Abstract
BACKGROUND
MicroRNAs (miRNAs) are key regulators of gene expression in the human genome and may contribute to risk for neuropsychiatric disorders. miRNAs play an acknowledged role in the strongest of genetic risk factors for schizophrenia, 22q11.2 deletions. We hypothesized that in schizophrenia there would be an enrichment of other rare copy number variants (CNVs) that overlap miRNAs.
METHODS
Using high-resolution genome-wide microarrays and rigorous methods, we compared the miRNA content of rare CNVs in well-characterized cohorts of schizophrenia cases (n = 420) and comparison subjects, excluding 22q11.2 CNVs. We also performed a gene-set enrichment analysis of the predicted miRNA target genes.
RESULTS
The schizophrenia group was enriched for the proportion of individuals with a rare CNV overlapping a miRNA (3.29-fold increase over comparison subjects, p < .0001). The presence of a rare CNV overlapping a miRNA remained a significant predictor of schizophrenia case status (p = .0072) in a multivariate logistic regression model correcting for total CNV size. In contrast, comparable analyses correcting for CNV size showed no enrichment of rare CNVs overlapping protein-coding genes. A gene-set enrichment analysis indicated that predicted target genes of recurrent CNV-overlapped miRNAs in schizophrenia may be functionally enriched for neurodevelopmental processes, including axonogenesis and neuron projection development. Predicted gene targets driving these results included CAPRIN1, NEDD4, NTRK2, PAK2, RHOA, and SYNGAP1.
CONCLUSIONS
These data are the first to demonstrate a genome-wide role for CNVs overlapping miRNAs in the genetic risk for schizophrenia. The results provide support for an expanded multihit model of causation, with potential implications for miRNA-based therapeutics.
Keywords: CAPRIN1, Copy number variation, MicroRNA, NEDD4, NTRK2, PAK2, RHOA, Schizophrenia, SYNGAP1, 16p13.11
Rare structural genetic changes (copy number variants [CNVs]) contribute to genetic risk for neurodevelopmental/neuropsychiatric disorders such as schizophrenia (1). In particular, several large, rare, recurrent CNVs are established risk factors of moderate effect for this genetically complex disease (2–5). Large CNVs are more likely to disrupt multiple genes simultaneously, supporting a “multiple-hit” hypothesis in schizophrenia; further evidence has shown significant enrichment in schizophrenia of individuals with two or more rare CNVs of any size that overlap coding sequences (3). Another potential mechanism for disruption of multiple gene targets involves changes in expression of regulatory elements such as microRNAs (miRNAs) (6,7). miRNAs are small noncoding RNAs that bind to the 3′-UTR (untranslated region) of usually many messenger RNAs (8). Through multiple mechanisms affecting transcription and translation, miRNAs can regulate the expression of suites of genes important for development and lifelong cellular functioning (9).
Several lines of evidence support a role for miRNAs in the etiology of schizophrenia, including the elevated (~25-fold) risk imparted by the most common of the pathogenic recurrent CNVs, a 22q11.2 deletion (10–12). The microRNA miR-185 at the 22q11.2 locus and its downstream pathways have been implicated in schizophrenia (12). In addition, a gene disrupted by typical 22q11.2 deletions, DGCR8, is a key component of the microprocessor complex involved in miRNA biogenesis (8). Reduced dosage of this gene causes global alterations in the miRNA complement, a potential mechanism for the high risk of schizophrenia with 22q11.2 deletions that is supported by mouse models (13–15). Postmortem brain (10,16,17) and association (18,19) studies provide further support implicating miRNAs in the pathogenesis of schizophrenia.
In the present study, we directly investigate for the first time the genome-wide miRNA content of rare CNVs in schizophrenia. We hypothesized that, excluding the known effect of rare 22q11.2 CNVs, individuals with schizophrenia would be enriched for rare CNVs that overlap miRNAs, even when accounting for CNV size. We also examined whether the target genes of the miRNAs overlapped by rare CNVs in schizophrenia are more likely to be involved in neurodevelopmental processes.
METHODS AND MATERIALS
Schizophrenia Cases and Ontario Population Genomics Platform Comparison Samples
As described in detail elsewhere (2,3), we have studied structural variants in a prospectively recruited cohort of unrelated Canadian patients of European ancestry meeting DSM-IV criteria for chronic schizophrenia or schizoaffective disorder, herein termed schizophrenia cases. The study was approved by local research ethics boards, and written informed consent was obtained for each participant. Case subjects with 22q11.2 deletions were a priori excluded to prevent findings from being driven by these established risk variants (3,10–12). The comparison sample comprised unrelated adults of European ancestry who are members of the Ontario Population Genomics Platform (OPGP) genetic epidemiological project (Supplemental Methods in Supplement 1) (3,20) and independent of the 2357 controls used for adjudication of CNV rarity (see below). In the present study of miRNA content of rare CNVs, we considered only schizophrenia case and OPGP comparison subjects with at least one rare CNV and excluded one comparison subject with a 22q11.2 duplication that was reciprocal to the 22q11.2 deletions (3).
