Abstract
Purpose
We hypothesized that common polymorphisms in excision repair cross-complementation group 1 (ERCC1), involved in nucleotide excision repair of platinum-induced damage, would be associated with progression-free survival (PFS) and overall survival (OS) in women with optimally resected, stage III epithelial ovarian cancer (EOC) treated with cisplatin and paclitaxel (C + P).
Patients and Methods
Single nucleotide polymorphism analysis was carried out by direct pyrosequencing at two sites (codon 118 and C8092A) in ERCC1 in leukocyte DNA from women who participated in the Gynecologic Oncology Group (GOG) phase III protocol-172 and were randomly assigned to intraperitoneal or intravenous C + P
Results
ERCC1 genotyping was performed in 233 of the 429 women who participated in GOG-172. The genotype distribution at codon 118 was 17% with C/C, 43% with C/T, and 40% with T/T, and the genotype distribution at C8092A was 56% with C/C, 37% with C/A, and 7% with A/A. Adjusted Cox regression analysis revealed that the codon 118 polymorphism in ERCC1 was not significantly associated with disease progression or death. Women with the C8092A C/A or A/A genotypes compared with the C/C genotype had an increased risk of disease progression (hazard ratio [HR] = 1.44; 95% CI, 1.06 to 1.94; P = .018) and death (HR = 1.50; 95% CI, 1.07 to 2.09; P = .018). Median PFS and OS were 6 and 17 months shorter for women with the C8092A C/A or A/A genotypes versus the C/C genotype, respectively.
Conclusion
Although the ERCC1 codon 118 polymorphism does not seem to be associated with clinical outcome, the C8092A polymorphism was an independent predictor of PFS and OS in women with optimally resected EOC.
INTRODUCTION
Epithelial ovarian cancer (EOC) is the leading cause of death in the United States in women diagnosed with gynecologic malignancies, with 21,650 new cases and 15,520 women estimated to die of ovarian cancer in 2008.1 The standard treatment for EOC includes staging laparotomy with cytoreduction followed by platinum/taxane-based chemotherapy.2–14 Despite impressive initial response rates, 5-year survival for this patient population remains approximately 30% to 50%.1,8–11 Patients with either platinum-refractory EOC who do not respond to initial cytotoxic chemotherapy or platinumresistant EOC who develop recurrent disease within 6 months of completion of adjuvant therapy have the worst prognosis. Identification of patients who are less responsive to platinum-based chemotherapy would permit treatment decisions tailored to the individual and allow for selection of novel agents and drug combinations that would hopefully have increased efficacy, reduced adverse effects, better quality of life, and long-term benefit. Therefore, any biologic or genetic markers that could identify women at risk for platinum-refractory or platinum-resistant disease would have immediate clinical utility.
Platinum agents induce formation of interstrand and intrastrand DNA cross-links. These adducts are recognized and repaired by the nucleotide excision repair pathway. Cells that have a robust nucleotide excision repair mechanism have a greater likelihood of repairing DNA lesions and surviving a platinum challenge. Thus, functional variants in genes involved in the DNA repair pathway may be important determinants of platinum response in women with platinum-sensitive, -resistant, or -refractory EOC. One of the genes in this pathway, excision repair cross-complementation group 1 (ERCC1), seems to play a significant role in platinum-DNA adduct repair. Expression levels of ERCC1 correlate strongly with response to platinum-based therapy.15–18 Increased ERCC1 mRNA levels in ovarian tumors resulted in decreased platinum sensitivity,19 and downregulation of ERCC1 expression, with antisense ERCC1 RNA, seemed to increase the sensitivity of the highly resistant ovarian cancer cell line OVCAR10 to cisplatin.20
Several common polymorphisms of the ERCC1 gene with proposed functional effects have been identified. The codon 118 C/T polymorphism is thought to affect mRNA levels21 and showed a significant association with overall survival (OS)22,23 and tumor response24 in advanced colorectal cancer patients. However, the codon 118 polymorphism was not associated with OS in melanoma,25 lung cancer,26 or ovarian cancer27 patients but was an independent predictor of reduced risk of platinum resistance, which was defined as disease recurrence within 6 months from the completion of chemotherapy in ovarian cancer.27 A second polymorphism, C8092A, located in the 3′ untranslated region, is thought to affect mRNA stability.28 The C8092A polymorphism was associated with more favorable outcomes in head and neck squamous cell carcinoma patients29 and better OS in advanced non–small-cell lung cancer patients.26
The use of ERCC1 genotyping to predict response to platinum-based chemotherapy and survival in epithelial cancers has not always produced consistent results. The aim of this current study was to evaluate associations between two common genetic variants in the ERCC1 gene and clinical outcomes in a phase III clinical trial of patients with optimally resected, stage III EOC treated with cisplatin and paclitaxel (C+P) administered via the intravenous (IV) versus intraperitoneal (IP) route conducted by the Gynecologic Oncology Group (GOG). By comparing the results between IV and IP groups in this relatively homogenous patient population who were consistently staged, treated, and evaluated, we could assess whether any observed associations were modified by the route of drug delivery. The influence of the route for chemotherapy administration is clinically relevant, and its relationship to genotype variations has never been studied.
PATIENTS AND METHODS
Study Population
Patients who participated in GOG-172 and provided blood specimens for translational research (TR) were included in this study. GOG-172 was a phase III randomized trial of IV versus IP C+P in patients with optimally resected, stage III EOC or primary peritoneal carcinoma. Details regarding eligibility criteria, treatment, and clinical outcomes have been previously published.14 In brief, patients with no residual mass ≥ 1.0 cm after surgery were randomly assigned to receive either 135 mg of IV paclitaxel per square meter (m2) of body-surface area over a 24-hour period followed by either 75 mg of IV cisplatin/m2 on day 2 (IV arm) or 100 mg of IP cisplatin/m2 on day 2 and 60 mg of IP paclitaxel/mm2 on day 8 (IP arm). Treatment was administered every 3 weeks for six cycles. Patients provided written informed consent to participate in GOG-172 and provided a blood specimen for TR consistent with all federal, state, and local requirements before enrollment onto the study.
