Abstract
The Wnts are secreted, lipidated glycoproteins that play a role in cellular processes of differentiation, proliferation, migration, survival, polarity and stem cell self-renewal. The majority of Wnts biological effects are through binding to specific frizzled (Fzd) receptor complexes leading to activation of downstream pathways. Secreted Frizzled-related proteins (sFRPs) were first identified as antagonists of Wnt signalling by binding directly to Wnts. They comprise two domains, a Fzd-like cysteine rich domain (CRD) and a netrin-like domain (NLD). Subsequently sFRPs have been shown to also interact with Fzd receptors and more diverse functions have been identified, including potentiation of Wnt signalling. Many aspects of the biology of this family remain to be elucidated. We used the number and brightness (N&B) method, a technique based on fluorescence fluctuation analysis, to characterise the intracellular aggregation and trafficking of sFRP4 domains. We expressed sFRP4 and its’ domains as EGFP fusions and then characterised the effect of endogenous Wnt3a by fluorescence confocal imaging. We observed vesicular trafficking of sFRP4 and that the NLD domain has a vesicular association signal. We found that sFRP4 and the CRD formed oligomeric aggregates in the perinuclear region while the NLD was distributed evenly throughout the cell with a larger proportion of aggregates. Most significantly we observed intracellular redistribution of sFRP4 in response to Wnt3a suggesting that Wnt3a can modulate intracellular localisation and secretion of sFRP4. Our results reveal a number of novel findings regarding sFRP4 which are likely to have relevance to this wider family.
Keywords: Wnt, sFRP, Cysteine-rich domain, Netrin-like domain, Protein aggregates, Live cell imaging
1. Introduction
The Wnts are secreted, lipidated glycoproteins (at least 19 in humans) that transduce signals by binding to specific frizzled (Fzd) receptor complexes (reviewed in Schulte, 2010) leading to activation of canonical or non-canonical pathways depending upon molecular context. Wnts are functionally integral to many processes during embryonic development and play an important role in homeostasis in adult tissues. The downstream pathways activated by Wnts play a role in a diverse array of cellular processes including differentiation, proliferation, migration, survival, polarity and stem cell self-renewal (Clevers and Nusse, 2012; Wang et al., 2012). Aberrant Wnt signalling is associated with several disorders, especially cancer (Anastas and Moon, 2013).
The mammalian secreted Frizzled-related proteins (sFRPs) are a family of five proteins which were first identified on the basis of their antagonistic effect upon Wnt signalling. Subsequent research has indicated more diverse functions for sFRPs, including potentiation of Wnt signalling in certain contexts (Uren et al., 2000; Kress et al., 2009; von Marschall and Fisher, 2010; Xavier et al., 2014), spacial diffusion of Wnts (Mii and Taira, 2009) or even Wnt independent effects (Martin-Manso et al., 2011). They contain two domains, a Fzd-like cysteine rich domain (CRD) and a netrin-like domain (NLD). The CRD of sFRPs was initially believed to play a role analogous to the CRD of Fzds as the principal mediators of Wnt binding (Lin et al., 1997). Although some studies have suggested that sFRPs interact with Wnts via their CRD domain (Lin et al., 1997) subsequent experiments have indicated that the NLD may play a more prominent role in this interaction (Bhat et al., 2007; Lopez-Rios et al., 2008). In addition it has been demonstrated that SFRPs and Fzds can also interact via their CRD domains to form heterodimers and homodimers suggesting an alternative mechanistic basis for sFRPs to influence Wnt signalling (Bafico et al., 1999; Rodriguez et al., 2005).
The biological significance of sFRPs is evinced by studies in developmental models and the frequent observation of aberrant expression in many cancers. Despite this evidence many aspects of the biology of this family remains to be elucidated. In malignant mesothelioma (MM) sFRP4 has been shown to be downregulated by promoter methylation and to function as a tumour suppressor (Lee et al., 2004; Kohno et al., 2010). Nearly all studies of sFRPs to date have focussed upon their extracellular interactions and very little is known about the intracellular trafficking, localization and behaviour of sFRP4 and its domains. Using a mesothelioma cell model which showed little or no sFRP4 expression (Fox et al., 2013) we undertook to characterise the intracellular trafficking of sFRP4 domains, particularly in cells exposed to endogenous Wnt3a.
