Abstract
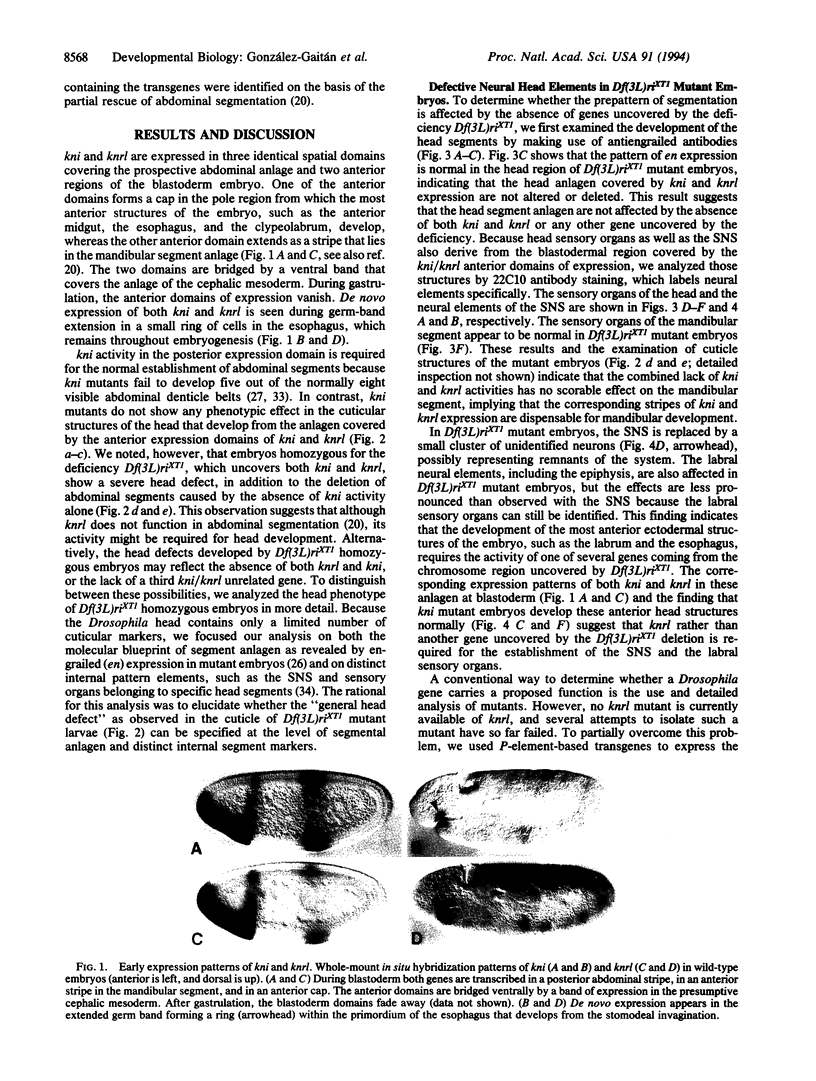
Developmental gene functions of Drosophila are typically characterized by a recognizable mutant phenotype. When molecular probes of such genes were used to isolate homologues, distinct spatially and temporally restricted expression patterns were observed in vertebrates as well. However, corresponding "gene knock-outs" often revealed subtle or no scorable phenotypes, a phenomenon attributed to redundant gene functions. We found that the evolutionarily related genes knirps (kni) and knirps-related (knrl) contribute to a similar phenomenon in Drosophila. The two closely situated genes show identical expression patterns in the developing embryo, including the posterior and anterior expression domains in the blastoderm. Here we show that the two biochemically equivalent gene products are both functional in the head anlage and that the lack of one gene activity can be overcome by the activity of the other. Whereas kni is also required for abdominal segmentation, knrl is nonfunctional in its posterior expression domain. Thus, the kni/knrl pair of genes provides a region-specific buffering system, rather than a case of global functional redundancy.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basson M. E., Thorsness M., Rine J. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5563–5567. doi: 10.1073/pnas.83.15.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner S., Bopp D., Burri M., Noll M. Structure of two genes at the gooseberry locus related to the paired gene and their spatial expression during Drosophila embryogenesis. Genes Dev. 1987 Dec;1(10):1247–1267. doi: 10.1101/gad.1.10.1247. [DOI] [PubMed] [Google Scholar]
- Cadigan K. M., Grossniklaus U., Gehring W. J. Functional redundancy: the respective roles of the two sloppy paired genes in Drosophila segmentation. Proc Natl Acad Sci U S A. 1994 Jul 5;91(14):6324–6328. doi: 10.1073/pnas.91.14.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman K. G., Poole S. J., Weir M. P., Soeller W. C., Kornberg T. The invected gene of Drosophila: sequence analysis and expression studies reveal a close kinship to the engrailed gene. Genes Dev. 1987 Mar;1(1):19–28. doi: 10.1101/gad.1.1.19. [DOI] [PubMed] [Google Scholar]
- Côté S., Preiss A., Haller J., Schuh R., Kienlin A., Seifert E., Jäckle H. The gooseberry-zipper region of Drosophila: five genes encode different spatially restricted transcripts in the embryo. EMBO J. 1987 Sep;6(9):2793–2801. doi: 10.1002/j.1460-2075.1987.tb02575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S. C., Zipursky S. L., Benzer S., Ferrús A., Shotwell S. L. Monoclonal antibodies against the Drosophila nervous system. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L. S. Functional redundancy in mitotic force generation. J Cell Biol. 1993 Jan;120(1):1–3. doi: 10.1083/jcb.120.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U., Pearson R. K., Gehring W. J. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 1992 Jun;6(6):1030–1051. doi: 10.1101/gad.6.6.1030. [DOI] [PubMed] [Google Scholar]
- Gutjahr T., Patel N. H., Li X., Goodman C. S., Noll M. Analysis of the gooseberry locus in Drosophila embryos: gooseberry determines the cuticular pattern and activates gooseberry neuro. Development. 1993 May;118(1):21–31. doi: 10.1242/dev.118.1.21. [DOI] [PubMed] [Google Scholar]