Isolation of DNA
DNA was extracted from WBCs recovered from whole blood using the Puregene DNA purification kit (GentraSystems Inc, Minneapolis, MN) or the ABI PRISM 6100 Nucleic Acid Prep Station (Applied Biosystems Inc, Foster City, CA).30
Genotyping
The ERCC1 codon 118 and C8092A polymorphisms were detected by polymerase chain reactions (PCR), followed by pyrosequencing. For codon 118, a 413-base pair region was amplified in a standard PCR mixture of template DNA, a biotin-labeled forward primer 5′/5Bio/GTG-CGA-GGA-GGC-AGG-AGG-TGT-GGG-3′, and the reverse primer 5′-TGT-TGC-ACT-GGG-CAC-CTC-CAG-GCC-3′ (IDT DNA, Coralville, IA). A 255-base pair region for C8092A was amplified in a PCR mixture of template DNA, forward primer 5′/TGA-GCC-AAT-TCA-GCC-ACT-3, and a biotin-labeled reverse primer 5′-/5Bio/TAG-TTC-CTC-AGT-TTC-CCG-3. The sequencing primer for codon 118 was 5′-ACG-TCG-CCA-AAT-TCC-CAG-GG-3′, and the primer for C8092A was 5′/AGG-CCG-GGA-CAA-GAA-GCG-GA-3. Pyrosequencing was completed using the PSQ96 MA and the SQA reagent kit (Biotage, Uppsala, Sweden).
Statistical Analysis
Clinical and follow-up data were prospectively collected as required by the protocol. Progression-free survival (PFS) was the time from study entry until disease recurrence or death, whichever came first. OS was the time from study entry until death regardless of cause. Associations between ERCC1 polymorphisms and clinical characteristics were evaluated using Pearson’s χ2 test or Fisher’s exact test. The Kaplan-Meier method was used to estimate PFS and OS by genotype, and the log-rank test was used to compare the survival distributions. Associations between ERCC1 polymorphisms and PFS and OS were evaluated using Cox proportional hazards analyses using reduced models adjusted for histology (clear cell/mucinous v other histologic subtypes), residual disease status (gross v none or microscopic tumor), and treatment arm (IP v IV). These covariates were chosen based on their documented prognostic relevance in a GOG meta-analysis in this patient population31 and univariate Cox modeling in this cohort. Additional Cox modeling was performed with adjustments for patient age, race, performance status, histologic cell type, tumor grade, residual disease status, and treatment regimen. The results and conclusions from the full models were similar to those obtained using the reduced models. Because of the small number of patients with A/A genotype for ERCC1 C8092A polymorphism, C/A and A/A genotypes were combined in the analysis, as suggested in other studies.26,32 The subgroup analysis in IV-versus IP-treated patients was exploratory. All statistical testing was two-sided and performed using SAS Version 9.1 (SAS Institute, Cary, NC).
RESULTS
Of the 429 women enrolled onto GOG-172, 371 provided a satisfactory blood specimen for TR. Leukocyte DNA was prepared and used to examine mutations in BRCA1 (unpublished data) and CHEK2.30 Only 233 women had sufficient DNA left over for genotyping codon 118 and the C8092A region of the ERCC1 gene. Patient characteristics for the 233 women in this cohort are listed in Table 1 and are representative of those observed in the entire GOG-172 cohort.14 Median age of the participants at enrollment was 56.9 years, most of the patients (91.9%) were white, and 93.2% of the patients had a GOG performance status of 0 to 1. The majority of tumors (76.8%) were serous histology, and most patients (58%) had gross residual disease at the completion of surgery. Women were randomly allocated to receive IV C+P (54.5%) or IP C+P (45.5%; Table 1). At the time of the analysis, the median follow-up time for those still alive was 75 months (range, 10 to 101 months); 57 women were alive with no evidence of disease, 34 women were alive with documented recurrence/disease progression, and 142 women died. The cause of death was disease progression in 113 patients, treatment in three patients, both disease progression and treatment in two patients, and other reasons in 24 patients.
Table 1.
Clinical Characteristics
| All Patients (N = 233)
|
Patients on IP Arm (n = 106)
|
Patients on IV Arm (n = 127)
|
||||
|---|---|---|---|---|---|---|
| Characteristic | No. | % | No. | % | No. | % |
| Age, years | ||||||
| ≤ 55 | 104 | 44.6 | 48 | 45.3 | 56 | 44.1 |
| 56–65 | 63 | 27.0 | 29 | 27.4 | 34 | 26.8 |
| > 65 | 66 | 28.3 | 29 | 27.4 | 37 | 29.1 |
| Median | 57 | 57 | 57 | |||
| Range | 32–83 | 32–83 | 32–78 | |||
|
| ||||||
| Race | ||||||
| White | 214 | 91.9 | 96 | 90.6 | 118 | 92.9 |
| Black | 7 | 3.0 | 4 | 5.7 | 3 | 2.4 |
| Other | 12 | 5.2 | 6 | 3.8 | 6 | 4.7 |
|
| ||||||
| GOG performance status | ||||||
| 0 | 102 | 43.8 | 45 | 42.5 | 57 | 44.9 |
| 1 | 115 | 49.4 | 49 | 46.2 | 66 | 52.0 |
| 2 | 16 | 6.9 | 12 | 11.3 | 4 | 3.2 |
|
| ||||||
| Histologic cell type | ||||||
| Serous adenocarcinoma | 179 | 76.8 | 79 | 74.5 | 100 | 78.7 |
| Endometrioid adenocarcinoma | 15 | 6.4 | 9 | 8.5 | 6 | 4.7 |
| Clear cell carcinoma | 14 | 6.0 | 7 | 6.6 | 7 | 5.5 |
| Mucinous adenocarcinoma | 1 | 0.4 | 1 | 0.9 | 0 | 0.0 |
| Other adenocarcinomas | 24 | 10.3 | 10 | 9.4 | 14 | 11.0 |
|
| ||||||
| Tumor grade | ||||||
| 1 | 23 | 9.9 | 13 | 12.3 | 10 | 7.9 |
| 2 | 93 | 39.9 | 43 | 40.6 | 50 | 39.4 |
| 3 or clear cell | 117 | 50.2 | 50 | 47.2 | 67 | 52.8 |
|
| ||||||
| Residual disease | ||||||
| Microscopic tumor | 97 | 41.6 | 44 | 41.5 | 53 | 41.7 |
| Gross tumor | 136 | 58.4 | 62 | 58.5 | 74 | 58.3 |
Abbreviations: IV, intravenous cisplatin and paclitaxel; IP, intraperitoneal cisplatin and paclitaxel; GOG, Gynecologic Oncology Group.
