Abstract
Nowadays, one can hardly imagine biology and medicine without flow cytometry to measure CD4 T cell counts in HIV, follow bone marrow transplant patients, characterize leukemias, etc. Similarly, without flow cytometry, there would be a bleak future for stem cell deployment, HIV drug development and full characterization of the cells and cell interactions in the immune system. But while flow instruments have improved markedly, the development of automated tools for processing and analyzing flow data has lagged sorely behind. To address this deficit, we have developed automated flow analysis software technology, provisionally named AutoComp and AutoGate. AutoComp acquires sample and reagent labels from users or flow data files, and uses this information to complete the flow data compensation task. AutoGate replaces the manual subsetting capabilities provided by current analysis packages with newly defined statistical algorithms that automatically and accurately detect, display and delineate subsets in well-labeled and well-recognized formats (histograms, contour and dot plots). Users guide analyses by successively specifying axes (flow parameters) for data subset displays and selecting statistically defined subsets to be used for the next analysis round. Ultimately, this process generates analysis “trees” that can be applied to automatically guide analyses for similar samples. The first AutoComp/AutoGate version is currently in the hands of a small group of users at Stanford, Emory and NIH. When this “early adopter” phase is complete, the authors expect to distribute the software free of charge to .edu, .org and .gov users.
Keywords: Multiparameter flow cytometry, Automating fluorescence compensation, Automatic cell subsets identification, Guiding gating strategy
Introduction
Flow cytometry is so deeply ingrained in the science and practice of medicine, and in basic studies that precede the work in the clinic that it is hard to imagine where many areas of biotechnology would be without appreciating what this widely used technology has contributed. However, anyone who has been faced with extracting meaningful data from multiparameter flow cytometry data sets is well aware of the skill required and the painstaking work needed, for this repetitively encountered task. For the novice, this can be a nerve-wracking and incredibly time-consuming experience. For the skilled practitioner, it is a labor whose recompense lies mainly in the value of the findings that may be had—the pot of gold at the end of the rainbow. It is a job that few love and fewer would claim to have fully mastered. Basically, while current reagents and instrumentation readily enable robust data collection, the data collection capabilities of flow hardware have well outstripped the ability of current flow software to enable users to fully or easily extract findings once data collection is complete.
To address this deficit, we have recently developed automated1 flow analysis software, provisionally named AutoComp and AutoGate. AutoComp replaces the manual fluorescence compensation software necessary to prepare flow data for analysis. AutoGate replaces the manual sub-setting capabilities provided by current analysis packages with newly defined statistical algorithms that automatically detect, visualize and compare subsets within a given sample. Users choose which of the visualized subsets to gate further and then choose which parameters to use for the next round of visualization and gating.
In essence, AutoGate obviates the need to draw arbitrary gates to define the subsets in a gating model. It defines these algorithmically instead. However, it keeps the user firmly in control of the analysis by enabling the user to choose the axes on which to display newly defined subsets and to choose which of the newly defined subsets to subset further. In addition, AutoGate also provides innovative statistical tools that enable users to determine whether subsets identified in a gating model built from data collected from a given sample are represented in data from similarly stained samples.
In the sections that follow, we present a bit of background information and then move to a description of AutoComp and AutoGate and the output this software generates. We focus here on fluorescence-based flow cytometry. However, except for details about fluorescence compensation, the discussion is equally applicable to CY-TOF data analysis or data collected with any other system where multiparameter data are collected for samples containing large numbers of items.
Results
Here, we present a novel package of innovative techniques called AutoGate to automate key flow data handling and data processing tasks:
Staining the samples
Subsets are defined by quantitative or qualitative co-expression of individual markers on/in cells. New researchers working with flow cytometry for the first time do not always appreciate that no matter how many subsets are identified within a given cell sample, more subsets may often be found when additional markers (more parameters) are used to analyze these or additional samples from the same or an appropriately related source. For these reasons, we usually advise colleagues to stain samples with as many fluorescence parameters as are compatible with their budgets and reagent availability, and to collect and store data for all of these markers.
Outside of cost and reagent availability, the broad approach of using more markers to better delineate cell subsets has been limited by the requirement for advanced fluorescence compensation and subset gating skills. However, the AutoComp and AutoGate combination provided by our new software largely removes these limitations and thus opens high-dimension fluorescence-based analysis to a much wider group of users.
