Summary
Cell division in all eukaryotes depends on function of the spindle, a microtubule-based structure that segregates chromosomes to generate daughter cells in mitosis or haploid gametes in meiosis. Spindle size adapts to changes in cell size and shape, which vary dramatically across species and within a multicellular organism, but the nature of scaling events and their underlying mechanisms are poorly understood. Cell size variations are most pronounced in early animal development, as egg diameters range from tens of microns up to millimeters across animal phyla, and decrease several orders of magnitude during rapid reductive divisions. During early embryogenesis in the model organisms X. laevis and C. elegans, the spindle scales with cell size [1,2], a phenomenon regulated by molecules that modulate microtubule dynamics [3–6], as well as by limiting cytoplasmic volume [7,8]. However, it is not known to what extent spindle scaling is conserved across organisms and among different cell types. Here we show that in a range of metazoan phyla, mitotic spindle length decreased with cell size across a ~30 fold difference in zygote size. Maximum spindle length varied, but linear spindle scaling occurred similarly in all species once embryonic cell diameter reduced to 140 μm. In contrast, we find that the female meiotic spindle does not scale as closely to egg size, adopting a more uniform size across species that likely reflects its specialized function. Our analysis reveals that spindle morphometrics change abruptly, within one cell cycle, at the transition from meiosis to mitosis in most animals.
Results and Discussion
Early embryo mitotic spindles scale to cell size across metazoans
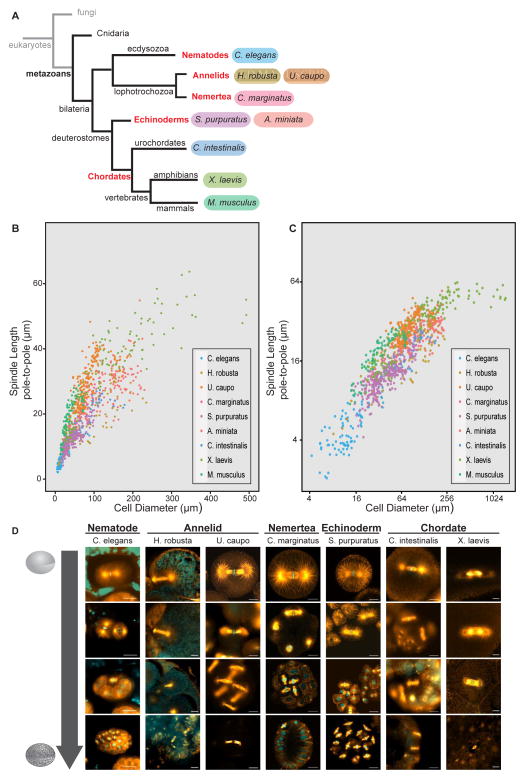
To evaluate scaling features among diverse animal species, we imaged embryos from eight different organisms representing five metazoan phyla and measured both cell diameter and several spindle size parameters (Figure 1). We also included published data from Mus musculus (Chordata) [7,9] in our analysis. The ‘pole-to-pole’ length was measured as the distance between the two positions where most interpolar spindle microtubules terminated (Figure S1A), a previously described measurement used to analyze spindle length scaling [1–3,7,8]. Many mitotic spindles contain centrosomes adjacent to the spindle poles that radiate microtubules and contribute to overall bipolar spindle structure. Therefore the ‘aster-to-aster’ length was also measured for each spindle as the distance between the centers of the two spindle asters emanating from presumed centrosomes as judged by tubulin immunofluorescence (Figure S1B). Cell size was measured as the longest cell diameter parallel to the spindle pole-to-pole axis. Metaphase mitotic spindle length scaled robustly with cell size in all embryos examined, with both the pole-to-pole and the aster-to-aster spindle lengths decreasing with cell diameter (Figure 1B, 1C, S1A and S1B).
Figure 1. Mitotic spindles scale to cell size across Metazoans.
(A) Phylogenetic tree depicting phyla (red) and species (colors) represented in our analysis. (B) Mitotic pole-to-pole spindle length versus cell diameter in early embryonic cells < 500 μm in diameter. (C) Mitotic pole-to-pole spindle length versus cell diameter in early embryos on a log2-log2 scale. Individual data points represent a single spindle measurement and different colors represent different species. (D) Images of fixed mitotic embryos at different early embryonic stages stained for tubulin (orange) and DNA (cyan). All scale bars = 20 μm.
