Abstract
Antibodies to human leukocyte antigens (HLAs) are a risk factor for acute renal allograft rejection and loss. The role of non-HLAs and their significance to allograft rejection have gained recent attention. Here, we applied protein microarray technology, with the capacity to simultaneously identify 5056 potential antigen targets, to assess non-HLA antibody formation in 15 pediatric renal transplant recipients during allograft rejection. Comparison of the pre- and post-transplant serum identified de novo antibodies to 229 non-HLA targets, 36 of which were present in multiple patients at allograft rejection. On the basis of its reactivity, protein kinase Cζ (PKCζ) was selected for confirmatory testing and clinical study. Immunohistochemical analysis found PKCζ both within the renal tissue and infiltrating lymphocytes at rejection. Patients who had an elevated anti-PKCζ titer developed rejection, which was significantly more likely to result in graft loss. The absence of C4d deposition in patients with high anti-PKCζ titers suggests that it is a marker of severe allograft injury rather than itself being pathogenic. Presumably, critical renal injury and inflammation associated with this rejection subtype lead to the immunological exposure of PKCζ with resultant antibody formation. Prospective assessment of serum anti-PKCζ levels at allograft rejection will be needed to confirm these results.
Keywords: acute rejection, pediatric kidney transplantation, renal transplantation
Although advances in allograft allocation and immunosuppression have reduced the incidence of acute rejection (AR) episodes after renal transplantation, AR remains a significant risk factor for allograft failure.1,2 Donor-specific antibodies (DSAs), are widely recognized as a risk factor, both for AR and for allograft loss.3 Recently, antibodies to non-human leukocyte antigens (non-HLAs) have been the subject of more intense scrutiny. The Collaborative Transplant Study described 4048 HLA-identical sibling transplants.4 In the course of 10 posttransplant years, a higher panel-reactive antibody was associated with significantly lower allograft survival. As these transplants involved HLA-identical siblings, the increase in allograft loss could not be attributed to DSAs. This study did not specifically detect non-HLA antibodies nor did it show causality, but it clearly established the negative impact of ‘non-HLA immunity’ on allograft survival and function. Collins et al.5 described C4d deposition in the absence of DSAs in HLA-identical, ABO-compatible renal allograft recipients who had experienced allograft failure. Although they were unable to investigate or identify non-HLA antibodies in these patients, the occurrence of presumed antibody-mediated rejection in these HLA-identical patients was thought to be caused by non-HLA alloantibody production.
Thus far, only a few non-HLA antibodies have been identified in humans.6-10 In addition, the absence of commercially available, validated detection strategies has hampered our ability to determine their clinical relevance and ascertain whether these antibodies are truly pathogenic.11,12 Protein microarrays offer a novel technique for the identification of patient-specific serum antibodies to non-HLA immunological targets, allowing simultaneous detection of antibodies to thousands of potential antigens. Although this technique has been applied to human autoimmune and oncological disease, our study represents the first use in the field of solid organ transplantation.13,14
We applied protein microarray technology to 15 pediatric patients who had experienced AR after renal transplantation. By paired comparative analysis using both pre-transplant and posttransplant serum samples,15 the protein microarray was able to identify 36 de novo antibody targets that were present in at least two patients at AR. In addition, a high antibody titer to one of these targets, protein kinase C-ζ (PKCζ), was associated with a recalcitrant subtype of AR and a significantly greater risk of allograft loss.
RESULTS
Antigen discovery using protein microarray
A total of 15 pediatric (mean age at transplantation 12.4±5.2 years) kidney transplant patients, with a mean HLA mismatch score of 4.1, were examined in our antigen discovery phase (Table 1). In total, 12 patients received steroid-free maintenance immunosuppression consisting of tacrolimus and mycophenolate mofetil, whereas the three remaining patients received steroid-based maintenance immunosuppression consisting of tacrolimus, mycophenolate mofetil, and prednisone. The patients developed AR at a mean of 22.3±20.7 months posttransplant. All patients experienced acute cellular rejection of whom four patients had Banff 1a rejection, eight patients had Banff 1b rejection, and three patients had Banff 2a rejection. Of the 15 patients with cellular rejection, only 4 (27%) had additional evidence of antibody-mediated rejection, based on positive C4d staining and the presence of DSAs. However, 53% (8/15) had at least one DSA at AR, whereas an additional 27% (4/15) had at least one non-DSA HLA antibody detected at AR.
Table 1.