Genotyping, CNV Determination, and Validation
Detailed methods are presented elsewhere (3) and in Supplemental Methods in Supplement 1. Briefly, genotyping and CNV analyses for all schizophrenia case and OPGP comparison samples were performed at the same laboratory using identical protocols. The Centre for Applied Genomics in Toronto, Canada, genotyped high-quality genomic DNA using the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Inc, Santa Clara, California). Only arrays that met the Affymetrix recommended quality control guideline of contrast quality control >.4 were used for further analysis. In this study, we consider only “stringent” CNVs: those detected by at least two of three CNV calling algorithms [Birdsuite (21), iPattern (22), and Affymetrix Genotyping Console] and spanning at least 10 kb in length and five or more consecutive array probes (3,20,23). Arrays with number of CNVs exceeding three times the standard deviation from the mean number of CNVs across the data set were excluded (23). To adjudicate the rarity of CNVs identified in schizophrenia case and OPGP comparison subjects, we used the CNVs identified using the same platform in independent population-based control cohorts comprising 2357 individuals of European ancestry (3,20,23). “Rare” and “very rare” CNVs were defined as those present in <.1% and 0% of these population-based controls, respectively, using 50% reciprocal overlap criteria (3,20). These methods showed a high rate of CNV validation (3,20,23): of 58 rare CNVs in this data set tested using another method, all 58 (100%) were validated, and these spanned various sizes, including 17 (30%) <50 kb. There were 376 (of 420) case and 371 (of 415) comparison subjects with at least one rare CNV (3).
As described previously (3), each large (>500 kb) rare CNV in a schizophrenia case or OPGP comparison subject was assessed independently by two experienced clinical cytogenetic laboratory directors, blind to schizophrenia case status. The CNVs deemed to be clinically “Pathogenic” or of “Uncertain clinical significance; likely pathogenic” per established American College of Medical Genetics criteria (24) were collectively termed “pathogenic,” and CNVs of “Uncertain clinical significance (no subclassification)” were termed “variants of unknown significance”; the remainder were termed “benign.”
miRNA Annotation and Burden Analyses
We previously published all rare CNVs in both the case and the comparison cohorts used (3). For the present study, we updated and reannotated the genomic content of these CNVs, using RefSeq (25) for genes and miRBase (release 20, accessed in August 2013; www.mirbase.org/) for the 2578 mature miRNAs annotated in this version of miRBase (26).
To investigate the relative miRNA content of rare CNVs in case and comparison subjects, we used two complementary CNV burden analysis methods, as before restricting to autosomal CNVs (3). First, we compared the proportions of subjects with one or more rare CNVs that overlapped at least one miRNA. Second, we compared the proportions of all rare CNVs in case (n = 1052; 56.2% losses; 57.0% genic) and comparison (n = 954; 58.3% losses; 54.7% genic) subjects that overlapped at least one miRNA. Pearson χ2 tests, odds ratios (ORs), and 95% confidence intervals (CIs) were calculated using standard methods. We also constructed multivariate logistic regression models (Supplemental Methods in Supplement 1) to investigate potential confounding variables in these two burden analyses. The dependent variable was schizophrenia case status. In the first model, predictor variables were the presence of a rare CNV overlapping a miRNA in the individual (binary indicator variable) and either the total number of rare CNVs or the total size of the rare CNVs (in base pairs) in the individual. In the second model, predictor variables were the presence of a miRNA within the genomic extent of a rare CNV (binary indicator variable) and size of the rare CNV. All tests were two-sided, with statistical significance defined as p < .05. All statistical analyses were performed using R 3.0.1 software.
miRNA Target Prediction and Overrepresentation Analyses
We used a conservative strategy to examine predicted target genes of miRNAs overlapped by rare CNVs in case and comparison subjects, similar to the strategy used in other studies (27,28). Targets considered were those identified by at least two of three established target prediction tools: TargetScan 6.2 (29), DIANA microT-CDS (30), and miRanda (31). These prediction tools employ varying score thresholds to produce target gene lists; we chose a stringent 97th percentile score for our analyses. We then investigated whether the predicted targets of miRNAs overlapped by rare CNVs in schizophrenia cases were enriched for genes involved in neurodevelopmental pathways, relative to the predicted target genes of miRNAs overlapped by rare CNVs in comparison subjects. For these analyses, we considered only targets of miRNAs that were overlapped by a rare CNV in two or more unrelated subjects with schizophrenia (“recurrent” case miRNAs). In OPGP comparison subjects, there were no such recurrent miRNAs, and we used the gene targets of all miRNAs overlapped by rare CNVs in these comparison subjects as the comparison group. We derived all functional “gene sets” from Gene Ontology (32), Kyoto Encyclopedia of Genes and Genomes (33), Reactome (34), National Cancer Institute (35), and BioCarta (http://www.biocarta.com). We compared the enrichment of all the functional gene sets in the predicted target genes for case and comparison subjects using Fisher exact tests, with statistical significance defined as p < .05. Because this analysis compares functional enrichment between schizophrenia case and OPGP comparison subject miRNA target genes, it is robust to functional biases in miRNA targets overall. To control the false discovery rate, we used the Benjamini-Hochberg correction.
RESULTS
Increased Burden of CNVs Overlapping miRNAs in Schizophrenia Cases
In subjects with at least one rare autosomal CNV, the proportion with a rare CNV that overlapped a miRNA was significantly greater in schizophrenia cases than in comparison subjects (62 of 376 vs. 21 of 371; p < .0001; OR, 3.29 [95% CI, 1.96–5.52]) (Table 1). In a multivariate logistic regression model that included the total number of rare CNVs per subject as an additional predictor variable, the presence of a rare CNV overlapping a miRNA was the sole significant predictor of schizophrenia case status (p < .0001; OR, 3.15 [95% CI, 1.90–5.42]). In a logistic regression model including total size, rather than total number, of rare CNVs per subject, the presence of a rare CNV overlapping a miRNA remained a significant predictor (p = .0072; OR, 2.18 [95% CI, 1.25–3.90]) (Table 1), although total size of rare CNVs was also significant. In contrast, comparisons using the proportion of individuals with a rare CNV that overlapped a protein-coding gene showed no significant difference after correcting for total number or total size of rare CNVs per subject (p = .452 and p = .925, respectively). As previously reported (3), the overall CNV profile (unrestricted, e.g., by rarity or size) was similar for case and comparison subjects.
