Bladder cancer is a smoking- and occupational exposure-related disease with a substantial genetic component (Boffetta, 2008[4]; Golka et al., 2012[17]; Roth et al., 2012[39]; Rushton et al., 2012[41]; Schwender et al., 2012[43]; Burger et al., 2013[7]). Approximately 30 % of all urinary bladder cancer cases can be attributed to genetic risk factors (Lichtenstein et al., 2000[30]; Selinski, 2012[44]; Hammad, 2013[21]). Both family studies and large genome-wide association analyses support a polygenetic basis for urinary bladder carcinomas, mainly because there is no evidence for a major gene (Aben et al., 2006[1]; Kiemeney, 2008[25]; Kiemeney et al., 2010[26]; Rafnar et al., 2011[38]; Stewart and Marchan, 2012[53]; Bolt, 2013[5][6]), and all known susceptibility variants show moderate risks (Grotenhuis et al., 2010[20]; Lehmann et al., 2010[29]; Golka et al., 2011[19]; Selinski et al., 2012[52][51]; Dudek et al., 2013[10]; Selinski, 2014[46]). Several of these moderate-risk variants, especially those categorized as phase II metabolism genes, have been shown to modulate bladder cancer risk depending on exposure to bladder carcinogens, in particular, aromatic amines and polycyclic aromatic hydrocarbons (Garcia-Closas et al., 2005[13], 2013[14]; Golka et al., 2009[15]; Rothman et al., 2010[40]; Moore et al., 2011[35]; Selinski et al., 2011[50], 2012[51]). These gene-environment interactions are well-investigated for several phase II genes, including the deletion variant of glutathione-S-transferase M1 (GSTM1) and the N-acetyltransferase 2 (NAT2) polymorphisms, both of which are particularly relevant in the presence of their carcinogenic substrates due to occupational or tobacco smoke exposure (Engel et al., 2002[11]; Golka et al., 2002[17], 2008[18], 2009[15]; Garcia-Closas et al., 2005[13]; Kopps et al., 2008[27]; Hengstler, 2010[22]; Moore et al., 2011[35]; Ovsiannikov et al., 2012[36]; Selinski, 2013[45], 2014[46]; Selinski et al., 2013[49][47], 2014[48]). Current studies focus on a broader range of polymorphisms identified by genome-wide association studies (GWAS) and the interaction of these polymorphisms with tobacco smoke exposure. Garcia-Closas et al. (2013[14]) investigated the interaction between smoking habits and the well-known panel of eleven single nucleotide polymorphisms (SNPs) from GWAS, in addition to GSTM1, in studies, which were all part of the NCI bladder cancer GWAS. The NCI bladder cancer GWAS led to the discovery of several of these bladder cancer susceptibility SNPs. The authors found additive interactions between exposure and six of the variants, in particular, rs1495741 (NAT2), rs17863783 (UDP glucuronosyltransferase 1 family, polypeptide A6 UGT1A6), GSTM1, rs2294008 (prostate stem cell antigen PSCA), rs9642880 (v-myc avian myelocytomatosis viral oncogene homolog MYC) and rs1014971 (chromobox homolog 6 CBX6, apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A APOBEC3A) (Garcia-Closas et al., 2013[14]). Figueroa et al. (2014[12]) searched genome-wide for SNP × smoking interactions in the same multicentric case-control series. Two novel SNPs could be validated in independent study groups: the non-smoker SNP rs1711973 near forkhead box F2 (FOXF2) and the ever smoker SNP rs12216499 in an intergenic region between the radial spoke 3 homolog (Chlamydomonas) (RSPH3), T-cell activation RhoGTPase activating protein (TAGAP) and ezrin (EZR) genes (Figueroa et al., 2014[12]). Meanwhile, further studies focused on the common effects of several genetic variants on urinary bladder cancer risk instead of analysing single variants or their gene-environment interactions. The approaches used encompassed SNP-SNP and gene-gene interaction analysis (Andrew et al., 2012[2]; Binder et al., 2012[3]; Schwender et al., 2012[43]; Hu et al., 2013[23]), pathway analysis (Menashe et al., 2012[33]; Pan et al., 2014[37]) and polygenetic scores (Garcia-Closas et al., 2013[14]; Wang et al., 2014[54][55]). Results from recent genetic interaction studies are summarised in Table 1(Tab. 1). Generally, SNP-SNP or gene-gene interaction analyses aim to identify single genetic variants that interact in an additive or multiplicative way to modify the outcome of interest, e. g. bladder cancer risk. Pathway analyses consider sets of variants associated with genes that belong to the same biological or artificial pathway. The association with a phenotype of interest is often tested via enrichment analysis, i. e., a significant overrepresentation of variants of a particular pathway. Polygenic risk scores are calculated as weighted or unweighted sums of risks alleles of a set of risk variants. The unweighted variant usually sums up all risk alleles of the SNP set whereas, the weighted variant uses the individual variant odds ratio (OR) to account for higher or lower impact of each polymorphism. Usually, higher versus the lowest quartiles are compared but thresholds are also common.
