Abstract
CUG triplet repeat-binding protein 2 (CUGBP2) is a RNA-binding protein that regulates mRNA translation and modulates apoptosis. Here, we report the identification of two splice variants (termed variants 2 and 3) in cultured human intestinal epithelial cells and in mouse gastrointestinal tract. The variants are generated from alternative upstream promoters resulting in the inclusion of additional NH2-terminal residues. Although variant 2 is the predominant isoform in normal intestine, its expression is reduced, whereas variant 1 is overexpressed following γ-irradiation. All three variants bind cyclooxygenase-2 (COX-2) mRNA. However, only variant 1 inhibits the translation of the endogenous COX-2 mRNA and a chimeric luciferase mRNA containing the COX-2 3′ untranslated region. Furthermore, whereas variant 1 is predominantly nuclear, variants 2 and 3 are predominantly cytoplasmic. These data imply that the additional amino acids affect CUGBP2 function. Previous studies have demonstrated that variant 1 induces intestinal epithelial cells to undergo apoptosis. However, in contrast to variant 1, the two novel variants do not affect proliferation or apoptosis of HCT116 cells. In addition, only variant 1 induced G2/M cell cycle arrest, which was overcome by prostaglandin E2. Moreover, variant 1 increased cellular levels of phosphorylated p53 and Bax and decreased Bcl2. Caspase−3 and −9 were also activated, suggesting the initiation of the intrinsic apoptotic pathway. Furthermore, increased phosphorylation of checkpoint kinase (Chk)1 and Chk2 kinases and increased nuclear localization of Cdc2 and cyclin B1 suggested that cells were in mitotic transition. Taken together, these data demonstrate that cells expressing CUGBP2 variant 1 undergo apoptosis during mitosis, suggesting mitotic catastrophe.
Keywords: cell cycle, apoptosis, G2/M arrest, checkpoint kinases
cugbp2 (CUG triplet repeat-binding protein 2, also known as ETR3, NAPOR2, and BRUNOL2) is an ubiquitously expressed RNA-binding protein that was originally identified in skeletal muscle through its interaction with CUG triplet repeats (11, 28, 30). CUGBP2 is a member of a family of proteins, which have been collectively termed CELF (CUGBP1, CUGBP2, and CELF3 through CELF6) (23). All the members of this family contain two RNA recognition motifs (RRMs) near the NH2 terminus and a third RRM near the COOH terminus (11, 23, 30). However, an important difference between the members of this family is the presence of a divergent domain of 160 to 230 amino acids between the second and third RRMs.
The CELF family members have been demonstrated to regulate multiple RNA pathways including splicing, editing, and translation. In muscle cells, CUGBP2 is involved in alternative splicing of muscle-specific genes including exon 5 of cardiac troponin T, exon 11 of insulin receptor, intron 2 of chloride channel 1, exons 5 and 21 of 1N-methyl-D-aspartate receptor-1, and the muscle-specific exon of α-actinin (3, 16, 19, 23, 27). A second function for CUGBP2 relates to its ability to bind to AU-rich sequences in 3’untranslated region (3’UTR) of the target mRNAs (34, 35). Upon binding to AU-rich sequences in cyclooxygenase-2 (COX-2) 3’UTR, CUGBP2 induces the stability of COX-2 mRNA. However, CUGBP2 binding also results in the inhibition of COX-2 mRNA translation. In addition, CUGBP2 can interact with HuR, a key inducer of RNA stability and translation, and competitively inhibit HuR function (46). More recently, plate-let-derived growth factor was shown to enhance CUGBP2 binding to COX-2 mRNA through increased phosphorylation of a tyrosine residue at position 39 in the protein (53). These data suggest that posttranscriptional control mechanisms are in place to modulate the CUGBP2 function as a regulator of stability and translation of AU-rich transcripts.
CUGBP2 expression is relatively lower in the intestinal epithelial cells compared with the skeletal or cardiac muscle (30, 36). Furthermore, overexpression of CUGBP2 results in HT-29 colon cancer cells undergoing apoptosis-mediated cell death (34). The mechanism for this function was correlated to the loss of COX-2 protein expression, thereby decreasing prostaglandin E2 (PGE2) production. CUGBP2 was also observed to regulate COX-2 mRNA translation in response to γ-irradiation (36). COX-2 mRNA has been shown to be significantly upregulated in the intestinal epithelial cells of mice within minutes of exposure to whole-body γ-irradiation (21, 36, 48). However, protein levels do not increase, due to the concomitant increase in CUGBP2 protein levels (36). The consequence of this is that there is a significant increase in radiation-mediated damage to the intestine. In contrast, administration of lipopolysaccharide to the animal radioprotects the intestinal epithelial cells, in part due to suppression of CUGBP2 expression and induction of COX-2-mediated PGE2 production (4, 37, 41, 42). Taken together, these data demonstrate that CUGBP2 is a potent inducer of cell death although the mechanism involving translational inhibition of COX-2 mRNA has not been described.
One intriguing point relates to the role of CUGBP2 in the normal intestine since it does not seem to cause apoptosis of the intestinal epithelial cells. This has led us to determine whether there is a difference in the CUGBP2 that is expressed under normal conditions and following radiation. In this regard, we now report that CUGBP2 is expressed from three distinct promoters and that the protein expressed from the proximal promoter is the prototype CUGBP2 protein that is 490 amino acids long. We have identified that there are two additional variants (termed variants 2 and 3) that are expressed in cultured intestinal epithelial cells, which arise as a result of transcription driven by two independent upstream promoters. The consequence of this is that the translation occurs from upstream sites, resulting in the presence of additional amino acids in the amino-terminus of the protein. These three variants were also expressed throughout the gastrointestinal tract in the mouse. Furthermore, under normal conditions, variant 2 is the predominant isoform. However, following exposure to 12 Gy γ-irradiation, there is a transcriptional switch, and variant 1 is the predominant isoform. The two novel variants neither affect COX-2 mRNA translation nor cell viability. We have also determined that CUGBP2 variant 1 (the prototype protein) causes cells to undergo mitotic catastrophe. These data suggest that, although variants 2 and 3 might have a physiological role in the normal intestine related to its splicing regulatory function, variant 1 regulates cell viability in response to injury.
MATERIALS AND METHODS
Animals
Wild-type C57BL/6 mice were obtained from Jackson Laboratories (Bar Harbor, ME). The mice were maintained on a 12 h:12 h light-dark schedule and fed standard laboratory chow. Mice were irradiated at 8 wk in a Gammacel 40-cesium irradiator at 0.96 cGy/min and allowed to recover. The proximal and distal jejunum were isolated and snap frozen in liquid nitrogen for RNA isolation. The animal protocol used in these studies was approved by the University of Oklahoma Institutional Animal Care and Use Committee.
Cells
HCT116, SW480, and HT-29 colon adenocarcinoma cells were obtained from American Type Culture Center and cultured in DMEM containing 10% heat-inactivated fetal bovine serum (Sigma Chemical, St. Louis, MO) and standard antibiotics in a carbon dioxide incubator with 5% CO2 at 37°C.
Plasmids
The full-length coding region of human CUGBP2 variants were amplified by RT-PCR and cloned in pGEM-T EZ (Promega, Madison, WI). After sequencing, the product was subcloned into pCMV-Tag2B (a cytomegalovirus immediate early promoter-driven expression vector) (Stratagene, La Jolla, CA) at the HindIII and XhoI restriction sites and expressed as amino-terminal Flag epitope-tagged proteins. Primers used for the cloning are: variant 1 5′-AAGCTTATGAACGGAGCTTTGGAT-3′ and 5′- CTCGAGT-CAGTAAGGTTTGCTGTCGTT-3′, variant 2 5′- AAGCTTATGAT-GGTCGAGGGCCGCCT-3 ′ and 5′-CTCGAGTCAGTAAGGTTTG-CTGTCGT-3′, and variant 3 5′-AAGCTTATGCGCTGTCCCAAATC-CGCT-3′ and 5′-CTCGAGTCAGTAAGGTTTGCTGTCGTT-3′. The luciferase expression vector containing the full-length murine COX-2 3 ′ UTR has been previously published (12).
Cell proliferation
HCT116 cells were seeded on 96-well plates at a density of 1 × 103 cells/well and allowed to adhere. The cells were also simultaneously transfected with plasmid pCMV-Tag2B–CUGBP2 variant 1, 2, or 3, or just the vector (as controls). Following 48-h incubation, proliferation was determined by hexosaminidase enzyme assay as previously described (26). Results were further confirmed by manual cell counts.
Apoptosis
HCT116 cells were grown in 96-well black plates. As mentioned above, the cells were also simultaneously transfected with plasmid pCMV-Tag2B–CUGBP2 variant 1, 2, or 3, or just the vector (as controls). After 48 h, caspase-3 and caspase-7 activity was measured with the Apo-one Homogeneous caspase-3 and caspase-7 Assay per the manufacturer’s instructions (Promega).
