Summary
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that mediate excitatory neurotransmission crucial for brain development and function including learning and memory formation. Recently a wealth of structural studies on iGluRs, including AMPA receptors (AMPARs), kainate receptors, and NMDA receptors (NMDARs) became available.. These studies showed structures of non-NMDARs including AMPAR and kainate receptor in various functional states, thereby providing the first visual sense of how non-NMDAR iGluRs may function in the context of homotetramers. Furthermore, they provided the first view of heterotetrameric NMDAR ion channels, which illuminated the similarities with and differences from non-NMDARs, thus raising a mechanistic distinction between the two groups of iGluRs. Here we review mechanistic insights into iGluR functions gained through structural studies of multiple groups.
Keywords: ionotropic glutamate receptors, structural biology, pharmacology
A brief history of structural studies on iGluRs
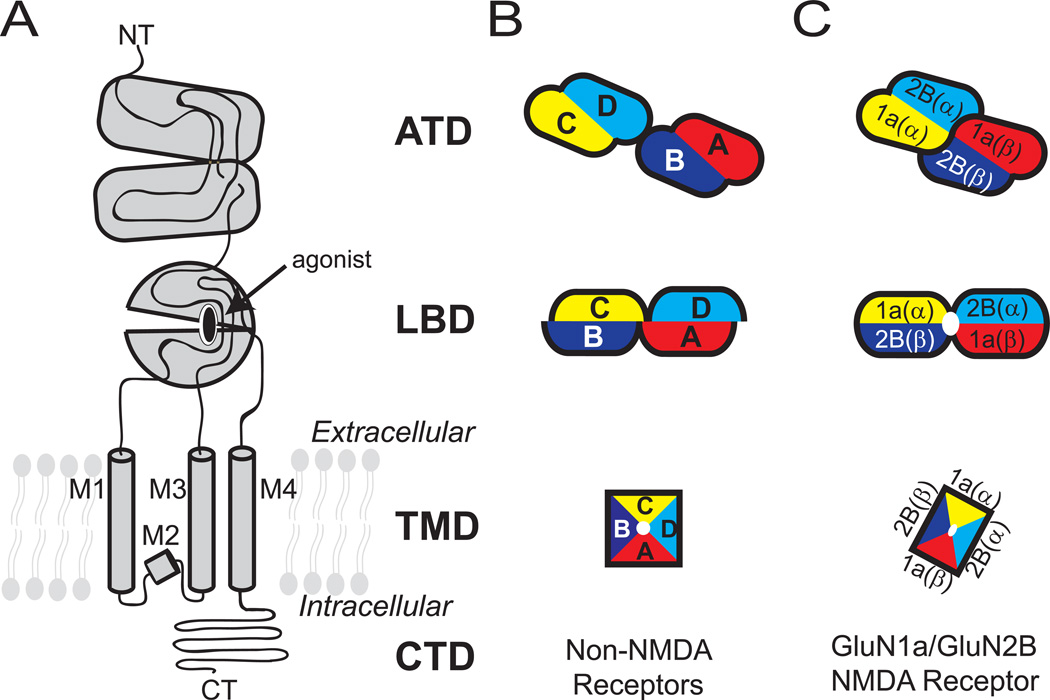
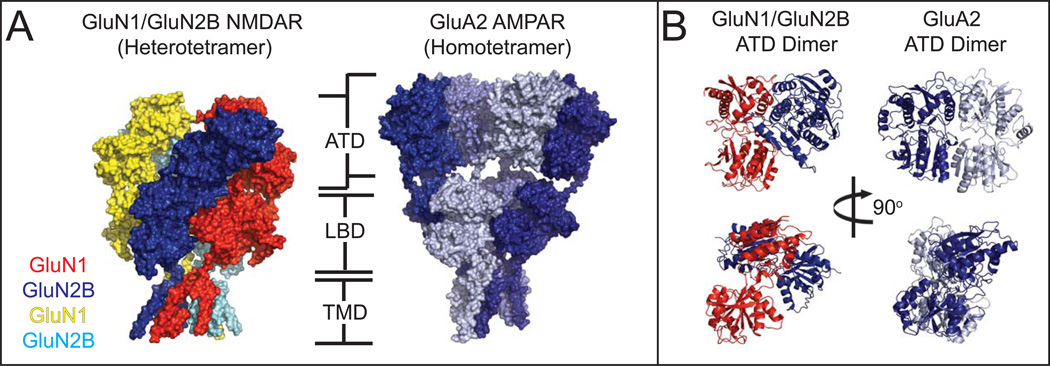
Glutamate, a simple amino acid, is the major excitatory neurotransmitter in mammalian brains [1]. The presynaptically released glutamate binds different subclasses of ionotropic glutamate receptors (iGluRs) including α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors (AMPARs), kainate receptors, and N-methyl-D-aspartate (NMDA) receptors (NMDARs). Glutamate binding mediates opening of their cationic ion channels to generate synaptic current pivotal to brain function. The iGluR subunits contain modular domains including the amino terminal domain (ATD), ligand-binding domain (LBD), transmembrane domain (TMD), and carboxy terminal domain (CTD), and assemble into tetramers within respective subclasses to form ligand-gated ion channels (Figure. 1). While NMDARs form functional ion channels only as heterotetramers of at least two distinct subunits, non-NMDARs do so by forming either homo or heterotetramers, Enthusiasm to understand the structure and function of iGluRs has been enormous over several decades due to the involvement of iGluRs in various aspects of neuronal functions as well as neurological diseases and disorders such as depression, schizophrenia, Alzheimer’s and Parkinson’s diseases, autism, seizure, and stroke [2].
Figure 1. Domain organization and subunit arrangement of iGluRs.
(A) The ionotropic glutamate receptor subunits are composed of distinct domains including the amino terminal domain (ATD), ligand-binding domain (LBD), transmembrane domain (TMD), and carboxyl terminal domain (CTD). The TMD of one subunit is composed of M1-M4 helices. Schematic representation of tetrameric subunit organization of non-NMDARs (B) and NMDARs (C) at ATD, LBD and TMD layers are shown. Dimer pairs at ATD and LBD are indicated by lack of black lines at the interface. The four subunits in non-NMDARs are noted as A-D.