In addition, we observed characteristic changes in spindle morphology during embryogenesis. In large cells of X. laevis early embryos, centrosomes appear detached from spindle poles with a region of low microtubule density between the spindle pole and centrosome aster [1]. Similarly, centrosome asters were separated from spindle poles in the largest cells of other animal embryos, with the average distance between the centrosome aster and spindle pole ranging from 2.3 μm ± 0.8 in U. caupo to 11.5 μm ± 4.5 in C. intestnalis (top row Figure 1D), resulting in an aster-to-aster spindle length greater than the pole-to-pole spindle length (Figure S1C). The difference between pole-to-pole and aster-to-aster spindle lengths in the earliest embryonic cells varied from organism to organism, with an average difference ranging from 4 to 45 μm that correlated with zygote size (r= 0.76, p=0.01; Figure S1D). Thus, organisms with larger embryos displayed a greater difference between aster-to-aster and pole-to-pole spindle lengths. Centrosome size, as measured by the diameter of bright and uniform tubulin fluorescence in the center of each aster, also scaled with cell size such that organisms with larger embryos contained larger centrosomes (Figure 1D and S1E). In C. elegans, centrosome size scales to cell volume and is limited by amount of centrosomal components [10]. We observed similar scaling of centrosome diameter to cell size in early embryos of all organisms (Figure S1F). As cells decreased in size, centrosomes not only decreased in size but also converged with spindle poles, leading to similar aster-to-aster and pole-to-pole lengths (Figure 1D, S1C and S1F). The separation of centrosome asters from spindle poles may serve as a second length scaling mechanism to efficiently segregate chromosomes across long distances, properly position centrosomes, and induce cleavage plane formation in large cells [11,12].
Conserved features of mitotic spindle scaling across metazoans
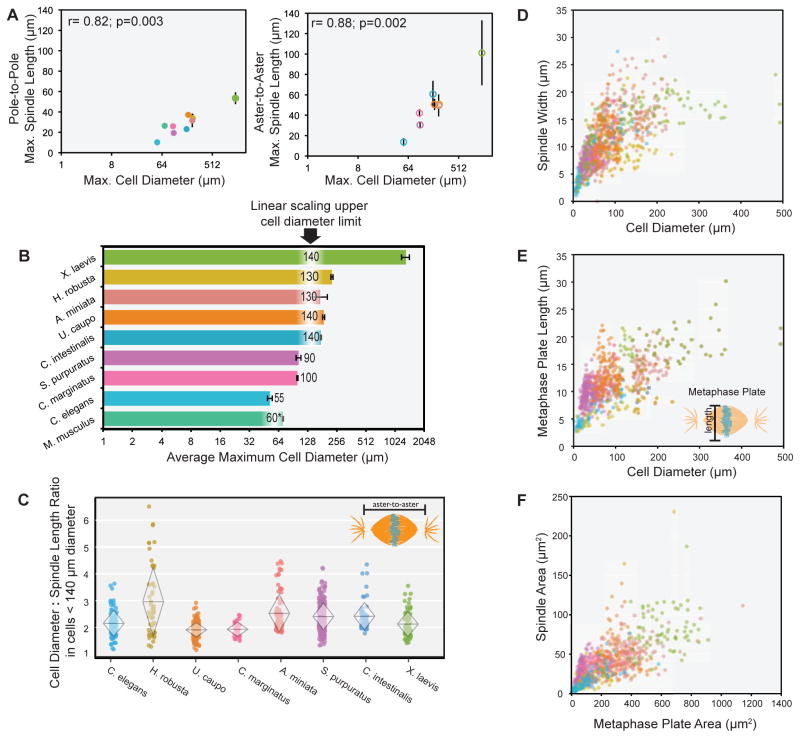
We next compared specific features of spindle scaling across animals. During the first several rounds of division in Xenopus embryos, mitotic spindles exhibit an upper size limit that can be recapitulated in cytoplasmic extracts prepared from embryos [1,7]. Likewise, pole-to-pole spindle length plateaued during the first divisions of large early embryos including A. miniata (30.9 μm ± 5.6), U. caupo (36.4 μm ± 2.5), C. intestinalis (23.7 μm ± 1.4), and H. robusta (27.7 μm ± 6.0) (Figure S1C). Across phyla, the maximum spindle length during the earliest mitotic divisions varied over five-fold, ranging in pole-to-pole distance from 10.2 μm ± 2.9 in C. elegans to 53.5 μm ± 5.9 in X. laevis. Remarkably, across this range the maximum pole-to-pole (r=0.82, p=0.003) and aster-to-aster (r=0.88, p=0.002) mitotic spindle lengths correlated with embryo size (Figure 2A). Recently it was observed that among 97 separate isolates of C. elegans from around the world, variation in cell size at the first embryonic division was the primary driver of variation in spindle size [13]. Taken together, these results indicate that cell size correlates with maximum mitotic spindle length both within a single species during embryo development and across metazoan phyla.
Figure 2. Linear size scaling relationships during animal development.