Cell cycle analyses
HCT116 cells transiently expressing Flag-tagged CUGBP2 were harvested by trypsinization and suspended in PBS. The single-cell suspensions were fixed with 70% ethanol for 2 h and subsequently permeabilized with PBS containing 1 mg/ml propidium iodide (Sigma-Aldrich), 0.1% vol/vol Triton X-100 (Sigma-Aldrich) and 2 mg DNase-free RNase (Sigma-Aldrich) at room temperature. Flow cytometry was done with a FACSCalibur analyzer 3-color (Becton Dickinson, Mountain View, CA), capturing 50,000 events for each sample; results were analyzed with ModFit LT software (Verity Software House, Topsham, ME).
Colony formation assay
Six-well dishes were seeded with 1,000 viable cells and simultaneously transfected with plasmid expressing the three CUGBP2 variants in complete medium. After 48 h, the cells were washed in PBS and incubated for an additional 15 days in complete medium. Each treatment was done in triplicate. The colonies obtained were washed with PBS and fixed in 10% formalin for 10 min at room temperature and were then washed with PBS followed by staining with hematoxylin. The colonies were counted and compared with untreated cells.
RT-PCR analyses
Total RNA was isolated with Trizol reagent (Invitrogen, Carlsbad, CA). Complementary DNAs were prepared using random hexamer oligonucleotides and used for semiquantitative polymerase chain reaction analysis using Jumpstart Taq DNA polymerase (Sigma) and α32P-dCTP (Perkin Elmer, Boston, MA). The products were resolved in a 5% native PAGE, 0.5X Tris-borate EDTA and autoradiographed. Radioactive products were also quantified with a Storm Phosphorimager apparatus (Bio-Rad Laboratories, Hercules, CA). Primers used in this study were as follows: β-actin, 5′-GAGT-GCTGTCTCCATGTTTGATG-3′ and 5′-CTCTAAGTTGCCAGC-CCTCCT-3′; COX-2, 5′-GAATCATTCACCAGGCAAA-TTG-3′ and 5′-TCTGTACTGCGGGTGGAACA-3′, mouse variant 1 5′- AGAATGGTCCCAGCATGCTGG-3′ and 5′-CTTGTTGGCT-GTGCCGTTAC-3′, human variant 1 5′-AGCTCTGCCTTTCTTTC-CCAG-3′ and the same reverse primer from mouse variant 1, mouse and human variant 2 5′-ATGATGGTCGAGGGCCGCCT-3′ and 5′-CGGAGGATCCGGCATTCTTC-3′, mouse and human variant 3 5′-ATGCGCTGTCCCAAATCCGCT-3′ and 5′-CGGAGGATCCG-GCATTCTTC-3′.
Immunoprecipitation coupled RT-PCR analyses
Cells were subjected to treatment with 1% formaldehyde for 10 min, and cell lysates were prepared by sonication. CUGBP2 variants were immunoprecipitated using an anti-Flag antibody. Total RNA was isolated from the precipitate and supernatant and subjected to RT-PCR analyses for COX-2 and β-actin as mentioned above.
Western blot analyses
HCT116 cells transfected with the indicated CUGBP2 isoform were allowed to grow for 48 h. Cell lysates were prepared and subjected to polyacrylamide gel electrophoresis and blotted onto Immobilon polyvinylidene difluoride membranes (Milli-pore, Bedford, MA). Antibodies were purchased from either Santa Cruz Biotechnology (Santa Cruz, CA) or Cell Signaling (Bedford, MA). Specific proteins were detected by an enhanced chemiluminescence system (Amersham Pharmacia Biotech, Piscataway, NJ). Spot densitometry on an Alpha Innotech Flurochem 8000 (San Leandro, CA) was used to obtain a density value for each spot.
Luciferase assay
Cells were transfected with plasmids pGL3-control plasmid or pGL3-Luc-COX plasmid encoding the murine COX-2 3′UTR using FuGENE 6 (Roche, Indianapolis, IN). Cells were cotransfected with plasmid pCMV-Tag2B encoding NH2-terminal Flag-tag CUGBP2 variants, and cells were allowed to grow for 24 h. As a transfection control, test plasmids were cotransfected with the plasmid pRL-TK (Clontech). Renilla luciferase was expressed under the control of the RSV thymidine kinase promoter. Lysates were prepared, and luciferase levels were determined with the use of the Dual-Luciferase Reporter Assay System (Promega).
Immunocytochemistry and immunofluorescence
Cells were transfected with plasmids encoding NH2-terminal Flag-tagged CUGBP2 variants and allowed to grow for 48 h. The cells were fixed with 10% buffer for 10 min at room temperature and subsequently washed three times with PBS. The cells were then permeabilized with PBS containing 0.5% Triton X-100 for 10 min at room temperature. For immunocytochemistry analyses of cyclin B1 and Cdc2, the cells were incubated with rabbit anticyclin B1 (Santa Cruz) and rabbit anti-Cdc2 antibodies (AbCam, Cambridge, MA), followed by biotinylated anti-rabbit IgG. The slides were further processed using Vectastain ABC Kit (Vector Laboratories, Burlingame, CA) followed by diamino-benzidine staining. For immunofluorescence, the cells were incubated with rabbit anti-Flag antibody (Affinity Bioreagents) followed by FITC-conjugated anti-rabbit IgG. The slides were mounted and examined with the use of Zeiss Axioskop 2 MOT plus microscope (Carl Zeiss, Thornwood, NY).
Statistical analysis
The values are expressed as the means ± SE. Data were analyzed using an unpaired Student’s t-test. We considered a P value of less than 0.05 to be statistically significant.
RESULTS
Differential promoter usage governs CUGBP2 expression in the mouse gastrointestinal tract
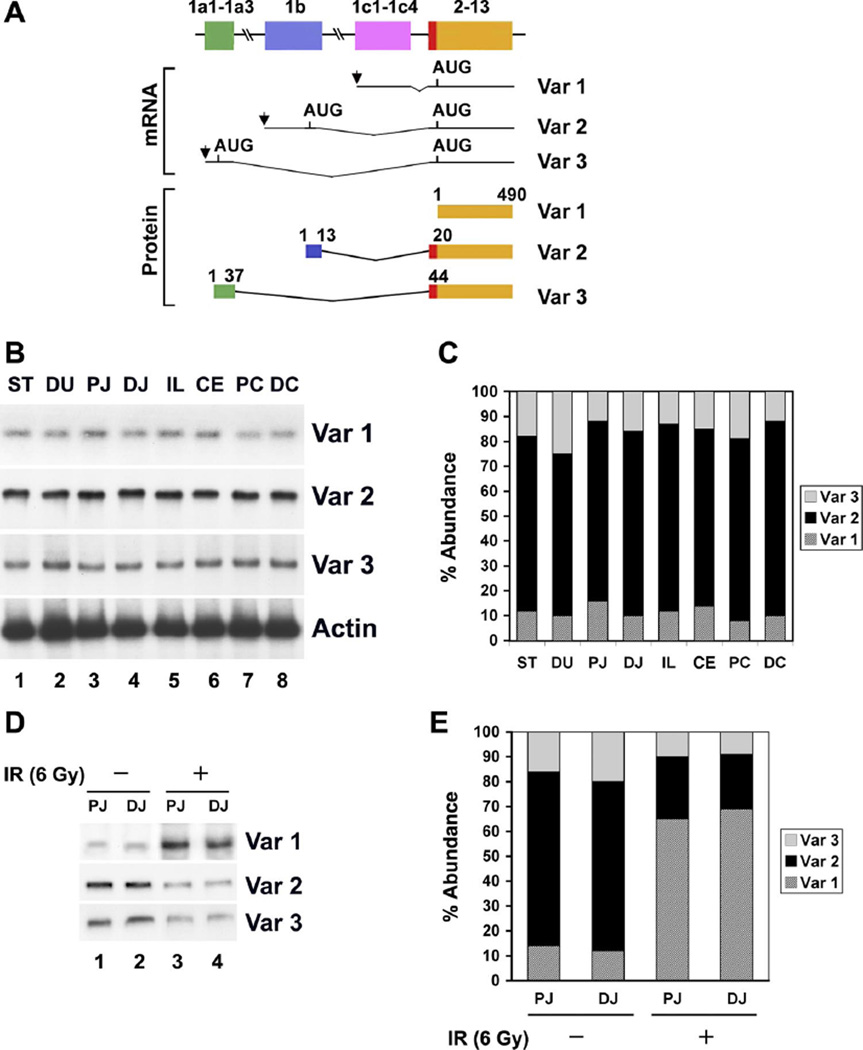
Mouse CUGBP2 gene spans 350 kb and consists of 20 exons in total. The expression is controlled by three distinct promoters located upstream of exons 1a, 1b, and 1c (Fig. 1A). All three transcripts splice into exon 2. Variant 1 is translated from the first AUG located in exon 2, whereas variants 2 and 3 are translated from an upstream AUG that is in frame with the AUG in exon 2. As a result, variants 2 and 3 have additional 20 and 44 amino acids added to the NH2 terminus (Fig. 1A). We first determined the expression of the three variants in the different regions of the gastrointestinal tract of mice. Semiquantitative radioactive PCR demonstrated that all three variants are expressed throughout the GI tract from the stomach to the distal colon (Fig. 1B). Quantitation of the expression demonstrated that variant 2 is expressed at the highest levels in all areas (Fig. 1C). More importantly, variant 1 was expressed at very low levels, with highest expression seen in the duodenum.
Fig. 1.