The structural biology of iGluRs started in the late 1990s after the molecular cloning of various iGluR genes [3–5]. Taking advantage of the modular nature of iGluR subunit proteins, structural determination of individual water-soluble domains such as the ATD and LBD was pursued first (primarily by x-ray crystallography) because it was substantially less technically demanding than solving the entire tetrameric transmembrane ion channels [6]. As of 2015, over 268 crystal structures of LBDs of various subunits from all of the major iGluR subclasses in complex with agonists, antagonists, partial agonists, and allosteric modulators are available. Despite low sequence identity between NMDAR and non-NMDAR subunits (~20%), the LBD structures, have similar bi-lobed clamshell-like architectures that undergo opening and closing upon unbinding and binding of ligands, respectively [7–14]. The structural mobility of LBD was also extensively tested and elaborated by nuclear magnetic resonance [15], molecular dynamics free energy simulation [16], and single molecular fluorescence resonance energy transfer [17]. Meanwhile, ATD structures for non-NMDARs [18–24] and NMDARs [25–27] became available but in limited numbers compared to LBD structures. ATDs from both NMDARs and non-NMDARs have bi-lobed structures but completely unrelated in primary sequence and overall architecture to LBDs. However, consistent with low sequence identity (<10%), conformations of bi-lobed architectures are highly distinct between NMDARs and non-NMDARs [28, 29]. In NMDARs, allosteric sites, not present in non-NMDARs, were observed at the heterodimeric subunit interface between GluN1 and GluN2B for ifenprodil [27] and within GluN2B ATDs for zinc [26]. In both studies of ATD and LBD, homodimeric and heterodimeric assemblies were observed in non-NMDARs and NMDARs, respectively. This led to the speculation that the basic assembly pattern of an iGluR subunit is a dimer of dimers instead of a bona fide four-fold symmetrical tetramer as in other tetrameric ion channels such as potassium and sodium channels [30–32]. However, integration of fragments of knowledge about the structure and function of individual extracellular domains required an overall view of the intact iGluR ion channels. This was an enormous challenge in x-ray crystallography because iGluR channels are tetramers of large transmembrane proteins that are heavily glycosylated. Attempts were made to observe intact AMPAR structures in the early 2000s by cryo-electron microscopy (cryo-EM), however, the overall resolution of the structure and thus, the structural interpretation was limited by the technical restriction of the time [33–36]. The first clear-cut structure of the intact iGluR channel became available when Gouaux and colleagues solved the structure of homotetrameric GluA2 AMPARs in complex with an antagonist by x-ray crystallography thereby revealing the pattern of subunit and domain arrangement and opening the new era of structural studies for intact iGluRs. [37]. Today, the iGluR field has multiple structures of the intact GluA2 AMPAR representing different functional states, the GluK2 kainate receptor, and even intact heterotetrameric NMDARs, which were once considered overwhelmingly challenging.
Structural biology of intact non-NMDARs – a giant step toward mechanistic understanding
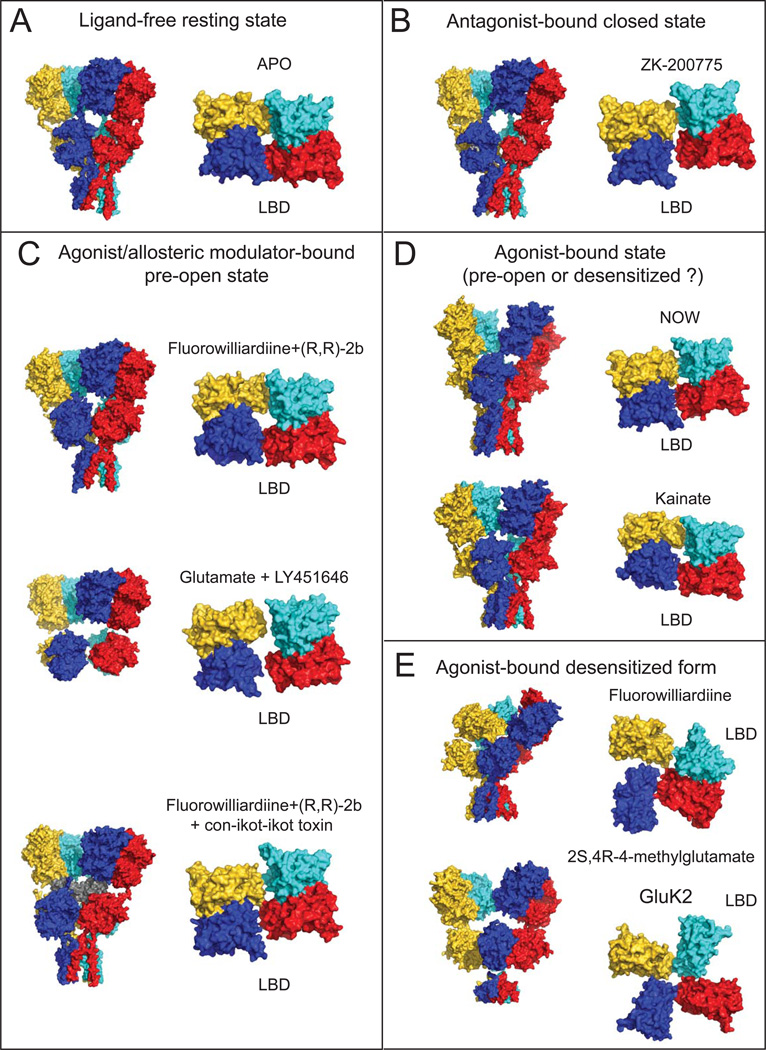
Recently a number of intact non-NMDAR structures have been published by multiple groups, providing solid molecular templates that permit a mechanistic understanding of their activation and desensitization. Specifically, in addition to the original GluA2 AMPAR structure in the antagonist-bound form, GluA2 AMPAR structures in the pre-activated (or pre-open) state and structures of the desensitized state of GluA2 AMPAR and GluK2 kainate receptor (obtained by both x-ray crystallography and cryo-EM) have become available. These studies provide clear ‘snap-shots’ for making valid predictions of how domains and subunits move upon entry into different functional states (Figure 2).
Figure 2. Structures of GluA2 AMPAR and GluK2 kainate receptors in various functional states.
Surface representation of the structures in ligand-free (A), antagonist-bound closed (B), agonist and allosteric modulator-bound pre-open (C) and agonist-bound (D) and agonist-bound desensitized (E) states are shown as viewed from the side for the whole receptor (left) and from top for the LBD tetramer (right). Ligand combinations used and PDB IDs for the structures are shown for each structure (Table 1). Subunits are colored as yellow, cyan, magenta and blue for the subunits A,B,C and D, respectively. The con-ikot-ikot toxin molecule in panel C is colored in grey.
GluA2 AMPARs in apo, antagonist-bound, and agonist-bound states have an overall Y-shaped architecture formed by staggered layers of ATD, LBD and TMD (Figure 1 and 2) [37]. The structure has a symmetry mismatch between the extracellular domains (ATDs and LBDs) and TMDs, which are arranged as a dimer of dimers with 2-fold symmetry and a tetramer with 4-fold rotational symmetry, respectively. The subunit and domain assembly is complex as the extracellular ATD and LBD have swapping of dimer pairs (i.e. Homotetrameric subunits assigned with letters A through D form AB and CD dimers at ATD and AD and BC dimers at LBD; Figure. 1 and 2).