(A) Average pole-to-pole mitotic spindle length (left) and aster-to-aster mitotic spindle length (right) versus cell diameter (log2 scale) during the first and second embryonic divisions of various species, r=0.82; p=0.003 (pole-to-pole) and r=0.88, p=0.002 (aster-to-aster). (B) Histogram of average maximum cell diameter of different species on a log2 scale. The largest cell diameter when linear scaling of spindle length is observed is indicated for each species. (C) Cell diameter : aster-to-aster spindle length ratio for cells < 140 μm in diameter. Overlaid diamonds indicate mean cell diameter : spindle length ratio (center horizontal line) and standard deviation (height of vertices). (D) Mitotic spindle width versus cell diameter in cells < 500 μm in diameter. For cells < 140 μm in diameter, r= 0.67, p<0.001. (E) Mitotic metaphase plate length versus cell diameter in cells < 500 μm in diameter. For cells < 140 μm in diameter, r= 0.62, p<0.001. (F) Spindle area (pole-to-pole spindle length x spindle width) versus metaphase plate area (metaphase plate length x width), r=0.73, p<0.001. (C – F) Individual points represent a single spindle measurement. Error bars represent standard deviation of the mean. Different colors represent different species as indicated in Figure 1A key.
The observation that spindles scale to cell size across species, but that mitotic spindle length reaches organism-specific plateaus, prompted us to ask whether there is a conserved range of cell sizes within which mitotic spindles scale linearly and above which mitotic spindles do not scale linearly to cell size. For spindle length versus cell diameter plots, we tracked the goodness-of-fit to a linear model (R2) over cell diameter bins, identifying the cell diameter range where spindle length scales linearly with cell size as having the highest R2 value. This analysis revealed that across organisms both the pole-to-pole and aster-to-aster mitotic spindle lengths scaled linearly with cell size in cells less than 140 μm in diameter (Figure 2B). Organisms with embryos smaller than 140 μm, including S. purpuratus, C. marginatus, and C. elegans, displayed linear mitotic spindle scaling to cell size from the very first division (see Figure S1C), with the exception of M. musculus. In mice, mitotic spindles did not begin scaling linearly to cell size until the second division, when cell diameter was less than 60 μm. One distinguishing feature of early mouse embryos is their lack of a paternally contributed centrosome at fertilization [9,14,15], a unique feature of rodent zygotes that might contribute to differences in spindle scaling. To test the validity of this approach, we compared the R2 analysis of all species, which shows peak values at 110–140 μm, with a piecewise regression analysis that plots a two component linear model to describe the scaling trends (linear scaling regime and plateau). This analysis revealed an optimal break point between two linear models at cell diameters of 110 – 120 μm, further supporting a linear spindle scaling regime at cell diameters below 140 μm (Figure S2A and S2B).
Within the linear scaling range, we observed a consistent cell diameter to aster-to-aster mitotic spindle length ratio of ~2.5 among organisms, varying between 1.9 ± 0.3 in C. marginatus to 3.0 ± 1.3 in H. robusta (Figure 2C). The ratio of cell diameter to pole-to-pole spindle length was also consistent at ~ 3.0 (Figure S2C). Therefore, the ratio of spindle length to cell diameter is conserved and maintained in early development across organisms in cells smaller than 140 μm. In addition, within this regime spindle width (Figure 2D and S2D) and metaphase plate length (Figure 2E and S2E) correlated with cell diameter, showing a linear relationship when plotted on a log2 - log2 scale (Figure S2F and S2G). Moreover, across organisms the area of the spindle correlated linearly with the area of the metaphase plate (r= 0.73, p<0.001; Figure 2F and S2H). In conjunction with previous work demonstrating that mitotic spindle assembly and shape is dependent on chromatin-derived signals [18,19] and that mitotic chromosomes scale in size during development [20,21], these data support the hypothesis that mechanisms regulating spindle size and mitotic chromosome condensation are conserved and coupled. Physical constraints dictated by cell volume that limit the amount of cellular material, for example decreasing levels of tubulin and other factors, may act as a general mechanism to reduce the size of intracellular structures as cells divide rapidly in the absence of growth during early embryogenesis [7,8]. In addition, highly conserved regulatory factors may function to coordinate subcellular scaling during early embryogenesis. For example, in X. laevis decreasing cytoplasmic levels of the transport factor importin α during development modulates both nuclear size and spindle size through regulation of nuclear import and microtubule stability, respectively [3,22].