We used a recently described fluorescence microscopy technique (Ossato et al., 2010), Number and Brightness (N&B) fluctuation spectroscopy analysis, to study intracellular localisation and aggregation of sFRP4, sFRP4 CRD and sFRP4 NLD. The basis for the N&B method is the analysis of fluorescence intensity distributions and permits measurement of both the number (N) and brightness (B) at each pixel in a stack of images. Since the brightness is directly related to the number of molecules aggregated, the method has been previously applied to live cell image analysis of intracellular protein aggregation (Ossato et al., 2010; Vetri et al., 2011). Our imaging and image analysis studies suggest that sFRP4 is localized in the perinuclear region and this localisation is likely to be a property of the CRD domain. We found that the NLD was likely to have a vesicle association signal involved in secretion of sFRP4. Notably we found that intracellular localisation and trafficking of sFRP4 domains could be modified in response to Wnt3a.
2. Materials and methods
2.1 Cell culture
The malignant mesothelioma cell line JU77 was used in this study (Manning et al., 1991). JU77 cells were cultured in RPMI 1640 supplemented with 5% foetal bovine serum, 2mM glutamine, penicillin (100IU/ml), and streptomycin (100ug/ml) (all from Thermofisher Hyclone, Vic., Aust).
2.2 Plasmid Constructs
Expression vectors for the human sFRP4 gene and the CRD and NLD domains were a kind gift from Prof. Roberts Friis, University of Bern, Switzerland. The sFRP4 constructs were prepared by PCR and cloning into the pEGFP-N1 vector (Clontech, CA, USA) so that the full length sFRP4, the CRD domain or the netrin C terminal domain were expressed in frame as amino terminal fusions to the GFP. The sFRP4, CRD and Netrin constructs retained the Kozak and signal sequences of sFRP4. The parental pEGFP-N1 vector expressing GFP was used as a control. Plasmid DNA for transfection was prepared using a HiSpeed Plasmid Midi Kit (Qiagen, Vic., Australia).
2.3 Transfection
Transient transfections were performed on JU77 cells using FUGENE® HD reagent and the pEGFP-N1 plasmid vector constructs. Cells were seeded at a density of 80% confluency in 250ul RPMI growth medium in 8 well Lab Tek chambered #1.0 Borosilicate cover glass system w/cvr (ThermoScientific) on the day of transfection. Following transfection with Reagent:DNA in a ratio of 3:1 for 24 hrs, a further 48 hrs of incubation was allowed for the protein to be expressed. The transfection reagent was removed from the cells and replaced with standard complete RPMI medium after 48 hrs. For the Wnt3a treatment group, 48 hrs post transfected cells in chambers were replenished with recombinant human Wnt3a (R&D systems) 250pg/mL in RPMI complete medium for 6 hrs and imaged under confocal microscope. The GFP-only vector was also expressed to act as a control. Transfected JU77 cells expressing GFP-fusion protein fluorescence were measured using a Nikon-A1+ confocal microscope (Nikon, Tokyo, Japan with 488nm laser point scanning)
2.4 Confocal live cell imaging and Number and Brightness analysis
Number and brightness (NB) analysis was performed on the transfected samples and measured by the time series of 100 frames of 256 × 256 pixels, obtained with a HV 96, offset-8 and laser 1.0 setting was used to obtain barely bright enough, best focus, and visible images to see the same GFP fluorescence for all the groups using a scan speed of 1/2. The pixel size was 50 nm with a pixel dwell time of 23.5 μs, and the pinhole was set at 33.0 μm. These image stacks were analyzed using SimFCS software (Laboratory for Fluorescence Dynamics, University of California, and Irvine CA). The detector calibration was obtained from 100 frames of background image taken before and after the experimental image stacks using the exact same settings but with the laser turned off as described previously (Dalal et al., 2008). Brightness (B) of fluorescence particle, B= 1 values represent the immobile fraction of the image, and B> 1 values represent the mobile fraction (Dalal et al., 2008). To obtain the molecular brightness in photons/molecule/s, the B= 1 value was divided by the pixel dwell time. The B vs intensity plot was analysed using two cursors. A red cursor selects all the pixels of the image that have B values between 1 and 1.5 and paints the pixels of the image in red. A second green cursor was used to select these pixels that have B values between 1.5 and 4 which were painted in green. There are essentially no pixels with B values above 4. Therefore the B images show in green the regions of the cell where larger aggregates form (larger B values). For each image the fraction of green and red pixels was also calculated. Changes upon stimulation of Wnt3a were also calculated by normalizing the difference of green pixels in the image before and after Wnt3a activation by the total number of pixels above a minimum of intensity threshold.