How ligand-gating occurs in non-NMDARs is a fundamental question that began to be addressed by extensive observation of conformational changes in isolated LBDs, for which binding of an agonist or antagonist stabilized the closed or open conformations, respectively. [6, 7, 13, 14]. This major conformational change in the LBD has been suggested to create tensions in the LBD-TMD linker leading to the opening of the channel [38, 39]; however, discerning what this actually means in the context of intact tetrameric iGluRs has been a challenging task that required intensive structural studies of full-length receptors. Recently, three independent studies attempted to trap the presumably active state of the GluA2 AMPAR by binding an allosteric modulator or a Conus striatus cone snail toxin, con-ikot-ikot, both of which block entry of AMPAR into the desensitized state [40]. These studies solved structures of trapped receptors by x-ray crystallography and cryo-EM [41–43] (Table 1). In the agonist/modulator-bound form, the receptor retains its Y-shaped architecture, but the vertical dimension of the receptor is shortened by 7-10 Å, bringing the ATD and LBD tetramers closer together compared to apo and antagonist-bound forms. Conversely, in the toxin-bound structure, the ATD and LBD are further separated by 10 Å because the toxin intercalates into the space between these domains and interacts with both ATDs and LBDs (Figure. 2C). Consistent with the studies on isolated LBDs, the LBD in the intact AMPAR bound to agonist/modulator had a closed cleft conformation whereas that in the antagonist-bound or apo state has an open cleft conformation. The extent of closure, however, is smaller in intact AMPARs compare to that in isolated LBDs possibly due to the fact that LBDs in intact receptors are tethered to the closed ion channel, thus, introducing some restrains in LBD motion [42]. However, this structural change appears to be insufficient to open the channel at the TMD – that is, the TMD in all of the crystal structures of the agonist/modulator-bound GluA2 adopts a closed channel similar to the apo and antagonist-bound forms. Therefore, these structures are currently proposed to represent an activated pre-open state as reasoned in the structural studies of the G-protein-regulated inward-rectifier potassium channel 2 channel complexed to the βγ G protein [44]. Other than the speculation that the closed channel may be energetically favored in general, it is unclear why the channels stay closed in many crystal structures of ion channels in “activating” conditions. The usual (but rather obscure) suspects include crystallization conditions containing high salt or high precipitant concentrations as well as crystal packing. In this regard, a comparison with the cryo-EM structure – in physiological buffer with no crystal packing – would have answered the above question, but unfortunately, the TMD was not resolvable in the recent study [41]. Other problems may be the absence of lipids or the presence of detergents in the samples and lack of voltage control. Application of new techniques such as visualization of liposome-reconstituted ion channels using cryo-EM as done in the study of the Big potassium (BK) channel might help resolve this issue since the liposome can provide a more physiological lipidic environment and even a voltage control [45, 46].
Table 1.
Structures of intact non-NMDA ionotropic glutamate receptors
| PDB ID | Constructsa | Agonists/ antagonists |
Allosteric modulator |
Technique | Resolution (Å) |
AC pair distancesb (Å) |
BD pair distancesc (Å) |
Reference | |
|---|---|---|---|---|---|---|---|---|---|
| Apo form | 4U2P | GluA25M | - | - | X-RAY | 3.24 | 32.0 | 21.9 | [1] |
| Antagonist –bound form |
3KG2 | GluA2cryst | ZK-200775 | - | X-RAY | 3.60 | 23.9 | 24.1 | [2] |
| 4U4G | GluA2* | ZK-200775 | - | X-RAY | 4.49 | 25.3 | 24.5 | [3] | |
| 4UQJ | GluA2em | ZK-200775 | - | Cryo-EM | ~10.0 | 27.2 | 28.0 | [4] | |
| Agonist- bound form with allosteric modulators |
4U1W | GluA25M | Kainate | (R,R)-2b | X-RAY | 3.25 | 39.8 | 25.7 | [1] |
| 4U1X | GluA210M | Kainate | (R,R)-2b | X-RAY | 3.30 | 38.5 | 25.4 | [1] | |
| 4U1Y | GluA210MDel | Fluorowilliardiine | (R,R)-2b | X-RAY | 3.90 | 39.9 | 25.8 | [1] | |
| 4U5D | GluA2cryst1 | Kainate | (R,R)-2b +toxind |
X-RAY | 3.58 | 28.1 | 21.7 | [5] | |
| 4U5F | GluA2cryst2 | Kainate | (R,R)-2b +toxind |
X-RAY | 3.70 | 27.8 | 21.7 | [5] | |
| 4U5C | GluA2cryst1 | Fluorowilliardiine | (R,R)-2b +toxind |
X-RAY | 3.69 | 27.4 | 22.0 | [5] | |
| 4U5E | GluA2cryst1 T625G |
Kainate | (R,R)-2b +toxind |
X-RAY | 3.51 | 27.9 | 21.8 | [5] | |
| 4U5B | GluA2cryst1 A622T |
Kainate | (R,R)-2b +toxind |
X-RAY | 3.50 | 27.9 | 21.8 | [5] | |
| 4UQ6 | GluA2em | Glutamate | LY451646 | Cryo-EM | ~12.0 | 40.2 | 30.5 | [4] | |
| 4UQK | GluA2em | Quisqualate | LY451646 | Cryo-EM | ~16.0 | 41.6 | 33.0 | [4] | |
| Agonist- bound form |
4U4F | GluA2* | (S)-5 -nitrowillardiine (NOW) |
- | X-RAY | 4.79 | 29.3 | 22.3 | [3] |
| 4U2Q | GluA25M | Kainate | - | X-RAY | 3.53 | 39.3 | 25.1 | [1] | |
| - | GluA25M | Fluorowilliardiine | - | X-RAY | 7.94 | 33.9 | 55.8 | [1] | |
| 4UQQ | GluK2em | 2S,4R-4- methylglutamate |
- | Cryo-EM | ~7.6 | 32.8 | 98.5 | [4] |
Constructs are labeled as described by the authors. All genes are from Rattus novergicus.
AC pair distances are measured between the Cα atoms of R660 for GluA2, K664 for GluK2.
BD pair distances are measured between the Cα atoms of Q756 for GluA2, E757 for GluK2.
Toxin: a Conus striatus cone snail toxin, con-ikot-ikot [6].
References:
Durr, K.L. et al. (2014) Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell 158, 778-792.
Sobolevsky, A.I. et al. (2009) X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745-756.
Yelshanskaya, M.V. et al. (2014) Structure of an agonist-bound ionotropic glutamate receptor. Science 345, 1070-1074.
Meyerson, J.R. et al. (2014) Structural mechanism of glutamate receptor activation and desensitization. Nature 514, 328-334.
Chen, L. et al. (2014) X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science 345, 1021-1026.
Walker, C.S. et al. (2009) A novel Conus snail polypeptide causes excitotoxicity by blocking desensitization of AMPA receptors. Current biology : CB 19, 900-908.