Among the 233 eligible patients, the genotype distribution at codon 118 was 17.2% with C/C, 43.4% with C/T, and 39.5% with T/T, and the genotype distribution at C8092A was 56.2% with C/C, 36.9% with C/A, and 6.9% with A/A. Both distributions were in Hardy-Weinberg equilibrium. There were no associations between codon 118 or C8092A polymorphisms in ERCC1 and patient age, tumor grade, histology, tumor residual volume, or treatment regimen (Table 2). Although only a limited number of African American women were enrolled onto GOG-172 and provided specimens for this project (n = 7), all of these patients had the C/C genotype for codon 118 (P < .001), and five of them had the C/A or A/A genotype in the C8092A polymorphism in the ERCC1 gene (P = .08). Table 2 illustrates the strong association between the codon 118 genotypes in ERCC1 and the C8092A genotypes in ERCC1 (P < .001). Specifically, 85% of the women with a C/C genotype in codon 118 exhibited the C/A or A/A genotypes in C8092A, and 97% of the women with the T/T genotype in codon 118 displayed the C/C genotype in C8092A.
Table 2.
Association Between Clinical Characteristics and ERCC1 Polymorphisms
| Codon 118 in ERCC1
|
C8092A in ERCC1
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C/C
|
C/T
|
T/T
|
C/C
|
C/A or A/A
|
||||||||
| Characteristic | No. of Patients | % | No. of Patients | % | No. of Patients | % | P | No. of Patients | % | No. of Patients | % | P |
| Age | .576 | .985 | ||||||||||
| ≤ 55 years | 17 | 16.4 | 44 | 42.3 | 43 | 41.4 | 58 | 55.8 | 46 | 44.2 | ||
| 56–65 years | 8 | 12.7 | 31 | 49.2 | 24 | 38.1 | 36 | 57.1 | 27 | 42.9 | ||
| > 65 years | 15 | 22.7 | 26 | 39.4 | 25 | 37.9 | 37 | 56.1 | 29 | 43.9 | ||
|
| ||||||||||||
| Race | < .001 | .080 | ||||||||||
| White | 30 | 14.0 | 93 | 43.5 | 91 | 42.5 | 125 | 58.4 | 89 | 41.6 | ||
| Black | 7 | 100.0 | 0 | 0.0 | 0 | 0.0 | 2 | 28.6 | 5 | 71.40 | ||
| Other | 3 | 25.0 | 8 | 66.7 | 1 | 18.3 | 4 | 33.3 | 8 | 66.7 | ||
|
| ||||||||||||
| GOG performance status | .678 | .736 | ||||||||||
| 0 | 15 | 14.7 | 44 | 43.1 | 43 | 42.2 | 59 | 57.8 | 43 | 42.2 | ||
| 1 | 23 | 20.0 | 51 | 44.4 | 41 | 35.7 | 62 | 53.9 | 53 | 46.1 | ||
| 2 | 2 | 12.5 | 6 | 37.5 | 8 | 50.0 | 10 | 62.5 | 6 | 37.5 | ||
|
| ||||||||||||
| Histologic cell type | .407 | .466 | ||||||||||
| Serous adenocarcinoma | 29 | 16.2 | 76 | 42.5 | 74 | 41.3 | 98 | 54.8 | 81 | 45.3 | ||
| Endometrioid adenocarcinoma | 5 | 33.3 | 8 | 53.3 | 2 | 13.3 | 8 | 53.3 | 7 | 46.7 | ||
| Clear cell carcinoma | 2 | 14.3 | 7 | 50.0 | 5 | 35.7 | 8 | 57.1 | 6 | 42.9 | ||
| Mucinous adenocarcinoma | 0 | 0.0 | 1 | 100.0 | 0 | 0.0 | 0 | 0.0 | 1 | 100.0 | ||
| Other adenocarcinomas | 4 | 16.7 | 9 | 37.5 | 11 | 45.8 | 17 | 70.8 | 7 | 29.2 | ||
|
| ||||||||||||
| Tumor grade | .788 | .997 | ||||||||||
| 1 | 4 | 17.4 | 9 | 39.1 | 10 | 43.5 | 13 | 56.5 | 10 | 43.5 | ||
| 2 | 17 | 18.3 | 44 | 47.3 | 32 | 34.4 | 52 | 55.9 | 41 | 44.1 | ||
| 3 or clear cell | 19 | 16.2 | 48 | 41.0 | 50 | 42.7 | 66 | 56.4 | 51 | 43.6 | ||
|
| ||||||||||||
| Residual disease | .961 | .695 | ||||||||||
| Microscopic tumor | 17 | 17.5 | 41 | 42.3 | 39 | 40.2 | 56 | 57.7 | 41 | 42.3 | ||
| Gross tumor | 23 | 16.9 | 60 | 44.1 | 53 | 39.0 | 75 | 55.2 | 61 | 44.9 | ||
|
| ||||||||||||
| Treatment regimen | .404 | .710 | ||||||||||
| IV arm | 23 | 18.1 | 50 | 39.4 | 54 | 42.5 | 70 | 55.1 | 57 | 44.9 | ||
| IP arm | 17 | 16.0 | 51 | 48.1 | 38 | 35.9 | 61 | 57.6 | 45 | 42.5 | ||
|
| ||||||||||||
| Codon 118 in ERCC1 | < .001 | |||||||||||
| C/C | 6 | 15.0 | 34 | 85.0 | ||||||||
| C/T | 36 | 35.6 | 65 | 64.4 | ||||||||
| T/T | 89 | 96.7 | 3 | 3.3 | ||||||||
|
| ||||||||||||
| C8092A in ERCC1 | < .001 | |||||||||||
| C/C | 6 | 4.6 | 36 | 27.5 | 89 | 67.9 | ||||||
| C/A or A/A | 34 | 33.3 | 65 | 63.7 | 3 | 2.9 | ||||||
NOTE. Fisher’s exact test for data in 2 × 2 tables and Pearson χ2 test for data in R × C tables.