Patient demographics
| Characteristic | Patients with acute rejection |
Stable posttransplant patients |
P-value |
|---|---|---|---|
| Number of patients | 15 | 28 | |
| Age at transplant (years) | 12.4±5.2 | 13.4±5.7 | 0.6 |
| HLA mismatch score | 4.1±1.9 | 4.2±1.5 | 0.9 |
| Gender | 0.9 | ||
| Male | 67% | 68% | |
| Female | 33% | 32% | |
| Allograft donor | 0.4 | ||
| Living donor | 53% | 39% | |
| Deceased donor | 47% | 61% | |
| Maintenance immunosuppression | 0.7 | ||
| Steroid free | 80% | 75% | |
| Steroid based | 20% | 25% | |
| Months posttransplant | < 0.005 | ||
| At acute rejection | 22.3±20.7 | ||
| At stable biopsy | 6.6±3.4 | ||
HLA, human leukocyte antigen.
Association of clinical variables between posttransplant patients who developed acute allograft rejection and posttransplant patients who did not develop acute allograft rejection. P-values <0.05 represent a significant difference between the two groups (by independent t-test for continuous variables and by χ2-test for categorical variables).
The values are expressed as means, standard deviations, and percentages.
At AR, de novo, serological, non-HLA responses were detected against 4.5% of the protein microarray targets (229/5056). At least one target was recognized in all patients, 36 targets were identified in at least two patients at AR. The mean protein microarray delta signal intensity of these targets in their respective patients was 1390±1061 intensity units compared with the mean delta signal intensity for all 5056 targets across all of the 15 patients, which was 7.6±198.3 (standard error, 0.7) intensity units. Patients with detectable anti-HLA recognized a mean of 24.4±15.4 non-HLA antigen targets. Patients without evidence of anti-HLA recognized a mean of 79.3±108.9 non-HLA antigen targets. This difference was not statistically significant (P=0.47); the greater mean number and larger standard deviation of non-HLA antigen targets recognized in patients without anti-HLA reactivity was primarily due to the fact that one of the three patients in this group recognized substantially more non-HLA antigens (205).
As this was a pilot study designed to assess the utility of the protein microarray technique in pediatric renal transplant recipients, we chose to focus our analysis on a single target, PKCζ, which had the highest mean signal intensity (6408 intensity units) of all 36 targets that were identified in two or more patients. In addition to having the strongest mean ProtoArray (Invitrogen, Carlsbad, CA, USA) signal, PKCζ was known to be expressed within renal parenchymal tissue, and has been shown to be actively involved in regulation of inflammation, cell survival, and apoptosis.16-24
Antigen validation of protein microarray results by enzyme-linked immunosorbent assay
Protein kinase C-ζ was analyzed by enzyme-linked immunosorbent assay (ELISA) across all 15 AR patients of the study set; ELISA showed a significant positive correlation with the protein microarray results (R2=0.84, P-value<0.001). Confirmation of ProtoArray-detected antibody presence and signal intensity, to our knowledge, has been validated for the first time in this study by ELISA. ELISA-determined at-event serum anti-PKCζ levels were plotted for the pre-transplant and the at-AR samples for each of the 15 patients, as well as for the posttransplant samples of 28 stable posttransplant patients who served as controls (Figure 1). The clinical characteristics of these control patients were similar to those of the 15 patients experiencing AR, with the exception of event time posttransplant (Table 1). AR occurred, on average, 22.3±20.7 months after transplant, whereas the biopsy showing the absence of AR occurred, on average, 6.6±3.4 months after transplant in the control patients (P<0.005). The mean anti-PKCζ serum levels for the pre-transplant, at-AR, and posttransplant stable control samples were 30.9±5.1 pg/μl, 46.7±34.9 pg/μl, and 34.8±8.6 pg/μl, respectively. Although there was a slight trend toward higher anti-PKCζ levels in the at-AR samples, this failed to reach statistical significance (P=0.07).
Figure 1. Enzyme-linked immunosorbent assay (ELISA) analysis of 15 patients with acute rejection (AR) and 28 stable posttransplant patients.
