Abstract
The cancer profile in the Indian state of Uttarakhand reveals that the breast cancer is the most prevalent type of cancers in females followed by cervical and ovarian type. Literature survey shows that the E6 protein of Human Papilloma Virus-16 (HPV-16) is responsible for causing several forms of cancer in human. Therefore, it is of interest to screen HPV-16 E6 target protein with known natural compounds using computer aided molecular modeling and docking tools. The complete structure of E6 is unknown. Hence, the E6 structure model was constructed using different online servers followed by molecular docking of Colchine, Curcumin, Daphnoretin, Ellipticine and Epigallocatechin-3-gallate; five known natural compounds with best E6 protein model predicted by Phyre2 server. The screening exercise shows that Daphnoretin (with binding free energy of -8.3 kcal/mol), a natural compound derived from Wikstroemia indica has the top binding properties. Thus, it is of interest to consider the compound for further validation.
Keywords: Human Papilloma Virus, E6 Protein, Daphnoretin, Wikstroemia indica, Molecular Docking
Background
Cervical cancer is one of the leading causes of deaths worldwide. The cancer profile of the Indian state of Uttarakhand reveals that the Breast cancer was most prevalent in female followed by cervical and ovarian cancer [1]. Human Papillomaviruses (HPVs) are the major cause of cervical cancer in the world, involved in 90% of all Cervical Cancers. The HPVs is divided into two groups, the ‘low risk’ such as (type 6 and 11) and the ‘igh risk’ (type 16 and 18) [2]. Although there are more than 200 types of HPV identified, the most commonly associated with cervical cancer are ‘high-risk’ HPV-16 and HPV-18 [3] HPV-16 and HPV-18 remain the primary drug targets for the development of anticancer drug. The two HPV genes, E6 and E7, are known to play crucial role in development of cancer. Both in vitro and in vivo experimental studies show that the function of E6 and E7 proteins, particularly of the ‘high risk types’, are essential for tumor formation [4, 5]. These oncogenes from the HPV-16 and HPV-18 have been showed to alter gene regulatory pathways involved in cell cycle control, interacting with and neutralizing the regulatory functions of two important tumor suppressor proteins, p53 and pRb (Retinoblastoma Protein) and deregulating key signal transduction pathways [6, 7]. HPV has been known to be play a major role in cervical cancer but effective treatment against HPV infection is still unavailable [8], Plant-derived product have currently become of great interest due to their medicinal values for prevention and treatment of cancer.
Plants secondary metabolites are known as magnificent accumulators of medicinal compounds. More than 50% of all the drug forms used in the clinical fields around the world have originated from the compounds extracted from plants. In our study, five different natural compounds from different plant sources were collected viz., Colchine, Curcumin, Daphnoretin, Ellipticine, and Epigallocatechine-3-gallate. Colchicine is a plant secondary metabolite extracted from Colchicum autumnale and Gloriosa superba L [9, 10]. Curcumin, a polyphenolic compound isolated from Curcuma longa, (commonly called turmeric), now finds its application as potential anti-cancer compound. It has been observed to induce apoptosis of various cancer cells and cause the modulation of the cell cycle pathway. However the exact mechanism is yet to be studied. To study its effect on colorectal cancer, multiple myeloma and pancreatic cancer, the compound is under Phase I/II trials. Curcumin is reported safe by phase I clinical trials, even when used at a high dosage level [11]. Daphnoretin, a bis-coumarin derivative, extracted from the root bark of Wikstroemia indica (Thymelaeaceae) in good amounts was found to have anti-cancer activity [12]. It is responsible for the suppression of protein and DNA synthesis in Ehrlich ascites carcinomas. Ellipticine, which is a plant alkaloid (5, 11-dimethyl-6H-pyrido [4, 3-b] carbazole) and its derivatives were isolated from Apocynaceae plant species. It intercalates with DNA and inhibits the Topoisomerase II activity. Apart from inhibiting the cell growth, it also leads to the apoptosis of human hepatocellular carcinoma HepG2 cells [13]. Epigallocatechin-3-gallate is a stable and water soluble compound and belongs to the group of flavonoids referred to as flavan-3-ols. Flavan-3-ols are mainly found in tea (black, green,) (Camellia sinensis), red wine (Vitis vinifera), strawberry (Fragaria ananassa) and cocao (Theobroma cacao) products [14, 15]. In order to understand the potential role of natural compounds as anticancer molecules, there is a need of computational drug designing tools that can identify and analyze protein-ligand interactions with respect to their binding affinity for investigation of novel drug molecule against HPV.
