Abstract
The purpose of the current study was to investigate the 11 bestrophin-1 (BEST1) exons in patients with best vitelliform macular dystrophy (BVMD), and to characterize the associated clinical features. Complete ophthalmic examinations were conducted on two families, and two family members were diagnosed with BVMD. Genomic DNA was extracted from the leukocytes of peripheral blood collected from the patients and their family members, in addition to 100 unrelated control subjects recruited from the same population. The polymerase chain reaction was used to amplify a total of 11 exons of the BEST1 gene, which were directly sequenced. Ophthalmic examinations, including best-corrected visual acuity, slit-lamp examination, fundus examination, fundus photography and fluorescein angiography imaging, as well as anterior segment analysis with Pentacam and optical coherence tomography, were conducted. The patients exhibited yellowish lesions in the macular area. A heterozygous mutation c.910_912delGAT (p.304del Asp) in exon 7 was identified in Case 1. A heterozygous BEST1 missense mutation c.685T>G (p.Trp229Gly) in exon 5 was identified in Case 2, but not in any of the unaffected family members or normal controls. Although BEST1 gene mutations and polymorphisms have previously been reported in various ethnic groups, the current study identified, for the first time to the best of our knowledge, two novel BEST1 gene mutations in patients with BVMD.
Keywords: best vitelliform macular dystrophy, bestrophin-1 gene, mutation
Introduction
Best vitelliform macular dystrophy (BVMD) is one of the most common forms of autosomal dominant macular dystrophy, characterized by the presence of yellowish lesions in the macular area (1–4).
BVMD has been classified into five phenotypic stages: The previtelliform, vitelliform, pseudohypopyon, vitelliruptive and atrophic stages. Not all patients will progress through all five stages, and the stages do not always occur consecutively. Furthermore, BVMD lesions may simultaneously display characteristics of various BVMD stages (5–8).
BVMD is associated with mutations in the bestrophin 1 gene (BEST1), on chromosome 11q12, which encodes a 585 amino acid transmembrane protein and is selectively expressed in the retinal pigment epithelium (RPE) (9–11). The BEST1 gene is the founding member of a family of four paralogs, which also includes BEST2, BEST3 and BEST4. BEST1 is expressed in the RPE cells of the developing and adult eye cells (1,9).
Greater than 300 distinct BEST1 mutations have been identified in families or sporadic patients affected by BVMD (12–17). It has been hypothesized that mutations in BEST1 may result in channel dysfunction which leads to abnormal fluid and ion transport through the RPE, resulting in defects during ocular growth and later-onset retinal dystrophy (18,19).
Mutations in the BEST1 gene are detected in the majority of cases of BVMD with a positive family history. The current study aimed to conduct mutational analysis of two Chinese families with BVMD at the gene level, in addition to analyzing the associated clinical features.
Patients and methods
Ethical approval
Approval for the current study was provided by the ethics committee of Zhongshan Ophthalmic Center, Sun Yat-Sen University (Guangzhou, China). Two patients were diagnosed with BVMD at the Zhongshan Ophthalmic Center.
Ophthalmic examinations
A variety of techniques were used for the ophthalmic examinations, which are outlined as follows: Visual acuity was examined using the Early Treatment Diabetic Retinopathy Study chart (Precision Vision, LaSalle, IL, USA). Anterior segment photographs were captured using a BX 900 Slit Lamp (Haag-Streit AG, Köniz, Switzerland). Anterior segment measurements were taken with Pentacam® HR version 70700 (OCULUS Optikgeräte GmbH, Wetzlar, Germany). Fundus photography and fundus fluorescein angiography (FFA) imaging was performed using a Heidelberg Retina Angiograph (Heidelberg Engineering GmbG, Heidelberg, Germany). In addition, physical examinations, including blood examination, a urine test, electrocardiogram, chest X-ray, blood biochemistry test, blood lipid and blood coagulation tests were conducted to exclude systemic diseases.