Table 1. Genetic interactions and pathways that confer urinary bladder cancer in recent studies.
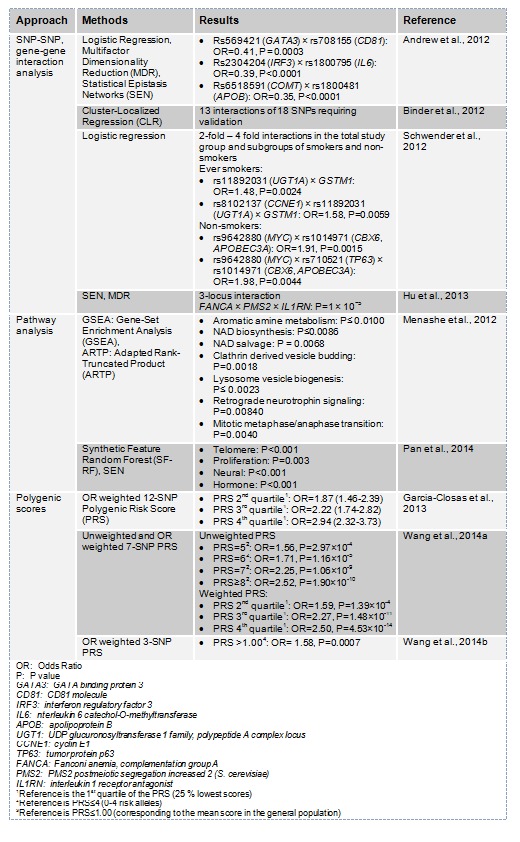
Genetic interaction studies are currently an important issue in cancer research. A number of approaches aim to elucidate the complex processes and interactions that lead to tumor development and progression, which has also recently been intensively studied in breast cancer (Chuang et al., 2013[9]; Sapkota et al., 2013[42]; Milne et al., 2014[34]; Yang et al., 2014[56]), prostate cancer (Lin et al., 2008[32], 2013[31]; Lavender et al., 2012[28]), lung cancer (Chu et al., 2014[8]) and colorectal cancer (Jiao et al., 2012[24]). Therefore, a new era has begun after successful identification of the most influential genetic variants. One of the goals of the post GWAS era is to understand and quantify SNP × SNP and SNP × environment interactions. The discussion on the most adequate techniques is still ongoing. A relatively easy and straight forward method is to sum up all risk alleles of relevant SNPs and study the association of the sum ('risk score') with cancer risk. A more challenging strategy is to calculate odds ratios for all combinations of variants and identify the most powerful interactions of high risk alleles. Although this approach is theoretically superior to simple 'risk score' approaches, it requires high computing capacity and very high case numbers. Currently, only few studies are available and the most critical interactions have most probably not yet been identified. However, the post GWAS era has only just begun.