Flow data collection and storage
The mechanics of flow cytometry data collection have been explained in greater or lesser detail in many publications. Briefly, cells are stained with antibodies and sometimes other reagents that are each associated with a distinctive fluorochrome. FACS instruments collect and record data for two to twenty measurements per cell for up to several million cells in a given sample. Results for each sample are stored separately in lengthy data files that may, for example, wind up containing measurements for each of 500,000 cells in the sample.
In the Stanford Shared FACS Facility (and now several other flow facilities at Stanford), the data files collected for a given flow experiment are usually packaged into a dataset and automatically stored centrally in a common data archive (built explicitly by Wayne Moore in our laboratory in 2003 to enable automatic long term, secure storage of FACS data). Still providing similar (albeit upgraded) functions today, users need only click once or twice to download previously collected datasets in a form that can directly be used by AutoGate, FlowJo or other co-operating data analysis programs.
Automating fluorescence compensation (AutoComp): supervised automation of the preparation of flow data for analysis
Data analysis for a flow cytometry dataset begins with a computation, referred to as fluorescence compensation that corrects for fluorescence overlap and reveals the actual amounts of fluorescence emitted by each of the fluoro-chromes associated with each of the reagents bound to each of the cells in the sample. This computation is usually necessary in multiparameter flow fluorescence experiments, even when only two or three colors are used, since the fluorescence (light) emitted by available fluorochromes commonly overlaps to some extent onto detector(s) for other fluorochromes and therefore spuriously contributes to the total fluorescence recorded by that detector. (We discuss this in detail in a 2006 review in Nature Immunology [1], which also provides key background for other instrumentation issues discussed here.)
At present, a variety of fluorescence compensation implementations are offered by current flow analysis packages. The well-known FlowJo data analysis package (TreeStar.com), for example, provides a reasonably well-automated compensation utility that applies compensation corrections to the appropriate data sets prior to analysis (although additional corrections sometimes need to be applied, as well). However, this and other current compensation methods all require human intervention and do not provide any sort of quality control information to detect compensation errors or identify inadequate samples [2–4]. Therefore, there is a need to create a new compensation data model to address these human errors and to deal with instrumentation and other types of errors that bedevil compensation. The fully automated fluorescence compensation computations provided by AutoComp serves these purposes by automatically computing the compensation values and requiring user input when necessary to resolve problems if they occur.
Thus, using advanced and often newly developed statistical procedures, AutoComp
prunes the data to remove spurious values that can skew computation of compensation corrections;
computes compensation corrections based on all of the data collected for each compensation sample, rather than just the upper and lower data clusters used by current methods;
corrects for high-end and low-end errors in data collection, which occur because some values “pile up” on the upper and lower bounds of the data set;
computes compensation corrections and reports these together with error estimations that indicate the reliability of the correction; and,
provides graphical representations and error estimates of the computed data for each measured parameter and, finally,
provides clear advice as to whether it is safe to proceed with applying the compensation values to the experiment data or whether significant errors were encountered and need user attention before proceeding (or aborting).
Once the user chooses to proceed, AutoComp applies the appropriate compensation corrections to the data for each of the samples, thus readying them without further user attention for AutoGate analysis.
Automation applied to gating for flow data
Once the compensation corrections are computed and applied, flow datasets are ready for analysis procedures aimed at identifying subsets of cells and at determining the frequency and/or the level(s) of marker representation on the identified subsets. With fluorescence-based (FACS) analyses, instruments report the amounts of individual fluorescent-labeled antibodies or other fluorescent markers associated with individual cells. Currently available flow instruments offer a variety of live display options to monitor and interpret the fluorescence measurements as they are acquired. However, in most cases, the data are written to (FCS standard) files that are stored for subsequent analysis.
During data analysis, cells that display similar combinations of markers or levels of markers are defined as belonging the same subset. A variety of software packages are available for visualizing and delineating such subsets. Some of these have (semi) automated components for computing and applying compensation corrections and/or for putting gates around subsets. Most enable application of the gates to data sets for other samples and provide for manual adjustment when necessary to better fit the for these samples. AutoGate also provides these capabilities. However, more importantly, AutoGate provides the user with the ability to automatically delineate subsets of cells, to select individual subsets or combine subsets for further subsetting, to apply the same subsetting procedure to similar samples and to automatically compare samples for the presence of absence of individual subsets.