Differential expression of CUG triplet repeat-binding protein 2 (CUGBP2) variants in mouse gastrointestinal tissues. A: schematic representation of the structure of the three CUGBP2 variants (var). Alternative promoter usage of the CUGBP2 gene generated 3 transcript variants, resulting in the presence of distinct exon 1 (1a–1c). Exons are depicted by boxes and introns by horizontal lines. The arrows indicate transcription start sites, and the location of AUGs encoding CUGBP2 variants are shown. For the sake of simplicity, the four exons that are included within the coding region (exons 2–13) are grouped together. Similarly, the 5′ exons that make up variant exons 1a and 1c are shown as one exon. The CUGBP2 variant 1 is 490 amino acids long starting from the AUG in exon 2. However, variants 2 and 3 have extra NH2-terminal 19 and 43 amino acids due to the presence of an additional AUG start codon in exon 1b and 1a, respectively. B: expression of the 3 variants in mouse gastrointestinal tissues. Representative autoradiograph of a semiquantitative RT-PCR was performed in the presence of 32P-CTP using variant-specific primers. All 3 isoforms are expressed. ST, stomach; DU, duodenum; PJ, proximal jejunum; DJ, distal jejunum; IL, ileum; CE, cecum; PC, proximal colon; DC, distal colon. C: abundance of the 3 variants. Data demonstrates that var 2 transcript is the predominant isoform in normal tissues, whereas var 1 is expressed at the lowest level. Data from 5 mice. D: differential variant expression in the small intestine following γ-irradiation. Total RNA from the proximal (PJ) and distal (DJ) jejunum from mice exposed to 6 Gy γ-irradiation were subjected to semiquantitative RT-PCR. Whereas var 1 expression is significantly upregulated, both var 2 and var 3 transcripts are downregulated compared with the same tissues from unirradiated animals. E: abundance of variants following γ-irradiation. Data demonstrates the significant down-regulation of var 2 and 3 and upregulation of var 1 transcript. Data from 3 mice.
Switch in CUGBP2 variant expression following radiation injury
In previous studies (34), we have demonstrated that CUGBP2 expression is rapidly induced in the mouse small intestine following exposure to γ-irradiation. Furthermore, administration of bacterial lipopolysaccharide before γ-irradiation protected the intestinal cells, in part, through inhibition of CUGBP2 expression in a prostaglandin-dependent mechanism (34). Therefore, we determined the expression of the three CUGBP2 variants following whole-body administration of 6 Gy γ-irradiation to the mice. This dose was chosen because previous studies (14, 33) have demonstrated that there is a marked increase in the number of epithelial cells in the intestinal crypts undergoing apoptosis at 24 h. There was a significant upregulation in variant 1 expression in proximal and distal jejunum 24 h after 6 Gy γ-irradiation (Fig. 1, D and E). In contrast, there was a reduction in the expression of both variants 2 and 3 (Fig. 1, D and E). These data suggest that there is a switch in the promoter usage following radiation, resulting in the expression of variant 1. Given that CUGBP2 overexpression after γ-irradiation results in inhibition of COX-2 mRNA, these data imply that CUGBP2 variant 1 might be the dominant inhibitor of COX-2 expression.
Differential CUGBP2 variant expression also occurs in human colon cancer cells
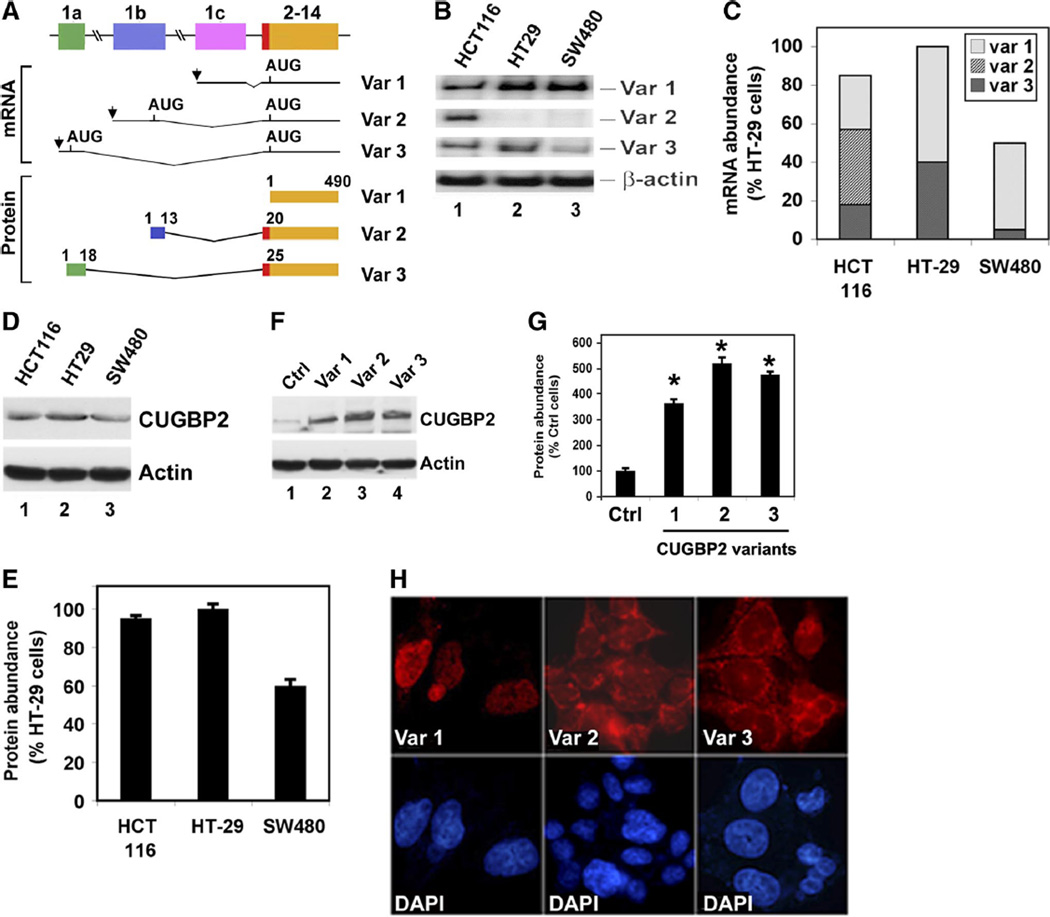
Three species of CUGBP2 are also present in human tissues, again resulting from differential promoter usage (Fig. 2A). Transcripts arising from exon 1c result in translation start from the AUG codon located in exon 2 leading to the generation of variant 1 CUGBP2 protein. In contrast, transcription start from the upstream promoters results in the inclusion of distinct exon 1 sequences, each of which also encodes an in-frame AUG codon. Translation from these AUGs results in the inclusion of additional 19 and 24 amino acids in variants 2 and 3, respectively. The distribution pattern of the three CUGBP2 variants was determined by semiquantitative RT-PCR analysis. In colon cancer cells, there were differences in the expression pattern of the three variants. Whereas HCT116 cells demonstrated the presence of all three transcripts, HT-29 and SW480 cells lacked variant 2 (Fig. 2, B and C). Furthermore, HT-29 had higher levels of variant 3 compared with SW480 cells. To confirm that the protein is expressed in the three colon cancer cell lines, Western blot analysis was performed. CUGBP2 expression was observed in all three cell lines (Fig. 2, D and E). We next compared the localization of the two novel protein variants to the prototype CUGBP2 variant 1 protein. For this, the cDNAs of the three variants were inserted into the mammalian expression vector pCMV-Tag2B and transfected into HCT116 cells. Western blot analysis confirmed the increased expression of the proteins in the transfected cells (Fig. 2, F and G). To determine whether the presence of the additional amino acids affects cellular localization of the variants, transfected cells were immunolabeled with anti-Flag antibody. Variant 1 protein was strongly enriched in the nucleus, which was somewhat diffuse but not present in the nucleolus (Fig. 2H). This is similar to the pattern previously demonstrated for the endogenous protein (34, 46). In contrast, both variants 2 and 3 were predominantly localized to the cytoplasm that was somewhat diffuse (Fig. 2H). However, there was an enrichment of variant 2 in distinct regions of the cytoplasm. In addition, variant 3 was predominantly localized to the perinuclear region of the cells.
Fig. 2.
Differential expression of CUGBP2 mRNA in human colorectal cancer cells. A: schematic representation of the origin and structure of the 3 CUGBP2 variants in 3 human cell lines. Like the mouse CUGBP2, alternative promoter usage generates 3 transcript variants in the human cells, resulting in the presence of distinct exon 1 (1a–1c). Human CUGBP2 variant 1 is also 490 amino acids long starting from the AUG in exon 2. Variant 2 is similar to that seen in mouse, with addition of NH2-terminal 19 amino acids. However, variant 3 in the human is not as long as that observed in the mouse, with the addition of only 24 NH2-terminal amino acids. B: RT-PCR analyses from 3 human colon cancer cell lines, HCT116, HT-29, and SW480, show that variant 1 is the predominant isoform in all the cell lines. Moreover, HCT116 has higher levels of expression of all 3 variants. β-actin was used as internal control for loading. C: distribution and abundance of CUGBP2 variant mRNAs in 3 colon cancer cell lines. The percentage was calculated considering CUGBP2 mRNA expression in HT29 as 100%. Data from 3 independent RT-PCR experiments. D: Western blot analysis for CUGBP2 expression in colon cancer cell lines. HT29 has the highest level of protein expression compared with HCT116 and SW480 cells. E: abundance of the CUGBP2 protein. Again, percentage was calculated using expression in HT-29 cells as 100%. Data from 3 independent experiments. F: Western blot analysis for CUGBP2 expression after transient transfection. Plasmids encoding Flag-tagged CUGBP2 variants were transiently transfected in HCT116. Extracts were tested for expression of the transfected proteins. Data shows the higher levels of transfected CUGBP2 variant proteins in the cells. G: abundance of CUGBP2 protein after transient expression of individual isoforms. Variant 1 is least expressed of the 3 variants. Data from 3 independent experiments. *P < 0.05 H: immunocytochemistry analysis for localization of the CUGBP2 variants following transient transfection. Whereas variant 1 is nuclear, variants 2 and 3 are predominantly cytoplasmic. 4′-6-Diamidino-2-phenylindole (DAPI) is used to stain the nucleus.