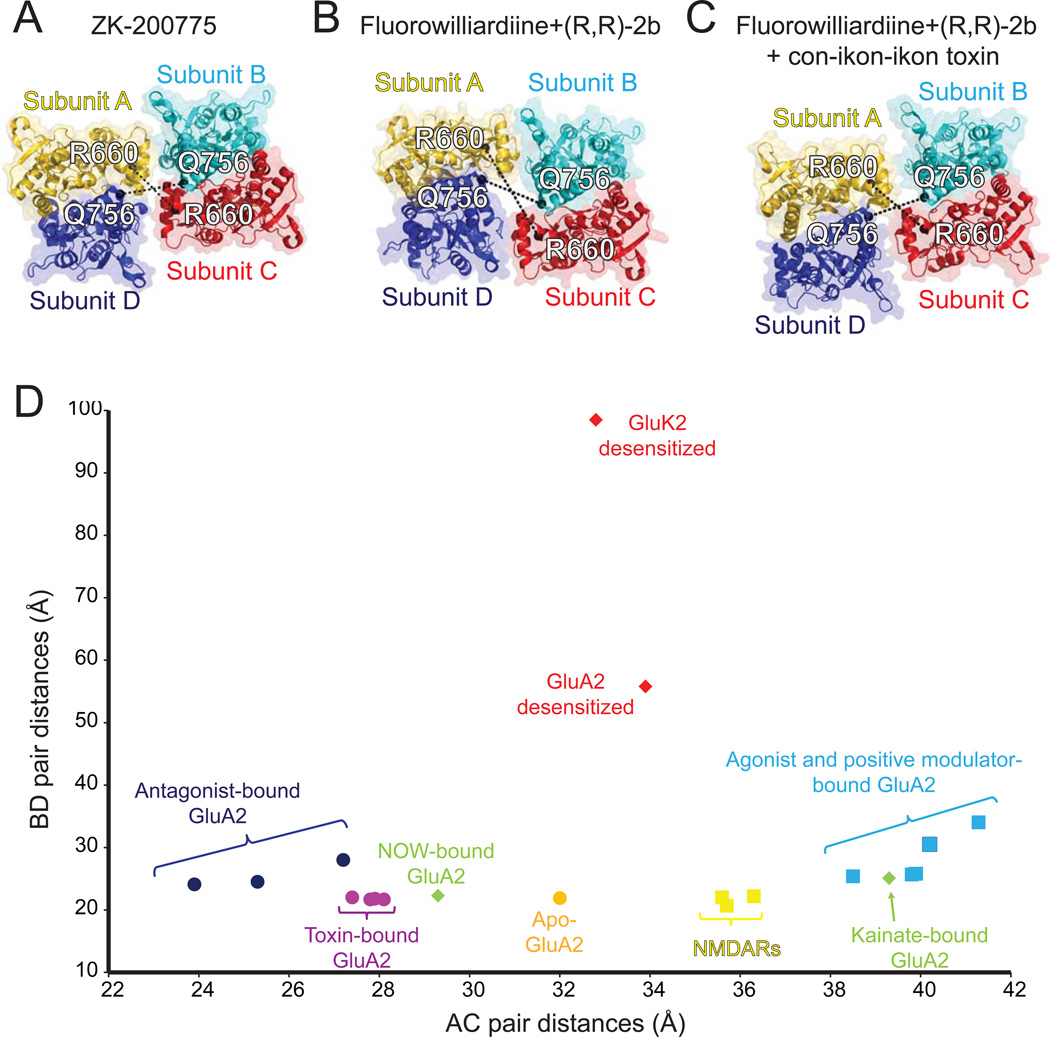
Another central question addressed in recent studies is what happens to the LBD in the pre-open state. An enlargement of the central opening of the LBD dimers observed in the structures of the agonist/modulator-bound GluA2 AMPAR is reminiscent of the ‘gating ring’ in the Ca2+-activated BK channel, which widens upon Ca2+ binding [42, 47]. In the AMPAR, the degree of gating ring enlargement is quantified by measuring intersubunit distances between Cα atoms of Arg 660 and Gln756 for the AC and BD pairs, respectively (Figure 3). In the partial agonist/modulator bound structures, the distance for AC and BD pairs increases by ~7 Å and ~4 Å, respectively, compared to the apo-state structure. The enlargement is more profound in the cryo-EM structures of GluA2 in the full agonist (i.e. glutamate)/modulator bound structure where the increase is about 8 Å for both AC and BD pairs compared to the apo-state structure. Thus, based on this structural comparison, it may be speculated that enlargement of the gating ring is correlated with the degree of receptor activation. However, the mechanism may not be this simple because in the toxin-bound structure which is likely representing an activated state, a decrease rather than an increase in the AC distance was observed while the distance for the BD pair remained similar. In addition, the angle between the two local two-fold rotation axes of the LBD dimers in the toxin-bound structures remained highly similar to that in the apo-state structure, whereas 18-20° increase is observed in the agonist/modulator bound structure (Chen at al, 2014 and Durr et al 2014). Therefore, despite extensive structural information, it is difficult to definitively conclude which conformations of the gating ring and the LBD tetramer facilitate opening of ion channels in non-NMDARs. One possible explanation for this observation is that binding of toxin favors the AMPAR to enter into one subconductance state over the others or creates unique conformations of the gating ring that enhance channel opening. Extensive single channel analyses of AMPARs in the presence of the toxin should answer this question and explain more clearly what functional state the crystal structure actually represents.
Figure 3. Structural changes in the gating ring during receptor activation of non-NMDARs.
LBD tetramers of antagonist-bound (PDB ID: 3KG2) (A), FW/(R,R)-2b-bound (PDB ID: 4U1Y) (B) and, FW/(R,R)-2b-bound and con-ikot-ikot toxin-bound (PDB ID: 4U5C) (C) GluA2 receptors are shown in ribbon representation and viewed from the top of the receptor. Distances between Cα atoms (black spheres) of Arg660 from A/C subunits, and Gln756 from B/D subunits are measured and plotted (D) along with the distances for GluA2 structures in other conformations. The distances for NMDARs are measured between Arg694 for GluN1 (chain A/C) and Asp786 for GluN2B (chain B/D). The distances for kainate receptors are measured between Lys664 for A/C subunits and Glu757 for B/D subunits.