Collectively, these and other statistical and computation advances in the methods built into AutoComp largely result in stable axes display that fit the data very well and do not call out for the “re-transformation” that often is needed for data display with current methods. In fact, our experience to date indicates that AutoComp performs well on various types of data set and seldom reports errors when the appropriate compensation controls are present and correctly labeled in the data set for a given experiment.
AutoComp and AutoGate software are both amenable for use at the time data are being collected, for example, during sorting or to enable monitoring during data collection. However, cooperation from the flow instrument manufacturers will be required to enable this “on line” use.
Basically, AutoGate uses mathematical procedures to rapidly delineate and present the user with statistically valid subsets drawn from the available data at each analysis round. Users simply select the subsets and select the axes (fluorescence markers) they want to use for the next round they want to use to display the output for the next analysis round. They then continue this rapid iterative procedure until they have created a gating model that captures the subsets of interest for the model datasets (see Fig. 1). To further facilitate the definition of this gating model, AutoGate ranks the axis choices at each iteration according to the probability of being useful (see Fig. 2). Thus, this new software system sequentially guides the iterative selection of axes to create lineal or branched analysis trees (aka gating trees or gating models).
Fig. 1.
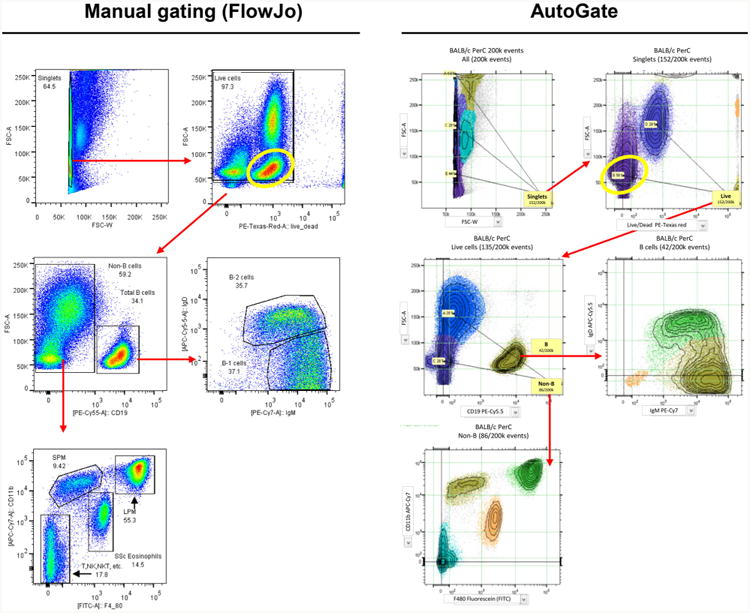
Comparison of gating using FlowJo software (manual) and AutoGate. The researcher in this example (Eliver Ghosn, Genetics Department, Stanford) initially used FlowJo to draw gates (left panel) that delineate macrophage and other subsets (clusters) revealed by FACS analysis of cells isolated from the peritoneal cavity (PerC) of unimmunized (naïve) mice [5]. At successive steps in this recursive procedure, the user locates and draws gates based on the color density plot representation of regions containing data of interest. The panel on the right shows that corresponding subsets automatically identified by AutoGate, which uses a unique color to identify the cells in distinct subsets. The yellow oval defines the location of the B cells in the sample. The X axis shows data for cells stained with a viability dye that is excluded from live cells and brightly stains dead cells (which mainly fall above 104 fluorescence units on the X axis). The position of the B cells (yellow ovals) differs between the FlowJo and AutoGate analysis because AutoGate automatically applies compensation corrections to all channels, including the B cell channel in this case. FlowJo, in contrast, allowed the user in this case to omit the compensation correction for this channel, leaving the B cells and the other cells in the sample with uncompensated signals that raised their fluorescence values to the second decade. Thus, the yellow oval and the B cells within it appear in different locations in the two plots, as do the other cells in the sample
Fig. 2.