Female meiotic spindles do not scale to egg size
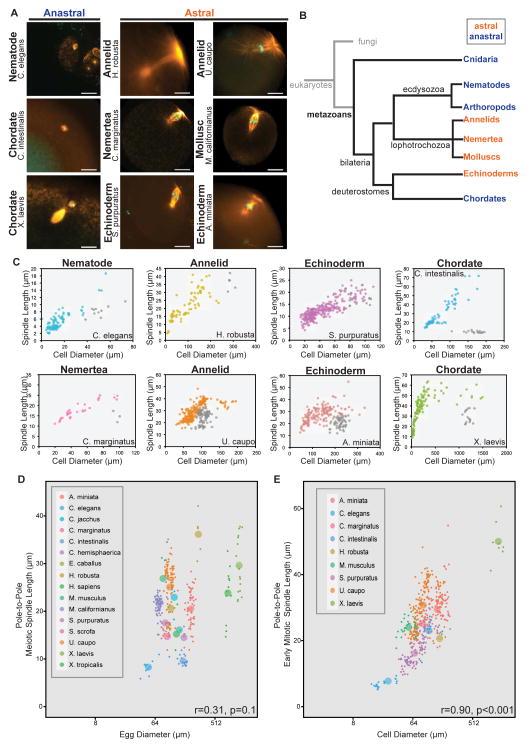
To date, all spindle scaling data is derived from mitotic spindle measurements [1–4] with the exception of data from male meiosis in Drosophila [23] and studies using unfertilized X. laevis egg cytoplasm, which recapitulates meiotic spindle assembly in vitro using demembranated sperm nuclei [2,7,24]. To examine female meiotic spindle features more broadly we fixed, stained and imaged eggs from ten organisms representing six different phyla (Figure 3A). In addition, published images of fixed female meiotic spindles from several organisms were included in the analysis (Figure 3D; [9,25–38]).
Figure 3. Meiotic spindles do not scale to egg size.
(A) Images of female meiotic spindles stained for tubulin (orange) and DNA (cyan). Scale bars = 20 μm, MI=meiosis I, MII= meiosis II. (B) Phylogenetic tree indicating phyla with astral (orange) or anastral (blue) female meiotic spindles. (C) Individual species plots of pole-to-pole mitotic (color) and meiotic (grey) spindle length versus cell diameter. (D) Female meiotic pole-to-pole spindle length versus egg diameter (log2 scale), r=0.31, p= 0.1. (E) Average mitotic pole-to-pole spindle length versus cell diameter (log2 scale) from 1- to 4- cell embryos, r= 0.90, p< 0.001. (C–E) Individual data points represent a single spindle measurement and different colors represent different species as indicated. (D and E) Larger points represent averages for each species.
As for mitotic spindles, great diversity in meiotic spindle morphology was observed among metazoans. Meiotic spindles were smaller than the largest mitotic spindles of the same species, and while mitotic spindle poles were associated with astral microtubules, meiotic spindles could be divided into two categories. Species from phyla including Cnidaria, Nematoda, Arthropoda, and Chordata possessed anastral female meiotic spindles that lacked apparent microtubule organizing centers at the poles (Figure 3A; [26,29]). These species represent the major animal model organisms used for studying mechanisms of meiotic spindle positioning and dynamics, namely frogs, mice, flies, and worms. In contrast, we observed that organisms of several animal phyla, including Echinodermata (S. purpuratus, A. miniata), Annelida (H. robusta, U. caupo), Mollusca (M. californianus), and Nemertea (C. marginatus), possessed female meiotic spindles with astral microtubules, indicating the presence of centrosomes (Figure 3A). In animals, the maternal centrosome is degraded or eliminated prior to the first embryonic division [39], but the timing of its demise differs, and our results indicate a role for the centrosome in female meiotic spindle assembly and function in some species. The evolutionary relationships among phyla with astral or anastral female meiotic spindles indicates that loss of centrosome function during female meiosis has evolved dynamically among metazoa (Figure 3B). Centrosome loss or maintenance during oogenesis is a major source of diversity among female animal meiotic spindles, but the underlying mechanisms are poorly understood.
In comparing female meiotic spindles to early embryo mitotic spindles we found that, with the exception of H. robusta, meiotic spindle lengths did not scale to cell size in a manner similar to mitotic spindle length scaling (Figure 3C). In each organism, meiotic spindle lengths were significantly shorter than mitotic spindle lengths in the first embryonic division, most dramatically in species in which the anastral meiotic spindle inhabits a large oocyte (C. intestinalis and X. laevis). Both the pole-to-pole (r=0.31, p= 0.1) and aster-to-aster (r=0.18, p=0.2) female meiotic spindle lengths did not correlate with egg size in either astral or anastral spindles (Figure 3D, S3A and S3B), which is in stark contrast to the strong correlation between first mitotic spindle length and zygote size (r=0.90, p<0.001; Figure 3E; [13]). In search of other parameters that correlate with female meiotic spindle length, we analyzed polar body size (r=0.29, p=0.1; Figure S3C), genome size (r=0.25, p=0.2; Figure S3D), diploid chromosome number (r=0.15, p=0.3; Figure S3E), and genome size normalized to diploid chromosome number (r=0.12, p=0.3; Figure S3F), but did not identify obvious scaling relationships. However, across a ~25-fold range in egg sizes (50 μm – 1300 μm) we observed a relatively small ~3.5-fold range in female meiotic spindle lengths (10 μm – 35 μm), significantly smaller than the ~6-fold range of early mitotic spindle lengths (10 μm – 60 μm). These results suggest that female meiotic spindle length is under different regulation than mitotic spindle length, which is not surprising considering the divergent roles of mitotic versus female meiotic spindles. Whereas mitotic spindles serve to equatorially segregate replicated chromosomes so that each of the newly formed daughter cells inherits identical genomic content from the parent cell, female meiotic spindles reductionally segregate chromosomes so that half of the genomic content is retained in the egg and the other half discarded. Furthermore, aster separation is needed to direct cytokinetic furrow formation in mitotic cells [40], while extreme asymmetric spindle positioning obviates this relationship in female meiosis. Therefore, while the general function of both mitotic and female meiotic spindles is to segregate chromosomes, their structures are dissimilar and likely require distinct regulatory mechanisms.