Sample collection
Two affected families were identified as described. A total of 100 subjects who exhibited no diagnostic features of BVMD were recruited from the same population to serve as normal controls. Written informed consent was obtained from all participating individuals prior to the commencement of the current study. According to the principles of the Declaration of Helsinki, venous blood samples were collected from peripheral blood leukocytes for genomic DNA extraction using a DNA extraction kit (Qiagen, Hilden, Germany).
Mutation detection
Exons of the BEST1 gene were amplified with the polymerase chain reaction (PCR), using the primers (Beijing Genomics Institute, Guangzhou, China) presented in Table I (20). In brief, PCR was performed in a 50-µl total volume reaction. All reagents used for PCR were purchased from (Takara Bio, Inc., Tokyo, Japan). The PCR cycling profile was as follows: One cycle at 94°C for 5 min, followed by 40 cycles at 94°C for 45 sec, 52–66°C for 45 sec and 72°C for 45 sec, in addition to one cycle at 72°C for 10 min. The PCR products were sequenced from both directions using an ABI3730 Automated Sequencer (Applied Biosystems Life Technologies, Foster City, CA, USA). The sequencing results were analyzed using SeqMan, version 2.3 (Technelysium Pty Ltd., Brisbane, Australia) and they were compared with the reference sequences in the database at the National Center for Biotechnology Information (NC_000011.9; http://www.ncbi.nlm.nih.gov/nuccore/224589802).
Table I.
Primers used for the polymerase chain reaction.
| Exon | Forward (5′-3′) | Reverse (5′-3′) | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| 2 | AGTCTCAGCCATCTCCTCGC | TGGCCTGTCTGGAGCCTG | 212 | 61 |
| 3 | GGGACAGTCTCAGCC ATCTC | CAGCTCCTCGTGATCCTCC | 238 | 58 |
| 4 | AGAAAGCTGGAGGAGCCG | GCGGCAGCCCTGTCTGTAC | 1408 | 59 |
| 5 | GGGGCAGGTGGTGTTCAGA | GGCAGCCTCACCAGCCTAG | 150 | 59 |
| 6 | GGGCAGGTGGTGTTCAGA | CCTTGGTCCTTCTAGCCTCAG | 181 | 59 |
| 7 | CATCCTGATTTCAGGGTTCC | CTCTGGCCATGCCTCCAG | 257 | 59 |
| 8 | AGCTGAGGTTTAAAGGGGGA | TCTCTTTGGGTCCACTTTGG | 215 | 59 |
| 9 | ACATACAAGGTCCTGCCTGG | GCATTAACTAGTGCTATTCTAAGTTCC | 298 | 59 |
| 10A | GGTGTTGGTCCTTTGTCCAC | CTCTGGCATATCCGTCAGGT | 591 | 59 |
| 10B | CTTCAAGTCTGCCCCACTGT | TAGGCTCAGAGCAAGGGAAG | 457 | 59 |
| 11 | CATTTTGGTATTTGAAATGAAGG | CCATTTGATTCAGGCTGTTG | 216 | 59 |
Summary of the primers and product lengths used for the amplification of the 11 exons of BEST1. bp, base pairs.