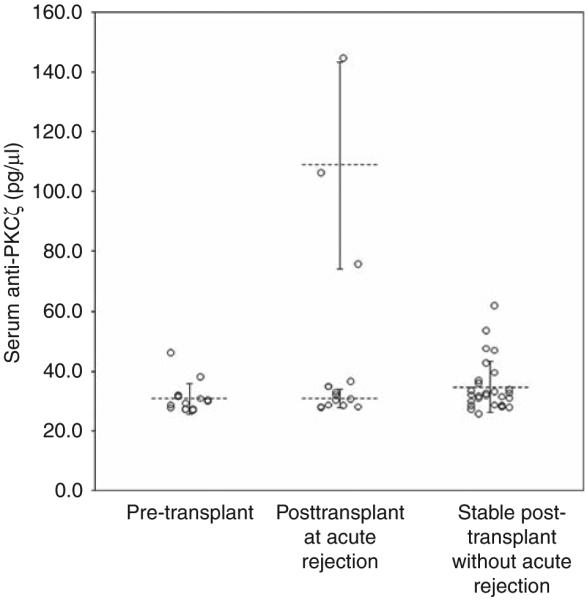
Among the three groups (pretransplant, posttransplant with AR, and posttransplant stable) there was a non-significant trend toward higher at-event anti-protein kinase C-ζ (anti-PKCζ) levels (P=0.07). The three patients with high anti-PKCζ levels had a mean concentration of 109±34.4 pg/μl. This was more than three times the mean concentration of the remaining 12 patients, that is, 31.1±3.1 pg/μl. This difference was statistically significant (P<0.001). The long horizontal bars represent the mean value for each group and the short horizontal bars represent one standard deviation. None of the patients had high anti-PKCζ levels pre-transplant (mean value 30.9±5.1 pg/μl). This suggests that the anti-PKCζ response in the three patients is de novo.
When the at-AR samples were further analyzed, the anti-PKCζ levels determined by ELISA were dramatically higher in 3 of the 15 AR patients, who all had values >75 pg/μl (Figure 1). The mean anti-PKCζ level in these three patients was 109±34.4 pg/μl. This was significantly greater than the mean anti-PKCζ level in the remaining 12 patients, 31.1±3.1 pg/μl (P<0.001). Comparative HLA and biopsy information for the patients with high anti-PKCζ levels and low anti-PKCζ levels is shown in Table 2. There was no association between the presence of high anti-PKCζ levels and the pathological severity of rejection, as graded by the Banff criteria (P=0.63), or a diagnosis concurrent to antibody-mediated rejection (P=0.24). In addition, there was no correlation between high anti-PKCζ levels and the presence of dense CD20-positive cell clusters (P=0.44). Finally, there was no association between high anti-PKCζ titers and development of antibodies to HLA targets. This held true both for DSA (P=0.60) and non-DSA HLA antibodies (P=0.44).
Table 2.
HLA antibody and biopsy data for patients with high and low anti-PKCζ levels
| Characteristic | Patients with high anti-PKCζ titers |
Patients with low anti-PKCζ titers |
P-value |
|---|---|---|---|
| Number of patients | 3 | 12 | |
| Allograft survival | 33.3% | 100% | 0.002 |
| Biopsy Banff score | 0.63 | ||
| 1a | 1 | 3 | |
| 1b | 2 | 6 | |
| 2a | 0 | 3 | |
| Antibody-mediated rejection | 0% | 33% | 0.24 |
| Dense CD20-positive clusters | 33.3% | 58.3% | 0.44 |
| Presence of donor-specific antibody at AR |
66.7% | 50% | 0.60 |
| Presence of non-donor-specific antibody HLA antibodies at AR |
66.7% | 41.7% | 0.44 |
| Presence of any HLA antibody at AR | 100% | 75% | 0.33 |
AR, acute rejection; HLA, human leukocyte antigen; PKCζ, protein kinase C-ζ Association of HLA antibody and biopsy factors with anti-PKCζ levels. P-values <0.05 represent a significant association between high anti-PKCζ and the respective variable.
Allograft survival analysis
When the high anti-PKCζ and the low anti-PKCζ patients were assessed by Kaplan–Meier analysis (Figure 2), at a mean follow-up of 4.5±0.5 years, the low anti-PKCζ patients had significantly better allograft survival than the patients with high anti-PKCζ levels (100% versus 33%; P=0.002). Although 4 of the 15 AR patients had C4d staining evident in their AR biopsy, none of the three patients with high anti-PKCζ levels had positive C4d staining.
Figure 2. Kaplan–Meier analysis of two subtypes of acute rejection (AR) based on serum anti-protein kinase C-ζ (anti-PKCζ) levels.
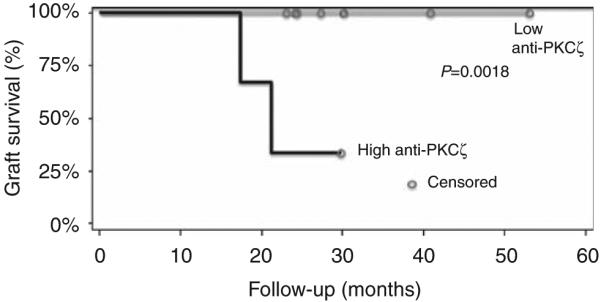
The gray line represents the 12 patients with low serum anti-PKCζ levels and the black line represents the 3 patients with high serum anti-PKCζ levels. Allograft survival for patients with high anti-PKCζ levels was lower (33%) than that for patients with low anti-PKCζ levels (100%). This was significantly different (P=0.002).