Spindle morphometrics differ between meiosis and mitosis
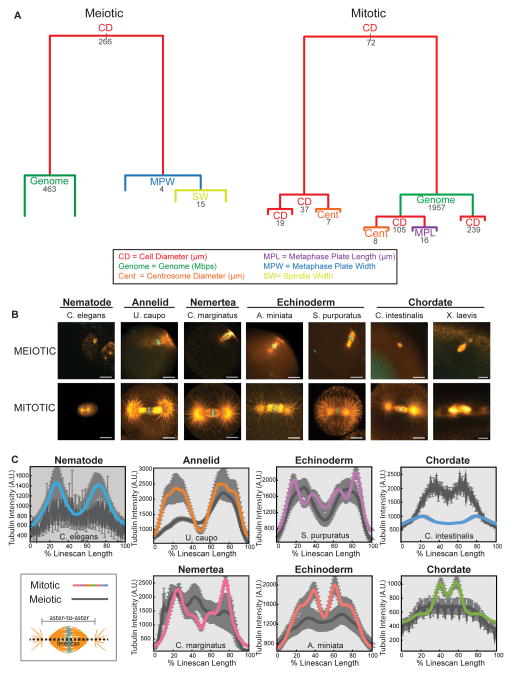
Our initial analysis revealed that most of the variation in mitotic spindle length could be ascribed to a direct linear relationship with cell size, but that a correlation between meiotic spindle size and cell size was not as apparent (Figure 3D and E). To more quantitatively define the morphometric and biological features that explain spindle size differences we applied a computational analysis using regression tree models (see Supplemental Experimental Procedures). The complete meiotic and mitotic data sets were analyzed to identify parameters that accounted for approximately 80% of variation in spindle size (Figure 4A). This analysis confirmed that meiotic and mitotic spindle sizes differ in the character of their dependence on the cell size. Interestingly, in both cases cell diameter accounted for the largest portion of variation in spindle size, although the fraction is larger for mitotic spindles (55% and 40% for mitotic and meiotic spindles, respectively). For meiotic spindles, genome size, metaphase plate width, and spindle width also contributed to variation in spindle length whereas genome size, centrosome diameter, and metaphase plate length contributed to variation in mitotic spindle length. Thus, although a direct relationship was not obvious between meiotic spindle length and cell size (Figure 3D), based on the regression tree analysis, cell size is the strongest predictor of meiotic spindle length. These findings highlight that different relationships exist between cell size and spindle size between meiosis and mitosis, and identify other parameters that differentially correlate with spindle variation.
Figure 4. Spindle morphometrics differ between meiotic and mitotic spindles.
(A) Regression tree models for meiotic and mitotic spindles obtained through recursive partitioning of continuous explanatory variables (x(i)) with meiotic or mitotic spindle size as response variable (y). Split corresponds to the biggest change in explained spindle size variation and the length of branches reflects the portion of variation explained. Numbers indicate explanatory variable values at each split. (B) Images of female meiotic spindles (top panel) and mitotic spindles from the first or second embryonic division (bottom panel) stained for tubulin (orange) and DNA (cyan). Scale bars = 20 μm. (C) Plots of integrated average microtubule intensity quantified from a 30 pixel-wide linescan that extended beyond the aster-to-aster spindle length. Intensity plots are scaled to 100% linescan length and averaged from metaphase mitotic spindles (color) from 1- to 4-cell embryos and female meiotic spindles (dark gray), error bars (light grey) = SEM.