Results
Clinical data
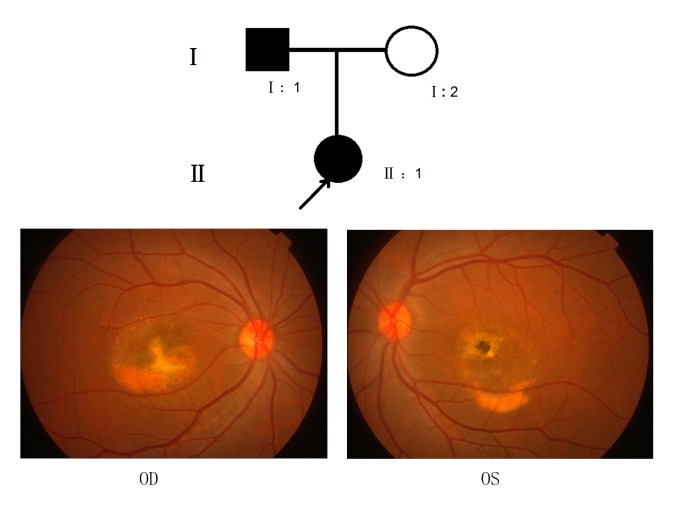
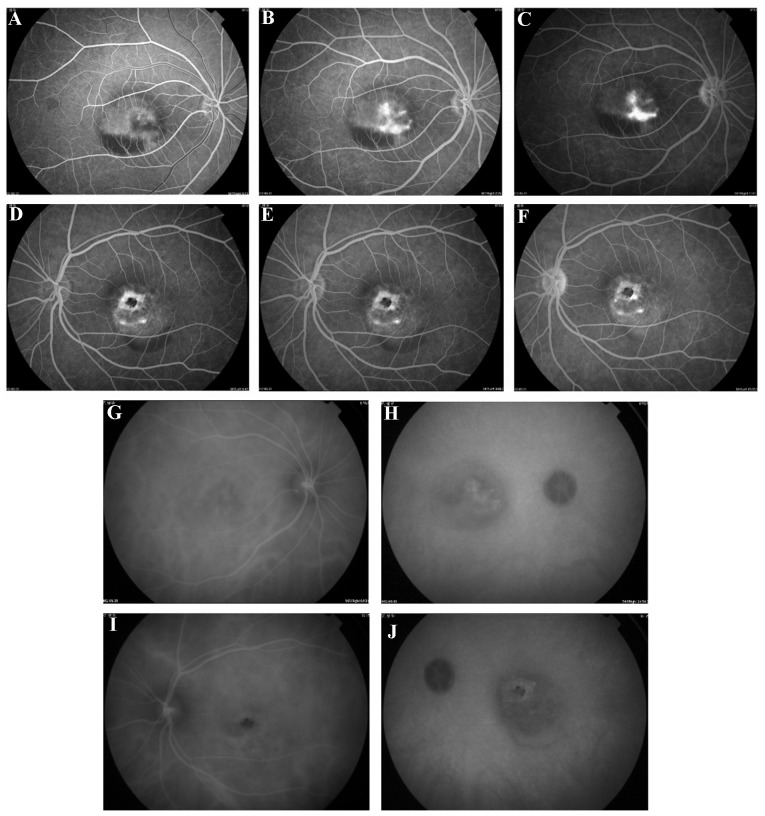
The patients evaluated in the current study were from the southern area of China. Case 1 (II:1) of Family 1 was a 23-year old female with a family history (Fig. 1) of ocular disease, diagnosed with BVMD at the age of 21 years. Her best-corrected visual acuity, as measured by the Logarithm of the Minimum Angle of Resolution (LogMAR) using the following formula: LogMAR visual acuity = 0.1 + LogMAR value of the best line read −0.02 X (number of letters read), was 0.1 in the right eye and 0.4 in the left eye. Examination of the fundus identified vitelliruptive lesions with a scrambled egg-like appearance and dispersion of the vitelliform material, including signs of atrophy, in the left eye (Fig. 1; OS). Fluorescein angiography (Fig. 2) demonstrated significant early hyperfluorescence (Fig. 2A) that increased in intensity at the late stage of the angiographic sequence, and was associated with moderate leakage (Fig. 2B and C) in the right eye, and macular lesions that simulated a pattern of dystrophy in the left eye (Fig. 2D–F). Indocyanine green chorioangiography (ICG; Fig. 2G–J) detected hypofluorescence in the early stages (Fig. 2G and I) of the angiographic sequence in the left and right eyes and subsequently detected little abnormal hyperfluorescence the late stages (Fig. 2H and J). The father of Case 1 was also diagnosed with ocular disease during his teenage years, and at the time of the present study, exhibited marked macular degeneration and cataracts.
Figure 1.