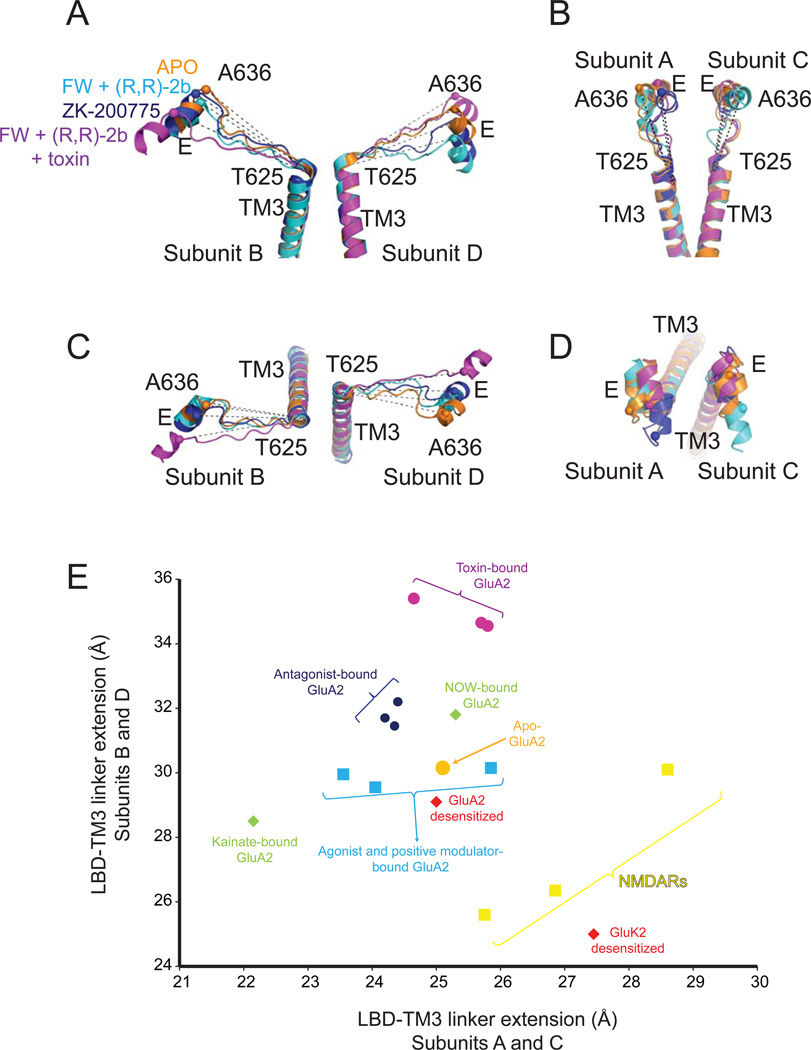
The LBD-TM3 linker connects the LBD to the TM3 gating helix, but how a conformational transition at the LBD tetramer leads to structural changes that create tension along the linker to facilitate ion channel opening, remains unknown. Here, we took the liberty of measuring a structural correlate of the LBD-TM3 linker tension by comparing the distances between the Cα of Ala636 on helix E to the Cα of Thr625 at the extracellular side of the TM3 gating helix (Figure 4). Comparison of the distances between several structures shows the level of linker extension due to the force applied by the LBD. The highest extension is observed in the toxin-bound structures where the distance is increased to ~35 Å for the B and D subunits from 30 Å in the apo-state structure, whereas no major change was observed for A and C subunits (Figure 4). Despite the 5 Å extension for the B and D subunits the channel remain closed implying that either further extension of the linker in the B and D subunits or extension of the linkers in A and C subunits is required to keep the channel open. The distance of the linker in the partial agonist/modulator bound structures is similar to that observed in the apo structure. There is currently no explanation to this unexpected result. The same distance comparison is not possible for the cryo-EM study of the full agonist/modulator bound structures due to the absence of EM-density in TMD.
Figure 4. Conformational changes at the LBD-TM3 linker during receptor activation.
Helices E on the LBD and TM3 at the TMD of B/D subunits (panels A and C) and A/C subunits (panels B and D) are viewed from side (panels A and B) and top (panels C and D). (E) Plot of the istances between Cα atoms (spheres) of Ala636 and Thr625 within the same subunits for apo, FW and (R,R)-2b bound, FW, (R,R)-2b and toxin bound, and ZK200775 bound structures along with the distances for GluA2 structures in other conformations. Distances between Val656 and Ile667 for NMDAR GluN1 subunit, Ile655 and Leu666 for NMDAR GluN2B subunit and Thr629 and Ala640 for kainate receptors are also plotted in (E). The values for the plot are averages of the distances of A/C subunits or B/D subunits.
An attempt to observe a structural representation of the desensitized state led three groups to solve GluA2 AMPAR [41, 42, 48] and GluK2 kainate receptor [41] structures in complex with partial and full agonists but without ‘desensitization blocking’ allosteric modulators or toxin (Figure 1 and Table). Cryo-EM studies of the AMPAR with the full agonist quisqualate, or partial agonist fluorowillardiine (FW), in the absence of allosteric modulators revealed that the desensitized receptors adopt variable conformations [41, 42]. The quisqualate-bound form was separated via 3D classification into three dominant classes with different degrees of separation between ATD dimers [41]. In these configurations, the LBD dimers separate into a quasi-four-fold arrangement. The ATD dimers appeared to be separated to varying extents in the ~8 Å low resolution crystal structure of the FW-bound GluA2 AMPAR, likely representing one of the many conformations observed in cryo-EM studies of the desensitized state (Figure 2E). In this structure, one LBD dimer pair stays intact whereas the other dissociates into two monomers. It is interesting to note that the pattern of subunit association in this dimer is similar to the one observed in the GluA2 Ser729Cys LBD structure, which was suggested to represent the desensitized state [49]. Taken together, the above desensitized structures all have dramatic rearrangements in their LBDs with the overall architecture deviating from the Y-shaped structure observed in apo, agonist/modulator-bound or antagonist-bound structures (Figure 2E) [42]. By contrast, the crystal structures of GluA2 in complex with kainate (KA) or (S)-5-nitrowillardiine (NOW) in the absence of any allosteric modulator (therefore thought to represent desensitized state) are Y-shaped with high similarity to agonist/modulator-bound structures (Figure 2D) [42, 48]. At this point, it is not clear if these structures represent the receptor trapped in one of the potential desensitized states or the agonist-bound pre-open state as above.
An intriguing case is the GluK2 kainate receptor structure in a desensitized state where dramatic rearrangement occurs in the LBD but not in the ATD. Unlike GluA2 AMPARs, GluK2 kainate receptors bound to the full agonist 2S,4R-4-methylglutamate adopt a more homogeneous conformation, perhaps consistent with the 100-fold greater stability of the GluK2 desensitized state revealed by electrophysiological studies [41, 50, 51]. The cryo-EM structure at ~7.6 Å revealed that the dimer of dimers arrangement of the ATD is preserved in the desensitized state while the LBD layer adopts a quasi-four fold symmetric arrangement (Figure 2E) [41]. The undisrupted arrangement of ATDs in GluK2 receptors compared to GluA2 receptors during desensitization is potentially due to tighter tetrameric arrangement of GluK2 ATDs, which is 1,000-fold stronger than the tetrameric arrangement of GluA2 ATDs [24, 52]. It is interesting that the subunit orientation of the disrupted LBD dimer in the crystal structure of FW-bound GluA2 AMPAR is roughly similar to that in the GluK2 structure (Figure 2E). Overall, the structures of non-NMDARs in different functional states agree with each other to a great extent but also disagree in many details. Further structural and functional studies will hopefully integrate the field and provide a consensus views on the mechanistic schemes.