3. Results
3.1 Establishment of B value for monomeric EGFP
In order to determine the brightness (B) of monomeric EGFP we transfected JU77 cells with the parental pEGFP-N1 vector and acquired images as described above. This enabled us to establish background correction and subsequently determine the B value due to monomeric EGFP expressed in JU77 cells essentially as previously described (Plotegher et al., 2014). As a result of this analysis we were able to determine that the brightess of monomeric EGFP was distributed from 1 to 1.5 with a mean value for B 1.25.
3.2 Peri-nuclear localization and vesicular trafficking of sFRP4
The nature of the intracellular localisation of sFRP4 (or indeed sFRPs in general) and whether it forms oligomeric aggregates is not known. Therefore JU77 cells were transiently transfected with sFRP4-EGFP and observed by live cell confocal imaging 72 hours after transfection. The sFRP4-EGFP was localized predominantly in the peri-nuclear region (Fig. 1A) and to a lesser extent in the cytoplasm, whereas there was a very low intensity in the nucleus. The images demonstrated (Fig. 1) that there are small sFRP4-EGFP containing vesicles with a size ranging from 250 to 300 nm (essentially diffraction-limited spots) diffusing in the cytoplasm with a characteristic vesicle transport-like movement (see movie in Supplementary file). These mobile vesicles observed in the cytosol are consistent with the secreted nature of this protein, where sFRP4 is synthesized, modified and accumulates in the endoplasmic reticulum in and around the peri-nuclear region, and then is transported via vesicles to the cell membrane from where they are secreted.
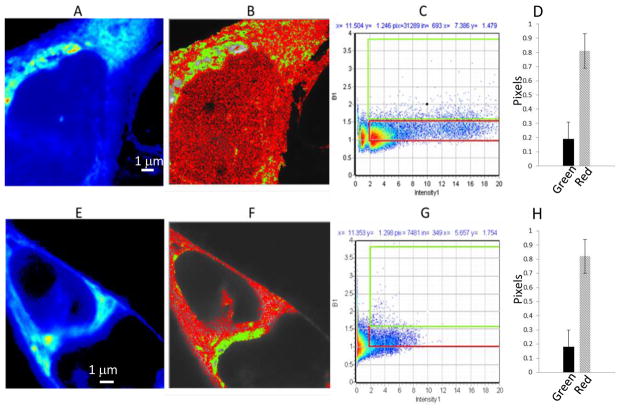
Fig. 1.
N&B analysis of JU77 cells expressing sFRP4-EGFP; (A–D) and (E–H) following Wnt3a treatment. A and E. Average intensity image of a single optical plane. B and F. Image with identification of pixels with brightness corresponding to monomers (red, B<1.5) or oligomers (green, B>1.5). C and G. Corresponding distribution of B values versus fluorescence intensity for the same cell and selection map for red (B= 1–1.5) and green (B>1.5) pixels. D and H. Fraction of pixels corresponding to oligomer (green) and monomer (red).
Interestingly, further analysis of these images using the number and brightness technique, did show that sFRP4-EGFP formed oligomeric aggregates and that these are particularly localised in the peri-nuclear region (Fig. 1B). The fraction of relatively larger aggregates was determined by selecting pixels with B values larger than 1.5 (green pixels in Fig. 1B, the cursor placement is shown in Fig. 1C), while values of B less than 1.5 (red pixels) were classified as small aggregates or protein monomers. This indicated that overall the monomeric form is predominant within the cell and in particular this is so outside the perinuclear region. We did observe that the vesicular sFRP4-EGFP particle brightness was less than we would otherwise expect given the vesicle size and this can be indicative of quenching through interaction with other molecules. However, it is not possible to establish this with certainty based upon N&B analysis and further investigation is required to determine if this is the case.