Immunohistochemical staining for PKCζ
To evaluate the localization of the PKCζ antigen in the transplant and the native kidney, immunohistochemical (IHC) staining was performed. PKCζ was shown in native and transplanted, non-rejecting kidney tissue, localizing both to the smooth muscle layer of arterioles and to the cytoplasmic domain of distal tubular cells (Figure 3a and b). IHC staining of renal allografts during AR shows the presence of PKCζ additionally in lymphocytes, both within lymphocyte aggregates and scattered throughout the tubulointerstitium (Figure 3c and d).
Figure 3. Immunohistochemical staining for PKCζ in normal renal tissue and renal parenchyma experiencing acute rejection.
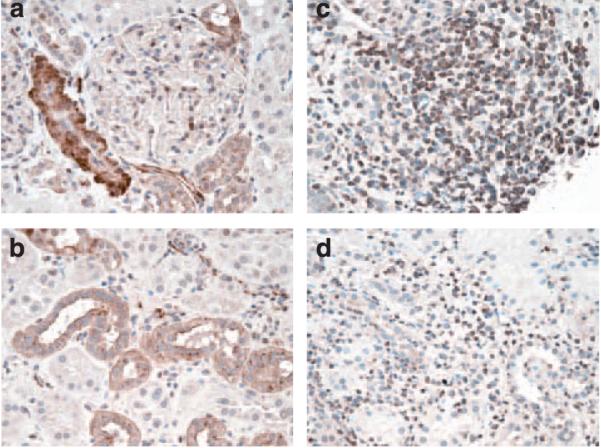
Within normal kidney (a and b), cytoplasmic granular staining for PKCζ is observed in a subset of tubules morphologically compatible with distal tubules (b) and the smooth muscle cells of the arteries (a). Patchy endothelial cell staining is observed in a few capillaries. No significant staining is observed in glomeruli except for an occasional infiltrating lymphocyte. In acute rejection (c and d), the tubular staining is less intense, but the infiltrating lymphocytes are PCKζ-positive, both when scattered (d) and when arranged in aggregates (c). Negative controls were run to identify non-specific anti-PKCζ staining. Tissue from non-rejecting allografts had a similar staining pattern to those of normal kidney (data not shown).
DISCUSSION
These results show the feasibility of applying protein microarrays to renal transplant recipients. It is a novel and emerging technology with the capacity to identify thousands of potential immunogenic non-HLA antigens. Previously, Robinson et al.14 fabricated an 1152-feature protein microarray that was used to show specific autoantibody binding and characterize sera in known autoimmune disease states. The currently used ProtoArray platform from Invitrogen offers a human protein microarray containing 5056 antigens. To date, there is a single publication using this technology to study human disease. In this study, the ProtoArray was probed with sera from patients with ovarian cancer.13 Although no target was identified universally in patients with ovarian cancer, autoantibodies to four antigens were found to have higher reactivity in patients with ovarian cancer when compared with healthy controls. Although not all patients with ovarian cancer formed detectable autoantibodies to these targets, combined immunostaining for two of the targets identified by protein microarray led to a highly sensitive and specific tissue diagnosis tool.
We have used the ProtoArray for the first time in solid organ transplantation to determine whether de novo, non-HLA targets, with clinical and prognostic relevance, can be identified in transplant patients experiencing AR. With this technology, we found biologically relevant antibody targets in multiple patients at AR. Interestingly, the repertoire of antigens recognized seems to be patient specific, with variable reactivity to the range of protein targets; the patients had antibody responses to between 0.1% and 4.1% of the possible antigens. In addition, in our small cohort, the number and specificity of antigen targets recognized did not seem to be associated with the development of HLA antibodies. In total, 36 of the 5056 antigens were recognized in at least 2 of the 15 AR patients. This seemingly low number is not surprising given that the ProtoArray was not designed to examine renal-related antigens or transplant-specific targets. It is likely that a protein microarray optimized for solid organ transplantation would have a higher net yield. Despite this, we were able to identify antibodies to numerous biologically relevant antigen targets, simultaneously using a single test and minimal patient serum. Given the preliminary nature of this study, we chose one such relevant target, PKCζ, for additional analysis. PKCζ was chosen because it had the strongest mean signal intensity of the 36 potential antigens, it is known to be present in renal tissue, and it is involved in inflammatory signal transduction pathways. Comprehensive analysis of other relevant targets will be the focus of future investigation.