Table 1.
Burden of Rare CNVs Overlapping miRNAs in Unrelated Adults of European Ancestry with Schizophrenia
| Schizophrenia Cases
|
OPGP Comparison Subjects
|
Analysis
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uncorrected
|
Correcteda
|
|||||||||||
| No. miRNA | No. Total | (%) | No. miRNA | No. Total | (%) | p | OR | (95% CI) | p | OR | (95% CI) | |
| All Rare CNVsb | 62 | 376 | (16.49) | 21 | 371 | (5.66) | <.0001 | 3.29 | (1.96) (5.52) | .0072 | 2.18 | (1.25) (3.90) |
|
| ||||||||||||
| Loss only | 24 | 306 | (7.84) | 7 | 307 | (2.28) | .0017 | 3.65 | (1.55) (8.60) | .0254 | 2.10 | (1.11) (4.09) |
|
| ||||||||||||
| Gain only | 41 | 264 | (15.53) | 14 | 239 | (5.86) | .0005 | 2.95 | (1.57) (5.57) | .0032 | 2.54 | (1.39) (4.83) |
|
| ||||||||||||
| Very rarec only | 50 | 334 | (14.97) | 13 | 316 | (4.11) | <.0001 | 4.10 | (2.18) (7.71) | .0166 | 2.05 | (1.15) (3.76) |
|
| ||||||||||||
| Small Rare CNVsb (<500 kb only) | 28 | 314 | (8.92) | 16 | 351 | (4.56) | .0240 | 2.05 | (1.09) (3.86) | .0258 | 2.10 | (1.11) (4.12) |
|
| ||||||||||||
| Very rarec only | 21 | 275 | (7.64) | 10 | 297 | (3.37) | .0242 | 2.37 | (1.10) (5.13) | .0598d | 1.93d | (.98) (3.91) |
CI, 95% confidence interval of odds ratio; CNV, copy number variant; Corrected p, two-sided Wald p value; miRNA, microRNA; OPGP, Ontario Population Genomics Platform; No. miRNA, number of individuals with at least one rare autosomal CNV overlapping an miRNA; No. total, total number of individuals with one or more rare autosomal CNVs; OR, odds ratio; Uncorrected p, two-sided Pearson χ2 p value.
Corrected for total CNV size through a logistic regression model including total size of rare CNVs per subject and presence of a rare CNV overlapping a miRNA as predictor variables.
Rare CNVs with <.1% prevalence in 2357 population controls of European ancestry (used to adjudicate CNVs in both schizophrenia cases and OPGP comparison subjects). Only CNVs >10 kb were considered (see Methods and Materials).
Very rare CNVs with 0% prevalence in 2357 population controls.
Trend only result; all other results shown in this table are significant (p < .05).
Results were similar using a different definition of the miRNA burden of rare CNVs. The proportion of rare CNVs that overlapped a miRNA was significantly greater in schizophrenia cases than in comparison subjects (67 of 1052 vs. 25 of 954; p < .0001; OR, 2.53 [95% CI, 1.58–4.04]) (Table S1 in Supplement 1). The presence of a miRNA within the genomic extent of a rare CNV remained a significant predictor of schizophrenia case status (p = .0189; OR, 1.83 [95% CI, 1.12–3.07]) (Table S1 in Supplement 1) in a multivariate logistic regression model that included rare CNV size as a significant predictor variable. Restricting to rare CNVs <500 kb in size, there was a nonsignificant trend toward an increased miRNA burden in cases after correcting for CNV size (p = .052) (Table S1 in Supplement 1). The proportion of rare CNVs that overlapped a protein-coding gene was not significantly different between case and comparison subjects after correcting for rare CNV size (p = .6156). These results were similar after restricting to rare CNVs <500 kb in size (data not shown).