We were interested to investigate the effect of the canonical ligand Wnt3a upon the intracellular behaviour of sFRP4 as we had previously found that sFRP4 can antagonize the effects of Wnt3a upon JU77 cells (manuscript in preparation). Therefore we treated sFRP4-EGFP expressing JU77 cells with Wnt3a and observed the effect by confocal fluorescent imaging. We did not observe significant changes in the area covered by oligomeric sFRP4-EGFP (ie B>1.5) upon treatment with Wnt3a (green regions in Fig. 1E and 1F). There was some indication that sFRP4-EGFP became more diffused intracellularly upon Wnt3a treatment, however, we did not observe a measurable effect upon intensity and brightness. There was little difference in the monomeric and oligomeric sFRP4-EGFP fractions as determined by the green and red selections (cursor positions shown in Fig. 1C and 1G) in response to Wnt3a (Fig. 1D vs 1H).
3.3 sFRP4 CRD localization, oligomerisation and response to Wnt3a
We next undertook to investigate the relative role of the CRD domain in the intracellular localisation and trafficking of sFRP4. The sFRP CRD domain is known to interact with other partners, specifically Wnts, Fzd receptors and may be capable of homodimerisation although most investigations have not identified the location of such interactions (Cruciat and Niehrs, 2013). We transfected JU77 cells with a CRD-EGFP expression construct and as before performed live cell confocal imaging after 72 hours. As with full length sFRP4 we found that CRD-EGFP was most concentrated in the peri-nuclear region, however, overall CRD-EGFP was more broadly distributed throughout the cell (Fig. 2A). In addition there are more pixels in the CRD-EGFP cells localized around the peri-nuclear region both with B<1.5 and B>1.5 (Fig. 2B) compared to sFRP4-EGFP (Fig. 1B) which may reflect higher expression of this construct. Notably we did not observe evidence of vesicular trafficking of CRD-EGFP.
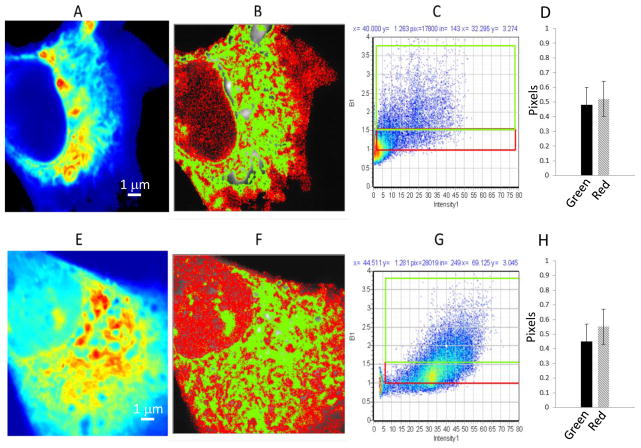
Fig. 2.
N&B analysis of JU77 cells expressing CRD-EGFP; (A–D) and (E–H) following Wnt3a treatment. A and E. Average intensity image of a single optical plane. B and F. Image with identification of pixels with brightness corresponding to monomers (red, B<1.5) or oligomers (green, B>1.5). C and G. Corresponding distribution of B values versus fluorescence intensity for the same cell and selection map for red (B= 1–1.5) and green (B>1.5) pixels. D and H. Fraction of pixels corresponding to oligomer (green) and monomer (red).
N & B analysis of these images demonstrated the CRD-EGFP behaved quite distinctly to sFRP4-EGFP with respect to aggregation properties and response to Wnt3a. The sizes of the oligomers (judged by the B value) are comparable to sFRP4 protein (Fig. 2B); however, the location and area where the oligomers are found are much larger in the CRD expressing cells (green area in Fig. 2B). In addition the distribution of aggregate sizes was different for CRD-EGFP with a much greater proportion of oligomers as shown in the graph (Fig. 2D).
When cells expressing CRD-GFP were exposed to Wnt3a, the distribution within the cell changed and fluorescence became more diffused (Fig. 2E). CRD-EGFP aggregates were not restricted to the peri-nuclear region; but seen in the cytosol as well as in the nucleus (Fig. 2E and F). Following Wnt3a treatment the actual area within the cell covered by the oligomers (green in Fig. 2F) remained constant although it became less localised (Fig. 2B and F). This is confirmed by the fact that the distribution of monomers/oligomers did not change (Fig. 2D and H). The CRD-EGFP protein must be bound to interacting partners for localisation to the peri-nuclear region and the effect of Wnt3a indicates that this interaction is affected through an undetermined mechanism. The nature and partners of this interaction with CRD and their significance to Wnt signalling of requires further investigation since such direct effects by Wnts upon sFRPs have not previously been described.