Protein Kinase C-ζ, which is expressed in a number of tissues, including brain, kidney, lung, and testes,25 is an atypical PKC which is an integral component of several pathways involved in cell survival, proliferation, and apoptosis17,20,21 Animal model data are concordant with available in vitro data, suggesting that PKCζ has an active, regulatory role in inflammation. PKCζ-deficient mice (PKCζ−) have reduced Peyer’s patch formation, a relative reduction of B cells in peripheral lymph nodes, and no B-cell follicle formation.16 In addition, they lack the anti-apoptotic signal mediated by tumor necrosis factor-a-activated NF-kB, which is present in normal mice. In a renal ischemia/reperfusion rat model, PKCζ had significantly upregulated expression during the first hour of reperfusion, at 1 day after reperfusion, and at days 5–7 after reperfusion.19 Human studies have been consistent with the in vitro and animal model data, establishing the active role PKCζ has in inflammatory cell signaling and cell survival. PKCζ is involved in intracellular signaling in human monocytes and macrophages, and mediates lipopolysaccharide-activated pro-inflammatory cytokine gene expression.23 In addition, PKCζ mediates regulation of the mitogen-activated protein kinase and mammalian target of rapamycin pathways in follicular lymphoma cells, and seems to exert a survival function in these cells. Administration of rituximab, a humanized anti-CD20 immunotherapy, led to reduced PKCζ activity and inhibited its survival effects.18 Finally, Zhao et al.22 recently showed increased PKCζ expression in psoriatic skin lesions compared with healthy skin. tumor necrosis factor-α, a well-described pathogenic factor in psoriasis, was found to be dependent on PKCζ for cell signaling and signal transduction. After tumor necrosis factor-α stimulation, cytoplasmic and nuclear staining for PKCζ was increased. Furthermore, activation of PKCζ was associated with an increased expression of CD1d, which interacts with natural killer T cells, and has an integral role in their cytokine production. Thus, PKCζ seems to have a significant role in inflammatory cell signaling and may be upregulated in inflammatory disease states, such as acute allograft rejection.
In our analysis, although there was a slight trend toward higher anti-PKCζ levels in the at-AR cohort compared with the pre-transplant and stable posttransplant cohorts, this trend failed to reach statistical significance. However, a subset of patients within the AR cohort had robust anti-PKCζ responses, suggesting the presence of an AR subtype. When allograft survival was assessed, the patients with elevated anti-PKCζ levels had significantly worse outcomes and anti-PKCζ levels were significantly associated with accelerated allograft loss at mean follow-up of 4.5±0.5 years.
It is important to interpret these results with caution; given the small size of our cohort, we cannot rule out that anti-PKCζ levels are elevated merely because of increased expression, abnormal splicing or protein folding, or polymorphism. In addition, although high anti-PKCζ titers were significantly associated with allograft loss in our study, there is no evidence of causality. In fact, given that none of the three patients with high anti-PKCζ titers had evidence of C4d deposition in their AR biopsies, it is likely that anti-PKCζ is a marker, or bystander molecule, related to cellular damage associated with severe AR, rather than being truly pathogenic. The fact that higher anti-PKCζ levels were not associated with development of HLA antibodies would also suggest a different mechanism than that seen with DSAs in antibody-mediated rejection. Our IHC results show that PKCζ is indeed present in renal parenchymal cells, localizing to smooth muscle and distal tubular cells in healthy renal allograft tissue. The presence of PKCζ within renal tubular cells is consistent with a recent study which showed that PKCζ is present in and regulates organic anion transporters in renal proximal tubular cells.26 Interestingly, in the setting of AR, our IHC staining also found PKCζ within infiltrating lymphocytes, suggesting either upregulation within the inflammatory cell or immunological exposure to the intracellular antigen. Our results are consistent with the premise that PKCζ is upregulated in the inflammation associated with AR; we hypothesize that in our subset of AR patients with high anti-PKCζ levels, severe renal injury and cell death led to immunological exposure of PKCζ with resultant antibody formation. In this setting, the elevated anti-PKCζ titer may be a marker for the damage associated with a more severe subtype of AR. Interestingly, in our small pilot study, there did not seem to be a specific histological feature that was associated with AR and the presence of higher anti-PKCζ levels; however, it is possible that in a larger patient cohort such a characteristic might be found.