Abbreviations: GOG, Gynecologic Oncology Group; IV, intravenous cisplatin and paclitaxel; IP, intraperitoneal cisplatin and paclitaxel.
There was no evidence of a statistically significant difference in either PFS or OS among women with the C/T or T/T genotype in codon 118 of the ERCC1 gene compared with women with the C/C genotype (Figs 1A and 1B; Table 3). These results were consistent with those obtained using a Cox regression model for PFS or OS with adjustments for the prognostic factors (histologic cell type, residual disease status, and treatment) in this patient population (Table 4). Similar results were obtained with the full Cox model described in Patients and Methods (Appendix Table A1, online only). There were no differences in risk of disease progression or death by codon 118 genotype for women treated with C+P administered IP or IV (Table 4).
Fig 1.
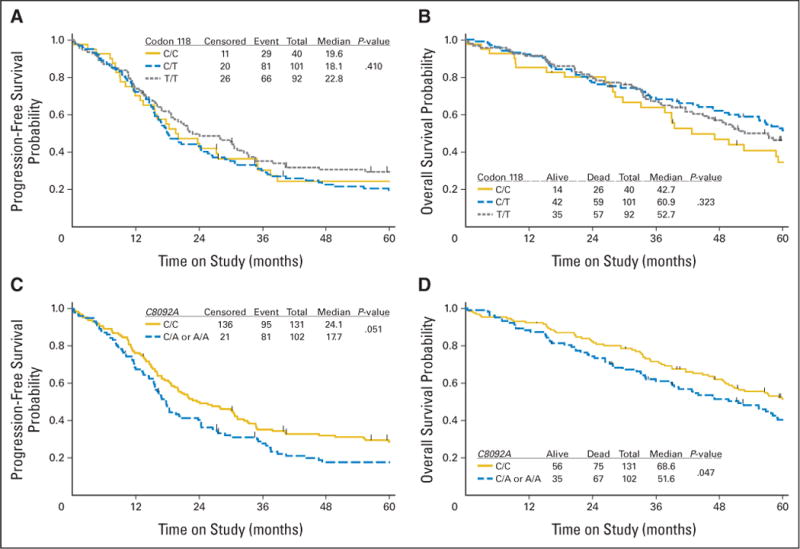
Kaplan-Meier estimates of (A, C) progression-free survival (PFS) and (B, D) overall survival (OS) in the entire cohort categorized by polymorphisms in (A, B) codon 118 and (C, D) C8092A in the ERCC1 gene. Censored indicates women who were alive with no evidence of disease progression at last contact, and event reflects women with documented recurrence/disease progression or death.
Table 3.
Median PFS and OS Time by ERCC1 Polymorphisms
| ERCC1 Genotype | No. of Patients | Median PFS (months) | P | Median OS (months) | P |
|---|---|---|---|---|---|
| All participants | |||||
| Codon 118 in ERCC1 | .410 | .323 | |||
| C/C | 40 | 19.6 | 42.7 | ||
| C/T | 101 | 18.1 | 60.9 | ||
| T/T | 92 | 22.8 | 52.7 | ||
| C8092A in ERCC1 | .051 | .047 | |||
| C/C | 131 | 24.1 | 68.6 | ||
| C/A or A/A | 102 | 17.7 | 51.6 | ||
|
| |||||
| IP arm subset | |||||
| Codon 118 in ERCC1 | .751 | .385 | |||
| C/C | 17 | 23.8 | 42.7 | ||
| C/T | 51 | 18.4 | 66.9 | ||
| T/T | 38 | 24.1 | 68.6 | ||
| C8092A in ERCC1 | .038 | .046 | |||
| C/C | 61 | 27.9 | 81.4 | ||
| C/A or A/A | 45 | 18.3 | 52.6 | ||
|
| |||||
| IV arm subset | |||||
| Codon 118 in ERCC1 | .440 | .725 | |||
| C/C | 23 | 17.8 | 47.1 | ||
| C/T | 50 | 16.9 | 59.9 | ||
| T/T | 54 | 20.9 | 50.4 | ||
| C8092A in ERCC1 | .465 | .333 | |||
| C/C | 70 | 20.3 | 56.5 | ||
| C/A or A/A | 57 | 17.7 | 49.7 | ||
NOTE. PFS and OS were estimated by Kaplan-Meier method, and median times are provided in months from enrollment. Log-rank test was used to compare survival distributions between groups.
Abbreviations: PFS, progression-free survival; OS, overall survival; IP, intraperitoneal cisplatin and paclitaxel; IV, intravenous cisplatin and paclitaxel.
Table 4.