Methodology
Sequence retrieval and phylogenetic analysis:
The 5 amino acid sequences of the E6 protein of HPV from different countries (Accession no-AAD33252.1, ABK32509.1, ACQ90215.1, AFF58869.1, AEN69303.1) was retrieved in FASTA format from National Centre for Biotechnology Information (http:// www.ncbi. nlm.nih.gov/) database for multiple sequence alignment. Phylogenetic tree was constructed using Neighbor-Joining method by MEGA6 [16]. E6 protein sequence (Accession no. AAD33252.1) was taken for protein 3D model construction.
Protein structure prediction and validation:
The 158 amino acids residue long E6 protein of HPV-16 was subjected to BLASTp (http: //blast. ncbi.nlm. nih. gov/Blast.cgi) analysis against PDB database (http://www.rcsb.org /) to identify suitable template for comparative protein modeling. Comparative homology modeling depends on a sequence alignment between target sequence and the template sequence whose 3D structure has been determined by experimental method. A target-template alignment was performed using ClustalX program [17] and ESPrint 3.0 (http://espript.ibcp.fr/ESPript/ESPript/) server was used to visualize the target-template alignment results (Figure 1). In addition to target-template alignment, the three-dimensional structure of E6 protein sequence were constructed using five different servers viz., Bhageerath (http: // www.scfbioiitd.res.in/bhageerath /index.jsp/), CPH Model (http: //www.cbs.dtu.dk / services /CPHmodels/), Geno3D (http://geno3dpbil.ibcp.fr), ITASSER(http://zhanglab.ccmb.med.umich.edu /I-TASSER/) and Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2 /html /page.cgi? id=index). The predicted models were subjected for further refinement in ModRefiner server (http://zhanglab.ccmb.med.umich.edu /ModRefiner /). The best model was selected on the basis of Ramachandran plot and protein stability analysis by Rampage (http: //mordred.bioc.cam.ac.uk /~rapper /rampage.php) and ProsaA (https: //prosa.services.came.sbg.ac.at/prosa.php). Discovery Studio 4.1 visualizer (http: // accelrys.com/products /discovery-studio/) was used for the visualization of modelled protein structure (Figure 2).
Figure 1.
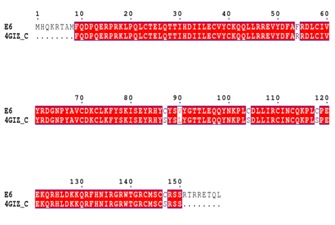
Pair-wise sequence alignment of the target protein E6 with C-chain of its template (PDB id: 4GIZ)
Figure 2.
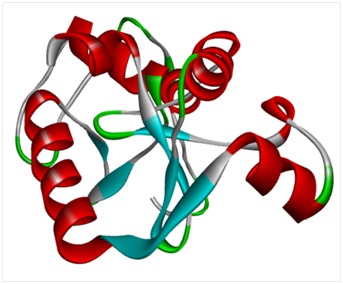
Homology model of E6 of HPV-16.
Ligand preparation:
The Structure of natural compounds derived from plants viz., Colchine (CID 2833), Curcumin (CID 969516), Daphnoretin (CID 5281406), Ellipticine (CID 3213) and Epigallocatechin-3- gallate (CID 65064) were retrieved from PubChem database at (http://www.ncbi.nlm.nih.gov/). These ligands were reported in literature. The three dimensional coordinates of ligands molecules were generated by Marvin Sketch (http://www.chemaxon.com /product /marvin /marvinsketch /) and saved in *.pdb files for molecular docking studies.
Molecular docking:
Molecular docking studies were performed using AutoDock Vina [18]. Preparation of required input files for AutoDock Vina was prepared using AutoDock 4.2 program. Preparation of files through AutoDock 4.2 involved addition of polar hydrogen atoms and gasteiger charges. The size of grid box was kept as 22, 20, 24 for X, Y, Z. The energy range was kept as 4 which is default setting. It is one of the most important highly cited molecular docking tools for the prediction of proteinligand interaction. It requires the three dimensional structure of both ligand and protein. The results with best conformation and energetic were selected for analysis. Ligplot (http://www.ebi.ac.uk/thornton-srv/software/LigPlus/) was used for visualization and analysis of protein-ligand complex [19].
Drug-likeness prediction:
OSIRIS Property Explorer (http: // www.organicchemistry. org/prog/peo/) was used to predict side effects, such as mutagenic, tumorigenic, irritant and reproductive effects which are also providing score for drug-relevant properties with respect to drug-likeness and overall drug-score [20].