References
- 1.Aben KKH, Baglietto L, Baffoe-Bonnie A, Coebergh JW, Bailey-Wilson JE, Trink B, et al. Segregation analysis of urothelial cell carcinoma. Eur J Cancer. 2006;42:1428–33. doi: 10.1016/j.ejca.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 2.Andrew AS, Hu T, Gu J, Gui J, Ye Y, Marsit CJ, et al. HSD3B and gene-gene interactions in a pathway-based analysis of genetic susceptibility to bladder cancer. PLoS One. 2012;7:e51301. doi: 10.1371/journal.pone.0051301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binder H, Muller T, Schwender H, Golka K, Steffens M, Hengstler JG, et al. Cluster-localized sparse logistic regression for SNP data. Stat Appl Gen Mol Biol. 2012;11(4):article 13. doi: 10.1515/1544-6115.1694. [DOI] [PubMed] [Google Scholar]
- 4.Boffetta P. Tobacco smoking and risk of bladder cancer. Scand J Urol Nephrol. 2008;Suppl(218):45–54. doi: 10.1080/03008880802283664. [DOI] [PubMed] [Google Scholar]
- 5.Bolt HM. Human bladder cancer risk calculation based on genome-wide analysis of genetic variants. Arch Toxicol. 2013;87:397–9. doi: 10.1007/s00204-013-1020-x. [DOI] [PubMed] [Google Scholar]
- 6.Bolt HM. Relevance of genetic disposition versus environmental exposure for cancer risk: an old controversy revisited with novel methods. EXCLI J. 2013;12:79–80. [PMC free article] [PubMed] [Google Scholar]
- 7.Burger M, Catto JWF, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63:234–41. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 8.Chu M, Zhang R, Zhao Y, Wu C, Guo H, Zhou B, et al. A genome-wide gene-gene interaction analysis identifies an epistatic gene pair for lung cancer susceptibility in Han Chinese. Carcinogenesis. 2014;35:572–7. doi: 10.1093/carcin/bgt400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuang L, Chang H, Lin M, Yang C. Improved branch and bound algorithm for detecting SNP-SNP interactions in breast cancer. J Clin Bioinforma. 2013;3(1):4. doi: 10.1186/2043-9113-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudek AM, Grotenhuis AJ, Vermeulen SH, Kiemeney, LALM, Verhaegh GW. Urinary bladder cancer susceptibility markers. [What do we know about functional mechanisms?]. Int J Mol Sci. 2013;14:12346–66. doi: 10.3390/ijms140612346. (Ger). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engel LS, Taioli E, Pfeiffer R, Garcia-Closas M, Marcus PM, Lan Q, et al. Pooled analysis and meta-analysis of glutathione S-transferase M1 and bladder cancer: a HuGE review. Am J Epidemiol. 2002;156:95–109. doi: 10.1093/aje/kwf018. [DOI] [PubMed] [Google Scholar]
- 12.Figueroa JD, Han SS, Garcia-Closas M, Baris D, Jacobs EJ, Kogevinas M, et al. Genome-wide interaction study of smoking and bladder cancer risk. Carcinogenesis. 2014;35:1737–44. doi: 10.1093/carcin/bgu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, Hein DW, et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366(9486):649–59. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Closas M, Rothman N, Figueroa JD, Prokunina-Olsson L, Han SS, Baris D, et al. Common genetic polymorphisms modify the effect of smoking on absolute risk of bladder cancer. Cancer Res. 2013;73:2211–20. doi: 10.1158/0008-5472.CAN-12-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golka K, Hermes M, Selinski S, Blaszkewicz M, Bolt HM, Roth G, et al. Susceptibility to urinary bladder cancer: relevance of rs9642880T, GSTM1 0/0 and occupational exposure. Pharmacogenet Genomics. 2009;19:903–6. doi: 10.1097/FPC.0b013e328331b554. [DOI] [PubMed] [Google Scholar]