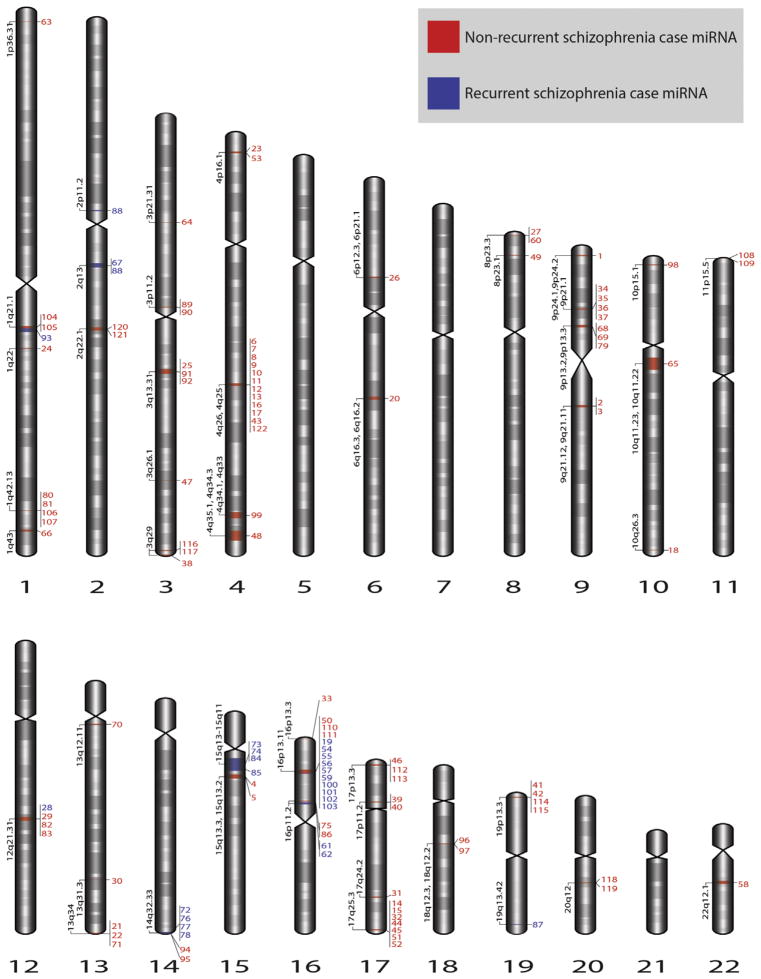
The 67 rare CNVs in 62 schizophrenia cases overlapped 122 distinct miRNAs, including 25 (20.5%) miRNAs that were implicated by rare CNVs in two or more unrelated subjects (Table 2, Figure 1, and Table S2 in Supplement 1). These 25 miRNAs, termed “recurrent” miRNAs, involved CNVs at eight loci: 1q21.1, 2q13, 12q21.31, 14q32.33, 15q11-15q13, 16p11.2, 16p13.11, and 19q13.42. To our knowledge, this is the first report implicating these 25 miRNAs in neuropsychiatric disease. Of the 67 rare CNVs, 20 were large (>500 kb) and were previously clinically classified (3,20,23); 7 (35%) as clinically pathogenic; 12 (60%) as variants of unknown significance; and 1 (5%), at 14q32.33, as benign. The number of protein-coding genes overlapped by these 20 large rare CNVs varied greatly (median, 5.5; range, 0–40). In contrast, in the OPGP comparison subjects, only 35 distinct miRNAs were overlapped by rare CNVs, none of which were recurrent (Table S2 in Supplement 1). The proportion of recurrent rare CNV-overlapped miRNAs was significantly greater in schizophrenia cases than in comparison subjects (25 of 122 vs. 0 of 35; p = .0013). None of the miRNAs implicated by rare CNVs in schizophrenia cases was the same as miRNAs in comparison subjects.
Table 2.
Recurrent miRNAs (n = 25) Overlapped by Rare CNVsa in Two or Moreb Unrelated Adults with Schizophrenia
| miRNA Name | Cytoband | Predicted Targetsc |
|---|---|---|
| hsa-miR-484 | 16p13.11 | ● |
| hsa-miR-617 | 12q21.31 | ● |
| hsa-miR-3179 | 16p13.11 | ● |
| hsa-miR-3180d | 16p13.11 | ● |
| hsa-miR-3180-3p, hsa-miR-3180-5pd | 16p13.11 | ● |
| hsa-miR-3670 | 16p13.11 | ● |
| hsa-miR-3680-3p, hsa-miR-3680-5p | 16p11.2 | |
| hsa-miR-4435 | 2q13 | ● |
| hsa-miR-4507 | 14q32.33 | |
| hsa-miR-4508 | 15q11-15q13 | |
| hsa-miR-4509 | 15q11-15q13 | ● |
| hsa-miR-4537 | 14q32.33 | |
| hsa-miR-4538 | 14q32.33 | ● |
| hsa-miR-4539 | 14q32.33 | ● |
| hsa-miR-4715-3p, hsa-miR-4715-5p | 15q11-15q13 | ● |
| hsa-miR-4752 | 19q13.42 | ● |
| hsa-miR-4771e | 2p11.2 and 2q13 | ● |
| hsa-miR-5087 | 1q21.1 | |
| hsa-miR-6506-3p, hsa-miR-6506-5p | 16p13.11 | |
| hsa-miR-6511a-3p, hsa-miR-6511a-5p | 16p13.11 |
CNV, copy number variant; miRNA, microRNA.
See Table S2 in Supplement 1 for additional details, including relevant CNV coordinates.
All miRNAs overlapped by CNVs in n = 2 unrelated probands except for hsa-miR-3680-3p, hsa-miR-3680-5p (n = 4); hsa-miR-4435 (n = 3); hsa-miR-4508 (n = 3); hsa-miR-4715-3p, hsa-miR-4715-5p (n=3); and hsa-miR-4771 (n = 4).
See Methods and Materials for details. Our stringent criteria (97th percentile target prediction score and with genes predicted by two of three established tools) and the lack of available data for many miRNAs (e.g., miRNAs numbered above 5000) resulted in many miRNAs not having a reliable list of target genes.
As detailed in miRBase, hsa-miR-3180, hsa-miR-3180-3p, and hsa-miR-3180-5p are distinct mature miRNAs.
As detailed in miRBase, hsa-miR-4771 is a mature miRNA located at two different cytobands: 2p11.2 and 2q13.
Figure 1.
Genomic location of miRNAs overlapped by CNVs in schizophrenia cases. The genomic locations of the 122 schizophrenia case miRNAs that are overlapped by rare autosomal CNVs are shown. Red labels indicate the 97 nonrecurrent and blue labels the 25 recurrent miRNAs overlapped by rare CNVs in schizophrenia cases. The miRNA numbers correspond to the miRNA identification numbers in Table S2 in Supplement 1. As detailed in miRBase, hsa-miR-4771 (no. 88) is a mature miRNA located at two different cytobands: 2p11.2 and 2q13. CNVs, copy number variants; miRNA, microRNA.