The first visualization of NMDAR heterotetramers
As several structural studies of NMDAR ATDs revealed a large difference in their architecture as well as the pattern of subunit interactions from non-NMDAR ATDs, it became clear that the mode of ATD and LBD interaction and subunit assembly in NMDARs would be distinct from that in non-NMDARs, and thus the AMPAR model cannot be extrapolated to study NMDARs [25–28]. In the wake of substantial enthusiasm to obtain the structure of the intact NMDAR, two very similar structures were published in 2014. The completion of the rat [53] and frog [54] GluN1a/GluN2B NMDAR structures, which have ~90% sequence identity, was a considerable step forward in understanding NMDAR function by putting the previously-solved structures of individual domains into the context of the intact heterotetramer. Despite differences in NMDAR orthologues, construct design, and protein production methods, both of the NMDAR structures, bound to agonists or partial agonists at the LBD and allosteric modulators at the ATD, proved to be similar with a root-mean-square deviation of of 2.1Å for the main chain carbons.
In general, the structures of the intact NMDARs confirmed several suspicions regarding the overall architecture of the protein (Figure. 5). The four subunits assemble in a 1-2-1-2 arrangement around a two-fold axis of symmetry as was predicted by earlier studies [37, 55, 56]. Similar to non-NMDARs, a domain-swap takes place between the ATD and LBD, wherein the GluN1/GluN2B heterodimers are staggered between the ATD and LBD layers [53, 54]. Meanwhile, the TMD exhibits a pseudo-fourfold symmetry resulting from a pseudo-symmetry mismatch between the extracellular domains and the TMD. Perhaps the most apparent difference between the NMDAR and AMPAR structures is the much closer association of the ATD with the LBD in the NMDAR, where substantially more extensive contacts between the ATD dimers and the LBD dimers are present. The distinct ATD-LBD domain arrangement stems from robust differences in the structure of the individual ATDs and in the pattern of the dimeric arrangement of ATDs in NMDARs compared to non-NMDARs (Figure. 5B). In contrast, patterns of inter-domain as well as inter-subunit arrangements appear to be roughly similar between non-NMDARs and NMDARs, perhaps indicating that the basic ligand-gating mechanism in the NMDAR may follow a similar structural changes as described in Figure 2.
Figure 5. Comparison of the intact GluN1/GluN2B NMDAR and GluA2 AMPAR structures.
(A). Both the NMDAR and non-NMDARs consist of four distinct domains: the extra cellular ATD and LBD, the TMD, and a long, largely unstructured Carboxyl Terminal Domain (not shown). The AMPAR assumes a domain-swapped assembly similar to the NMDAR, but with a more extended conformation leading to substantially less interaction between the ATD and LBD. (B). The ATD dimers in particular show dramatically different interactions between the subunits in the AMPAR (and Kainate receptor) vs NMDAR.
The tight packing between ATDs and LBDs in the NMDAR is consistent with existing functional knowledge that its channel activity, such as open channel probability, is regulated by the ATD [57, 58]. By contrast, little or no ATD-mediated regulation is observed in non-NMDARs [59]. That is, the close juxtaposition of the ATDs to the LBDs in the NMDAR likely allows for the efficient propagation of conformational alterations in the ATD to the LBD and vice versa, whereas in non-NMDARs the ATD is too distant from the LBD to affect conformation or subunit arrangement in the LBD, as is evident from crystallographic and cryo-EM studies. Indeed, bilobed structures of ATDs have been shown to undergo dynamic intrinsic conformational alterations, which are most likely regulated by ATD-targeting compounds that inhibit or potentiate channel activity [27, 60–62]. Thus far, only the closed conformation of the ATD bound to phenylethanolamines, representative of the “inhibited conformation,” has been observed by crystallography. Structurally capturing “active” conformations of the ATD and visuallizing changes in ATD conformations that affect the LBD conformation will be the key to understanding activation mechanism of NMDARs and perhaps bring us one step closer to deciphering complex and intriguing pattern of gating kinetics [63, 64].
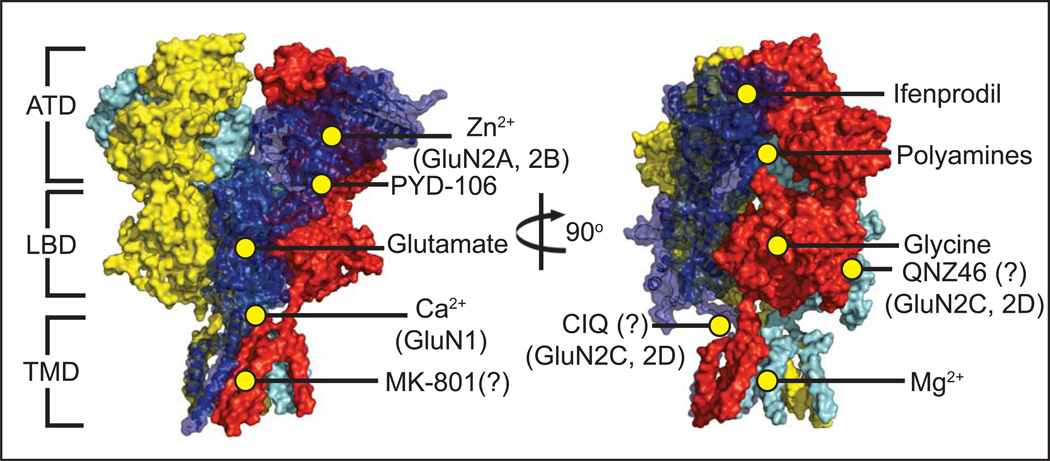
The local electron density in both the Karakas and Lee structures is limited within and around the TMD, but the two taken together can serve as complementary components to describe the finer features of this domain. The NMDAR has the same membrane topology as non-NMDARs, with the subunits arranged at the TMD in a pseudo four-fold symmetry consistent with its heterotetrameric assembly. Both of the recent NMDAR structures appear to have the closed channel consistent with the speculation that both structures likely captured the allosterically inhibited state. One key feature visible in the structure by Karakas & Furukawa is the DRPEER motif present in the GluN1 M3-S2 linker, named after the sequence of amino acids shown to regulate Ca2+ permeation and found exclusively in GluN1 subunits (and absent in the GluN2 and all non-NMDAR subunits) [65]. The lanthanides holmium and gadolinium, which can occupy Ca2+-binding sites [66, 67] and show strong anomalous signal in x-ray crystallography, were identified adjacent to the DRPEER motif, thus visually confirming the validity of previous electrophysiological data. Additionally, Lee, et al. reported an intriguing electron density near the center of the channelthat may represent the channel blocker MK-801 included in their crystallization condition. However, further work such as molecular docking and mutagenesis will be necessary to gain clear insights into the architecture of the blocker binding site. Another victim of reduced electron density at the TMD is the so-called S/L site in the GluN2 subunit. Recent work showed that subtype-specific sensitivity to Mg2+ block, single-channel conductance, and Ca2+ permeability were all closely linked to the identity of a single residue at the base of the GluN2 M3 helix and its interaction with an adjacent residue in the GluN1 M3 helix [68]. Unfortunately, the sidechains of those residues were not resolved in either of the crystallographic studies. Homology modelling suggests that the effect on channel properties propagated by this site may potentially function in a manner similar to the sodium/potassium channel and non-NMDAR iGluRs [68]. The development of a method to obtain a significantly higher resolution structure would be useful in resolving the chemistry unique to NMDARs, such as calcium permeability, magnesium block, and blocker binding. A method involving lipidic cubic phase (LCP) for crystallization has been instrumental in obtaining high-resolution structures of various membrane proteins, most notably G protein-coupled receptors in recent years [69]. Unfortunately, this method has not been successfully implemented on iGluR proteins, perhaps due to their large extracellular domains which exceed the predicted size limitation for the LCP matrix. Nevertheless, the crystal structures provide the long-awaited molecular templates to locate known pharmacological sites in the context of the heterotetramers and better predict potential binding sites for novel compounds, an example of which is the recently work on PYD-106 (Figure 6) [70].