It is remarkable that there are negligible changes in cell shape or size at the transition from meiotic to mitotic divisions that follow fertilization, yet these two spindle types vary substantially in structure and cell size scaling relationships. To investigate this dramatic shift in spindle morphometrics, we compared meiotic and early mitotic spindles from various organisms. In addition to an increase in spindle length, the meiotic-to-mitotic transition occurred with an abrupt change in spindle morphology and spindle positioning within the cell (Figure 4B), illustrated by comparing a variety of spindle measurements between female meiotic and early mitotic spindles (Figure S4A). Plotting the pattern of average microtubule intensity along the aster-to-aster spindle length revealed distinct profiles between meiotic and early mitotic spindles (Figure 4C). Early mitotic spindles showed distinct peaks of microtubule intensity in the interpolar region and at the asters that were not as apparent for meiotic spindles. Not surprisingly, differences in spindle parameters and microtubule intensity profiles between meiotic and mitotic spindles were most prominent in organisms with anastral female meiotic spindles.
The progression from meiotic egg to mitotic zygote is a fundamental transition in animal development. It was recently proposed that the transition from meiotic to mitotic spindle assembly occurs gradually over the first eight embryonic divisions during mammalian development [9]. However, this was based on studies in mouse development, in which there is no paternal contribution of centrioles to the zygote during fertilization [39]. Our data suggests that across animals, with the exception of rodents, the meiotic-to-mitotic transition catalyzes abrupt changes in spindle morphometrics.
In conclusion, we found that mitotic spindle scaling in the early embryo is not only conserved across metazoan phyla but that linear scaling occurs within a similar cell size range and that a consistent cell diameter to spindle length ratio is maintained in early development across organisms. In contrast, female meiotic spindles do not scale linearly to egg size, highlighting one of the many features that vary between female meiotic and mitotic spindles and change abruptly at fertilization. Our analysis demonstrates that features of mitotic and meiotic spindles are shared among animal phyla, which suggests conservation of mechanisms that determine spindle shape and size across metazoans. However, the molecular nature of these mechanisms is poorly understood. For example, why is the cell-to-spindle size ratio not maintained in larger cells where there are no constraints on abundance of cytoplasmic materials or geometric restrictions, and what factors initiate linear spindle scaling in cells less than 140 μm in diameter? What determines the narrow range of female meiotic spindle sizes, and what is the contribution of the centrosome if it is present? What molecular changes occur at the meiotic-to-mitotic transition that profoundly alter spindle architecture? These questions necessitate future studies in diverse model systems to elucidate conservation and deviations among mechanisms that regulate spindle structure and function across species and during development.
Supplementary Material
Highlights.
Early embryo mitotic spindles scale to cell size across metazoans
Linear mitotic spindle scaling occurs within a conserved regime of cell sizes
Across animals, female meiotic spindles do not scale to egg size
Spindle morphometrics change abruptly at the meiosis-to-mitosis transition
Acknowledgments
We thank University of California faculty David Weisblat (H. robusta), Michael Levine (C. intestinalis), Abby Dernburg (C. elegans), and Fred Wilt (S. purpuratus) for access to the indicated species, and for distribution of specific reagents and equipment needed to handle various organisms. We are also indebted to Dr. Chris Killian (University of California Berkeley), and to faculty and staff at the Bodega Marine Laboratory, specifically Dr. Gary Cherr (BML, University of California, Davis), Dr. Carol Vines (BML, University of California), and Karl Menard for generously providing guidance and assistance with handling various marine invertebrates as well as space for carrying out off-site experiments. We thank the Cancer Research Laboratory Molecular Imaging Center (University of California, Berkeley) for use of microscopy equipment and Dr. Andreas Ettinger for advice on statistical analysis and coding. We would also like to thank Dr. Zac Cande, Dr. Nicole King, Dr. Andy Lane, Dr. Kara Helmke, Dr. Chris Brownlee, Dr. Romain Gibeaux, Dr. Lauren Slevin, Andrew Grenfell and Kelly Miller for helpful suggestions and discussions. The CRL Molecular Imaging Center was supported by the NSF grant DBI-1041078. M.E.C. was supported by the National Institutes of Health (NIH) Ruth L. Kirschstein National Research Service Award 5F32 GM105199. This study was also supported by the NIH grant R01 GM057839 to R.H. and by NSF grant MCB-0917887 to G. von D.