Pedigree of Family 1. Square symbols denote males, and circular symbols denote females. The shaded symbols indicate ophthalmologist-confirmed best vitelliform macular dystrophy. The arrow indicates the proband (II:1). Fundus examination revealed vitelliruptive lesions with a scrambled egg-like appearance and dispersion of the vitelliform material, with signs of atrophy in the left eye of Case 1 (II:1) (magnification, ×50). OD, oculus dexter; OS, oculus sinister.
Figure 2.
Fundus photography, FFA imaging and ICG were performed using a retina angiograph. (A) FFA revealed significant early hyperfluorescence (B and C) that increased in intensity at the late stage of the angiographic sequence (D–F) and was associated with moderate leakage in the right eye and macular lesions that simulated a pattern of dystrophy in the left eye. (G and I) ICG detected hypofluorescence in the early stages of the angiographic sequence in the left and right eyes and subsequently detected (H and J) little abnormal hyperfluorescence at the late stages (magnification, ×50). ICG, indocyanine green chorioangiography; FFA, fundus fluorescein angiography.
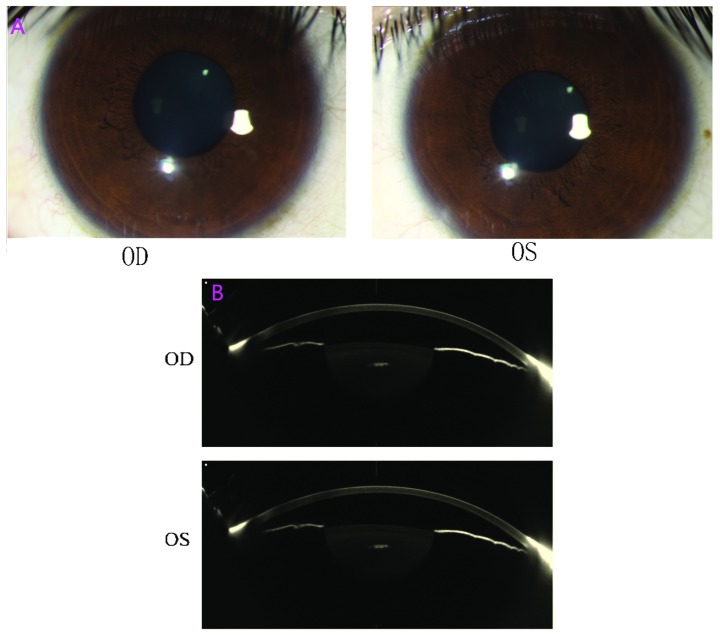
The anterior segment photograph captured using a BX 900 Slit Lamp is presented in Fig. 3A, and the images presented in Fig. 3B were captured using Pentacam. The anterior chamber depths were 2.09 mm [oculus dexter (OD)] and 2.12 mm [oculus sinister (OS)], and the central corneal thickness was identified to be 548 µm (OD) and 558 µm (OS).
Figure 3.
(A) Anterior segment photograph of Case 1 captured with a BX 900 Slit Lamp. (B) Anterior segment photograph of Case 1 captured with a Pentacam. The anterior chamber depths were 2.09 mm (OD) and 2.12 mm (OS), and the central corneal thickness was demonstrated to be 548 µm (OD) and 558 µm (OS) (magnification, ×50). OD, oculus dexter; OS, oculus sinister.