Figure 6. Heterotetrameric GluN1/GluN2B NMDAR and small molecule binding sites.
Numerous ligands have been identified that influence NMDAR activity. Co-agonists glycine [76] and glutamate bind at the LBD of the GluN1 and GluN2 subunits, respectively [77]. The small molecule PYD-106 is a GluN2C-specific allosteric activator which binds at the ATD/LBD interface [70]. Polyamines such as spermine and spermidine bind at the interface of two ATDs in GluN2B-containing receptors and potentiate channel activity [78], but only in GluN1 isoforms lacking Exon5 [79]. A number of NMDAR inhibitors have also been identified, including extracellular Zn2+ [53] binding at the hinge region of the GluN2 ATD [26] (and with a notably stronger effect on GluN2A-containing receptors over other types), and voltage-dependent block by Mg2+ at the TMD [68]. Phenylethanolamines, such as ifenprodil, bind at the interface of the GluN1/GluN2B ATDs [27], while the inhibitor MK-801 is purported to bind very near to the center of the ion channel [54]. A sequence of acidic residues in the LBD/TMD M3 linker in the GluN1 subunit known as the DRPEER motif is essential for calcium binding and conductance [53].
Concluding remarks
The structural studies of iGluRs are now entering a new, exciting phase in which the intact tetrameric structures of all of the major iGluR subclasses are revealed. Structural studies of iGluRs are not only important for the understanding of general mechanisms underlying functional regulation (ligand-gating, channel opening, allosteric regulation, etc.) but also to visualize pharmacological binding sites useful for the development of therapeutic compounds (Figure. 6). While significant insights into a functional mechanistic scheme can be gained at intermediate to low resolution (~3.5 -15 Å), pharmacological studies ideally require resolution higher than 2.5 Å, which permits visualization of ordered water molecules and ions. Due to the low-resolution nature of structural studies on the full-length iGluRs, it is still meaningful to work on isolated domains including the ATD and LBD to obtain high-resolution structures useful for such purposes. Nevertheless, with the availability of intact iGluR structures today, the structural information of isolated domains can be extrapolated reasonably well in order to speculate mechanisms in the context of the tetrameric ion channels. Undeniably, x-ray and EM structures are “snapshots” of protein architectures, thus, detailed functional analyses by well-established techniques such as electrophysiology, fast-advancing molecular dynamic studies [71–74], and fluorescent methods [17, 75] are necessary to gain deeper mechanistic insights into how different members of the large iGluR family function. Overall, the structural templates of intact iGluRs provided by the recent studies in combination with the isolated domain structures serve well to understand general functional mechanisms and to facilitate compound development for neurological disorders and diseases in an unprecedented manner.
Highlights.
-
5
Structures of the AMPAR and kainate receptor in multiple states are now available.
-
6
These structures show how non-NMDAR iGluRs move to mediate defined functions.
-
7
Structures of GluN1/GluN2B show NMDARs and non-NMDARs have distinct architectures.
-
8
Despite efforts, a bona fide “open channel” structure remains to be observed.
Acknowledgement
We thank Eric Gouaux for providing the structural coordinate for the desensitized AMPAR. Mark Mayer, Sasha Sobolevsky, Andrew Plested, Katharina Dürr, Lei Chen, Chia-Hsueh Lee, and Xianqiang Song are thanked for open discussion on the topics in this review at the 59th biophysical society meeting. This work was supported by National Institute of Health (MH085926 and GM105730 to HF), Stanley institute of cognitive genomics, and Robertson Research Fund of Cold Spring Harbor Laboratory (to HF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hayashi T. Effects of sodium glutamate on the nervous system. Keio J. Med. 1954;3:192–193. [Google Scholar]
- 2.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacological reviews. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moriyoshi K, et al. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- 4.Hollmann M, et al. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989;342:643–648. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- 5.Werner P, et al. Cloning of a putative high-affinity kainate receptor expressed predominantly in hippocampal CA3 cells. Nature. 1991;351:742–744. doi: 10.1038/351742a0. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong N, et al. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 8.Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa H, et al. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 10.Jespersen A, et al. Structural Insights into Competitive Antagonism in NMDA Receptors. Neuron. 2014;81:366–378. doi: 10.1016/j.neuron.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vance KM, et al. Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nature communications. 2011;2:294. doi: 10.1038/ncomms1295. doi:210.1038/ncomms1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albrecht T, et al. New developments in nanopore research-from fundamentals to applications. Journal of physics. Condensed matter : an Institute of Physics journal. 2010;22:450301. doi: 10.1088/0953-8984/22/45/450301. [DOI] [PubMed] [Google Scholar]
- 13.Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 14.Mayer ML, et al. Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McFeeters RL, Oswald RE. Structural mobility of the extracellular ligand-binding core of an ionotropic glutamate receptor. Analysis of NMR relaxation dynamics. Biochemistry. 2002;41:10472–10481. doi: 10.1021/bi026010p. [DOI] [PubMed] [Google Scholar]
- 16.Lau AY, Roux B. The hidden energetics of ligand binding and activation in a glutamate receptor. Nature structural & molecular biology. 2011;18:283–287. doi: 10.1038/nsmb.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landes CF, et al. Structural landscape of isolated agonist-binding domains from single AMPA receptors. Nature chemical biology. 2011;7:168–173. doi: 10.1038/nchembio.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin R, et al. Crystal structure and association behaviour of the GluR2 amino-terminal domain. Embo J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar J, et al. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clayton A, et al. Crystal structure of the GluR2 amino-terminal domain provides insights into the architecture and assembly of ionotropic glutamate receptors. J Mol Biol. 2009;392:1125–1132. doi: 10.1016/j.jmb.2009.07.082. [DOI] [PubMed] [Google Scholar]
- 21.Sukumaran M, et al. Dynamics and allosteric potential of the AMPA receptor N-terminal domain. The EMBO journal. 2011;30:972–982. doi: 10.1038/emboj.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao G, et al. Crystal structure of the glutamate receptor GluA1 N-terminal domain. The Biochemical journal. 2011;438:255–263. doi: 10.1042/BJ20110801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar J, Mayer ML. Crystal structures of the glutamate receptor ion channel GluK3 and GluK5 amino-terminal domains. Journal of molecular biology. 2010;404:680–696. doi: 10.1016/j.jmb.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar J, et al. Structure and assembly mechanism for heteromeric kainate receptors. Neuron. 2011;71:319–331. doi: 10.1016/j.neuron.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farina AN, et al. Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. The Journal of neuroscience. 2011;31:3565–3579. doi: 10.1523/JNEUROSCI.6041-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karakas E, et al. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karakas E, et al. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011;475:249–253. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furukawa H. Structure and function of glutamate receptor amino terminal domains. The Journal of physiology. 2012;590:63–72. doi: 10.1113/jphysiol.2011.213850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen KB, et al. Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol. 2010;78:535–549. doi: 10.1124/mol.110.067157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payandeh J, et al. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 32.Long SB, et al. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa T, et al. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 34.Safferling M, et al. First images of a glutamate receptor ion channel: oligomeric state and molecular dimensions of GluRB homomers. Biochemistry. 2001;40:13948–13953. doi: 10.1021/bi011143g. [DOI] [PubMed] [Google Scholar]