Footnotes
Author Contributions
M.E.C, J.D.W, and R.H. conceived the project. M.E.C. performed most experimental procedures, data collection, measuring, and analysis. M.S. performed all computational and statistical analysis, and created online repository for the project. J.D.W, M.C.G, and G. von D. contributed experimental data. M.E.C., M.S., and R.H. prepared the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wuhr M, Chen Y, Dumont S, Groen AC, Needleman DJ, Salic A, Mitchison TJ. Evidence for an upper limit to mitotic spindle length. Curr Biol. 2008;16:1256–1261. doi: 10.1016/j.cub.2008.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hara Y, Kimura A. Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr Biol. 2009;18:1549–1554. doi: 10.1016/j.cub.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 3.Wilbur JD, Heald R. Mitotic spindle scaling during Xenopus development by kif2a and importin alpha. Elife. 2013:e00290. doi: 10.7554/eLife.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hara Y, Kimura A. An allometric relationship between mitotic spindle width, spindle length, and ploidy in Caenorhabditis elegans embryos. Mol Biol Cell. 2013;9:1411–1419. doi: 10.1091/mbc.E12-07-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reber SB, Baumgart J, Widlund PO, Pozniakovsky A, Howard J, Hyman AA, Julicher F. XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat Cell Biol. 2013;9:1116–1122. doi: 10.1038/ncb2834. [DOI] [PubMed] [Google Scholar]
- 6.Young S, Besson S, Welburn JP. Length-dependent anisotropic scaling of spindle shape. Biol Open. 2014;12:1217–1223. doi: 10.1242/bio.201410363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. Cytoplasmic volume modulates spindle size during embryogenesis. Science. 2013;6160:856–860. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science. 2013;6160:853–856. doi: 10.1126/science.1243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courtois A, Schuh M, Ellenberg J, Hiiragi T. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol. 2012;3:357–370. doi: 10.1083/jcb.201202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decker M, Jaensch S, Pozniakovsky A, Zinke A, O’Connell KF, Zachariae W, Myers E, Hyman AA. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr Biol. 2011;15:1259–1267. doi: 10.1016/j.cub.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Ishihara K, Nguyen PA, Groen AC, Field CM, Mitchison TJ. Microtubule nucleation remote from centrosomes may explain how asters span large cells. Proc Natl Acad Sci U S A. 2014;50:17715–17722. doi: 10.1073/pnas.1418796111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wuhr M, Tan ES, Parker SK, Detrich HW, 3rd, Mitchison TJ. A model for cleavage plane determination in early amphibian and fish embryos. Curr Biol. 2010;22:2040–2045. doi: 10.1016/j.cub.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farhadifar R, Baer CF, Valfort AC, Andersen EC, Muller-Reichert T, Delattre M, Needleman DJ. Scaling, Selection, and Evolutionary Dynamics of the Mitotic Spindle. Curr Biol. 2015 doi: 10.1016/j.cub.2014.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manandhar G, Sutovsky P, Joshi HC, Stearns T, Schatten G. Centrosome reduction during mouse spermiogenesis. Dev Biol. 1998;2:424–434. doi: 10.1006/dbio.1998.8947. [DOI] [PubMed] [Google Scholar]
- 15.Schatten G. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol. 1994;2:299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- 16.Huang FZ, Kang D, Ramirez-Weber FA, Bissen ST, Weisblat DA. Micromere lineages in the glossiphoniid leech Helobdella. Development. 2002;3:719–732. doi: 10.1242/dev.129.3.719. [DOI] [PubMed] [Google Scholar]
- 17.Weisblat DA. Asymmetric cell divisions in the early embryo of the leech Helobdella robusta. Prog Mol Subcell Biol. 2007:79–95. doi: 10.1007/978-3-540-69161-7_4. [DOI] [PubMed] [Google Scholar]
- 18.Dinarina A, Pugieux C, Corral MM, Loose M, Spatz J, Karsenti E, Nedelec F. Chromatin shapes the mitotic spindle. Cell. 2009;3:502–513. doi: 10.1016/j.cell.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 19.O’Connell CB, Khodjakov AL. Cooperative mechanisms of mitotic spindle formation. J Cell Sci Pt. 2007;10:1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- 20.Kieserman EK, Heald R. Mitotic chromosome size scaling in Xenopus. Cell Cycle. 2011;22:3863–3870. doi: 10.4161/cc.10.22.17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara Y, Iwabuchi M, Ohsumi K, Kimura A. Intranuclear DNA density affects chromosome condensation in metazoans. Mol Biol Cell. 2013;15:2442–2453. doi: 10.1091/mbc.E13-01-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy DL, Heald R. Nuclear size is regulated by importin alpha and Ntf2 in Xenopus. Cell. 2010;2:288–298. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lattao R, Bonaccorsi S, Gatti M. Giant meiotic spindles in males from Drosophila species with giant sperm tails. J Cell Sci Pt. 2012;3:584–588. doi: 10.1242/jcs.101469. [DOI] [PubMed] [Google Scholar]