Reduced Multivariate Cox Regression Analysis for ERCC1 Polymorphisms and Clinical Outcome
| Polymorphism and Genotype | No. of Patients | PFS
|
OS
|
||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||
| All participants*† | 233 | ||||||
| Codon 118 in ERCC1 | .300 | .135 | |||||
| C/C | 40 | 1.00 | 1.00 | ||||
| C/T | 101 | 1.00 | 0.66 to 1.54 | 0.62 | 0.39 to 0.99 | ||
| T/T | 92 | 0.79 | 0.51 to 1.22 | 0.72 | 0.45 to 1.16 | ||
| C8092A in ERCC1 | .018 | .018 | |||||
| C/C | 131 | 1.00 | 1.00 | ||||
| C/A or A/A | 102 | 1.44 | 1.06 to 1.94 | 1.50 | 1.07 to 2.09 | ||
|
| |||||||
| IP arm subset†‡ | 106 | ||||||
| Codon 118 in ERCC1 | .713 | .139 | |||||
| C/C | 17 | 1.00 | 1.00 | ||||
| C/T | 51 | 0.84 | 0.43 to 1.63 | 0.49 | 0.24 to 1.00 | ||
| T/T | 38 | 0.75 | 0.37 to 1.50 | 0.55 | 0.26 to 1.16 | ||
| C8092A in ERCC1 | .012 | .011 | |||||
| C/C | 61 | 1.00 | 1.00 | ||||
| C/A or A/A | 45 | 1.81 | 1.14 to 2.88 | 1.96 | 1.17 to 3.30 | ||
|
| |||||||
| IV arm subset†‡ | 127 | ||||||
| Codon 118 in ERCC1 | .394 | .637 | |||||
| C/C | 23 | 1.00 | 1.00 | ||||
| C/T | 50 | 1.12 | 0.64 to 1.98 | 0.74 | 0.40 to 1.38 | ||
| T/T | 54 | 0.83 | 0.47 to 1.47 | 0.82 | 0.44 to 1.49 | ||
| C8092A in ERCC1 | .350 | .295 | |||||
| C/C | 70 | 1.00 | 1.00 | ||||
| C/A or A/A | 57 | 1.21 | 0.81 to 1.80 | 1.27 | 0.81 to 1.97 | ||
Abbreviations: PFS, progression-free survival; OS, overall survival; HR, hazard ratio; IP, intraperitoneal cisplatin and paclitaxel; IV, intravenous cisplatin and paclitaxel.
Adjustments were made for the prognostic factors including histology cell type (clear cell/mucinous v other histologic subtypes), residual disease status (macroscopic v none or microscopic), and treatment arm (IP v IV) in this patient population.
Similar results and conclusions were observed using full multivariate Cox models for PFS and OS, with adjustments for patient age, race, Gynecologic Oncology Group performance status, histologic cell type, tumor grade, residual disease status, and treatment arm, and reduced multivariate Cox models for PFS and OS with the two ERCC1 polymorphisms and adjustments for histologic cell type, residual disease status, and treatment arm.
Adjustments were made for histology cell type (clear cell/mucinous v other histologic subtypes) and residual disease status (macroscopic v none or microscopic) in women randomly assigned to the IP or IV treatment arm. Testing was performed to determine whether there was statistical evidence of an interaction between the codon 118 or the C8092A polymorphism in ERCC1 and treatment in this cohort (P > .05).
Analysis of the C8092A polymorphism in ERCC1 demonstrated an association with prognosis (Figs 1C and 1D; Tables 3 and 4). When compared with women exhibiting the C8092A C/C genotype, women carrying at least one A allele (C/A or AA) had a 6-month shorter median PFS time and a 17-month shorter median OS time (Table 3). Kaplan-Meier plots illustrate the differences in PFS (Fig 1C; P = .051) and OS (Fig 1D; P = .047) distributions for women categorized by C8092A polymorphisms in ERCC1. After adjusting for the prognostic factors in this patient population (histologic cell type, residual disease status, and treatment arm), women with the C/A or A/A genotype at C8092A had an increased risk of disease progression (hazard ratio [HR] = 1.44; 95% CI, 1.06 to 1.94; P = .018) and death (HR = 1.50; 95% CI, 1.07 to 2.09; P = .018) compared with women with the C/C genotype (Table 4). Similar results were obtained with the full Cox model described in Patients and Methods (Appendix Table A1) and when both ERCC1 polymorphisms were included in reduced multivariate Cox models (data not shown).
Next, we explored whether the association between C8092A genotype in the ERCC1 gene and clinical outcome was modified by the route of drug delivery. Of the 233 women in this cohort, 106 and 127 women were randomly assigned to the IP or IV arm, respectively. There were no differences in the clinical characteristics in these subgroups (Table 1). Subset analysis stratified by treatment regimen demonstrated a distinct PFS (Fig 2A) and OS (Fig 2B) advantage for women with the C8092A C/C genotype compared with women with either the C/A or A/A genotype in patients randomly allocated to the IP treatment arm (Figs 2C and 2D). Adjusted Cox regression analysis (Table 4) suggested that women with a C/A or A/A genotype, compared with a C/C genotype, had a significantly higher risk of disease progression (HR = 1.81; 95% CI, 1.14 to 2.88) and death (HR = 1.96; 95% CI, 1.17 to 3.30) when randomly assigned to the IP arm versus the IV arm (PFS: HR = 1.21; 95% CI, 0.81 to 1.80; OS: HR = 1.27; 95% CI, 0.81 to 1.97). Although the subset analysis suggested that the effect of the C/C genotype was more evident for IP patients, this study was underpowered to evaluate an interaction between genotype and treatment arm.
Fig 2.
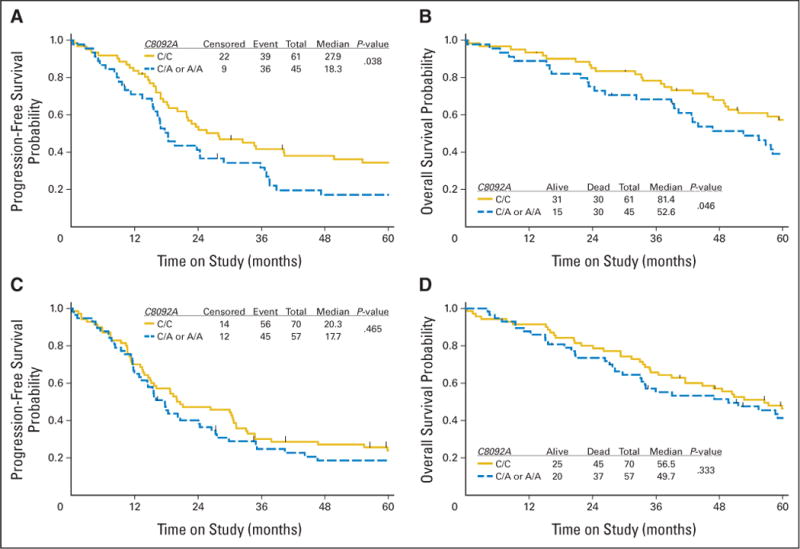
Kaplan-Meier estimates of (A, C) progression-free survival and (B, D) overall survival in the subset of women randomly assigned to the (A, B) intraperitoneal (IP) arm or (C, D) intravenous arm and categorized by polymorphisms in C8092A in the ERCC1 gene. Censored indicates women who were alive with no evidence of disease progression at last contact, and event reflects women with documented recurrence/disease progression or death.