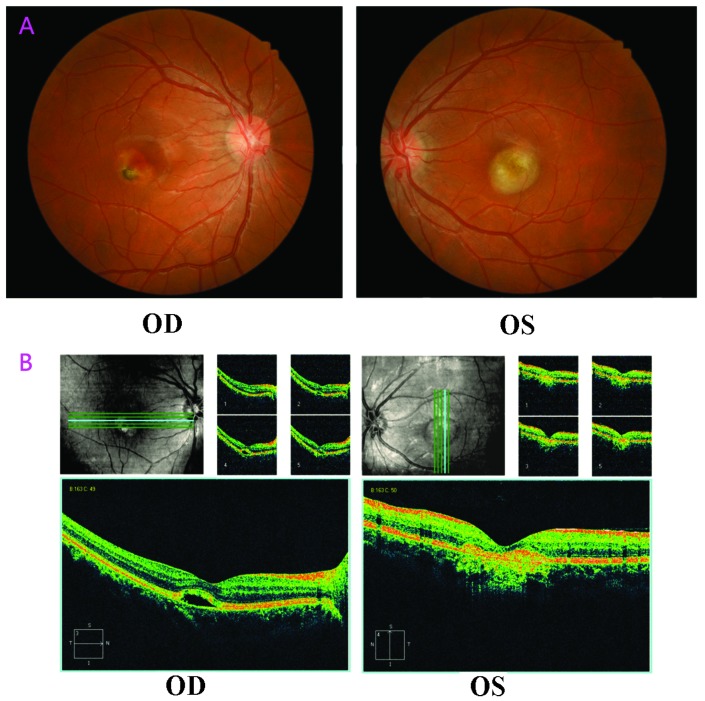
Case 2 of Family 2 was a 22 year-old male with no known familial history of ocular disease at the time of diagnosis of BVMD aged 19 years. His best-corrected visual acuity, as measured by LogMAR, was 0.7 in the right eye and 0.4 in the left eye. Fundus examination (Fig. 4A) indicated vitelliruptive lesions with a scrambled egg-like appearance and dispersion of the vitelliform material with signs of atrophy of the right eye. This patient was identified to be supersensitive to fluorescein, therefore no FFA or ICGA results were obtained.
Figure 4.
(A) Examination of the fundus of Case 2. Vitelliruptive lesions with a scrambled egg-like appearance were observed, in addition to dispersion of the vitelliform material with signs of atrophy in the right eye. (B) Spectral domain optical coherence tomography scans demonstrated clear abnormalities, including the absence of the foveal pit, serous retinal detachment, cystoid macular edema and interruption of the outer limiting membrane. The right eye exhibited marked yellow-white subretinal scarring with pigmented borders, surrounded by serous retinal detachment. In addition, the left eye exhibited an atrophic lesion (magnification, ×50). OD, oculus dexter; OS, oculus sinister.
The anterior chamber depths were 3.15 mm (OD) and 3.17 mm (OS), and the central corneal thickness was identified to be 564 µm (OD) and 568 µm (OS). Spectral domain optical coherence tomography (OCT) scans revealed clear abnormalities, including the absence of the foveal pit, serous retinal detachment, cystoid macular edema and interruption of the outer limiting membrane (Fig. 4B). The right eye was observed to exhibit prominent yellow-white subretinal scarring with pigmented borders, surrounded by a serous retinal detachment. Additionally, the left eye presented with an atrophic lesion.
OCT revealed that the foveal region of the right eye was unusually thick due to the presence of abnormal neuroretinal detachment from the RPE, which was suggested to have been triggered by an aberrant accumulation of fluid within the choriocapillaris and between the RPE and the fovea. In addition, OCT indicated that the foveal region of the left eye was abnormally thick due to irregularities in the deep retinal layers, with aberrant junctions between the inner and outer segents, as well as a prominent and hyperreflective structure in contact with the overlying retina, which was surrounded by elevated retina with underlying spots of increased reflectivity.