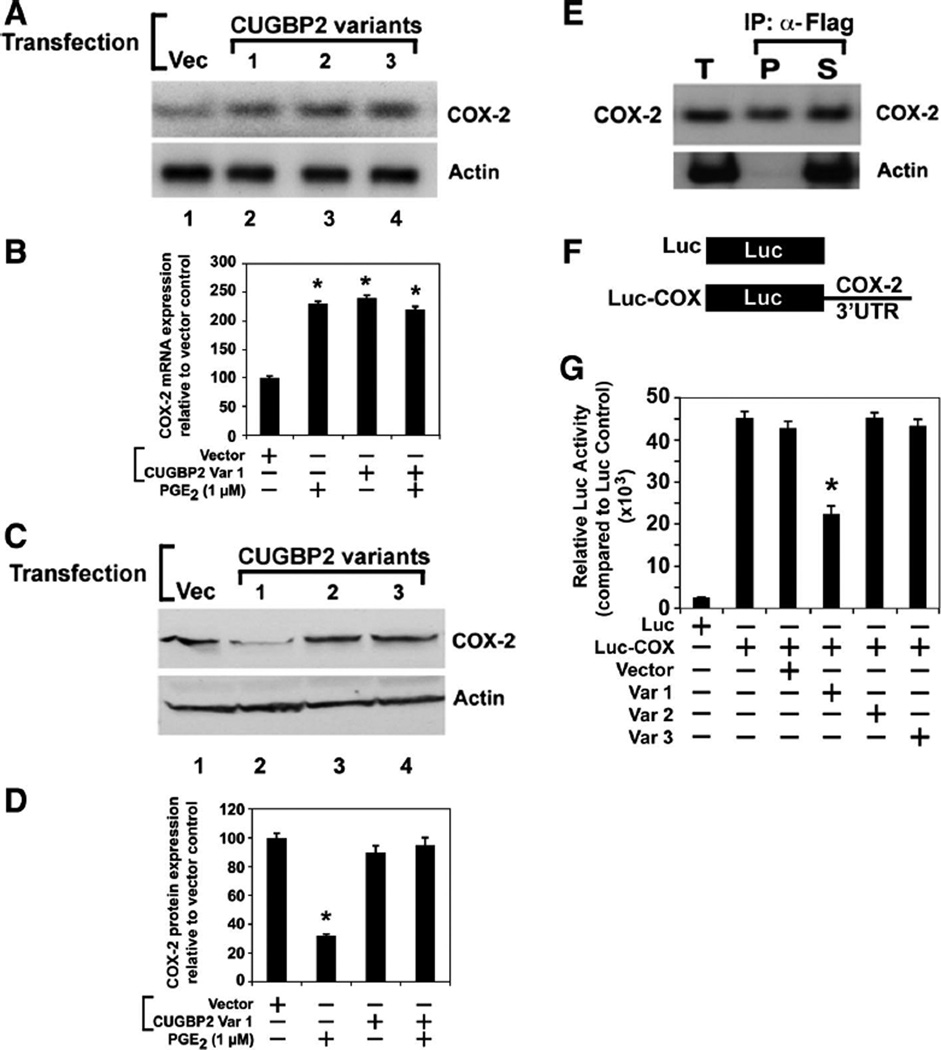
Only variant 1 inhibits COX-2 mRNA translation
Previous studies (34, 46) have demonstrated that CUGBP2 variant 1 binds to AU-rich sequences in the 3’UTR of COX-2 mRNA and stabilizes the mRNA but inhibits its translation. To determine whether the additional amino acids in the NH2 terminus of variants 2 and 3 affect the translation activity, we next transfected the plasmids into HCT116 cells and tested for COX-2 levels. Semiquantitative RT-PCR assays demonstrated that COX-2 mRNA levels were significantly upregulated following expression of all three CUGBP2 variants (Fig. 3, A and B). Furthermore, real-time RT-PCR demonstrated that there was a three- to fourfold increase in COX-2 mRNA levels due to the expression of CUGBP2 variants (data not shown). In previous studies, we have demonstrated that CUGBP2 variant 1 increased COX-2 mRNA stability but inhibits COX-2 mRNA translation (34, 46). To determine whether variants 2 and 3 also regulate COX-2 mRNA stability, cell lysates were subjected to Western blot analysis for COX-2 protein. CUGBP2 variant 1 significantly inhibited COX-2 protein levels (Fig. 3, C and D). In contrast, neither variant 2 nor variant 3 inhibited COX-2 protein levels. These data suggest that the additional amino acids present in the NH2 terminus of CUGBP2 variants 2 and 3 affect the ability of the protein to inhibit COX-2 mRNA translation. To confirm that this is not due to a loss in RNA-binding activity, a coupled immunoprecipitation-RT-PCR analysis was performed with cells in which CUGBP2 variant 2 was transfected. Extracts were immunoprecipitated for the transfected variant 2 with the use of anti-Flag antibody and subjected to radioactive RT-PCR. COX-2 mRNA was identified in both the precipitate and the supernatant, suggesting that the transiently expressed protein binds to the COX-2 mRNA (Fig. 3 E). Similar results were obtained with variants 1 and 3 (data not shown). In previous studies (34, 46), we have demonstrated that CUGBP2 mediated increased COX-2 mRNA stability and inhibition of translation occurs through the 3′UTR. To confirm the role of COX-2 3′UTR in CUGBP2 function, cells were transfected with a luciferase construct encoding the COX-2 3′UTR along with the CUGBP2 variants (Fig. 3F). Luciferase assays demonstrated that only variant 1 but not variants 2 and 3 inhibited luciferase mRNA translation (Fig. 3G). These data confirm the Western blot results that only CUGBP2 variant 1 inhibits COX-2 mRNA translation.
Fig. 3.
Only CUGBP2 variant 1 inhibits cyclooxygenase-2 (COX-2) mRNA translation. A: HCT116 cells were transiently transfected with plasmid-encoding Flag-tagged CUGBP2 variants, whereas the control cells were transfected with the empty vector (Vec). RT-PCR analyses showed that all the 3 variants increase COX-2 mRNA expression. B: abundance of COX-2 mRNA following transfection of the CUGBP2 variants. COX-2 mRNA levels increased following expression of all 3 variants. *P < 0.05 from 3 independent experiments. PGE2, prostaglandin E2. C: lysates from the CUGBP2 variant transfections were subjected to Western blot analysis for COX-2 protein. There was a marked decrease in COX-2 protein levels in the Flag-tagged CUGBP2 variant 1-transfected cells. D: abundance of COX-2 protein following expression of the CUGBP2 variants. COX-2 protein levels are significantly reduced following expression of variant 1. *P < 0.05 from 3 independent experiments. E: whole cell extracts (T) from Flag-tagged CUGBP2 variant 2-expressing cells were crosslinked and subjected to immunoprecipitation with anti-Flag antibody. RNA present in the immunoprecipitate (P) and supernatant (S) were isolated after reverse crosslink and subjected to RT-PCR for COX-2 mRNA. Data demonstrates the presence of COX-2 mRNA in the pellet. Actin was used as control to demonstrate specificity of CUGBP2 binding. F: schematic representation of control luciferase mRNA (Luc) and luciferase mRNA containing the full length mouse COX-2 3′untranslated region (3’UTR) (Luc-COX) that is encoded in the plasmid under the control of the SV40 promoter. G: relative expression of the firefly luciferase reporter activity in HCT116 cells following expression of the 3 CUGBP2 variants. Extracts were subjected to luciferase activity measurements. Data shows that over-expression of variant 1 resulted in significant reduction of luciferase expression. Data from 3 independent experiments. *P < 0.05.