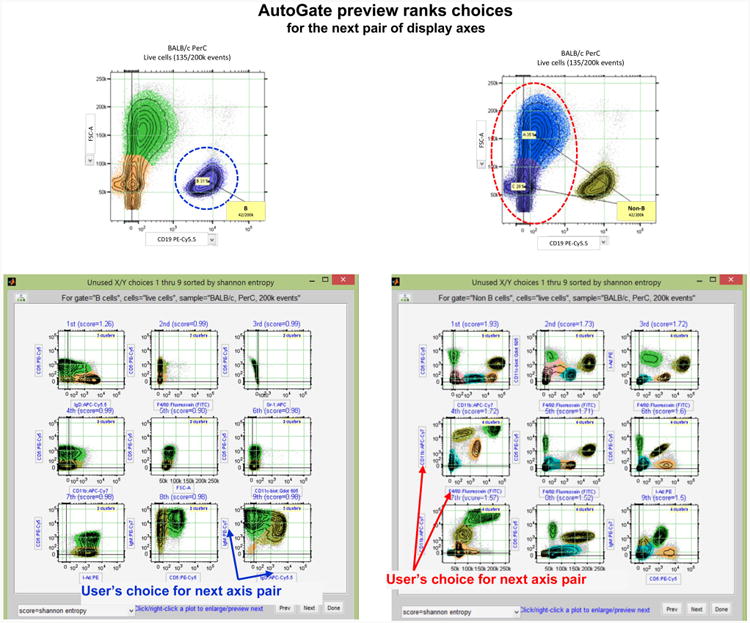
AutoGate preview ranks choices for the next pair of display axes. An example of the first 9 (out of 44 possible in this case) axis pairs which AutoGate advised as a next step in gating process. In both cases (B cells—highlighted with purple color and Non-B cells— highlighted with red color subsets—see Fig. 1 for details), user's choice of the axis pair among the first 9 axis pairs advised by AutoGate
Once a gating model is complete, the users simply select the additional datasets to which it should be applied and trigger the full analysis to complete automatically. For each sample, AutoGate automatically locates subsets defined in the model and creates a gating tree for the target sample. It fits the identified subsets with statistically defined bounds that approximate the bounds in the gating model but are appropriately modified to fit the data in the sample. In cases where a subset in the model is not discovered in the target sample, or where a subset is present in the sample but is not present in the model, AutoGate automatically displays a note to this effect in the appropriate location on the gating tree. Finally, AutoGate displays frequencies and other statistics for each subset it identifies.
In essence, AutoGate enables the sequential definition of subsets much the way current software does, but with certain practical differences. With current analyses software (e.g., FlowJo), users iteratively build a gating model by sequentially choosing sets of axes (staining parameters) to visualize the data, manually drawing boundaries (gates) around subsets of cells and then restricting the next visualization to the cells within a chosen gate (see Fig. 1). The series of specified gates for a given data set constitutes a gating model, which users can apply (with adjustments, when needed) to discover, visualize and quantitate similar subsets in other samples.
AutoGate similarly enables users to sequentially visualize data and select subsets, and to define and apply gating models. However, in addition to offering users traditional manual gating capabilities, AutoGate offers powerful statistical procedures that automatically locate and draw subset boundaries during the definition of a gating model. Furthermore, AutoGate's statistical arsenal offers powerful tools that can intelligently apply it to similarly stained samples to rapidly identify matching subsets, distinguish absent and additional subsets, and quantify differences between like subsets.
To date, we have developed and tested this method with FACS data sets that include up to 12 fluorescence and two light scatter measurements. However, we expect the method to be equally well usable for analysis of CYTOF and other very high-dimensional datasets, including those acquired for data outside the flow arena. The CYTOF instruments (http://www.dvssciences.com/mass-cytometry), which use mass spectrometry rather than fluorescence measurements to associate marker expression with cells, provide a creative way to relieve the need for complex compensation corrections. These instruments offer a much wider range of co-utilizable reagents on individual cells. However, limitations in the number of cells that can be analyzed per minute might restrict the routine use of these instruments to more highly represented subsets (or to very patient users). However, there clearly are situations where the featured high parameterization of CyTOF balances the benefits of speed on traditional fluorescence platforms. In any event, AutoGate can be expected to work equally well with data acquired with mass spectrometry and fluorescence-based reagents. Thus, the flow analysis automation tool discussed here (AutoGate) potentially expands the subset-defining capabilities of both types of instruments.