- 24.Loughlin R, Wilbur JD, McNally FJ, Nedelec FJ, Heald R. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 2011;6:1397–1407. doi: 10.1016/j.cell.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delimitreva S, Tkachenko OY, Berenson A, Nayudu PL. Variations of chromatin, tubulin and actin structures in primate oocytes arrested during in vitro maturation and fertilization--what is this telling us about the relationships between cytoskeletal and chromatin meiotic defects? Theriogenology. 2012;7:1297–1311. doi: 10.1016/j.theriogenology.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 26.Amiel A, Leclere L, Robert L, Chevalier S, Houliston E. Conserved functions for Mos in eumetazoan oocyte maturation revealed by studies in a cnidarian. Curr Biol. 2009;4:305–311. doi: 10.1016/j.cub.2008.12.054. [DOI] [PubMed] [Google Scholar]
- 27.Dekens MP, Pelegri FJ, Maischein HM, Nusslein-Volhard C. The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development. 2003;17:3907–3916. doi: 10.1242/dev.00606. [DOI] [PubMed] [Google Scholar]
- 28.Nair S, Marlow F, Abrams E, Kapp L, Mullins MC, Pelegri F. The chromosomal passenger protein birc5b organizes microfilaments and germ plasm in the zebrafish embryo. PLoS Genet. 2013;4:e1003448. doi: 10.1371/journal.pgen.1003448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radford SJ, Harrison AM, McKim KS. Microtubule-depolymerizing kinesin KLP10A restricts the length of the acentrosomal meiotic spindle in Drosophila females. Genetics. 2012;2:431–440. doi: 10.1534/genetics.112.143503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tremoleda JL, Van Haeften T, Stout TA, Colenbrander B, Bevers MM. Cytoskeleton and chromatin reorganization in horse oocytes following intracytoplasmic sperm injection: patterns associated with normal and defective fertilization. Biol Reprod. 2003;1:186–194. doi: 10.1095/biolreprod.102.012823. [DOI] [PubMed] [Google Scholar]
- 31.Coticchio G, Guglielmo MC, Dal Canto M, Fadini R, Mignini Renzini M, De Ponti E, Brambillasca F, Albertini DF. Mechanistic foundations of the metaphase II spindle of human oocytes matured in vivo and in vitro. Hum Reprod. 2013;12:3271–3282. doi: 10.1093/humrep/det381. [DOI] [PubMed] [Google Scholar]
- 32.Duncan FE, Hornick JE, Lampson MA, Schultz RM, Shea LD, Woodruff TK. Chromosome cohesion decreases in human eggs with advanced maternal age. Aging Cell. 2012;6:1121–1124. doi: 10.1111/j.1474-9726.2012.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim NH, Chung HM, Cha KY, Chung KS. Microtubule and microfilament organization in maturing human oocytes. Hum Reprod. 1998;8:2217–2222. doi: 10.1093/humrep/13.8.2217. [DOI] [PubMed] [Google Scholar]
- 34.Nichols SM, Gierbolini L, Gonzalez-Martinez JA, Bavister BD. Effects of in vitro maturation and age on oocyte quality in the rhesus macaque Macaca mulatta. Fertil Steril. 2010;5:1591–1600. doi: 10.1016/j.fertnstert.2008.12.141. [DOI] [PubMed] [Google Scholar]
- 35.Breed WG, Simerly C, Navara CS, VandeBerg JL, Schatten G. Microtubule configurations in oocytes, zygotes, and early embryos of a marsupial, Monodelphis domestica. Dev Biol. 1994;1:230–240. doi: 10.1006/dbio.1994.1194. [DOI] [PubMed] [Google Scholar]
- 36.Brunet S, Maria AS, Guillaud P, Dujardin D, Kubiak JZ, Maro B. Kinetochore fibers are not involved in the formation of the first meiotic spindle in mouse oocytes, but control the exit from the first meiotic M phase. J Cell Biol. 1999;1:1–12. doi: 10.1083/jcb.146.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunet S, Dumont J, Lee KW, Kinoshita K, Hikal P, Gruss OJ, Maro B, Verlhac MH. Meiotic regulation of TPX2 protein levels governs cell cycle progression in mouse oocytes. PLoS One. 2008;10:e3338. doi: 10.1371/journal.pone.0003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueno S, Kurome M, Ueda H, Tomii R, Hiruma K, Nagashima H. Effects of maturation conditions on spindle morphology in porcine MII oocytes. J Reprod Dev. 2005;3:405–410. doi: 10.1262/jrd.16091. [DOI] [PubMed] [Google Scholar]
- 39.Manandhar G, Schatten H, Sutovsky P. Centrosome reduction during gametogenesis and its significance. Biol Reprod. 2005;1:2–13. doi: 10.1095/biolreprod.104.031245. [DOI] [PubMed] [Google Scholar]
- 40.Lewellyn L, Dumont J, Desai A, Oegema K. Analyzing the effects of delaying aster separation on furrow formation during cytokinesis in the Caenorhabditis elegans embryo. Mol Biol Cell. 2010;1:50–62. doi: 10.1091/mbc.E09-01-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.