Mutation screening
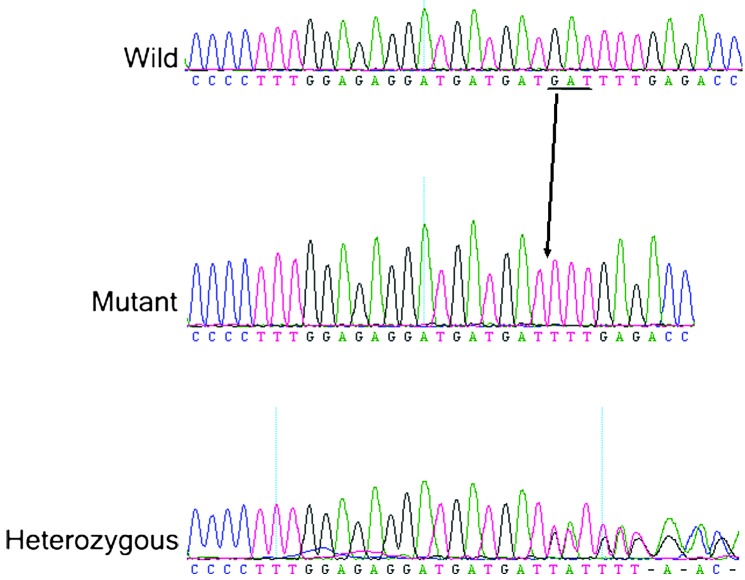
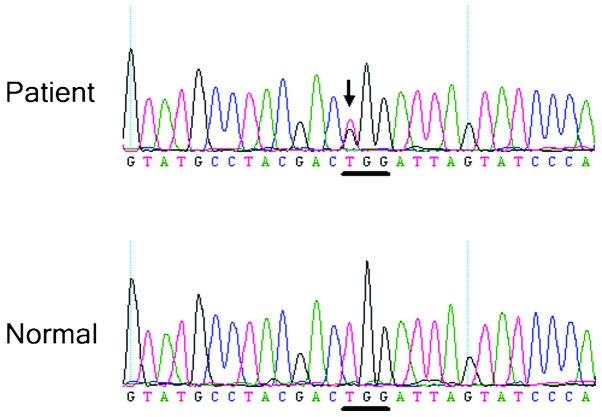
A heterozygous mutation c.910_912delGAT (p.304del Asp) was identified in exon 7 in the affected family members of Family 1 (Fig. 5). A heterozygous BEST1 missense mutation c.685T>G (p.Trp229Gly) in exon 5 was identified in the affected Case 2 (Fig. 6), but not the unaffected members of Family 2 or the normal controls. These identified mutations had not previously been reported.
Figure 5.
A heterozygous mutation c.910_912delGAT (p.304del Asp) in exon 7 of BEST1 was identified in the affected members of Family 1.
Figure 6.
A heterozygous BEST1 missense mutation c.685T>G (p.Trp229Gly) in exon 5 was identified in the affected Case 2 (Family 2).
Discussion
BVMD is associated with mutations in the BEST1 gene, which was formerly known as the VMD2 gene. The BEST1 gene encodes bestrophin-1, a transmembrane protein located in the basolateral membrane of the RPE (21–23).
A large number of allelic variants of the BEST1 gene have previously been identified and are recorded in the Regensburg University (Bavaria, Germany) database (24).
In the current study, two mutations were identified in the BEST-1 gene, which are associated with Best Syndrome: c.910_912delGAT (p.304del Asp) and c.685T>G (p.Trp229Gly). These mutations, rather than a rare polymorphism in the normal population, are suggested to be the causative mutations in the two sporadic BVMD patients.
Although >300 distinct BEST1 allelic variants have been identified in BVMD, a comprehensive database summarizing the known associations between BEST1 mutations and the prognosis of patients with BVMD worldwide remains to be developed. This is due to the fact that systematic whole BEST1 gene sequencing and clinical examinations have not been completed in a sufficiently large cohort of affected patients.
BVMD is a bilateral, symmetric, progressive disease of the macular area, associated with long-term loss of vision. Onset of the disease is predominantly early in life, often occurring by 10 years of age (7,25,26). Amongst certain patients, BVMD rapidly progresses towards the loss of central vision (22,26); thus, a thorough investigation of the age of onset of this disease may aid the development of valuable diagnostic/prognostic techniques. Conducting studies on independent cohorts of children belonging to families with a known history of BVMD may also be beneficial.
In conclusion, the current study identified two novel mutations of the BEST1 gene in two Chinese families with BVMD. The results of the current study expanded the library of known mutations of BEST1 and are valuable for the development of genetic counseling and prenatal diagnosis in families with BVMD.