Predicted Targets of miRNAs Implicated in Schizophrenia Cases Are Enriched for Neurodevelopmental Genes
There were 145 predicted target genes of the 25 recurrent rare CNV-overlapped miRNAs (Table S3 in Supplement 1) in schizophrenia cases and 387 predicted target genes of the 35 comparison subject miRNAs, including 9 genes common to both case and comparison subjects. Of the 145 case miRNA targets, three genes (SYNGAP1, GLT8D2, and GOLGA6L6) were predicted targets of ≥2 of the 25 miRNAs involved, excluding the overlap of target genes of hsa-miR-3180 and hsa-miR-3180-3p (Table S3 in Supplement 1). With our stringent cutoff and data currently available, there were no predicted target genes for the miRNAs overlapped by two recurrent pathogenic CNVs, 16p11.2 duplications and 1q21.1 duplications (Table 2).
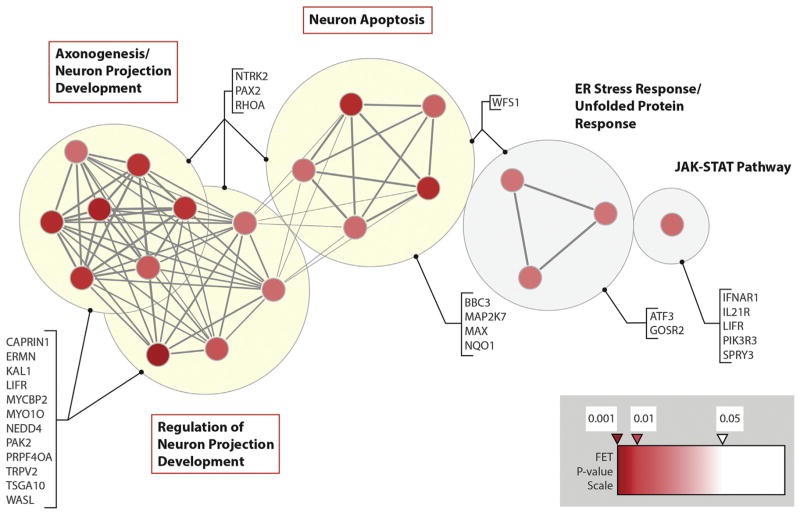
The gene-set overrepresentation analysis comparing case with comparison subjects showed that 16 (80%) of the top 20 gene sets (each with nominally significant enrichment in schizophrenia) could be considered related to neurodevelopmental functions, including axonogenesis, neuron projection development, and neuron death (Figure 2; Table S4 in Supplement 1). Predicted gene targets driving these results included CAPRIN1 (36,37), MYO10 (38), NEDD4 (39), NTRK2 (40), PAK2 at 3q29 (41), and RHOA (3). After correcting for multiple testing, none of the overrepresentation analyses had significant p values.
Figure 2.
Top 20 functional gene sets enriched in predicted targets of recurrent miRNAs overlapped by rare CNVs in schizophrenia. A functional map is shown of recurrent schizophrenia miRNAs overlapped by rare CNVs using results of the gene-set association analysis of predicted miRNA gene targets, displayed as a network of 20 gene sets (circles) related by mutual overlap (lines). The map was created using the Cytoscape plugin Enrichment Map (version 1.2) (70), the gene-set enrichment table was loaded using the generic format interface, and gene-set overlaps were quantified using the Jaccard+Overlap combined coefficient with the filtering threshold set at .3. Circle color is proportional to the total number of “support” genes in each gene set based on the functional enrichment p value (inset), and line thickness represents the number of genes in common between two gene sets. Each support gene is a predicted target of a recurrent miRNA overlapped by more than one CNV in unrelated schizophrenia cases and not found as a miRNA target in Ontario Population Genomics Platform comparison subjects. Groups of functionally related gene sets are highlighted by large background circles and respective labels; three major neurodevelopmentally relevant functional groups are further highlighted by yellow filled circles. Support genes for schizophrenia are shown. CNVs, copy number variants; ER, endoplasmic reticulum; FET, the Fisher exact test; JAK-STAT, Janus kinase–signal transducer and activator of transcription; miRNA, microRNA.
DISCUSSION
To our knowledge, this is the first genome-wide study investigating CNVs that overlap miRNAs in schizophrenia. Consistent with preliminary findings in other diseases (28,42–44), the results showed both quantitative and qualitative CNV-related miRNA differences between schizophrenia cases and comparison subjects. There was a significant enrichment in schizophrenia cases of rare CNVs that overlap miRNAs. Also, the predicted target genes of the 25 CNV-overlapped miRNAs that were recurrent in unrelated subjects with schizophrenia tended to be involved in neurodevelopmental processes. These data provide further support for a model of causation in which simultaneous disruption of multiple genetic pathways may be necessary for expression of schizophrenia within an individual.
Delineating a miRNA Target Gene Network in Schizophrenia
Our results indicate that rare CNVs that disrupt DNA encoding a miRNA may contribute to causing schizophrenia in a substantial minority of individuals. Such disruption is predicted to affect the functioning of the miRNAs with respect to their target genes (12). Several of the top 20 miRNA target gene sets (Figure 2) show an overlap with the most relevant functional gene-set clusters identified previously for this cohort using the protein-coding genetic content of rare deletions (3). These gene sets include cell projection, axonogenesis, and neuron development. A gene represented in 10 of the top 20 recurrent CNV-overlapped miRNA target gene sets, CAPRIN1, is relevant to autism (36) and is involved in dendritic spine morphogenesis (45). Of the 26 predicted target genes of recurrent schizophrenia-related miRNAs identified in this study that were overrepresented in the top 20 gene sets (Figure 2), 8—CAPRIN1, KAL1, MAP2K7, MYO10, NTRK2, PRPF40A, SPRY3, and TSGA10—are differentially expressed in human dorsolateral prefrontal cortex (27). NTRK2 (known sometimes as TrkB), encoding a brain-derived neurotrophic factor receptor, also has altered levels in brains with schizophrenia (46) and is involved in dendritic spine morphogenesis (40). Additionally, one of the three genes predicted to be targeted by two miRNAs, hsa-miR-3179 and hsa-miR-3180-3p overlapped by 16p13.11 CNVs (47), was SYNGAP1, a gene implicated in autism, intellectual disability, and schizophrenia (48–50). The laminin gene LAMB3, a target of the miRNA hsa-miR-484, is another promising candidate for schizophrenia, with other laminin genes previously implicated by rare CNVs and point mutations in schizophrenia (50–56).