Thus, we see AutoGate, together with CytoGenie and AutoComp, as opening complex high-dimensional flow analysis to a broad group of users, most of whom currently have a much better understanding of how flow data inform biomedical studies than to use current technology to extract or evaluate high-dimensional flow data.
Discussion
Experience has shown that the greater the number of reagents used in a staining panel, the better the chance of resolving the subsets in a targeted sample and of determining the expression levels of various determinants on the cells within the subsets. In today's flow world, even average users working on typical projects with cells from human or animal sources have come to recognize that the complexity of the cell populations with which they work requires them to deal at least with 6-12 color data—and upwards of that if their work requires data collection with today's high-powered FACS and CyTOF instruments. However, as we and others have recognized for some time now, the manual analysis packages that have served us so well for many years are simply not up to enabling facile handling of data acquired with these modern high-powered instruments.
AutoGate, in contrast, provides the analysis automation necessary to meet the needs of users working with the large, high-dimensional datasets that modern flow instruments (including CYTOF) generate. Basically, with Aut-oGate, users no longer have to manually draw and apply arbitrary gates to delineate subsets, nor do they have to manually apply and adjust these gates to extract subset data from like samples. Instead, AutoGate provides an automated iterative procedure that guides the users through successively refined gating decisions to generate a gating “tree” that can then be applied in automated analyses of samples for which comparable datasets are available.
Our laboratory wrote some of the earliest flow data display packages, and has contributed to this area over the years. AutoGate now brings us back into this arena, this time with a package that significantly simplifies the mechanics of flow data analysis and significantly speeds up the process. The work is a further contribution to provide users tools that allow them to focus on the meaning of the data than on the mechanics of producing it.
Since most of the work on this project was done at Stanford University, principally in the Departments of Genetics and Statistics, we plan to secure agreement to distribute the AutoComp and AutoGate packages at minimal or no cost to flow users based in academic or government institutions (i.e., those meriting the.edu, .gov or .org designation). Negotiations toward this end are near completion.
Acknowledgments
Work described in this article is sponsored in part by the NIH Grant number R01AI098519.
Footnotes
This software in its entirety is appropriately designated “semi-automated” since the user remains in control and is required to “supervise” the process by making choices and providing other input at key points during the process. Once the user provides this input, the compensation, clustering and other statistical operations are triggered to perform as automated tasks that return to the user for inspection of output and for input of further guidance. We use the term “automated” here for textual simplicity, not to imply full automation of the analysis process, which (in our view) would not be an acceptable in a tool intended for the complex processes involved in flow data analysis.
Contributor Information
Stephen Meehan, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA, Department of Statistics, Stanford University, Stanford, CA 94305, USA.
Guenther Walther, Department of Statistics, Stanford University, Stanford, CA 94305, USA.
Wayne Moore, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
Darya Orlova, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA, Institute of Chemical Kinetics and Combustion, Novosibirsk 630090, Russia.
Connor Meehan, Department of Mathematics, California Institute of Technology, Pasadena, CA 91125, USA.
David Parks, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
Eliver Ghosn, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
Megan Philips, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
Erin Mitsunaga, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
Jeffrey Waters, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
Aaron Kantor, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
Ross Okamura, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
Solomon Owumi, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
Yang Yang, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
Leonard A. Herzenberg, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA
Leonore A. Herzenberg, Email: LeeHerz@Darwin.stanford.edu, Department of Genetics, Stanford University School of, Medicine, Stanford, CA 94305, USA.
References
- 1.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–5. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 2.Bagwell CB, Adams EG. Fluorescence spectral overlap compensation for any number of flow cytometry parameters. Ann NY Acad Sci. 1993;677:167–84. doi: 10.1111/j.1749-6632.1993.tb38775.x. [DOI] [PubMed] [Google Scholar]
- 3.Hahne F, et al. FlowCore: a bioconductor package for high throughput flow cytometry. BMC Bioinform. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roederer M. Compensation in flow cytometry. In: Paul Robinson J, et al., editors. Current protocols in cytometry/editorial board. Chapter 1. 2002. p. 14. Unit 1. [DOI] [PubMed] [Google Scholar]
- 5.Ghosn EE, et al. Two physically, functionally, and developmen-tally distinct peritoneal macrophage subsets. Proc Natl Acad Sci USA. 2010;107:2568–73. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]