- 16.Golka K, Kopps S, Prager HM, Mende Sv, Thiel R, Jungmann O, et al. Bladder cancer in crack testers applying azo dye-based sprays to metal bodies. J Toxicol Environ Health. 2012;75:566–71. doi: 10.1080/15287394.2012.675309. [DOI] [PubMed] [Google Scholar]
- 17.Golka K, Prior V, Blaszkewicz M, Bolt HM. The enhanced bladder cancer susceptibility of NAT2 slow acetylators towards aromatic amines: a review considering ethnic differences. Toxicol Lett. 2002;128:229–41. doi: 10.1016/s0378-4274(01)00544-6. [DOI] [PubMed] [Google Scholar]
- 18.Golka K, Schmidt T, Seidel T, Dietrich H, Roemer HC, Lohlein D, et al. The influence of polymorphisms of glutathione S-transferases M1 and M3 on the development of human urothelial cancer. J Toxicol Environ Health A. 2008;71:881–6. doi: 10.1080/15287390801988087. [DOI] [PubMed] [Google Scholar]
- 19.Golka K, Selinski S, Lehmann M, Blaszkewicz M, Marchan R, Ickstadt K, et al. Genetic variants in urinary bladder cancer: collective power of the "wimp SNPs". Arch Toxicol. 2011;85:539–54. doi: 10.1007/s00204-011-0676-3. [DOI] [PubMed] [Google Scholar]
- 20.Grotenhuis AJ, Vermeulen SH, Kiemeney LA. Germline genetic markers for urinary bladder cancer risk, prognosis and treatment response. Future Oncol. 2010;6:1433–60. doi: 10.2217/fon.10.109. [DOI] [PubMed] [Google Scholar]
- 21.Hammad S. Interaction of genetic variants towards increased cancer risk. EXCLI J. 2013;12:625–627. [PMC free article] [PubMed] [Google Scholar]
- 22.Hengstler JG. Experimental and Clinical Sciences: at interface to toxicology. EXCLI J. 2010;9:141–3. [Google Scholar]
- 23.Hu T, Andrew AS, Karagas MR, Moore JH. Statistical epistasis networks reduce the computational complexity of searching three-locus genetic models. Pac Symp Biocomput. 2013;2013:397–408. [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao S, Hsu L, Berndt S, Bézieau S, Brenner H, Buchanan D, et al. Genome-wide search for gene-gene interactions in colorectal cancer. PLoS One. 2012;7(12):e52535. doi: 10.1371/journal.pone.0052535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiemeney LA. Hereditary bladder cancer. Scand J Urol Nephrol. 2008;Suppl(218):110–5. doi: 10.1080/03008880802283755. [DOI] [PubMed] [Google Scholar]
- 26.Kiemeney LA, Sulem P, Besenbacher S, Vermeulen SH, Sigurdsson A, Thorleifsson G, et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat Genet. 2010;42:415–9. doi: 10.1038/ng.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopps S, Angeli-Greaves M, Blaszkewicz M, Prager H, Roemer HC, Lohlein D, et al. Glutathione S-transferase P1 ILE105Val polymorphism in occupationally exposed bladder cancer cases. J Toxicol Environ Health Part A. 2008;71:898–901. doi: 10.1080/15287390801988483. [DOI] [PubMed] [Google Scholar]
- 28.Lavender NA, Rogers EN, Yeyeodu S, Rudd J, Hu T, Zhang J, et al. Interaction among apoptosis-associated sequence variants and joint effects on aggressive prostate cancer. BMC Med Genomics. 2012;5:11. doi: 10.1186/1755-8794-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehmann ML, Selinski S, Blaszkewicz M, Orlich M, Ovsiannikov D, Moormann O, et al. Rs710521[A] on chromosome 3q28 close to TP63 is associated with increased urinary bladder cancer risk. Arch Toxicol. 2010;84:967–78. doi: 10.1007/s00204-010-0617-6. [DOI] [PubMed] [Google Scholar]