Rare CNVs in Schizophrenia Are Enriched for miRNAs, Providing Further Insight into the Complex Genetic Architecture of Schizophrenia
Previous research has shown a significant enrichment of large multigenic rare CNVs in schizophrenia (2–5). The increased burden of rare CNVs that overlap miRNAs in schizophrenia provides an alternative or enhanced explanation for the genetic disruption associated with rare CNVs because individual miRNAs can target diverse networks of genes. The fact that these genes may be anywhere in the rest of the genome and each subject to common and rare variation could help to explain the variability of expression of recurrent CNVs [e.g., 16p13.11 duplications (3,47,57–59)]. Also, in the schizophrenia cases, there were rare CNVs that overlapped miRNAs but no protein-coding genes located at eight loci: 3p11.2, 3q26.1, 9p21.1, 10p15.1, 10q26.3, 13q31.3, 14q32.33, and 18q12.2-18q12.3 (Table S2 in Supplement 1). Although such miRNA-related CNVs may confer risk for schizophrenia, they would not be reliably identified in exome sequencing studies.
Advantages and Limitations
As before (3,20), in the present study, we applied identical molecular and conservative analytic methods to unrelated cases and to a similar-sized Canadian sample of epidemiologic comparison subjects. Advantages derived from our sampling strategy are detailed elsewhere (2,3). With respect to the CNVs themselves, our strategy emphasizes CNVs likely to have an enhanced effect size because we use a stringent level to define rarity (<.1%). Although this strategy could mean missing more common variants with a lower effect size, using an independent population-based control set to adjudicate rarity ensured equal effects for both cases and OPGP comparison subjects and similar expected rates of type 1 and 2 errors. Our methods show high validation rates for rare CNVs (3,20,23), including 100% (58 of 58) in this data set. Most case and comparison subject rare CNVs that overlapped miRNAs (79.1% of case subjects and 84.0% of comparison subjects) were >100 kb in size, further increasing confidence in the CNV results. We ensured that the miRNA-related results were not driven by effects of 22q11.2 deletions by excluding a priori subjects with these CNVs (3), known to have high penetrance for schizophrenia and important miRNA-related mechanisms (12–15). Although outside the scope of this initial genome-wide case-control study, it would be important to investigate specifically the potential role of miRNAs in other established large rare CNVs associated with schizophrenia. However, the a priori exclusion of all such CNVs would reduce the power of the analyses undertaken and be premature at this point given the lack of data on effects of these CNVs related to their miRNA content. A related issue is that there may be different risks for neuropsychiatric disorders when considering losses and gains (deletions and duplications) at the same genomic locus separately (60). However, predicting the related effects of loss or gain CNVs on expression of protein-coding genes is not straightforward (e.g., a deletion does not always mean reduced expression of overlapped genes) (61), and there are as yet limited data about CNV effects on miRNA expression in humans, even at a well-studied locus such as 22q11.2 (11).
A limitation faced by all current miRNA studies is that one must rely on target gene prediction tools, given that there are limited validated gene target data available, and even then the miRNA expression studies that are the gold standard may be imperfect (62,63). Only 2 of the 25 recurrent case miRNAs (hsa-miR-484 and hsa-miR-617) in this study had validated targets annotated using TarBase v6.0 (64). Also, miRNAs are being identified daily, and the prediction tools are limited by the data available on these miRNAs and their gene targets. To minimize false positive targets, we adopted a high level of stringency and considered only gene targets predicted by at least two of three established prediction tools, treating case and comparison cohorts equally. Although imperfect, the integration of more than one prediction method tends to balance out the precision and recall, resulting in better accuracy and coverage of predictions (40). As a result of these factors, true target genes of the miRNAs observed were likely missed. No predicted target genes were found for 34.4% of case and 28.6% of comparison subject miRNAs, including none for more recently discovered miRNAs numbered above 5000 (Table S2 in Supplement 1). In time, improved annotation of experimentally validated miRNA target genes may facilitate the reanalysis of these CNV data and could minimize both false-positive and false-negative targets and potentially assess protein translation effects. Although the sample size of this study had sufficient power to show significant results for the main burden analyses, a larger cohort is required to refine further the set of candidate miRNAs recurrent in schizophrenia but not comparison subjects for gene-set enrichment analyses. Nevertheless, the gene-set enrichment results align with previous reports for protein-coding genes (3,50,53,54). Definitively proving causality of specific genetic variants for schizophrenia is beyond the scope of this and most other studies (3,48,53,54,59,65–67).
In conclusion, these findings provide further support for a model of the genetic causation of schizophrenia that extends beyond protein-coding genes. The apparent significance of the miRNA content of rare CNVs has implications for the interpretation of rare structural variants in schizophrenia and the current and potential future yield of clinical microarray testing (3,68). Rare single nucleotide variants in miRNAs and their target genes may similarly play an important role in schizophrenia. Whole-genome sequencing in this cohort is a logical future consideration. Lastly, in light of developments regarding miRNA-based therapeutics in other diseases such as cancer (69), these results suggest that a similar approach to novel treatment design could hold promise in schizophrenia.