3.4 sFRP4 NLD has a vesicular association signal
In order to characterise the NLD domain we similarly transfected JU77 cells with a NLD-EGFP expression construct followed by live cell confocal imaging at 72 hours. Our observations for the localisation of NLD-EGFP were quite different to those of either the full length sFRP4 or the CRD construct. The NLD-EGFP protein was more or less uniformly distributed across the cell (Fig. 3A). This indicates that the peri-nuclear localisation described above for the previous constructs is due to an inherent property of the CRD domain. It should be noted that all of the constructs used in the present study were engineered to contain the sFRP4 signal peptide and there is evidence that they are secreted (Longmann et al., 2012).
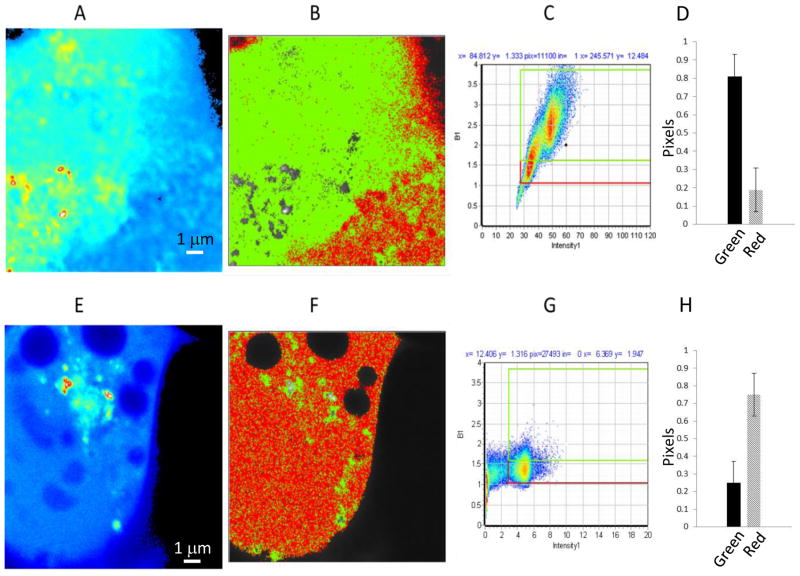
Fig. 3.
N&B analysis on JU77 cells expressing the NLD domain; (A–D) and (E–H) following Wnt3a treatment. A and E. Average intensity image of a single optical plane. B and F. Image with identification of pixels with brightness corresponding to monomers (red, B<1.5) or oligomers (green, B>1.5). C and G. Corresponding distribution of B values versus fluorescence intensity for the same cell and selection map for red (B= 1–1.5) and green (B>1.5) pixels. D and H. Fraction of pixels corresponding to oligomer (green) and monomer (red).
In addition we observed bright particle-like aggregates formed by NLD-EGFP which demonstrate apparent associated with vesicles (Supplementary data: video). The size of these particles ranges from 250 nm to 300 nm and were similar in size to those seen in sFRP4-EGFP expressing cells. As can be seen in Fig 3B the larger oligomeric aggregates were distributed throughout the cell and significantly, there was a much higher proportion of oligomeric aggregates for NLD-EGFP (Fig. 3D) than for the other fusions used.
When cells expressing NLD-EGFP were treated with exogenous Wnt3a there were profound changes in the overall intracellular fluorescence intensity (Fig. 3E) and in the relative proportion of oligomeric aggregates (Fig. 3H), Some particle-like aggregates remained in the perinuclear region but were strongly reduced as judged by the area in which they can be found (green areas in Fig. 3F, selection graph in Fig. 3G). This strong reduction in vesicular fluorescence in response to Wnt3a suggests that the NLD plays a role in secretion of sFRP4. Most significantly, it supports a role for Wnt3a in regulation of sFRP4 trafficking and secretion which has not previously been described in sFRPs generally.