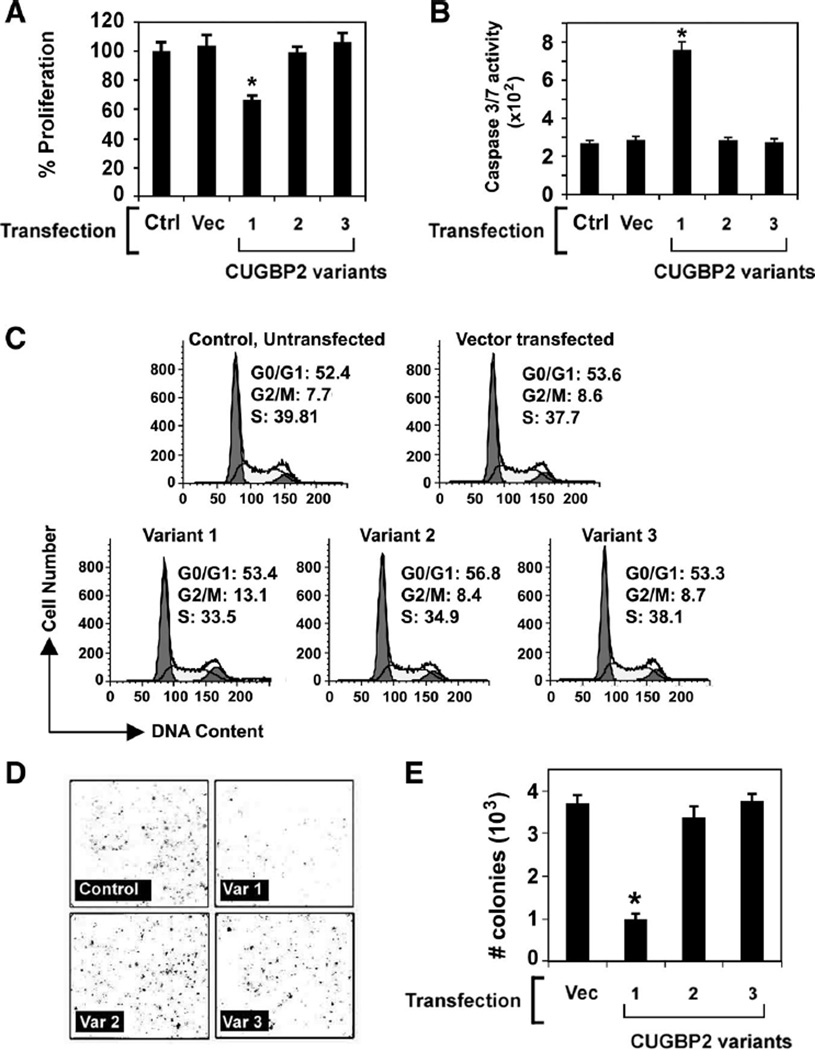
CUGBP2 variant 1 induces cells to undergo G2/M arrest
Stable cell lines of CUGBP2 variant 1 have been problematic (34). To determine the cell fate after expression of the CUGBP2 variants, HCT116 cells were transfected with the pCMV-Tag2B plasmids expressing Flag-tagged CUGBP2 variants, and growth of the cells was monitored. Cell proliferation was not affected by variants 2 and 3, whereas variant 1 significantly retarded the growth of the cells in culture (Fig. 4A). Furthermore, analysis for cell death using the caspase-3/7 activity assay demonstrated that only the cells expressing CUGBP2 variant 1, but not with variant 2 or variant 3, undergo apoptosis (Fig. 4B). To confirm these results, cell cycle analysis was performed following propidium iodide staining. There were increased numbers of cells in G2/M phase of the cell cycle in the presence of variant 1, but not in the presence of variants 2 and 3, suggesting that the decreased proliferation of the cells in the presence of variant 1 is due to an arrest in the G2/M phase of the cell cycle (Fig. 4C). The effect of CUGBP2 variant 1 on G2/M phase arrest in the cells was largely at the expense of the S phase, with a nonsignificant change in G0/G1 phase when compared with untransfected or vector-transfected cells. Given the effect on proliferation and cell cycle, we next investigated whether CUGBP2 variants affect colony formation. Whereas expression of variant 1 significantly decreased the number of HCT116 colonies formed compared with the control group, there was no effect observed when the cells expressed variants 2 or 3 (Fig. 4, D and E). These data suggest that variant 1, but not variants 2 and 3, inhibits cell proliferation and induces cell death at the G2/M stage of cell cycle.
Fig. 4.
CUGBP2 variant 1 inhibits the growth of colon cancer cells. A: CUGBP2 variant 1 inhibited the proliferation of HCT116 cells. The cells were transiently transfected with plasmid-expressing Flag-tagged CUGBP2 variants for 48 h and subsequently analyzed for proliferation based on hexosaminidase enzyme activity. Data shows only variant 1 inhibited the proliferation of HCT116 cells. Data from 3 independent experiments. *P< 0.05. B: CUGBP2 variant 1 increases apoptosis of HCT116 cells. Cells were analyzed for caspase-3/7 activity 48 h following transient expression of Flag-tagged CUGBP2 variants. CUGBP2 variant 1 overexpression resulted in increased caspase activity. *P < 0.01. C: CUGBP2 variant 1 overexpression induces G2/M arrest. Cell-cycle profiles were analyzed for HCT116 cells transiently transfected with plasmid-expressing Flag-tagged CUGBP2 variants for 48 h by fluorescence-activated cell sorting using propidium iodide staining for DNA content. The percentage of cells in the G2/M phase following variant 1 overexpression was increased compared with either the control or vector-transfected cells. Data from 3 independent experiments. D: CUGBP2 variant 1 induces cell death. HCT116 cells transiently transfected with plasmid-expressing Flag-tagged CUGBP2 variants were allowed to grow for 15 days. Number of colonies was identified by staining with hematoxylin. The data demonstrates that variant 1 expression induces cell death. E: number of colonies was counted and the quantitative estimation on number of colonies was presented. The graph shows variant 1 overexpression resulted in significantly lower number of colonies, whereas variants 2 and 3 had near normal values. Data from 3 independent experiments. *P < 0.01.
Cell death mediated by CUGBP2 variant 1 is due to mitotic catastrophe that occurs in part through loss of PGE2
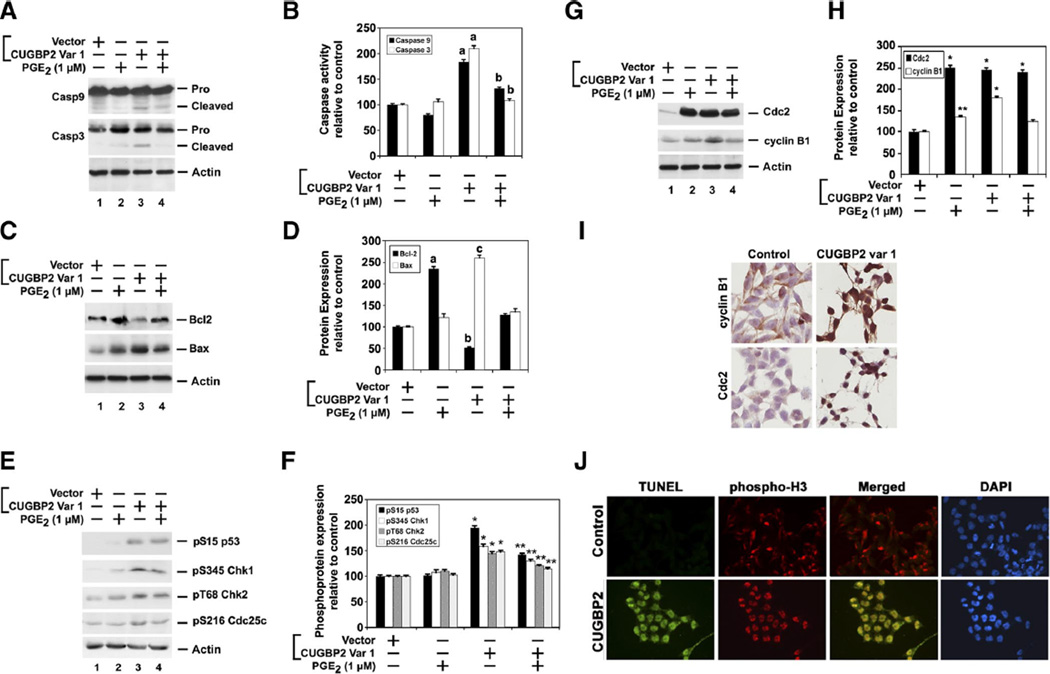
The results from above demonstrate that cells expressing CUGBP2 variant 1 undergo G2/M arrest and have decreased colonies, suggesting that cell death occurs during the G2/M stage of cell cycle. In addition, there was an increase in effector caspase activity. To determine whether expression of CUGBP2 variant 1 resulted in increased intrinsic caspase pathway, Western blot analyses were performed. Confirmation of caspase-3 activation was first obtained. Activated caspase-3 was observed in cells transfected with CUGBP2 variant 1 (Fig. 5, A and B). Furthermore, caspase-9 was also activated in cells expressing variant 1. Since variant 1 inhibited the expression of COX-2 protein, we determined whether the activation of the two caspases occurred due to loss of prostaglandins, the product of COX-2 enzyme activity. Hence, we also treated cells with PGE2, a potent prostaglandin that inhibits apoptosis through inhibition of caspase activation (38, 47, 50, 52). When the cells expressing CUGBP2 variant 1 were also treated with PGE2, the activation of both caspase-9 and caspase-3 was inhibited (Fig. 5, A and B). Caspase-9 activation typically occurs following stimulation of the intrinsic or mitochondrial pathway of apoptotic cell death (20, 22). Bcl2, an integral membrane protein, is a potent inhibitor of apoptosis, and inactivation of the protein augments apoptosis (5, 54, 55). On the other hand, Bax is a cytosolic monomer protein in healthy cells that changes conformation during apoptosis and integrates into the outer mitochondrial membrane (5, 54, 55). Bax then oligomerizes, which contributes to the permeabilization of the outer mitochondrial membrane, thereby allowing the efflux of proteins involved in apoptosis (5, 54, 55). Western blot analyses demonstrated that, at baseline, Bcl2 expression was higher than Bax (Fig. 5, C and D). In contrast, cells expressing CUGBP2 variant 1 contained higher Bax (eightfold) and lower Bcl2 (two- to threefold). This was somewhat abrogated when the cells were also treated with PGE2 (Fig. 5, C and D). There was also a small increase in Bax expression following PGE2 treatment. However, given the significant increase in Bcl2, the ratio of Bax:Bcl2 suggests that this increase in Bax expression does not affect the cell. These data suggest that CUGBP2 variant 1-mediated apoptosis occurs in part through increasing the proapoptotic Bax-Bcl2 ratio in the cells.