- Higashijima S., Michiue T., Emori Y., Saigo K. Subtype determination of Drosophila embryonic external sensory organs by redundant homeo box genes BarH1 and BarH2. Genes Dev. 1992 Jun;6(6):1005–1018. doi: 10.1101/gad.6.6.1005. [DOI] [PubMed] [Google Scholar]
- Joyner A. L., Herrup K., Auerbach B. A., Davis C. A., Rossant J. Subtle cerebellar phenotype in mice homozygous for a targeted deletion of the En-2 homeobox. Science. 1991 Mar 8;251(4998):1239–1243. doi: 10.1126/science.1672471. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., McGill C., Fasano O., Strathern J., Broach J., Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984 Jun;37(2):437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Lohnes D., Kastner P., Dierich A., Mark M., LeMeur M., Chambon P. Function of retinoic acid receptor gamma in the mouse. Cell. 1993 May 21;73(4):643–658. doi: 10.1016/0092-8674(93)90246-m. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986 Dec 11;324(6097):537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- McMahon A. P., Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990 Sep 21;62(6):1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Nauber U., Pankratz M. J., Kienlin A., Seifert E., Klemm U., Jäckle H. Abdominal segmentation of the Drosophila embryo requires a hormone receptor-like protein encoded by the gap gene knirps. Nature. 1988 Dec 1;336(6198):489–492. doi: 10.1038/336489a0. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980 Oct 30;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Oro A. E., Ong E. S., Margolis J. S., Posakony J. W., McKeown M., Evans R. M. The Drosophila gene knirps-related is a member of the steroid-receptor gene superfamily. Nature. 1988 Dec 1;336(6198):493–496. doi: 10.1038/336493a0. [DOI] [PubMed] [Google Scholar]
- Pankratz M. J., Busch M., Hoch M., Seifert E., Jäckle H. Spatial control of the gap gene knirps in the Drosophila embryo by posterior morphogen system. Science. 1992 Feb 21;255(5047):986–989. doi: 10.1126/science.1546296. [DOI] [PubMed] [Google Scholar]
- Patel N. H., Martin-Blanco E., Coleman K. G., Poole S. J., Ellis M. C., Kornberg T. B., Goodman C. S. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell. 1989 Sep 8;58(5):955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Rothe M., Nauber U., Jäckle H. Three hormone receptor-like Drosophila genes encode an identical DNA-binding finger. EMBO J. 1989 Oct;8(10):3087–3094. doi: 10.1002/j.1460-2075.1989.tb08460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M., Pehl M., Taubert H., Jäckle H. Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature. 1992 Sep 10;359(6391):156–159. doi: 10.1038/359156a0. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 1983 Sep 24;11(18):6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ott U., González-Gaitán M., Jäckle H., Technau G. M. Number, identity, and sequence of the Drosophila head segments as revealed by neural elements and their deletion patterns in mutants. Proc Natl Acad Sci U S A. 1994 Aug 30;91(18):8363–8367. doi: 10.1073/pnas.91.18.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ott U., Technau G. M. Expression of en and wg in the embryonic head and brain of Drosophila indicates a refolded band of seven segment remnants. Development. 1992 Sep;116(1):111–125. doi: 10.1242/dev.116.1.111. [DOI] [PubMed] [Google Scholar]
- Tautz D., Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989 Aug;98(2):81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature. 1990 Aug 30;346(6287):847–850. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]