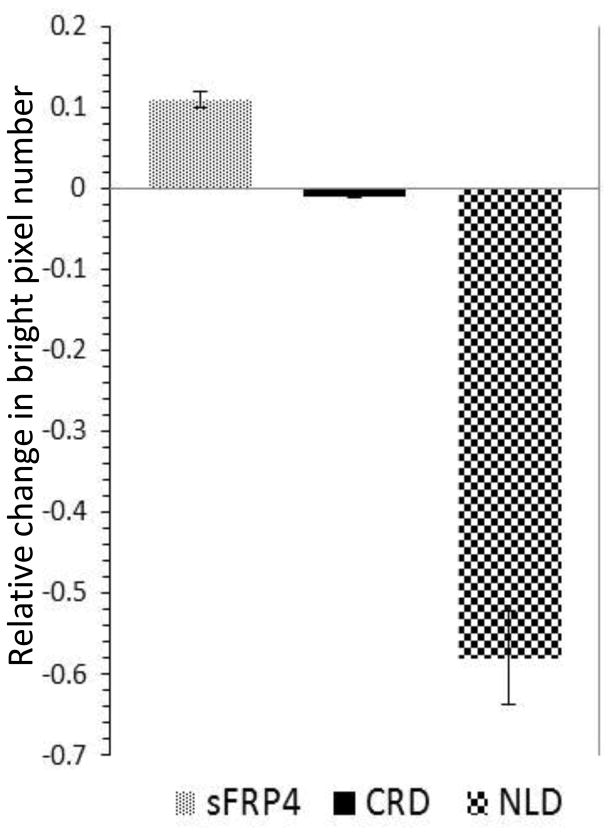
If we consider the relative changes of the number of pixels in the B values > 1.5, normalized to the total number of pixels, we found that activation of Wnt3a produces a larger change in the NLD expressing cells (Fig. 4). Therefore we concluded that the NLD domain must have a strong vesicle associated signal since the changes in aggregation are much larger for this construct.
Fig. 4.
Relative change in number of bright pixels with Wnt3a activation. Changes in the fraction (relative to total) of pixels corresponding to oligomers (green, B>1.5) upon activation of Wnt3a
4. Discussion
This study reports new evidence regarding the biology of sFRP4 which is likely to have relevance to other members of this family and their role in regulation of Wnt signalling. While recent studies have improved our understanding of the intracellular production and trafficking of Wnts, very little is known regarding this aspect of sFRPs. We were able to identify vesicular trafficking of sFRP4 and we observed that the NLD domain has a vesicular association signal. Most notably our results suggest that Wnt3a can modulate intracellular localisation and secretion of sFRP4. Recent evidence has shown that the NLD domain of sFRPs interacts strongly with Wnts (Lopez-Rios et al., 2008) while the CRD most likely acts through interactions with Fzd receptors (Rodriguez et al., 2005). This is consistent with our unpublished observations that the NLD domain exerts the same biological effects as sFRP4 in mesothelioma cells. Interestingly, our results suggest that both the NLD and CRD exist in oligomeric form within the cell. Dimerization of sFRP and Fzd CRDs has been widely reported (Dann et al., 2001; Bafico et al., 1999; Rodriguez et al., 2005), however, insofar as we are aware there is only one report of netrin dimerization (Koch et al., 2000). How this influences the interaction of sFRP4 with other binding partners remains to be investigated, however, it has been suggested that the dimerisation of the CRD domain affects its activity (Xavier et al., 2014).
The observation that Wnt3a may modulate intracellular localisation and secretion of sFRP4 is an unexpected finding of our study which requires further confirmatory investigation. Wnt signalling is tightly regulated through numerous feedback mechanisms (Logan and Nusse, 2004), primarily through gene expression of Wnt pathway components. Negative regulators are often induced by the signals that they control including the secreted regulators Dkk (Gonzalez-Sancho et al., 2005) and sFRP-1 (Gibb et al., 2013). In terms of protein trafficking, we know from recent evidence that Wnt signalling can induce ubiquitination and endocytosis of Fzd receptors as a mechanism of feedback regulation (Hao et al., 2012; Koo et al., 2012). Feedback by regulated secretion of sFRPs may provide additional fine adjustment of Wnt signalling activity at the cell membrane through interactions with ligands and receptors. In summary, our results provide evidence regarding the intracellular localisation, aggregation, trafficking and secretion of sFRP4 not previously reported. Further investigation of these findings are likely to provide insight into the function of sFRPs in Wnt regulation.