Supplementary Material
Acknowledgments
This work was supported by the Canadian Institutes of Health Research Grant Nos. MOP-89066 and MOP-111238 (to ASB) as well as grants from the University of Toronto McLaughlin Centre, NeuroDevNet, Genome Canada and the Ontario Genomics Institute, the Canadian Institutes of Health Research, the Canadian Institute for Advanced Research, the Canada Foundation for Innovation, the government of Ontario, Autism Speaks, and The Hospital for Sick Children Foundation (to SWS). SWS holds the GlaxoSmithKline–Canadian Institutes of Health Research Chair in Genome Sciences at the University of Toronto and The Hospital for Sick Children. ASB holds the Canada Research Chair in Schizophrenia Genetics and Genomic Disorders and the Dalglish Chair in 22q11.2 Deletion Syndrome.
We thank the patients and their families for their participation; colleagues for referring patients; research assistants; staff at Saint John Community Mental Health Services, Saint John Regional Hospital, Hillsborough Hospital, and Hamilton clinic; staff at The Centre for Applied Genomics; and fellows and students who assisted in the collection and analysis of data for the study. We thank A. Fiebig, A. Franke, and S. Schreiber at PopGen (University of Kiel, Germany) and A. Stewart, R. McPherson, and R. Roberts of the University of Ottawa Heart Institute (University of Ottawa, Canada) for providing comparison subject data. We thank S. Bekeschus for figure design and formatting and A. Lionel (The Centre for Applied Genomics) for critical discussion.
Footnotes
DISCLOSURES
SWS is on the Scientific Advisory Board of Population Diagnostics, Inc., and is a cofounder of YouNique Genomics. All other authors report no biomedical financial interests or potential conflicts of interest.
Supplementary material cited in this article is available online at http://dx.doi.org/10.1016/j.biopsych.2014.05.011.
References
- 1.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 2.Bassett AS, Costain G, Fung WLA, Russell KJ, Pierce L, Kapadia R, et al. Clinically detectable copy number variations in a Canadian catchment population of schizophrenia. J Psychiatr Res. 2010;44:1005–1009. doi: 10.1016/j.jpsychires.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costain G, Lionel AC, Merico D, Forsythe P, Russell K, Lowther C, et al. Pathogenic rare copy number variants in community-based schizophrenia suggest a potential role for clinical microarrays. Hum Mol Genet. 2013;22:4485–4501. doi: 10.1093/hmg/ddt297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costain G, Bassett AS. Clinical applications of schizophrenia genetics: Genetic diagnosis, risk, and counseling in the molecular era. Appl Clin Genet. 2012;5:1–18. doi: 10.2147/TACG.S21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- 7.De Smaele E, Ferretti E, Gulino A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010;1338:100–111. doi: 10.1016/j.brainres.2010.03.103. [DOI] [PubMed] [Google Scholar]
- 8.Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 9.Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: A developing story. Curr Opin Genet Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. 2012;46:263–271. doi: 10.1016/j.nbd.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 11.Brzustowicz LM, Bassett AS. miRNA-mediated risk for schizophrenia in 22q11.2 deletion syndrome. Front Genet. 2012;3:291. doi: 10.3389/fgene.2012.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forstner AJ, Degenhardt F, Schratt G, Nothen MM. MicroRNAs as the cause of schizophrenia in 22q11.2 deletion carriers, and possible implications for idiopathic disease: A mini-review. Front Mol Neurosci. 2013;6:47. doi: 10.3389/fnmol.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15:1176–1189. doi: 10.1038/mp.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 15.Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry. 2011;69:188–193. doi: 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smalheiser NR, Lugli G, Zhang H, Rizavi H, Cook EH, Dwivedi Y. Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PLoS One. 2014;9:e86469. doi: 10.1371/journal.pone.0086469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. 2013;18:11–12. doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- 20.Silversides CK, Lionel AC, Costain G, Merico D, Migita O, Liu B, et al. Rare copy number variations in adults with tetralogy of Fallot implicate novel risk gene pathways. PLoS Genet. 2012;8:e1002843. doi: 10.1371/journal.pgen.1002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29:512–520. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lionel AC, Crosbie J, Barbosa N, Goodale T, Thiruvahindrapuram B, Rickaby J, et al. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med. 2011;3:95ra75. doi: 10.1126/scitranslmed.3002464. [DOI] [PubMed] [Google Scholar]
- 24.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 25.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: Current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziats MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.93. published online ahead of print Aug 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaishnavi V, Manikandan M, Tiwary BK, Munirajan AK. Insights on the functional impact of microRNAs present in autism-associated copy number variants. PLoS One. 2013;8:e56781. doi: 10.1371/journal.pone.0056781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, et al. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, et al. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang YH, Yuen RK, Jin X, Wang M, Chen N, Wu X, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Fatimy R, Tremblay S, Dury AY, Solomon S, De Koninck P, Schrader JW, et al. Fragile X mental retardation protein interacts with the RNA-binding protein Caprin1 in neuronal RiboNucleoProtein complexes [corrected] PLoS One. 2012;7:e39338. doi: 10.1371/journal.pone.0039338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin WH, Hurley JT, Raines AN, Cheney RE, Webb DJ. Myosin X and its motorless isoform differentially modulate dendritic spine development by regulating trafficking and retention of vasodilator-stimulated phosphoprotein. J Cell Sci. 2013;126:4756–4768. doi: 10.1242/jcs.132969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gautam V, Trinidad JC, Rimerman RA, Costa BM, Burlingame AL, Monaghan DT. Nedd4 is a specific E3 ubiquitin ligase for the NMDA receptor subunit GluN2D. Neuropharmacology. 