Results and Discussion
Multiple sequence alignment of five E6 protein sequences from India, USA, China, Canada and Spain were performed using ClustalX [17] and Phylogenetic tree was constructed using Neighbor-Joining method in MEGA6 (Figure 3) [16]. Phylogenetic analysis clearly revealed that the E6 protein of Human Papilloma Virus of USA origin is closely related to the Indian and it is experimentally verified, therefore E6 protein sequence from USA (Accession no. AAD33252.1) was preferred for the modeling of 3D structure. BLASTp search revealed one putative template (PDB id: 4GIZ, Chain C) of high-label identity and query coverage with the target sequence, as the best template for comparative modeling. The pair-wise sequence alignment of HPV-16 E6 and template was generated using ClustalX, and alignment was showed in ESPrint 3.0 (Figure 1). Based on target-template alignment, five different server viz. I-TASSER, Geno-3D, Phyre 2, CPH model and Bhageerath generated protein 3D models of E6.
Figure 3.
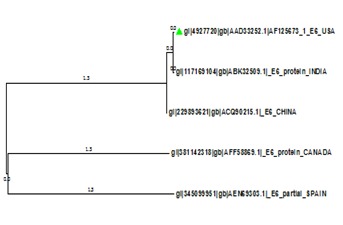
Neighbor-Joining tree of E6 protein sequences of five countries constructed in MEGA6. E6 protein from India and USA is closely related.
Structure refinements through energy minimization were performed using ModRefiner server ( http://zhanglab. ccmb.med. umich.edu /ModRefiner /). The minimized structures were finally saved as Hpv.pdb. The best model was selected based on of its stability assessment by RamPage and ProsaA. The model which had highest percentage of amino acid residues in the favoured region was chosen. Phyre2 Model, having 97.1% of residues in the most favoured region, 2.9% residues was found in additional allowed region and no residue observed in the generously allowed and disallowed region was selected Table 1 (see supplementary material). The energy profile of the selected model and the Z-score value (a measure of model quality) were obtained using ProsA program that calculates the interaction energy per residue using a distance-based pair potential. Based on ProsA analysis, the model constructed by Phyre2 showed a Z score of -4.46 and that of template as -4.91 which represent the reliability of predicted model Table 2 (see supplementary material), indicating that this model is suitable for wide spectrum of applications.
The binding sites predicted by the I-TASSER during modeling (135,137,138, 139,111,112,140) were chosen for molecular docking studies [21]. The grid box was prepared by AutoDock 4.2 program as 22, 20, and 24 for X, Y, Z coordinates of E6. The energy range was kept as 4 which is default parameter.
Molecular docking was carried out with each of the natural compounds viz.,colchine, curcumin, Daphnoretin, Ellipticine, Epigallocatechin-3-gallate at the binding site grid generated by AutoDock [18]. For each compound, out of the many docking poses, only those that possessed the highest docking score and relatively good hydrogen bond interaction were chosen. Colchine was docked with E6 protein of HPV showing docking energy -6.8 kcal/mol with three hydrogen bonds, Curcumin was docked with E6 protein of HPV showing docking energy - 6.3 kcal/mol with three hydrogen bonds, Daphnoretin was docked with E6 protein of HPV showing docking energy -8.3 kcal/mol with five hydrogen bonds, Ellipticine was docked with E6 protein of HPV showing docking energy -7.4 kcal/mol with one hydrogen bond and Epigallocatechin -3-gallate was docked with E6 protein of HPV showing docking energy -6.6 kcal/mol with two hydrogen bonds Table 3 (see supplementary material).
Ligplot were used to study the interaction site analysis [19]. The interaction analysis for binding site of natural compound with E6 protein has been done to find out the residues that play an important role in binding. The Daphnoretin shows highest affinity to bind with E6 protein with energy value -8.3 kcal/mol. It interacts with E6 amino acid residues Tyr39, Cys58, Ser78, Gln114, Arg138 with 5 hydrogen bonds (Figure 4). The hydrogen bonding is of considerable importance in the interaction of molecules. On the basis of docking energies natural inhibitor Daphnoretin may be lead compound for prevention of various form of cervical cancer caused by HPV. Hence this information would prove to be important for desiging of novel drug molecules against E6 protein of HPV.
Figure 4.