Acknowledgments
The authors would like to thank all patients, families and normal control volunteers for their participation in the present study. The current study was supported by the National Natural Science Foundation of China (grant no. 30973277); the Science and Technology Planning Project of Guangdong Province, China (grant nos. 2010B090400416 and 2011B080701033); the Medical Scientific Research Foundation of Guangdong Province (grant no. B2012126); the Key Clinical Program of the Ministry of Health (grant no. 2010.439); the Youth Project of Fundamental Research Funds of the State Key Laboratory of Ophthalmology (grant no. 2011Q09) and the Project of Fundamental Research Funds of the State Key Laboratory of Ophthalmology (grant no. 2012KF03).
References
- 1.White K, Marquardt A, Weber BH. VMD2 mutations in vitelliform macular dystrophy (Best disease) and other maculopathies. Hum Mutat. 2000;15:301–308. doi: 10.1002/(SICI)1098-1004(200004)15:4<301::AID-HUMU1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 2.Sun H, Tsunenari T, Yau KW, Nathans J. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc Natl Acad Sci USA. 2002;99:4008–4013. doi: 10.1073/pnas.052692999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sodi A, Passerini I, Murro V, Caputo R, Bacci GM, et al. BEST1 sequence variants in Italian patients with vitelliform macular dystrophy. Mol Vis. 2012;18:2736–2748. [PMC free article] [PubMed] [Google Scholar]
- 4.MacDonald IM, Lee T. Best vitelliform macular dystrophy. In: Pagon RA, Adam MP, Ardinger HH, Bird TD, Dolan CR, et al., editors. GeneReviews® (Internet) University of Washington; Seattle, WA: 2003. [Google Scholar]
- 5.Hayami M, Decock C, Brabant P, Van Kerckhoven W, Lafaut BA, et al. Optical coherence tomography of adult-onset vitelliform dystrophy. Bull Soc Belge Ophtalmol. 2003;289:53–61. [PubMed] [Google Scholar]
- 6.Eksandh L, Bakall B, Bauer B, Wadelius C, Andréasson S. Best’s vitelliform macular dystrophy caused by a new mutation (Val89Ala) in the VMD2 gene. Ophthalmic Genet. 2001;22:107–115. doi: 10.1076/opge.22.2.107.2226. [DOI] [PubMed] [Google Scholar]
- 7.Boon CJ, Theelen T, Hoefsloot EH, van Schooneveld MJ, Keunen JE, et al. Clinical and molecular genetic analysis of best vitelliform macular dystrophy. Retina. 2009;29:835–847. doi: 10.1097/IAE.0b013e31819d4fda. [DOI] [PubMed] [Google Scholar]
- 8.Furino C, Boscia F, Cardascia N, Sborgia L, Sborgia C. Fundus autofluorescence, optical coherence tomography and visual acuity in adult-onset foveomacular dystrophy. Ophthalmologica. 2008;222:240–244. doi: 10.1159/000130427. [DOI] [PubMed] [Google Scholar]
- 9.Caldwell GM, Kakuk LE, Griesinger IB, Simpson SA, Nowak NJ, et al. Bestrophin gene mutations in patients with best vitelliform macular dystrophy. Genomics. 1999;58:98–101. doi: 10.1006/geno.1999.5808. [DOI] [PubMed] [Google Scholar]
- 10.Krämer F, Mohr N, Kellner U, Rudolph G, Weber BH. Ten novel mutations in VMD2 associated with best macular dystrophy (BMD) Hum Mutat. 2003;22:418. doi: 10.1002/humu.9189. [DOI] [PubMed] [Google Scholar]
- 11.Krämer F, White K, Pauleikhoff D, Gehrig A, Passmore L, et al. Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. Eur J Hum Genet. 2000;8:286–292. doi: 10.1038/sj.ejhg.5200447. [DOI] [PubMed] [Google Scholar]
- 12.MacDonald IM, Gudiseva HV, Villanueva A, Greve M, Caruso R, et al. Phenotype and genotype of patients with autosomal recessive bestrophinopathy. Ophthalmic Genet. 2012;33:123–129. doi: 10.3109/13816810.2011.592172. [DOI] [PubMed] [Google Scholar]