Fig. 5.
CUGBP2 variant 1 overexpression results in mitotic catastrophe. A: CUGBP2 variant 1 overexpression resulted in activation of proapoptotic caspases. Lysates from HCT116 cells transiently expressing Flag-tagged CUGBP2 variant 1 and treated with PGE2 (1 µM)were subjected to Western blot analysis for caspase-3 and caspase-9. Both caspases were activated in the presence of CUGBP2 variant 1, which was partially suppressed when cells were also incubated with PGE2. β-actin was used as internal control for loading. B: abundance of caspase activity. Chemiluminescence data from 3 independent experiments was graphed. Expression of variant 1 significantly upregulated caspase-3 and caspase-9. a, P < 0.001 compared with control. b, P < 0.001 compared with CUGBP2 var 1 alone. C: proapoptotic Bax and antiapoptotic Bcl2 protein expression were analyzed in the lysates from HCT116 cells. Flag-tagged CUGBP2 variant 1 induced Bax expression but inhibited Bcl2. D: abundance of Bcl2 and Bax proteins. Chemiluminescence levels from 3 independent experiments were compared with levels observed in control, vector transfected cells. PGE2 significantly induced, whereas CUGBP2 variant 1 inhibited expression of Bcl2. On the other hand, CUGBP2 induced expression of Bax protein. Data from 3 independent experiments. a, P < 0.001 compared with Bcl2 expression in control cells; b, P < 0.05 compared with control cells; c, P < 0.001 compared with Bax expression in control cells. E: CUGBP2 variant 1 overexpression induces checkpoint proteins. Western blot analysis of lysates from HCT116 cells expressing Flag-tagged CUGBP2 variant 1 and treated with PGE2 (1 µM) demonstrated increased phosphorylation of Chk1 and Chk2 at Ser345 and Thr68, respectively. In addition, Cdc25c and p53 were phosphorylated at Ser216 and Ser15, respectively. F: levels of phosphorylated protein expression. Data from 3 independent experiments were plotted comparing the expression to that observed in control, vector transfected cells. *P <0.05 compared with control, vector transfected cells, and **P < 0.05 compared with CUGBP2 variant 1 alone. G: CUGBP2 variant 1 increases cyclin B1 and Cdc2 expression. Western blot analysis demonstrates increased levels of cyclin B1 and Cdc2 in cells expressing CUGBP2 variant 1. Cdc2 was also significantly upregulated in cells treated with PGE2. H: levels of Cdc2 and cyclin B1 protein. Chemiluminescence data from 3 independent experiments demonstrates that both proteins are upregulated following expression of CUGBP2 variant 1 or treatment with PGE2 *P < 0.001, **P < 0.05, both compared with control, vector transfected cells. I: CUGBP2 variant induces the nuclear localization of cyclin B1 and Cdc2. Although cyclin B1 and Cdc2 are in the cytoplasm of control cells, there are increased levels of both proteins in the nucleus of variant 1-expressing cells. J: CUGBP2 overexpression leads to mitotic catastrophe. HCT116 cells transiently transfected with plasmid-expressing Flag-tagged CUGBP2 variant 1 were stained for TUNEL (green) and phosphorylated histone H3 (red). The merged image shows cells were yellow stain positive for colocalization. DAPI is used to stain the nucleus.
Presence of increased numbers of cells in the G2/M stage of cell cycle suggested that CUGBP2 might affect mitosis (1, 13, 32). Checkpoint failure increases genomic instability, which, in turn, could lead to cell death. Chk1 and Chk2 are regulated by phosphorylation and play an important role in mediating G2/M DNA damage checkpoint that prevents mitosis when DNA is being repaired (31, 40, 51). To determine whether Chk1 and Chk2 are activated in CUGBP2 variant 1-expressing cells, Western blot analysis was performed. Both Chk1 and Chk2 were phosphorylated in Ser345 and Thr68, respectively, in response to CUGBP2 variant 1 overexpression (Fig. 5, E and F). Furthermore, treatment with PGE2 only partially suppressed the phosphorylation of the two checkpoint proteins. Chk1 is known to phosphorylate Cdc25c on Ser216 (8, 18, 25). Since both Chk1 and Chk2 are activated in the CUGBP2 variant 1-overexpressing cells, we also determined the phosphorylation status of Cdc25c. In CUGBP2 variant 1-overexpressing cells, there was a significant amount of Cdc25c that was phosphorylated at Ser216 residue (Fig. 5, E and F). p53-induced growth arrest occurs during G1/S or G2/M stages and is required for apoptosis (6, 29). Phosphorylation of p53 on Ser15 is known to lead to its stabilization and subsequent activation as a transcription factor (49). Here we observed that in cells overexpressing CUGBP2 variant 1, there was increased phosphorylation of p53 (Fig. 5, E and F). Cyclin-dependent kinase Cdc2 is an important activator of mitosis that associates with its regulatory subunit, cyclin B1, whose levels rise during S phase and G2 and peak in mitosis (2, 15, 45). They are kept in the inactive state by phosphorylation at Thr14 and Tyr15 through the actions of wee1 protein kinase (7). To determine whether Cdc2 and cyclin B1 are affected, Western blot analyses were performed and total levels of the two proteins were determined. Cdc2 expression was significantly upregulated by both PGE2 treatment and CUGBP2 variant 1 overexpression. However, cyclin B1 expression was upregulated only in the presence of CUGBP2 variant 1 (Fig. 5, G and H). At the end of G2, rapid dephosphorylation of Cdc2 by the Cdc25c phosphatase triggers the Cdc2-cyclin B1 complex to enter the nucleus and trigger mitosis. Therefore, we performed immunocytochemistry analysis to determine the cellular localization of Cdc2 and cyclin B1. Although both Cdc2 and cyclin B1 are present in the cytoplasm of control cells, they accumulate significantly in the nucleus in the CUGBP2 variant 1-overexpressing cells (Fig. 5I). This suggests that the CUGBP2 variant 1-overexpressing cells are undergoing mitosis. All the data implies that cells undergoing mitosis are also undergoing apoptosis, suggesting that the cells are undergoing mitotic catastrophe (9, 44). To confirm that the cells are indeed undergoing mitosis and apoptosis at the same time, we performed immunocytochemistry analysis. In CUGBP2 variant 1-overexpressing cells, there were high levels of nuclear-phosphorylated histone H3 (signaling active cell division) and terminal deoxynucleotidyl transferase dUTP-mediated nick-end labeling (TUNEL) positivity (signaling apoptosis) (9, 39) (Fig. 5J). Furthermore, there was colocalization of the phosphorylated histone 3 with TUNEL staining. In contrast, the control cells neither demonstrate any nuclear-phosphorylated histone H3 nor were positive for TUNEL. Taken together, these data demonstrate that cells in which CUGBP2 variant 1 is overexpressed are driven to undergo mitotic catastrophe.
DISCUSSION
CUGBP2 is a protein with many functions in RNA metabolism including modulation of splicing, editing, stability, and translation. Its expression is highest in skeletal and cardiac muscle cells where it has been shown to bind to specific sequences in the intron and oppose hnRNP-I/PTB activity to induce splicing of muscle-specific transcripts such as troponin T and α-actinin (10, 19). CUGBP2 is also expressed in other cell types, and one physiological function is in regulation of apolipoprotein B mRNA editing in hepatocytes (3). However, its function in other cells, including the intestinal epithelial cells, is presently unknown. Given its ability to regulate alternative splicing in the muscle cells, the possibility exists that the protein also regulates such transcripts in the epithelial cells. In this regard, it was shown that CUGBP2 can activate alternative splicing of the transcript encoding cystic fibrosis transmembrane conductance regulator (CFTR) in in vitro studies (16). However, in vivo studies are required to demonstrate this function in intestinal epithelial cells. If CUGBP2 does regulate CFTR splicing in vivo, it would be interesting to determine whether CFTR splicing is altered following radiation.