In summary, protein microarrays were able to successfully identify AR-specific antigenic targets in a high throughput manner and represent an appealing technology to better assess alloimmunity in solid organ transplantation. In addition, based on our results, PKCζ is a potential non-HLA antigen target recognized in pediatric renal transplant patients experiencing AR. It is not a target in all AR episodes, but there seems to be a subtype of AR, characterized by exposure of and antibody formation against PKCζ, which is associated with poor allograft survival. Our results suggest that anti-PKCζ is a marker, rather than a truly pathogenic antibody and further research is necessary to accurately define the role that PKCζ has in AR.
METHODS
Patient selection
A review of our pediatric transplant database identified patients who had undergone renal allograft transplantation and experienced at least one episode of acute allograft rejection. A total of 15 patients were selected based on availability of serum samples, both before transplantation and at the time of AR. All transplant allograft biopsies were graded on the basis of the Banff classification.27,28 Pre-transplant serum samples were obtained within 48 h before allograft placement. The at-AR serum samples were obtained concurrently with the biopsy showing AR and before initiation of anti-rejection therapy. No patients received antibody therapy, including intravenous immunoglobulin, before the sample being obtained. Anti-HLA testing was performed as standard posttransplant care and the results were obtained from our histocompatibility laboratory. Pre- and posttransplant serum samples from all 15 AR patients were processed for ProtoArray and ELISA experiments. An additional 28 stable, posttransplant pediatric renal allograft recipients were selected as controls for the ELISA analysis using our validated PKCζ ELISA. These 28 patients were chosen based on clinical similarity to the 15 AR patients and the presence of a posttransplant surveillance biopsy showing the absence of AR. Serum samples for these patients were obtained concurrently with the biopsy showing the absence of AR. Pre-transplant serum samples were not available for these 28 allograft recipients. These serum samples were processed for ELISA experiments. All serum samples were available under a previously institutional review board approved protocol (no 13443).
Identification of autoantibody targets using protein microarray
A total of 30 protein microarrays (ProtoArray V3; Invitrogen, Carlsbad, CA, USA) were used for this study, one each for the pre-transplant and the at-AR serum samples of the 15 patients with AR. The ProtoArrays were blocked with blocking buffer for 1 h followed by application of plasma sample (1:150) for 90 min. After washing the protein microarray four times for 10 min each, the protein microarrays were probed with secondary antibody (goat anti-human Alexa 647, Molecular Probes, Eugene, OR, USA) for 90 min. After washing the slides, the protein microarrays were dried and scanned using a fluorescent microarray scanner (GSI Luminoics, Perkin-Elmer scanner, Waltham, MA, USA). All steps were carried out on a rotating platform and at 4°C. The slides were scanned at a photomultiplier gain of 60% with a laser power of 90% and a focus point of 0 mm. The ‘.gal’ files were obtained from a ProtoArray central portal on the Invitrogen website (www.invitrogen.com/ProtoArray) by submitting the barcode of each protein microarray. Data was obtained using GenePix software (Version 6, Molecular Devices, Sunnyvale, CA, USA). Using the appropriate ‘.gal’ file and the respective microarray image obtained from the scanners. Novel alloimmune antibody responses are identified by subtracting the pre-transplant data set from the posttransplant data set (delta); all reported ProtoArray signal intensities represent the delta intensity (signal at AR–signal pre-transplant). A target response was considered positive, and indicative of de novo antibody formation, if the response delta, defined as the response intensity at AR subtracting the pre-transplant response intensity, was arbitrarily 500 or greater. Positive antibody responses were arranged according to occurrence frequency, and all targets identified in at least two patients were reviewed with specific attention directed at the strength of the antibody response, human tissue expression data, gene ontology of the target, and the relevance to immunological function. Given the preliminary nature of this study, a single target, PKCζ, was selected as a candidate target for further analysis on the basis of the aforementioned factors.
ELISA validation of PKCζ protein microarray results
Both the pre-transplant and the at-AR serum samples from the 15 AR patients and the posttransplant serum samples from the 28 stable kidney transplant recipients were analyzed by ELISA. Insect cell-expressed human recombinant protein, PKCζ was obtained from Invitrogen. The 96-well microwell ELISA plate was coated with 0.27 μg PKCζ protein in 50 μl coating buffer (15 mm Na2CO3, 30 mm NaHCO3, 0.02% NaN3, pH 9.6) and incubating overnight at 4°C. The standard curve was generated using rabbit polyclonal antibody to PKCζ (Abcam, Cambridge, MA, USA), and Zymax-grade AP-conjugated goat anti-rabbit IgG (Invitrogen). After washing the plate with tris-buffered saline tween 20 buffer five times, the non-specific protein binding was blocked by 100 μl, 2% dry milk in tris-buffered saline tween 20 buffer for 1 h at room temperature. After the blocking step, 50 μl serum samples (40-fold diluted with 2% milk in tris-buffered saline tween 20 buffer) were incubated on the wells for 1 h at room temperature. The plate was washed five times with tris-buffered saline tween 20 buffer and incubated in 50 μl AP-conjugated AffiniPure Mouse anti-human IgG (Jackson ImmunoResearch, West Grove, PA, USA). The color was developed by using AP-pNPP liquid substrate system for ELISA (Sigma-Aldrich, St Louis, MO, USA). Absorption was measured at 405 nm with a SPECTRAMax 190 microplate reader (Molecular Devices, Sunnyvale, CA, USA). Serum PKCζ antibody concentrations were determined from the standard curve.