Supplementary Material
Acknowledgments
The authors would like to express their gratitude to Prof. Robert Friis, University of Bern, Switzerland for the kind gift of the sFRP4 and domain plasmid constructs used in this study. The authors acknowledge Dr. Paul Rigby, Centre for Microscopy Characterisation & Analysis, The University of Western Australia, Dr. Connie Jackaman and Mr Benjamin Dwyer, CHIRI, Biomedical Sciences, Curtin University for their help with confocal microscopy. V.P. was recipient of a Curtin International Postgraduate Research Scholarship. This work was in part funded by grants from the US National Institute of Health NIH P41-GM103540 and NIH P50-GM076516 [EG]. AD supported by strategic research funds from School of Biomedical Sciences, Commercialisation Advisory Board of Curtin University, Cancer Council, WA and Actinogen Ltd, Perth
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- Bafico A, Gazit A, Pramila T, Finch PW, Yaniv A, Aaronson SA. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J Biol Chem. 1999;274:16180–7. doi: 10.1074/jbc.274.23.16180. [DOI] [PubMed] [Google Scholar]
- Bhat RA, Stauffer B, Komm BS, Bodine PVN. Structure-function analysis of secreted frizzled-related protein-1 for its Wnt antagonist function. J Cell Biochem. 2007;102:1519–28. doi: 10.1002/jcb.21372. [DOI] [PubMed] [Google Scholar]
- Cruciat C-M, Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb Perspect Biol. 2013;5:a015081. doi: 10.1101/cshperspect.a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Dalal RB, Digman MA, Horwitz AF, Vetri V, Gratton E. Determination of particle number and brightness using a laser scanning confocal microscope operating in the analog mode. Microsc Res Tech. 2008;71:69–81. doi: 10.1002/jemt.20526. [DOI] [PubMed] [Google Scholar]
- Fox SA, Richards AK, Kusumah I, Perumal V, Bolitho EM, Mutsaers SE, et al. Expression profile and function of Wnt signaling mechanisms in malignant mesothelioma cells. Biochem Biophys Res Commun. 2013;440:82–7. doi: 10.1016/j.bbrc.2013.09.025. [DOI] [PubMed] [Google Scholar]
- Gibb N, Lavery DL, Hoppler S. sfrp1 promotes cardiomyocyte differentiation in Xenopus via negative-feedback regulation of Wnt signalling. Dev Camb Engl. 2013;140:1537–49. doi: 10.1242/dev.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Sancho JM, Aguilera O, García JM, Pendás-Franco N, Peña C, Cal S, et al. The Wnt antagonist DICKKOPF-1 gene is a downstream target of beta-catenin/TCF and is downregulated in human colon cancer. Oncogene. 2005;24:1098–103. doi: 10.1038/sj.onc.1208303. [DOI] [PubMed] [Google Scholar]
- Hao H-X, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- Koch M, Murrell JR, Hunter DD, Olson PF, Jin W, Keene DR, et al. A novel member of the netrin family, beta-netrin, shares homology with the beta chain of laminin: identification, expression, and functional characterization. J Cell Biol. 2000;151:221–34. doi: 10.1083/jcb.151.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H, Amatya VJ, Takeshima Y, Kushitani K, Hattori N, Kohno N, et al. Aberrant promoter methylation of WIF-1 and SFRP1, 2, 4 genes in mesothelioma. Oncol Rep. 2010;24:423–31. doi: 10.3892/or_00000875. [DOI] [PubMed] [Google Scholar]
- Koo B-K, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–9. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- Kress E, Rezza A, Nadjar J, Samarut J, Plateroti M. The frizzled-related sFRP2 gene is a target of thyroid hormone receptor alpha1 and activates beta-catenin signaling in mouse intestine. J Biol Chem. 2009;284:1234–41. doi: 10.1074/jbc.M806548200. [DOI] [PubMed] [Google Scholar]
- Lee AY, He B, You L, Dadfarmay S, Xu Z, Mazieres J, et al. Expression of the secreted frizzled-related protein gene family is downregulated in human mesothelioma. Oncogene. 2004;23:6672–6. doi: 10.1038/sj.onc.1207881. [DOI] [PubMed] [Google Scholar]