Here, we have identified that the two novel splice variants of CUGBP2 that result from alternative transcription from upstream promoters resulting in additional NH2-terminal amino acids. These splice variants are expressed in the intestinal epithelial cells in the mouse. As mentioned above, one physiological function for the three variants might be in regulating alternative splicing of distinct transcripts. In previous studies, we have demonstrated that CUGBP2, which we now call variant 1, is predominantly nuclear in intestinal epithelial cells and is significantly upregulated when the cells are exposed to γ-irradiation (34). Furthermore, CUGBP2 binds to AU-rich sequences in the 3′UTR of COX-2 mRNA and increases the stability of the mRNA but inhibits translation, resulting in decreased PGE2 production, thereby leading to cell death (34). However, the primers used for the identification of the transcript were located within the coding region that would not differentiate between the three variants. On the basis of the data in Fig. 1, it is now apparent that the variant that was observed before radiation was predominantly variant 2, whereas seen after radiation was variant 1. We believe this is significant because all three variants increased the steady-state levels of COX-2 mRNA; however, only variant 1 inhibited COX-2 mRNA translation. Furthermore, overexpression of variant 1, but not variants 2 or 3, resulted in cells undergoing mitotic catastrophe. Indeed, in normal tissue, variant 2 is the predominant isoform that is expressed, whereas variant 1 is very low. This suggests that CUGBP2 expression is tightly regulated with distinct function for the variants.
An interesting point to note is that the presence of additional amino acids in the NH2 terminus of variants 2 and 3 results in altered localization of the protein. Whereas variant 1 is nuclear, the two novel variants are predominantly localized in the cytoplasm. This change in localization is somewhat perplexing because previous studies have demonstrated that the nuclear localization signals are located within the divergent region and third RRM that are present in the carboxyl terminus of the protein (24). However, the first two RRMs located near the amino-terminus have been shown to potentially include signals for cytoplasmic localization (24). The possibility therefore exists that the presence of the additional amino acids in the amino-terminus of CUGBP2 might unmask these domains, resulting in increased cytoplasmic localization of the protein. An additional possibility is that the additional amino acids themselves encode cytoplasmic localization signals for which additional studies are necessary. However, database searches did not reveal the presence of any of the known signals that direct a protein to the cytoplasm.
We have observed that there is a differential expression pattern for the three splice variants in the three colon cancer cell lines tested. Furthermore, variant 1 was demonstrated to affect COX-2 mRNA translation, which was confirmed in this manuscript. However, the two novel splice variants did not have any effect on COX-2 mRNA translation, although they also increased steady-state COX-2 mRNA levels. These data suggest that, although the two variants retain the ability to stabilize COX-2 mRNA, they have lost the translation inhibitory function. Once additional mRNA targets for CUGBP2 are identified, it would be interesting to determine whether the loss of translation inhibitory function is a general phenomenon or specific to COX-2 mRNA.
Mitotic catastrophe is a unique form of cell death that occurs either during mitosis or resulting from failed mitosis, the molecular mechanisms involving premature activation of Chk1 and Chk2 (9, 44). Under normal conditions, Chk1 and Chk2 are supposed to prevent the activation of Cdc2-cyclin B1 complex, thereby arresting the cells in G2 phase of the cell cycle through the activation of Cdc25c (17, 43). These are the first studies demonstrating the ability of CUGBP2 variant 1 to induce mitotic catastrophe. We demonstrate that overexpression of CUGBP2 variant 1 results in activation of Chk1 and Chk2 and activation of Cdc25c. As would be predicted from this, Cdc2 and cyclin B1 are localized to the nucleus, suggesting that they, too, have been activated, their function in the nucleus being to induce mitosis. However, it is not clear how p53 is being phosphorylated and whether this is a major cause of the mitotic catastrophe that is observed following the overexpression of CUGBP2 variant 1. One p53 transcriptional target gene is Bax, which induces mitochondrial membrane permeabilization with subsequent cytochrome c-mediated caspase activation (5, 55). Indeed, our results demonstrate induction of Bax expression. Further studies are required to determine whether this is a major pathway for CUGBP2 variant 1-mediated increased cell death. Studies are also required to determine whether inhibition of Chk1 and Chk2 with several pharmacological inhibitors or by transfecting dominant negative constructs to gain further insights into whether CUGBP2 affects centrosomes or cyclin-dependent kinases such as Cdk1. We believe these ongoing studies will help in identifying whether CUGBP2 variant 1 drives premature induction of mitosis before the completion of S or G2 phase of the cell cycle.
ACKNOWLEDGMENTS
The authors thank James Wyche for insightful comments during the course of these studies.
GRANTS
The studies were supported by National Institutes of Health Grants DK-62265 and CA-109269.
REFERENCES
- 1.Abraham RT. Checkpoint signaling: epigenetic events sound the DNA strand-breaks alarm to the ATM protein kinase. Bioessays. 2003;25:627–630. doi: 10.1002/bies.10310. [DOI] [PubMed] [Google Scholar]
- 2.Abrieu A, Doree M, Fisher D. The interplay between cyclin-B-Cdc2 kinase (MPF) and MAP kinase during maturation of oocytes. J Cell Sci. 2001;114:257–267. doi: 10.1242/jcs.114.2.257. [DOI] [PubMed] [Google Scholar]
- 3.Anant S, Henderson JO, Mukhopadhyay D, Navaratnam N, Kennedy S, Min J, Davidson NO. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J Biol Chem. 2001;276:47338–47351. doi: 10.1074/jbc.M104911200. [DOI] [PubMed] [Google Scholar]
- 4.Anant S, Murmu N, Houchen CW, Mukhopadhyay D, Riehl TE, Young SG, Morrison AR, Stenson WF, Davidson NO. Apobec-1 protects intestine from radiation injury through posttranscriptional regulation of cyclooxygenase-2 expression. Gastroenterology. 2004;127:1139–1149. doi: 10.1053/j.gastro.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 5.Basu A, Haldar S. The relationship between BcI2, Bax and p53: consequences for cell cycle progression and cell death. Mol Hum Reprod. 1998;4:1099–1109. doi: 10.1093/molehr/4.12.1099. [DOI] [PubMed] [Google Scholar]
- 6.Bates S, Vousden KH. p53 in signaling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:12–18. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 7.Berry LD, Gould KL. Regulation of Cdc2 activity by phosphorylation at T14/Y15. Prog Cell Cycle Res. 1996;2:99–105. doi: 10.1007/978-1-4615-5873-6_10. [DOI] [PubMed] [Google Scholar]
- 8.Bulavin DV, Amundson SA, Fornace AJ. p38 and Chk1 kinases: different conductors for the G(2)/M checkpoint symphony. Curr Opin Genet Dev. 2002;12:92–97. doi: 10.1016/s0959-437x(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 9.Castedo M, Perfettini JL, Roumier T, Kroemer G. Cyclin-dependent kinase-1: linking apoptosis to cell cycle and mitotic catastrophe. Cell Death Differ. 2002;9:1287–1293. doi: 10.1038/sj.cdd.4401130. [DOI] [PubMed] [Google Scholar]
- 10.Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 11.Choi DK, Ito T, Tsukahara F, Hirai M, Sakaki Y. Developmentally-regulated expression of mNapor encoding an apoptosis-induced ELAV-type RNA binding protein. Gene. 1999;237:135–142. doi: 10.1016/s0378-1119(99)00312-1. [DOI] [PubMed] [Google Scholar]
- 12.Cok SJ, Morrison AR. The 3′-untranslated region of murine cyclooxygenase-2 contains multiple regulatory elements that alter message stability and translational efficiency. J Biol Chem. 2001;276:23179–23185. doi: 10.1074/jbc.M008461200. [DOI] [PubMed] [Google Scholar]
- 13.Collins I, Garrett MD. Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Curr Opin Pharmacol. 2005;5:366–373. doi: 10.1016/j.coph.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Coopersmith CM, Gordon JI. gamma-Ray-induced apoptosis in transgenic mice with proliferative abnormalities in their intestinal epithelium: re-entry of villus enterocytes into the cell cycle does not affect their radioresistance but enhances the radiosensitivity of the crypt by inducing p53. Oncogene. 1997;15:131–141. doi: 10.1038/sj.onc.1201176. [DOI] [PubMed] [Google Scholar]