Longitudinal allograft survival analysis
Allograft survival was assessed in the 15 patients with AR in the study set. Patients were divided into AR subtypes based on their serum anti-PKCζ levels at AR: 3 with high serum anti-PKCζ levels and 12 with low serum anti-PKCζ levels. Follow-up commenced at the time of the initial AR event. Follow-up was continued until allograft loss occurred or until the time of most recent assessment of allograft function. Allograft loss was defined as a return to dialysis.
IHC staining for PKCζ in renal parenchyma
Immunohistochemical staining was performed using antibodies directed against PKCζ (GeneTex, San Antonio, TX, USA catalog no GTX40214). Formalin-fixed, paraffin-embedded tissue were pretreated with citrate and stained with polyclonal antiserum to PKCζ (dilution 1:2000 for 18 h). A rabbit ABC detection kit (Vector Labs, Burlingame, CA, USA) was used (PK-6101). Negative controls were run to assess for non-specific anti-PKCζ staining.
Statistical analysis
t-Test, ANOVA (analysis of variance), and χ2-test were used for analysis of continuous or categorical types of data. Correlation analysis was performed for antigens detected by ProtoArray and ELISA. Graft survival rate was based on Kaplan–Meier survival analysis at current follow-up. P-values ≤0.05 were considered statistically significant. Results are reported as mean±standard deviation. All statistical analyses were performed using SAS 9.1.3 (SAS Institute, Cary, NC, USA).
ACKNOWLEDGMENTS
We thank the following people for their invaluable assistance with this project: Sue Hsieh, Poonam Sansanwal, Neeraja Kambham, and Lauren Weintraub. This work was supported by grant NIH/NIAID R01 AI 61739-01 (to MS).
Footnotes
DISCLOSURE
All the authors declared no competing of interests.
REFERENCES
- 1.Wissing KM, Fomegné G, Broeders N, et al. HLA mismatches remain risk factors for acute kidney allograft rejection in patients receiving quadruple immunosuppression with anti-interleukin-2 receptor antibodies. Transplantation. 2008;85:411–416. doi: 10.1097/TP.0b013e31816349b5. [DOI] [PubMed] [Google Scholar]
- 2.Hariharan S, Johnson CP, Bresnahan BA, et al. Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med. 2000;342:605–612. doi: 10.1056/NEJM200003023420901. [DOI] [PubMed] [Google Scholar]
- 3.Mao Q, Terasaki PI, Cai J, et al. Extremely high association between appearance of HLA antibodies and failure of kidney grafts in a five-year longitudinal study. Am J Transplant. 2007;7:864–871. doi: 10.1111/j.1600-6143.2006.01711.x. [DOI] [PubMed] [Google Scholar]
- 4.Opelz G. Non-HLA transplantation immunity revealed by lymphocytotoxic antibodies. Lancet. 2005;365:1570–1576. doi: 10.1016/S0140-6736(05)66458-6. [DOI] [PubMed] [Google Scholar]
- 5.Collins AB, Chicano SL, Cornell LD, et al. Putative antibody-mediated rejection with C4d deposition in HLA-identical, ABO-compatible renal allografts. Transplant Proc. 2006;38:3427–3429. doi: 10.1016/j.transproceed.2006.10.159. [DOI] [PubMed] [Google Scholar]
- 6.Carter V, Shenton BK, Jaques B, et al. Vimentin antibodies: a non-HLA antibody as a potential risk factor in renal transplantation. Transplant Proc. 2005;37:654–657. doi: 10.1016/j.transproceed.2004.12.043. [DOI] [PubMed] [Google Scholar]
- 7.Dragun D, Müller DN, Bräsen JH, et al. Angiotensin II type 1-receptor activating antibodies in renal-allograft rejection. N Engl J Med. 2005;352:558–569. doi: 10.1056/NEJMoa035717. [DOI] [PubMed] [Google Scholar]
- 8.Zou Y, Stastny P, Süsal C, et al. Antibodies against MICA antigens and kidney-transplant rejection. N Engl J Med. 2007;357:1293–1300. doi: 10.1056/NEJMoa067160. [DOI] [PubMed] [Google Scholar]
- 9.Sun Q, Liu Z, Yin G, et al. Detectable circulating antiendothelial cell antibodies in renal allograft recipients with C4d-positive acute rejection: a report of three cases. Transplantation. 2005;79:1759–1762. doi: 10.1097/01.tp.0000163290.19788.e7. [DOI] [PubMed] [Google Scholar]