- Lin K, Wang S, Julius MA, Kitajewski J, Moos M, Luyten FP. The cysteine-rich frizzled domain of Frzb-1 is required and sufficient for modulation of Wnt signaling. Proc Natl Acad Sci. 1997;94:11196–200. doi: 10.1073/pnas.94.21.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Longman D, Arfuso F, Viola HM, Hool LC, Dharmarajan AM. The role of the cysteine-rich domain and netrin-like domain of secreted frizzled-related protein 4 in angiogenesis inhibition in vitro. Oncol Res. 2012;20:1–6. doi: 10.3727/096504012x13425470196010. [DOI] [PubMed] [Google Scholar]
- Lopez-Rios J, Esteve P, Ruiz JM, Bovolenta P. The Netrin-related domain of Sfrp1 interacts with Wnt ligands and antagonizes their activity in the anterior neural plate. Neural Develop. 2008;3:19. doi: 10.1186/1749-8104-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning LS, Whitaker D, Murch AR, Garlepp MJ, Davis MR, Musk AW, et al. Establishment and characterization of five human malignant mesothelioma cell lines derived from pleural effusions. Int J Cancer J Int Cancer. 1991;47:285–90. doi: 10.1002/ijc.2910470219. [DOI] [PubMed] [Google Scholar]
- Von Marschall Z, Fisher LW. Secreted Frizzled-related protein-2 (sFRP2) augments canonical Wnt3a-induced signaling. Biochem Biophys Res Commun. 2010;400:299–304. doi: 10.1016/j.bbrc.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Manso G, Calzada MJ, Chuman Y, Sipes JM, Xavier CP, Wolf V, et al. sFRP-1 binds via its netrin-related motif to the N-module of thrombospondin-1 and blocks thrombospondin-1 stimulation of MDA-MB-231 breast carcinoma cell adhesion and migration. Arch Biochem Biophys. 2011;509:147–56. doi: 10.1016/j.abb.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mii Y, Taira M. Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Dev Camb Engl. 2009;136:4083–8. doi: 10.1242/dev.032524. [DOI] [PubMed] [Google Scholar]
- Ossato G, Digman MA, Aiken C, Lukacsovich T, Marsh JL, Gratton E. A two-step path to inclusion formation of huntingtin peptides revealed by number and brightness analysis. Biophys J. 2010;98:3078–85. doi: 10.1016/j.bpj.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotegher N, Gratton E, Bubacco L. Number and Brightness analysis of alpha-synuclein oligomerization and the associated mitochondrial morphology alterations in live cells. Biochim Biophys Acta. 2014;1840:2014–24. doi: 10.1016/j.bbagen.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Esteve P, Weinl C, Ruiz JM, Fermin Y, Trousse F, et al. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat Neurosci. 2005;8:1301–9. doi: 10.1038/nn1547. [DOI] [PubMed] [Google Scholar]
- Schulte G International Union of Basic and Clinical Pharmacology. LXXX. The class Frizzled receptors. Pharmacol Rev. 2010;62:632–67. doi: 10.1124/pr.110.002931. [DOI] [PubMed] [Google Scholar]
- Uren A, Reichsman F, Anest V, Taylor WG, Muraiso K, Bottaro DP, et al. Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J Biol Chem. 2000;275:4374–82. doi: 10.1074/jbc.275.6.4374. [DOI] [PubMed] [Google Scholar]
- Vetri V, Ossato G, Militello V, Digman MA, Leone M, Gratton E. Fluctuation methods to study protein aggregation in live cells: concanavalin A oligomers formation. Biophys J. 2011;100:774–83. doi: 10.1016/j.bpj.2010.11.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sinha T, Wynshaw-Boris A. Wnt Signaling in Mammalian Development: Lessons from Mouse Genetics. Cold Spring Harb Perspect Biol. 2012;4:a007963. doi: 10.1101/cshperspect.a007963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier CP, Melikova M, Chuman Y, Üren A, Baljinnyam B, Rubin JS. Secreted Frizzled-related protein potentiation versus inhibition of Wnt3a/β-catenin signaling. Cell Signal. 2014;26:94–101. doi: 10.1016/j.cellsig.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.