2013;74:96–107. doi: 10.1016/j.neuropharm.2013.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balu DT, Li Y, Puhl MD, Benneyworth MA, Basu AC, Takagi S, et al. Multiple risk pathways for schizophrenia converge in serine racemase knockout mice, a mouse model of NMDA receptor hypo-function. Proc Natl Acad Sci U S A. 2013;110:E2400–E2409. doi: 10.1073/pnas.1304308110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mills JD, Kavanagh T, Kim WS, Chen BJ, Kawahara Y, Halliday GM, et al. Unique transcriptome patterns of the white and grey matter corroborate structural and functional heterogeneity in the human frontal lobe. PLoS One. 2013;8:e78480. doi: 10.1371/journal.pone.0078480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiao Y, Badduke C, Mercier E, Lewis SM, Pavlidis P, Rajcan-Separovic E. miRNA and miRNA target genes in copy number variations occurring in individuals with intellectual disability. BMC Genomics. 2013;14:544. doi: 10.1186/1471-2164-14-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xing HJ, Li YJ, Ma QM, Wang AM, Wang JL, Sun M, et al. Identification of microRNAs present in congenital heart disease associated copy number variants. Eur Rev Med Pharmacol Sci. 2013;17:2114–2120. [PubMed] [Google Scholar]
- 44.Serrano NA, Xu C, Liu Y, Wang P, Fan W, Upton MP, et al. Integrative analysis in oral squamous cell carcinoma reveals DNA copy number-associated miRNAs dysregulating target genes. Otolaryngol Head Neck Surg. 2012;147:501–508. doi: 10.1177/0194599812442490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiina N, Tokunaga M. RNA granule protein 140 (RNG140), a paralog of RNG105 localized to distinct RNA granules in neuronal dendrites in the adult vertebrate brain. J Biol Chem. 2010;285:24260–24269. doi: 10.1074/jbc.M110.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong J, Rothmond DA, Webster MJ, Weickert CS. Increases in two truncated TrkB isoforms in the prefrontal cortex of people with schizophrenia. Schizophr Bull. 2013;39:130–140. doi: 10.1093/schbul/sbr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramalingam A, Zhou XG, Fiedler SD, Brawner SJ, Joyce JM, Liu HY, et al. 16p13.11 duplication is a risk factor for a wide spectrum of neuropsychiatric disorders. J Hum Genet. 2011;56:541–544. doi: 10.1038/jhg.2011.42. [DOI] [PubMed] [Google Scholar]
- 48.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sodhi MS, Simmons M, McCullumsmith R, Haroutunian V, Meador-Woodruff JH. Glutamatergic gene expression is specifically reduced in thalamocortical projecting relay neurons in schizophrenia. Biol Psychiatry. 2011;70:646–654. doi: 10.1016/j.biopsych.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 52.Need AC, McEvoy JP, Gennarelli M, Heinzen EL, Ge D, Maia JM, et al. Exome sequencing followed by large-scale genotyping suggests a limited role for moderately rare risk factors of strong effect in schizophrenia. Am J Hum Genet. 2012;91:303–312. doi: 10.1016/j.ajhg.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 55.Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radmanesh F, Caglayan AO, Silhavy JL, Yilmaz C, Cantagrel V, Omar T, et al. Mutations in LAMB1 cause cobblestone brain malformation without muscular or ocular abnormalities. Am J Hum Genet. 2013;92:468–474. doi: 10.1016/j.ajhg.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagamani SC, Erez A, Bader P, Lalani SR, Scott DA, Scaglia F, et al. Phenotypic manifestations of copy number variation in chromosome 16p13.11. Eur J Hum Genet. 2011;19:280–286. doi: 10.1038/ejhg.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tropeano M, Ahn JW, Dobson RJ, Breen G, Rucker J, Dixit A, et al. Male-biased autosomal effect of 16p13.11 copy number variation in neurodevelopmental disorders. PLoS One. 2013;8:e61365. doi: 10.1371/journal.pone.0061365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costain G, Lionel AC, Fu F, Stavropoulos DJ, Gazzellone MJ, Marshall CR, et al. Adult neuropsychiatric expression and familial segregation of 2q13 duplications. Am J Med Genet B Neuropsychiatr Genet. 2014;165:337–344. doi: 10.1002/ajmg.b.32236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bassett AS, Scherer SW, Brzustowicz LM. Copy number variations in schizophrenia: Critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry. 2010;167:899–914. doi: 10.1176/appi.ajp.2009.09071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet. 2009;18:R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 62.Sorefan K, Pais H, Hall AE, Kozomara A, Griffiths-Jones S, Moulton V, et al. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence. 2012;3:4. doi: 10.1186/1758-907X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marco A, Griffiths-Jones S. Detection of microRNAs in color space. Bioinformatics. 2012;28:318–323. doi: 10.1093/bioinformatics/btr686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: A functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, et al. Copy number variants in schizophrenia: Confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costain G, McDonald-McGinn DM, Bassett AS. Prenatal genetic testing with chromosomal microarray analysis identifies major risk variants for schizophrenia and other later-onset disorders. Am J Psychiatry. 2013;170:1498. doi: 10.1176/appi.ajp.2013.13070880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xi JJ. MicroRNAs in cancer. Cancer Treat Res. 2013;158:119–137. doi: 10.1007/978-3-642-31659-3_5. [DOI] [PubMed] [Google Scholar]
- 70.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.