- 30.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer - analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 31.Lin H, Amankwah EK, Tseng T, Qu X, Chen D, Park JY. SNP-SNP interaction network in angiogenesis genes associated with prostate cancer aggressiveness. PLoS One. 2013;8(4):e59688. doi: 10.1371/journal.pone.0059688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin H, Wang W, Liu Y, Soong S, York TP, Myers L, et al. Comparison of multivariate adaptive regression splines and logistic regression in detecting SNP-SNP interactions and their application in prostate cancer. J Hum Genet. 2008;53:802–11. doi: 10.1007/s10038-008-0313-z. [DOI] [PubMed] [Google Scholar]
- 33.Menashe I, Figueroa JD, Garcia-Closas M, Chatterjee N, Malats N, Picornell A, et al. Large-scale pathway-based analysis of bladder cancer genome-wide association data from five studies of European background. PLoS One. 2012;7(1):e29396. doi: 10.1371/journal.pone.0029396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milne RL, Herranz J, Michailidou K, Dennis J, Tyrer JP, Zamora MP, et al. A large-scale assessment of two-way SNP interactions in breast cancer susceptibility using 46,450 cases and 42,461 controls from the breast cancer association consortium. Hum Mol Genet. 2014;23:1934–46. doi: 10.1093/hmg/ddt581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore LE, Baris DR, Figueroa JD, Garcia-Closas M, Karagas MR, Schwenn MR, et al. GSTM1 null and NAT2 slow acetylation genotypes, smoking intensity and bladder cancer risk: results from the New England bladder cancer study and NAT2 meta-analysis. Carcinogenesis. 2011;32:182–9. doi: 10.1093/carcin/bgq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ovsiannikov D, Selinski S, Lehmann M, Blaszkewicz M, Moormann O, Haenel MW, et al. Polymorphic enzymes, urinary bladder cancer risk, and structural change in the local industry. J Toxicol Environ Health A. 2012;75:557–65. doi: 10.1080/15287394.2012.675308. [DOI] [PubMed] [Google Scholar]
- 37.Pan Q, Hu T, Malley JD, Andrew AS, Karagas MR, Moore JH. A system-level pathway-phenotype association analysis using synthetic feature random forest. Genet Epidemiol. 2014;38:209–19. doi: 10.1002/gepi.21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rafnar T, Vermeulen SH, Sulem P, Thorleifsson G, Aben KK, Witjes JA, et al. European genome-wide association study identifies SLC14A1 as a new urinary bladder cancer susceptibility gene. Hum Mol Genet. 2011;20:4268–81. doi: 10.1093/hmg/ddr303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth E, Selinski S, Schikowsky C, Seidel T, Volkert F, Blaszkewicz M, et al. Bladder cancer survival in a former industrial area in Saxony-Anhalt, Germany. J Toxicol Environ Health A. 2012;75:1216–25. doi: 10.1080/15287394.2012.709168. [DOI] [PubMed] [Google Scholar]
- 40.Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42:978–84. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rushton L, Hutchings SJ, Fortunato L, Young C, Evans GS, Brown T, et al. Occupational cancer burden in Great Britain. Br J Cancer. 2012;107(Suppl 1):S3–7. doi: 10.1038/bjc.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sapkota Y, Mackey JR, Lai R, Franco-Villalobos C, Lupichuk S, Robson PJ, et al. Assessing SNP-SNP interactions among DNA repair, modification and metabolism related pathway genes in breast cancer susceptibility. PLoS One. 2013;8(6):e64896. doi: 10.1371/journal.pone.0064896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwender H, Selinski S, Blaszkewicz M, Marchan R, Ickstadt K, Golka K, et al. Distinct SNP combinations confer susceptibility to urinary bladder cancer in smokers and non-smokers. PLoS One. 2012;7(12):e51880. doi: 10.1371/journal.pone.0051880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selinski S. Genetic variants confer susceptibility to urinary bladder cancer: an updated list of confirmed polymorphisms EXCLI J. 2012;11:743–7. [PMC free article] [PubMed] [Google Scholar]