DISCUSSION
Common functional genetic polymorphisms in ERCC1 exist in the population in exon sites (codon 118), as well as in the 3′ untranslated region (C8092A). Both variants are thought to affect ERCC1 levels and have been associated with clinical outcomes in patients treated with various platinum analogs. Polymorphisms at codon 118 have been studied extensively, with variable results. Our study indicates that the ERCC1 codon 118 C/T polymorphism was not associated with differences in PFS or OS in women with optimally resected stage III EOC. C/C genotype was associated with better outcome in patients with advanced colorectal cancer,23 patients with refractory colorectal cancer,33 and two of three studies in lung cancer patients,26,34,35 whereas the T/T genotype was associated with better tumor response in advanced colorectal carcinomas.24 In addition, melanoma patients treated with cisplatin had a less favorable response when carrying the C/C genotype.25 Kang et al27 studied a small series of ovarian cancer patients and reported that the codon 118 polymorphism in ERCC1 was an independent predictor of reduced risk of platinum resistance, which was defined as disease recurrence within 6 months from the completion of chemotherapy, but was not associated with OS. Smith et al36 demonstrated that the C/C genotype in codon 118 of ERCC1 was associated with an increased risk of disease progression (HR = 1.95, P = .051) and death (HR = 2.01, P = .033) in women with EOC treated with platinum without paclitaxel but not in EOC patients treated with platinum and paclitaxel, suggesting that the influence of the codon 118 genotype on the responsiveness of platinum therapy may be minimized by the addition of a taxane, which targets tubulin rather than DNA. The 233 women in our cohort were all treated with C+P, which may explain why we did not see a significant association between the codon 118 polymorphisms in ERCC1 and either PFS or OS. Finally, Marsh et al37 did not find any evidence of an association between the codon 118 polymorphism in ERCC1 and PFS, CA125 response, or clinical/radiographic response in women with EOC treated with carboplatin and paclitaxel or docetaxel.
The C8092A polymorphism in ERCC1 has not been studied as extensively as the codon 118 variant. In a study of 128 patients with advanced non–small-cell lung cancer, Zhou et al26 demonstrated a significant association between the C/C genotype and OS. Consistent with this finding, we observed a significant association between the C8092A polymorphism in ERCC1 and OS. We also observed a statistically significant association between the C8092A polymorphism in ERCC1 and PFS. Marsh et al37 did not find any statistical evidence of an association between this ERCC1 polymorphism and PFS, CA125 response, or clinical/radiographic response in EOC.
Inconsistencies in the reported data for the codon 118 and C8092A genotypes in ERCC1 could be attributable, at least in part, to differences in tumor biology, cancer types, stage of disease, responsiveness to C+P, study design, and sample size between the published studies. It is also possible that the ERCC1 polymorphisms do not directly affect treatment outcomes, but rather are in linkage disequilibrium with another causative locus. The possibility of increased chemotherapy toxicity leading to this inconsistency in outcomes was also evaluated. There was no association between ERCC1 polymorphisms and common grade 3 or 4 adverse effects (data not shown).
To our knowledge, this is the first study to demonstrate a 6-month and 17-month median PFS and OS advantage, respectively, in EOC patients with the C/C genotype compared with the C/A or A/A genotypes in the C8092A region of the ERCC1 gene and to demonstrate that the C8092A polymorphism is an independent prognostic factor for PFS and OS in optimally resected, stage III EOC. There was great interest in assessing whether the association between genotype and clinical outcome was modified by the route of drug delivery. Although this study was not powered to evaluate an interaction between ERCC1 genotype and treatment arm, exploratory analyses were performed to prioritize future studies. An exploratory subset analysis stratified by treatment provided suggestive evidence that the associations between the C8092A polymorphism and clinical outcome were most pronounced in the IP arm.
Median PFS and OS times for women with the C8092A C/C genotype were 8 and 25 months longer, respectively, for women on IP versus IV therapy (Table 3). In contrast, median PFS and OS times for women with at least one A allele in C8092A were similar for women on IP versus IV therapy (Table 3). A larger study is required to validate the observation that women with the C8092A C/C genotype had a significant PFS and OS advantage when treated with IP versus IV C+P, whereas women with the C/A or A/A genotype had similar risks for disease progression and death when treated with IP versus IV C+P. If these associations are confirmed, testing for the C/C genotype in the C8092A region of the ERCC1 gene may serve as a potential prescreening test for women contemplating IP therapy, enabling clinicians and patients to make more informed treatment management decisions. It may be that the effects of variant genotypes are mild, and only with differential drug distribution and higher levels of drug at the tumor site do you observe an association between the C/C genotype and phenotype (clinical outcome). It is also possible that the C8092A C/C genotype in ERCC1 may be in linkage disequilibrium with another causative locus. Alternatively, the effects of C/C genotype may be enhanced in women who have a better prognosis as a result of the treatment arm, and thus, their survival may be affected by factors other than treatment (eg, younger age and better performance status, which may improve patients’ tolerance of chemotherapy and the IP catheter). The results of the primary analysis in all participants and the exploratory subset analysis in women randomly assigned to IP versus IV C+P therapy are intriguing, but larger studies are required to validate these associations, and mechanistic studies are needed to ascertain the nature of this relationship (eg, whether the C8092A C/C genotype alters cisplatin sensitivity, is associated with another prognostic factor through linkage, or is preferentially observed in women who can tolerate six cycles of IP therapy).