- 35.Tichelaar W, et al. The Three-dimensional Structure of an Ionotropic Glutamate Receptor Reveals a Dimer-of-dimers Assembly. Journal of molecular biology. 2004;344:435–442. doi: 10.1016/j.jmb.2004.09.048. [DOI] [PubMed] [Google Scholar]
- 36.Nakagawa T, et al. Three-dimensional structure of an AMPA receptor without associated stargazin/TARP proteins. Biological chemistry. 2006;387:179–187. doi: 10.1515/BC.2006.024. [DOI] [PubMed] [Google Scholar]
- 37.Sobolevsky AI, et al. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kazi R, et al. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nature neuroscience. 2014;17:914–922. doi: 10.1038/nn.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, et al. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 40.Walker CS, et al. A novel Conus snail polypeptide causes excitotoxicity by blocking desensitization of AMPA receptors. Current biology : CB. 2009;19:900–908. doi: 10.1016/j.cub.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyerson JR, et al. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durr KL, et al. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, et al. X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science. 2014;345:1021–1026. doi: 10.1126/science.1258409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whorton MR, MacKinnon R. X-ray structure of the mammalian GIRK2-betagamma G-protein complex. Nature. 2013;498:190–197. doi: 10.1038/nature12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Sigworth FJ. Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature. 2009;461:292–295. doi: 10.1038/nature08291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Sigworth FJ. Automatic cryo-EM particle selection for membrane proteins in spherical liposomes. Journal of structural biology. 2014;185:295–302. doi: 10.1016/j.jsb.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan P, et al. Open structure of the Ca2+ gating ring in the high-conductance Ca2+-activated K+ channel. Nature. 2012;481:94–97. doi: 10.1038/nature10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yelshanskaya MV, et al. Structure of an agonist-bound ionotropic glutamate receptor. Science. 2014;345:1070–1074. doi: 10.1126/science.1256508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armstrong N, et al. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 50.Schauder DM, et al. Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5921–5926. doi: 10.1073/pnas.1217549110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carbone AL, Plested AJ. Coupled control of desensitization and gating by the ligand binding domain of glutamate receptors. Neuron. 2012;74:845–857. doi: 10.1016/j.neuron.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 52.Zhao H, et al. Analysis of high-affinity assembly for AMPA receptor amino-terminal domains. The Journal of general physiology. 2013;141:747–749. doi: 10.1085/jgp.20121077004292013c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee CH, et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salussolia CL, et al. Arrangement of subunits in functional NMDA receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:11295–11304. doi: 10.1523/JNEUROSCI.5612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riou M, et al. An alternating GluN1-2-1-2 subunit arrangement in mature NMDA receptors. PloS one. 2012;7:e35134. doi: 10.1371/journal.pone.0035134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan H, et al. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gielen M, et al. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moykkynen T, et al. The N-terminal domain modulates alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor desensitization. The Journal of biological chemistry. 2014;289:13197–13205. doi: 10.1074/jbc.M113.526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burger PB, et al. Mapping the binding of GluN2B-selective N-methyl-D-aspartate receptor negative allosteric modulators. Molecular pharmacology. 2012;82:344–359. doi: 10.1124/mol.112.078568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu S, et al. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nature structural & molecular biology. 2013;20:477–485. doi: 10.1038/nsmb.2522. [DOI] [PubMed] [Google Scholar]
- 62.Sirrieh RE, et al. Amino-terminal domain tetramer organization and structural effects of zinc binding in the N-methyl-D-aspartate (NMDA) receptor. The Journal of biological chemistry. 2013;288:22555–22564. doi: 10.1074/jbc.M113.482356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Popescu G, Auerbach A. The NMDA receptor gating machine: lessons from single channels. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2004;10:192–198. doi: 10.1177/1073858404263483. [DOI] [PubMed] [Google Scholar]
- 64.Popescu G, et al. Reaction mechanism determines NMDA receptor response to repetitive stimulation. Nature. 2004;430:790–793. doi: 10.1038/nature02775. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe J, et al. DRPEER: a motif in the extracellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. The Journal of Neuroscience. 2002;22:10209–10216. doi: 10.1523/JNEUROSCI.22-23-10209.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li GW, et al. EF hands at the N-lobe of calmodulin are required for both SK channel gating and stable SK-calmodulin interaction. The Journal of general physiology. 2009;134:281–293. doi: 10.1085/jgp.200910295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liddington RC, et al. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 68.Siegler Retchless B, et al. A single GluN2 subunit residue controls NMDA receptor channel properties via intersubunit interaction. Nature neuroscience. 2012;15:406–413. S401–S402. doi: 10.1038/nn.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nature protocols. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khatri A, et al. Structural determinants and mechanism of action of a GluN2C-selective NMDA receptor positive allosteric modulator. Molecular pharmacology. 2014;86:548–560. doi: 10.1124/mol.114.094516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao Y, et al. Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics. Structure. 2013 doi: 10.1016/j.str.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dawe GB, et al. Defining the structural relationship between kainate-receptor deactivation and desensitization. Nature structural & molecular biology. 2013;20:1054–1061. doi: 10.1038/nsmb.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dror RO, et al. Biomolecular simulation: a computational microscope for molecular biology. Annual review of biophysics. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 74.Dai J, Zhou HX. An NMDA receptor gating mechanism developed from MD simulations reveals molecular details underlying subunit-specific contributions. Biophysical journal. 2013;104:2170–2181. doi: 10.1016/j.bpj.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi UB, et al. Effect of Src kinase phosphorylation on disordered C-terminal domain of N-methyl-D-aspartic acid (NMDA) receptor subunit GluN2B protein. The Journal of biological chemistry. 2011;286:29904–29912. doi: 10.1074/jbc.M111.258897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson JW, Ascher P. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature. 1987;325:529–531. doi: 10.1038/325529a0. [DOI] [PubMed] [Google Scholar]
- 77.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mony L, et al. Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. The EMBO journal. 2011;30:3134–3146. doi: 10.1038/emboj.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Traynelis SF, et al. Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science. 1995;268:873–876. doi: 10.1126/science.7754371. [DOI] [PubMed] [Google Scholar]