- 13.Marchant D, Gogat K, Boutboul S, Pequignot M, Sternberg C, et al. Identification of novel VMD2 gene mutations in patients with best vitelliform macular dystrophy. Hum Mutat. 2001;17:235. doi: 10.1002/humu.9. [DOI] [PubMed] [Google Scholar]
- 14.Marchant D, Yu K, Bigot K, Roche O, Germain A, et al. New VMD2 gene mutations identified in patients affected by best vitelliform macular dystrophy. J Med Genet. 2007;44:e70. doi: 10.1136/jmg.2006.044511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musarella MA. Molecular genetics of macular degeneration. Doc Ophthalmol. 2001;102:165–177. doi: 10.1023/A:1017510515893. [DOI] [PubMed] [Google Scholar]
- 16.Sodi A, Passerini I, Simonelli F, Testa F, Menchini U, et al. A novel mutation in the VMD2 gene in an Italian family with best maculopathy. J Fr Ophtalmol. 2007;30:616–620. doi: 10.1016/S0181-5512(07)89667-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhao L, Grob S, Corey R, Krupa M, Luo J, et al. A novel compound heterozygous mutation in the BEST1 gene causes autosomal recessive best vitelliform macular dystrophy. Eye (Lond) 2012;26:866–871. doi: 10.1038/eye.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shamshinova AM. Local ERG for clinical examination of eye diseases. Doc Ophthalmol. 1990;76:1–11. doi: 10.1007/BF00140493. [DOI] [PubMed] [Google Scholar]
- 19.Querques G, Zerbib J, Santacroce R, Margaglione M, Delphin N, et al. Functional and clinical data of best vitelliform macular dystrophy patients with mutations in the BEST1 gene. Mol Vis. 2009;15:2960–2972. [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y, Liang X, Ai S, Chen C, Liu X, Luo L, et al. FGFR2 molecular analysis and related clinical findings in one Chinese family with Crouzon syndrome. Mol Vis. 2012;18:449–454. [PMC free article] [PubMed] [Google Scholar]
- 21.Allikmets R, Seddon JM, Bernstein PS, Hutchinson A, Atkinson A, et al. Evaluation of the best disease gene in patients with age-related macular degeneration and other maculopathies. Hum Genet. 1999;104:449–453. doi: 10.1007/s004390050986. [DOI] [PubMed] [Google Scholar]
- 22.Brecher R, Bird AC. Adult vitelliform macular dystrophy. Eye (Lond) 1990;4:210–215. doi: 10.1038/eye.1990.28. [DOI] [PubMed] [Google Scholar]
- 23.Preising MN, Pasquay C, Friedburg C, Bowl W, Jager M, et al. Autosomal recessive bestrophinopathy (ARB): a clinical and molecular description of two patients at childhood. Klin Monbl Augenheilkd. 2012;229:1009–1017. doi: 10.1055/s-0032-1327782. In German. [DOI] [PubMed] [Google Scholar]
- 24.Lacassagne E, Dhuez A, Rigaudière F, Dansault A, Vêtu C, Bigot K, Vieira V, Puech B, Defoort-Dhellemmes S, Abitbol M. Phenotypic variability in a French family with a novel mutation in the BEST1 gene causing multifocal best vitelliform macular dystrophy. Mol Vis. 2011;17:309–322. [PMC free article] [PubMed] [Google Scholar]
- 25.Booij JC, Boon CJ, van Schooneveld MJ, ten Brink JB, Bakker A, et al. Course of visual decline in relation to the Best1 genotype in vitelliform macular dystrophy. Ophthalmology. 2010;117:1415–1422. doi: 10.1016/j.ophtha.2009.11.044. [DOI] [PubMed] [Google Scholar]
- 26.Fishman GA, Baca W, Alexander KR, Derlacki DJ, Glenn AM, et al. Visual acuity in patients with best vitelliform macular dystrophy. Ophthalmology. 1993;100:1665–1670. doi: 10.1016/S0161-6420(93)31420-X. [DOI] [PubMed] [Google Scholar]