Supplementary Material
Acknowledgments
We are indebted to Mary Strange for excellent technical assistance and Anne Reardon for assistance in preparing this manuscript for publication. Special acknowledgments go to Brian Bundy, PhD, for his work on GOG-172 and to Suzanne Baskerville for coordinating the clinical data for GOG-172. Finally, we thank Heather Lankes, PhD, and the GOG Publications Subcommittee for their critical review of the manuscript and helpful suggestions.
Supported by National Cancer Institute Grant No. CA 27469 to the Gynecologic Oncology Group (GOG) Administrative Office and the GOG Tissue Bank and Grant No. CA 37517 to the GOG Statistical and Data Center, as well as by grants from the Gynecologic Oncology Group/Ovarian Cancer Research Fund New Investigator Award (T.C.K.), The Jennie K. Scaife Foundation (J.A.D.), and The Pittsburgh Foundation (J.A.D.).
Appendix
The Appendix is included in the full-text version of this article, available online at www.jco.org. It is not included in the PDF version (via Adobe® Reader®).
Footnotes
Presented in part at the 43rd Annual Meeting of the American Society of Clinical Oncology, June 1–5, 2007, Chicago, IL; at the 38th Annual Meeting of the Society of Gynecologic Oncologists, March 3–7, 2007, San Diego, CA; and the 39th Annual Meeting of the Society of Gynecologic Oncologists, March 9–12, 2008, Tampa, FL.
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO’s conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: None Stock Ownership: None Honoraria: None Research Funding: Kathleen M. Darcy, National Cancer Institute; Chunqiao Tian, National Cancer Institute Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Thomas C. Krivak, Kathleen M. Darcy, Chunqiao Tian, Holly Gallion, Julie A. DeLoia
Provision of study materials or patients: Deborah Armstrong, Bora E. Baysal, Holly Gallion
Collection and assembly of data: Chunqiao Tian, Julie A. DeLoia
Data analysis and interpretation: Thomas C. Krivak, Kathleen M. Darcy, Chunqiao Tian, Deborah Armstrong, Bora E. Baysal, Christine B. Ambrosone
Manuscript writing: Thomas C. Krivak, Kathleen M. Darcy, Chunqiao Tian, Deborah Armstrong, Christine B. Ambrosone, Julie A. DeLoia
Final approval of manuscript: Thomas C. Krivak, Kathleen M. Darcy, Chunqiao Tian, Deborah Armstrong, Bora E. Baysal, Holly Gallion, Julie A. DeLoia
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2007. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Colombo N, Van Gorp T, Parma G, et al. Ovarian cancer. Crit Rev Oncol Hematol. 2006;60:159–179. doi: 10.1016/j.critrevonc.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Bhoola S, Hoskins WJ. Diagnosis and management of epithelial ovarian cancer. Obstet Gynecol. 2006;107:1399–1410. doi: 10.1097/01.AOG.0000220516.34053.48. [DOI] [PubMed] [Google Scholar]
- 4.Bookman MA. Standard treatment in advanced ovarian cancer in 2005: The state of the art. Int J Gynecol Cancer. 2005;15(suppl 3):212–220. doi: 10.1111/j.1525-1438.2005.00444.x. [DOI] [PubMed] [Google Scholar]
- 5.Berkenblit A, Cannistra SA. Advances in the management of epithelial ovarian cancer. J Reprod Med. 2005;50:426–438. [PubMed] [Google Scholar]
- 6.Muggia FM. Relevance of chemotherapy dose and schedule to outcomes in ovarian cancer. Semin Oncol. 2004;31:19–24. doi: 10.1053/j.seminoncol.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Ozols RF. Update on Gynecologic Oncology Group (GOG) trials in ovarian cancer. Cancer Invest. 2004;22:11–20. doi: 10.1081/cnv-200030113. [DOI] [PubMed] [Google Scholar]
- 8.Hoskins WJ, Bundy BN, Thigpen JT, et al. The influence on cytoreductive surgery on recurrence free interval and survival in small volume stage III epithelial ovarian cancer: A Gynecologic Oncology group study. Gynecol Oncol. 1992;47:159–166. doi: 10.1016/0090-8258(92)90100-w. [DOI] [PubMed] [Google Scholar]
- 9.Hoskins WJ, McGuire WP, Brady MF, et al. The effect of largest residual disease on survival after primary cytoreductive surgery in patients with suboptimal residual ovarian carcinoma. Am J Obstet Gynecol. 1994;170:974–979. doi: 10.1016/s0002-9378(94)70090-7. [DOI] [PubMed] [Google Scholar]
- 10.McGuire V, Jesser CA, Whittmore AS. Survival among U.S. women with invasive epithelial ovarian cancer. Gynecol Oncol. 2002;84:399–403. doi: 10.1006/gyno.2001.6536. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong DK. Relapsed ovarian cancer: Challenges and management strategies for a chronic disease. Oncologist. 2002;7:20–28. doi: 10.1634/theoncologist.7-suppl_5-20. [DOI] [PubMed] [Google Scholar]
- 12.Alberts DS, Liu PY, Hannigan EV, et al. Intraperitoneal cisplatin plus intravenous cyclophosphamide versus intravenous cisplatin plus intravenous cyclophosphamide for stage III ovarian cancer. N Engl J Med. 1996;335:1950–1955. doi: 10.1056/NEJM199612263352603. [DOI] [PubMed] [Google Scholar]
- 13.Markman M, Bundy BN, Alberts DS, et al. Phase III trial of standard dose intravenous cisplatin plus paclitaxel versus moderately high dose carboplatin followed by intravenous paclitaxel and intraperitoneal cisplatin in small volume stage III ovarian carcinoma: An intergroup study of the GOG, SWOG, and ECOG. J Clin Oncol. 2001;19:1001–1007. doi: 10.1200/JCO.2001.19.4.1001. [DOI] [PubMed] [Google Scholar]
- 14.Armstrong DK, Bundy BN, Wenzel L, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 15.Lord RV, Brabender J, Gandara D, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res. 2002;8:2286–2291. [PubMed] [Google Scholar]