- 15.Doree M, Hunt T. From Cdc2 to Cdk1: when did the cell cycle kinase join its cyclin partner? J Cell Sci. 2002;115:2461–2464. doi: 10.1242/jcs.115.12.2461. [DOI] [PubMed] [Google Scholar]
- 16.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotoh T, Ohsumi K, Matsui T, Takisawa H, Kishimoto T. Inactivation of the checkpoint kinase Cds1 is dependent on cyclin B-Cdc2 kinase activation at the meiotic G(2)/M-phase transition in Xenopus oocytes. J Cell Sci. 2001;114:3397–3406. doi: 10.1242/jcs.114.18.3397. [DOI] [PubMed] [Google Scholar]
- 18.Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O’Connor PM, Piwnica-Worms H. The Chk1 protein kinase and the Cdc25c regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 19.Gromak N, Matlin AJ, Cooper TA, Smith CW. Antagonistic regulation of alpha-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA. 2003;9:443–456. doi: 10.1261/rna.2191903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson CR, Jarvis WD. Caspase-9 regulation: an update. Apoptosis. 2004;9:423–427. doi: 10.1023/B:APPT.0000031457.90890.13. [DOI] [PubMed] [Google Scholar]
- 21.Keskek M, Gocmen E, Kilic M, Gencturk S, Can B, Cengiz M, Okten RM, Koc M. Increased expression of cyclooxygenase-2 (COX-2) in radiation-induced small bowel injury in rats. J Surg Res. 2006;135:76–84. doi: 10.1016/j.jss.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 22.Kuida K. Caspase-9. Int J Biochem Cell Biol. 2000;32:121–124. doi: 10.1016/s1357-2725(99)00024-2. [DOI] [PubMed] [Google Scholar]
- 23.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladd AN, Cooper TA. Multiple domains control the subcellular localization and activity of ETR-3, a regulator of nuclear and cytoplasmic RNA processing events. J Cell Sci. 2004;117:3519–3529. doi: 10.1242/jcs.01194. [DOI] [PubMed] [Google Scholar]
- 25.Lam MH, Rosen JM. Chk1 versus Cdc25: chking one’s levels of cellular proliferation. Cell Cycle. 2004;3:1355–1357. doi: 10.4161/cc.3.11.1225. [DOI] [PubMed] [Google Scholar]
- 26.Landegren U. Measurement of cell numbers by means of the endogenous enzyme hexosaminidase. Applications to detection of lymphokines and cell surface antigens. J Immunol Methods. 1984;67:379–388. doi: 10.1016/0022-1759(84)90477-0. [DOI] [PubMed] [Google Scholar]
- 27.Leroy O, Dhaenens CM, Schraen-Maschke S, Belarbi K, Delacourte A, Andreadis A, Sablonniere B, Buee L, Sergeant N, Caillet-Boudin ML. ETR-3 represses Tau exons 2/3 inclusion, a splicing event abnormally enhanced in myotonic dystrophy type I. J Neurosci Res. 2006;84:852–859. doi: 10.1002/jnr.20980. [DOI] [PubMed] [Google Scholar]
- 28.Lichtner P, Attie-Bitach T, Schuffenhauer S, Henwood J, Bouvagnet P, Scambler PJ, Meitinger T, Vekemans M. Expression and mutation analysis of BRUNOL3, a candidate gene for heart and thymus developmental defects associated with partial monosomy 10p. J Mol Med. 2002;80:431–442. doi: 10.1007/s00109-002-0331-9. [DOI] [PubMed] [Google Scholar]
- 29.Liebermann DA, Hoffman B, Steinman RA. Molecular controls of growth arrest and apoptosis: p53-dependent and independent pathways. Oncogene. 1995;11:199–210. [PubMed] [Google Scholar]
- 30.Lu X, Timchenko NA, Timchenko LT. Cardiac elav-type RNA-binding protein (ETR-3) binds to RNA CUG repeats expanded in myotonic dystrophy. Hum Mol Genet. 1999;8:53–60. doi: 10.1093/hmg/8.1.53. [DOI] [PubMed] [Google Scholar]
- 31.Malmanche N, Maia A, Sunkel CE. The spindle assembly checkpoint: preventing chromosome missegregation during mitosis and meiosis. FEBS Lett. 2006;580:2888–2895. doi: 10.1016/j.febslet.2006.03.081. [DOI] [PubMed] [Google Scholar]
- 32.McGowan CH. Checking in on Cds1 (Chk2): a checkpoint kinase and tumor suppressor. Bioessays. 2002;24:502–511. doi: 10.1002/bies.10101. [DOI] [PubMed] [Google Scholar]
- 33.Merritt AJ, Allen TD, Potten CS, Hickman JA. Apoptosis in small intestinal epithelial from p53-null mice: evidence for a delayed, p53-independent G2/M-associated cell death after gamma-irradiation. Oncogene. 1997;14:2759–2766. doi: 10.1038/sj.onc.1201126. [DOI] [PubMed] [Google Scholar]
- 34.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 35.Mukhopadhyay D, Jung J, Murmu N, Houchen CW, Dieckgraefe BK, Anant S. CUGBP2 plays a critical role in apoptosis of breast cancer cells in response to genotoxic injury. Ann NY Acad Sci. 2003;1010:504–509. doi: 10.1196/annals.1299.093. [DOI] [PubMed] [Google Scholar]
- 36.Murmu N, Jung J, Mukhopadhyay D, Houchen CW, Riehl TE, Stenson WF, Morrison AR, Arumugam T, Dieckgraefe BK, Anant S. Dynamic antagonism between RNA-binding protein CUGBP2 and cyclooxygenase-2-mediated prostaglandin E2 in radiation damage. Proc Natl Acad Sci USA. 2004;101:13873–13878. doi: 10.1073/pnas.0406066101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Oehlert W, Oehlert M, Monig H, Konermann G. The effect of endotoxin on preirradiated mice. Strahlenther Onkol. 1992;168:716–727. [PubMed] [Google Scholar]
- 38.Papadimitriou A, King AJ, Jones PM, Persaud SJ. Anti-apoptotic effects of arachidonic acid and prostaglandin E2 in pancreatic beta-cells. Cell Physiol Biochem. 2007;20:607–616. doi: 10.1159/000107544. [DOI] [PubMed] [Google Scholar]
- 39.Pascreau G, Arlot-Bonnemains Y, Prigent C. Phosphorylation of his-tone and histone-like proteins by aurora kinases during mitosis. Prog Cell Cycle Res. 2003;5:369–374. [PubMed] [Google Scholar]
- 40.Rhind N, Baber-Furnari BA, Lopez-Girona A, Boddy MN, Brondello JM, Moser B, Shanahan P, Blasina A, McGowan C, Russell P. DNA damage checkpoint control of mitosis in fission yeast. Cold Spring Harb Symp Quant Biol. 2000;65:353–359. doi: 10.1101/sqb.2000.65.353. [DOI] [PubMed] [Google Scholar]
- 41.Riehl T, Cohn S, Tessner T, Schloemann S, Stenson WF. Lipopoly-saccharide is radioprotective in the mouse intestine through a prostaglandin-mediated mechanism. Gastroenterology. 2000;118:1106–1116. doi: 10.1016/s0016-5085(00)70363-5. [DOI] [PubMed] [Google Scholar]
- 42.Riehl TE, Newberry RD, Lorenz RG, Stenson WF. TNFR1 mediates the radioprotective effects of lipopolysaccharide in the mouse intestine. Am J Physiol Gastrointest Liver Physiol. 2004;286:G166–G173. doi: 10.1152/ajpgi.00537.2002. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez R, Meuth M. Chk1 and p21 cooperate to prevent apoptosis during DNA replication fork stress. Mol Biol Cell. 2006;17:402–412. doi: 10.1091/mbc.E05-07-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roninson IB, Broude EV, Chang BD. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist Updat. 2001;4:303–313. doi: 10.1054/drup.2001.0213. [DOI] [PubMed] [Google Scholar]
- 45.Solomon MJ. Activation of the various cyclin/Cdc2 protein kinases. Curr Opin Cell Biol. 1993;5:180–186. doi: 10.1016/0955-0674(93)90100-5. [DOI] [PubMed] [Google Scholar]
- 46.Sureban SM, Murmu N, Rodriguez P, May R, Maheshwari R, Dieckgraefe BK, Houchen CW, Anant S. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology. 2007;132:1055–1065. doi: 10.1053/j.gastro.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 47.Takadera T, Ohyashiki T. Prevention of rat cortical neurons from prostaglandin E2-induced apoptosis by glycogen synthase kinase-3 inhibitors. Neurosci Lett. 2006;400:105–109. doi: 10.1016/j.neulet.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 48.Tessner TG, Muhale F, Schloemann S, Cohn SM, Morrison AR, Stenson WF. Ionizing radiation up-regulates cyclooxygenase-2 in I407 cells through p38 mitogen-activated protein kinase. Carcinogenesis. 2004;25:37–45. doi: 10.1093/carcin/bgg183. [DOI] [PubMed] [Google Scholar]
- 49.Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vassiliou E, Sharma V, Jing H, Sheibanie F, Ganea D. Prostaglandin E2 promotes the survival of bone marrow-derived dendritic cells. J Immunol. 2004;173:6955–6964. doi: 10.4049/jimmunol.173.11.6955. [DOI] [PubMed] [Google Scholar]
- 51.Weaver BA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 52.Weinreb M, Shamir D, Machwate M, Rodan GA, Harada S, Keila S. Prostaglandin E2 (PGE2) increases the number of rat bone marrow osteogenic stromal cells (BMSC) via binding the EP4 receptor, activating sphingosine kinase and inhibiting caspase activity. Prostaglandins Leukot Essent Fatty Acids. 2006;75:81–90. doi: 10.1016/j.plefa.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 53.Xu K, Kitchen CM, Shu HK, Murphy TJ. Platelet-derived Growth Factor-induced Stabilization of Cyclooxygenase 2 mRNA in Rat Smooth Muscle Cells Requires the c-Src family of protein-tyrosine kinases. J Biol Chem. 2007;282:32699–32709. doi: 10.1074/jbc.M705272200. [DOI] [PubMed] [Google Scholar]
- 54.Yang E, Korsmeyer SJ. Molecular thanatopsis: a discourse on the BCL2 family and cell death. Blood. 1996;88:386–401. [PubMed] [Google Scholar]
- 55.Zinkel S, Gross A, Yang E. BCL2 family in DNA damage and cell cycle control. Cell Death Differ. 2006;13:1351–1359. doi: 10.1038/sj.cdd.4401987. [DOI] [PubMed] [Google Scholar]