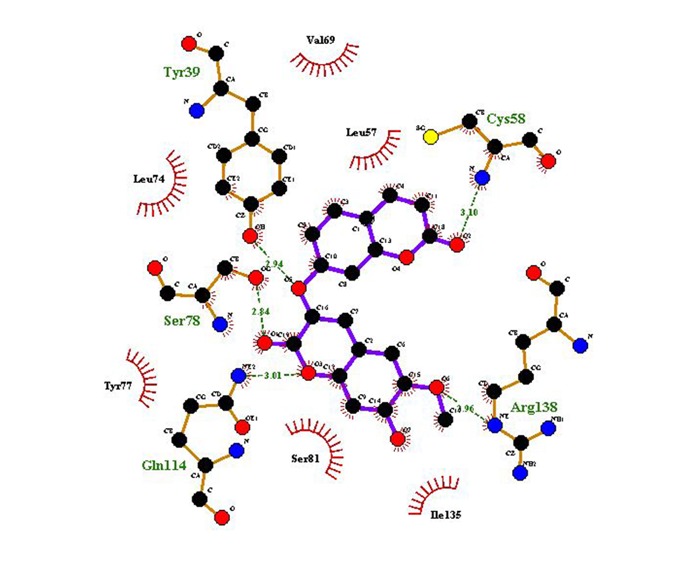
2D representation of 3D structure of Daphnoretin with E6 protein of HPV-16 created by Ligplot, showing H- bond interaction with CYS58, TYR39, SER78, GLN114, ARG138, CYS58 and Hydrophobic interactions with LEU74, VAL69, LEU57, TYR77, SER81, ILE135.
Conclusion
The inhibitory effect of natural compounds against E6 protein has been studied by few researchers. Advancements in computational chemistry and bioinformatics are very useful for the investigation of novel inhibitors from natural sources. The E6 protein of HPV-16 inactivates p53, therefore, the process of gene regulation is disturbed, which is a fundamental cause of cervical cancer. Thus, E6 of HPV-16 is of considerable interest for discovery and designing of novel molecule to overcome the challenges. A high-quality three-dimensional model of E6 obtained through in silico approach and molecular docking were performed using AutoDock vina. Daphnoretin was predicted to be the most potent inhibitor amongst all known selected natural compounds that may lead to the inhibition of E6 protein. In vitro and in vivo experimentaion are needed to confirm its efficacy and potency. Further it can be used for design candidate drug with desired biological properties by chemical modification in functional group at appropriate places.
Supplementary material
Acknowledgments
The authors would like to thank Department of Biotechnology, G. B. Pant Engineering College, Pauri Garhwal, Uttarakhand, India for providing all necessary facilities. One of the authors, Saril Mamgain gratefully acknowledges Technical Education Quality Improvement Programme (TEQIP), G. B. Pant Engineering College, Pauri Garhwal, India for fellowship.
Footnotes
Citation:Mamgain et al, Bioinformation 11(5): 236-242 (2015)
References
- 1.Arundhati B, et al. J Nat Sci Biol Med. 2012;3:186. [Google Scholar]
- 2.GY Ho, et al. J Natl Cancer Inst. 1995;87:1365. doi: 10.1093/jnci/87.18.1365. [DOI] [PubMed] [Google Scholar]
- 3.Guan P, et al. Int J Cancer. 2012;131:2349. doi: 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- 4.Matlashewski G, et al. EMBO J. 1987;6:1741. doi: 10.1002/j.1460-2075.1987.tb02426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vousden K. FASEB J. 1993;7:872. doi: 10.1096/fasebj.7.10.8393818. [DOI] [PubMed] [Google Scholar]
- 6.Rangarajan A, et al. Virology. 2001;286:23. doi: 10.1006/viro.2001.0867. [DOI] [PubMed] [Google Scholar]
- 7.Kiyono T, et al. Proc Natl Acad Sci U S A. 1997;94:11612. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bharti AC, et al. Indian J Med Res. 2009;130:296. [PubMed] [Google Scholar]
- 9.Dubey K, et al. Process Biochem. 2008;43:251. [Google Scholar]
- 10.Larkin MA, et al. Bioinformatics. 2007;23:2947. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 11.Goel A, et al. Biochem Pharmacol. 2008;75:787. doi: 10.1016/j.bcp.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Lu, et al. Food Chem. 2011;124:1500. [Google Scholar]
- 13.Kuo, et al. Life Sciences. 2006;78:2550. doi: 10.1016/j.lfs.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 14.Batra P, Sharma A. 3 Biotech. 2013;3:439. doi: 10.1007/s13205-013-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corcoran MP, et al. J Nutr Gerontol Geriatr. 2012;31:176. doi: 10.1080/21551197.2012.698219. [DOI] [PubMed] [Google Scholar]
- 16.Tamura K, et al. Mol Biol Evol. 2013;30:2725. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larkin MA, et al. Bioinformatics. 2007;23:2947. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 18.Trott O, et al. J Comput Chem. 2010;31:455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laskowski RA, et al. J Chem Inf Model. 2011;51:2778. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 20.Pathak RK, et al. Bioinformation. 2014;10:428. doi: 10.6026/97320630010428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ambrish R, et al. Nat Protoc. 2010;5:725. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