- 16.Joshi MBM, Shirota Y, Danenberg KD, et al. High gene expression of TS1, GSTP1, and ERCC1 are risk factors for survival in patients with trimodality therapy for esophageal cancer. Clin Cancer Res. 2005;11:2215–2221. doi: 10.1158/1078-0432.CCR-04-1387. [DOI] [PubMed] [Google Scholar]
- 17.Ceppi P, Volante M, Novello S, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17:1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 18.Bellmunt J, Paz-Ares L, Cuello M, et al. Gene expression of ERCC1 as a novel prognostic marker in advanced bladder cancer patients receiving cisplatin-based chemotherapy. Ann Oncol. 2007;18:522–528. doi: 10.1093/annonc/mdl435. [DOI] [PubMed] [Google Scholar]
- 19.Dabholkar M, Vionnet J, Bostick-Bruton F, et al. Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. J Clin Invest. 1994;94:703–708. doi: 10.1172/JCI117388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selvakumaran M, Pisarcik DA, Bao R, et al. Enhanced cisplatin cytotoxicity by disturbing the nucleotide excision repair pathway in ovarian cancer cell lines. Cancer Res. 2003;63:1311–1316. [PubMed] [Google Scholar]
- 21.Yu JJ, Lee KB, Mu C, et al. Comparison of two human ovarian carcinoma cell lines (A2780/CP70 and MCAS) that are equally resistant to platinum, but differ at codon 118 of the ERCC1 gene. Int J Oncol. 2000;16:555–560. doi: 10.3892/ijo.16.3.555. [DOI] [PubMed] [Google Scholar]
- 22.Moreno V, Gemignani F, Landi S, et al. Polymorphisms in genes of nucleotide and base excision repair: Risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–2108. doi: 10.1158/1078-0432.CCR-05-1363. [DOI] [PubMed] [Google Scholar]
- 23.Park DJ, Zhang W, Stoehlmacher J, et al. ERCC1 gene polymorphism as a predictor for clinical outcome in advanced colorectal cancer patients treated with platinum-based chemotherapy. Clin Adv Hematol Oncol. 2003;1:162–166. [PubMed] [Google Scholar]
- 24.Viguier J, Boige V, Miquel C, et al. ERCC1 codon 118 polymorphism is a predictive factor for the tumor response to oxaxiplatin/5-fluorouracil combination chemotherapy in patients with advanced colorectal cancer. Clin Cancer Res. 2005;11:6212–6217. doi: 10.1158/1078-0432.CCR-04-2216. [DOI] [PubMed] [Google Scholar]
- 25.Liu D, O’Day SJ, Yang D, et al. Impact of gene polymorphisms on clinical outcome for stage IV melanoma patients treated with biochemotherapy: An exploratory study. Clin Cancer Res. 2005;11:1237–1246. [PubMed] [Google Scholar]
- 26.Zhou W, Gurubhagavatula S, Liu G, et al. Excision repair cross-complementation group 1 polymorphism predicts overall survival in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res. 2004;10:4939–4943. doi: 10.1158/1078-0432.CCR-04-0247. [DOI] [PubMed] [Google Scholar]
- 27.Kang S, Ju W, Kim JW, et al. Association between excision repair cross-complementation group 1 polymorphism and clinical outcome of platinum-based chemotherapy in patients with epithelial ovarian cancer. Exp Mol Med. 2006;38:320–324. doi: 10.1038/emm.2006.38. [DOI] [PubMed] [Google Scholar]
- 28.Chen P, Wiencke J, Aldape K, et al. Association of an ERCC1 polymorphism with adult-onset glioma. Cancer Epidemiol Biomarkers Prev. 2000;9:843–847. [PubMed] [Google Scholar]
- 29.Quintela-Fandino M, Hitt R, Medina PP, et al. DNA-repair gene polymorphisms predict favorable clinical outcome among patients with advanced squamous cell carcinoma of the head and neck treated with cisplatin-based induction chemotherapy. J Clin Oncol. 2006;24:4333–4339. doi: 10.1200/JCO.2006.05.8768. [DOI] [PubMed] [Google Scholar]
- 30.Baysal BE, DeLoia JA, Willett-Brozick JE, et al. Analysis of CHEK2 gene for ovarian cancer susceptibility. Gynecol Oncol. 2004;95:62–69. doi: 10.1016/j.ygyno.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 31.Winter WE, Maxwell GL, Tian C, et al. Prognostic factors for stage III epithelial ovarian cancer: A Gynecologic Oncology Group study. J Clin Oncol. 2007;25:3621–3627. doi: 10.1200/JCO.2006.10.2517. [DOI] [PubMed] [Google Scholar]
- 32.Zhou W, Liu G, Park S, et al. Gene-smoking interaction associations for the ERCC1 polymorphisms in the risk of lung cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:491–496. doi: 10.1158/1055-9965.EPI-04-0612. [DOI] [PubMed] [Google Scholar]
- 33.Stoehlmacher J, Park DJ, Zhang W, et al. A multivariate analysis of genomic polymorphisms: Prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer. 2004;91:344–354. doi: 10.1038/sj.bjc.6601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryu JS, Hong YC, Han HS, et al. Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer. 2004;44:311–316. doi: 10.1016/j.lungcan.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Isla D, Sarries C, Rosell R, et al. Single nucleotide polymorphisms and outcome in docetaxelcisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15:1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 36.Smith S, Su D, Rigault de la Longrais IA, et al. ERCC1 genotype and phenotype in epithelial ovarian cancer identify patients likely to benefit from paclitaxel treatment in addition to platinum-based therapy. J Clin Oncol. 2007;25:5172–5179. doi: 10.1200/JCO.2007.11.8547. [DOI] [PubMed] [Google Scholar]
- 37.Marsh S, Paul J, King CR, et al. Pharmacogenetic assessment of toxicity and outcome after platinum plus taxane chemotherapy in ovarian cancer: The Scottish Randomised Trial in Ovarian Cancer. J Clin Oncol. 2007;25:4528–4535. doi: 10.1200/JCO.2006.10.4752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