- 10.Sun Q, Liu Z, Chen J, et al. Circulating anti-endothelial cell antibodies are associated with poor outcome in renal allograft recipients with acute rejection. Clin J Am Soc Nephrol. 2008;3:1479–1486. doi: 10.2215/CJN.04451007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dragun D. Humoral responses directed against non-human leukocyte antigens in solid-organ transplantation. Transplantation. 2008;86:1019–1025. doi: 10.1097/TP.0b013e3181889748. [DOI] [PubMed] [Google Scholar]
- 12.Tinckam KJ, Chandraker A. Mechanisms and role of HLA and non-HLA alloantibodies. Clin J Am Soc Nephrol. 2006;1:404–414. doi: 10.2215/CJN.00270106. [DOI] [PubMed] [Google Scholar]
- 13.Hudson ME, Pozdnyakova I, Haines K, et al. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci USA. 2007;104:17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson WH, DiGennaro C, Hueber W, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Wadia P, Chen R, et al. Identifying compartment-specific non-HLA targets after renal transplantation by integrating transcriptome and ““antibodyome”” measures. Proc Natl Acad Sci. 2009;106:4148–4153. doi: 10.1073/pnas.0900563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leitges M, Sanz L, Martin P, et al. Targeted disruption of the zetaPKC gene results in the impairment of the NF-kappaB pathway. Mol Cell. 2001;8:771–780. doi: 10.1016/s1097-2765(01)00361-6. [DOI] [PubMed] [Google Scholar]
- 17.Leroy I, de Thonel Al, Laurent G, et al. Protein kinase C zeta associates with death inducing signaling complex and regulates Fas ligand-induced apoptosis. Cell Signal. 2005;17:1149–1157. doi: 10.1016/j.cellsig.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Leseux L, Laurent G, Laurent C, et al. PKC zeta mTOR pathway: a new target for rituximab therapy in follicular lymphoma. Blood. 2008;111:285–291. doi: 10.1182/blood-2007-04-085092. [DOI] [PubMed] [Google Scholar]
- 19.Padanilam BJ. Induction and subcellular localization of protein kinase C isozymes following renal ischemia. Kidney Int. 2001;59:1789–1797. doi: 10.1046/j.1523-1755.2001.0590051789.x. [DOI] [PubMed] [Google Scholar]
- 20.San-Antonio B, Iñiguez MA, Fresno M. Protein kinase Czeta phosphorylates nuclear factor of activated T cells and regulates its transactivating activity. J Biol Chem. 2002;277:27073–27080. doi: 10.1074/jbc.M106983200. [DOI] [PubMed] [Google Scholar]
- 21.Xin M, Gao F, May WS, et al. Protein kinase Czeta abrogates the proapoptotic function of Bax through phosphorylation. J Biol Chem. 2007;282:21268–21277. doi: 10.1074/jbc.M701613200. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y, Fishelevich R, Petrali JP, et al. Activation of keratinocyte protein kinase C zeta in psoriasis plaques. J Invest Dermatol. 2008;128:2190–2197. doi: 10.1038/jid.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, Chen L-Y, Doerner A, et al. An atypical protein kinase C (PKC zeta) plays a critical role in lipopolysaccharide-activated NF-kappa B in human peripheral blood monocytes and macrophages. J Immunol. 2009;182:5810–5815. doi: 10.4049/jimmunol.0804073. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Johnston T, Jeon H, et al. Cyclosporin A up-regulates and activates protein kinase C-zeta in EBV-infected and EBV-transformed human B-cells. J Surg Res. 2009;153:156–161. doi: 10.1016/j.jss.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 25.SOURCE Search for PRKCZ http://smd.stanford.edu/cgi-bin/source/sourceResult. Accessed on June 2008.
- 26.Barros SA, Srimaroeng C, Perry JL, et al. Activation of protein kinase c{zeta} increases OAT1 (SLC22A6)- and OAT3 (SLC22A8)-mediated transport. J Biol Chem. 2009;284:2672–2679. doi: 10.1074/jbc.M808078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Racusen LC, Solez K, Colvin RB, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55:713–723. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 28.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8:753–760. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]