- 45.Selinski S. Functional consequences of urinary bladder cancer risk variants EXCLI J (Highlight report:) 2013;12:1017–9. [PMC free article] [PubMed] [Google Scholar]
- 46.Selinski S. Urinary bladder cancer risk variants: recent findings and new challenges of GWAS and confirmatory studies. Arch Toxicol. 2014;88:1469–75. doi: 10.1007/s00204-014-1297-4. [DOI] [PubMed] [Google Scholar]
- 47.Selinski S, Blaszkewicz M, Agundez JA, Martinez C, Garcia-Martin E, Hengstler JG, et al. Clarifying haplotype ambiguity of NAT2 in multi-national cohorts. Front Biosci (Schol Ed) 2013;5:672–684. doi: 10.2741/s399. [DOI] [PubMed] [Google Scholar]
- 48.Selinski S, Blaszkewicz M, Ickstadt K, Hengstler JG, Golka K. Improvements in algorithms for phenotype inference: the NAT2 example. Curr Drug Metab. 2014;15:233–49. doi: 10.2174/1389200215666140202215717. [DOI] [PubMed] [Google Scholar]
- 49.Selinski S, Blaszkewicz M, Ickstadt K, Hengstler JG, Golka K. Refinement of the prediction of N-acetyltransferase 2 (NAT2) phenotypes with respect to enzyme activity and urinary bladder cancer risk. Arch Toxicol. 2013;87:2129–39. doi: 10.1007/s00204-013-1157-7. [DOI] [PubMed] [Google Scholar]
- 50.Selinski S, Blaszkewicz M, Lehmann ML, Ovsiannikov D, Moormann O, Guballa C, et al. Genotyping NAT2 with only two SNPs (rs1041983 and rs1801280) outperforms the tagging SNP rs1495741 and is equivalent to the conventional 7-SNP NAT2 genotype. Pharmacogenet Genomics. 2011;21:673–8. doi: 10.1097/FPC.0b013e3283493a23. [DOI] [PubMed] [Google Scholar]
- 51.Selinski S, Lehmann ML, Blaszkewicz M, Ovsiannikov D, Moormann O, Guballa C, et al. Rs11892031[A] on chromosome 2q37 in an intronic region of the UGT1A locus is associated with urinary bladder cancer risk. Arch Toxicol. 2012;86:1369–78. doi: 10.1007/s00204-012-0854-y. [DOI] [PubMed] [Google Scholar]
- 52.Selinski S, Lehmann ML, Blaszkewicz M, Ovsiannikov D, Moormann O, Guballa C, et al. Urinary bladder cancer risk in relation to a single nucleotide polymorphism (rs2854744) in the insulin-like growth factor-binding protein-3 (IGFBP3) gene. Arch Toxicol. 2012;86:195–203. doi: 10.1007/s00204-011-0747-5. [DOI] [PubMed] [Google Scholar]
- 53.Stewart JD, Marchan R. Polymorphisms hit the headlines. Arch Toxicol. 2012;86:1799–801. doi: 10.1007/s00204-012-0973-5. [DOI] [PubMed] [Google Scholar]
- 54.Wang M, Chu H, Lv Q, Wang L, Yuan L, Fu G, et al. Cumulative effect of genome-wide association study-identified genetic variants for bladder cancer. Int J Cancer. 2014;35:2653–60. doi: 10.1002/ijc.28898. [DOI] [PubMed] [Google Scholar]
- 55.Wang P, Ye D, Guo J, Liu F, Jiang H, Gong J, et al. Genetic score of multiple risk-associated single nucleotide polymorphisms is a marker for genetic susceptibility to bladder cancer. Genes Chromosomes Cancer. 2014;53:98–105. doi: 10.1002/gcc.22121. [DOI] [PubMed] [Google Scholar]
- 56.Yang C, Lin Y, Chuang L, Chang H. Double-bottom chaotic map particle swarm optimization based on chi-square test to determine gene-gene interactions. Biomed Res Int. 2014;2014:172049. doi: 10.1155/2014/172049. [DOI] [PMC free article] [PubMed] [Google Scholar]


