Abstract
Localization to sites of DNA damage is a hallmark of DNA damage response (DDR) proteins. To identify new DDR factors, we screened epitope-tagged proteins for localization to sites of chromatin damaged by UV laser microirradiation and found >120 proteins that localize to damaged chromatin. These include the BAF tumor suppressor complex and the ALS candidate protein TAF15. TAF15 contains multiple domains that bind damaged chromatin in a PARP-dependent manner, suggesting a possible role as glue that tethers multiple PAR chains together. Many positives were transcription factors and >70% of randomly tested transcription factors localized to sites of DNA damage and approximately 90% were PARP-dependent for localization. Mutational analyses showed that localization to damaged chromatin is DNA-binding domain-dependent. By examining Hoechst staining patterns at damage sites, we see evidence of chromatin decompaction that is PARP-dependent. We propose that PARP-regulated chromatin remodeling at sites of damage allows transient accessibility of DNA-binding proteins.
Graphical abstract
INTRODUCTION
Each day cells are subjected to many different types of DNA damage including chemical adducts, depurinations, depyrimidinations, abasic lesions, double-strand breaks, single-strand breaks, and replication stress errors. The cell, however, has a myriad of enzymes that repair damaged DNA through chemical modification, including nucleases, kinases, helicases, phosphatases, recombinases, topoisomerases, ligases, demethylases and polymerases.
DNA remodeling enzymes must be carefully regulated both because they are potentially dangerous to genomic integrity and because carrying out the appropriate type of DNA repair necessitates that the right enzymes be activated in the right location at the right time. To facilitate this regulation, cells have evolved the DNA damage response (DDR) network, a sensory signal transduction pathway that recognizes different types of DNA lesions and provides information to the cell in order to allow proper orchestration of the myriad of responses that promote cell and organismal survival. A major mechanism by which the DDR accomplishes this is extensive proteomic remodeling through post-translational modification, including phosphorylation, ubiquitination, sumoylation and acetylation (Bennetzen et al., 2010; Lukas et al., 2011; Matsuoka et al., 2007). Additionally, the DDR coordinates mechanisms of spatial regulation through direct recruitment of specific factors to sites of DNA damage (Bekker-Jensen et al., 2006). To accomplish this, the DDR assembles platforms for recruitment, including the poly-(ADP-ribose) polymerase (PARP) enzymes, the RPA complex, the histone H2A family member H2AX, PCNA, and the FANCI-FANCD2 complex. Importantly, each of these mediates the recruitment of distinct repair factors corresponding to a distinct set of structural alterations in the DNA.
H2AX and RPA respond to double-strand breaks (DSBs) and stalled replication forks (Ciccia and Elledge, 2010; Rogakou et al., 1998). RPA is recruited to these lesions through recognition of extensive ssDNA that is formed nearby, and once bound, facilitates localized accumulation of several important repair complexes, including ATRIP/ATR, the RAD17/RFC2-5 clamp loader, and the 9-1-1 heterotrimer bound to TOPBP1, as well as SMARCAL1, PRP19 (Marechal et al., 2014), RHNO1 (Cotta-Ramusino et al., 2011), and other proteins. H2AX recruits repair factors to DSBs through a distinct feed-forward signaling mechanism. Specifically, H2AX is phosphorylated on Ser139 in response to DNA damage and acts as a scaffold to recruit MDC1, which in turn, binds the ATM kinase to further propagate phosphorylation of H2AX for up to 2 megabases of adjacent chromatin, generating a visible focus of phosphorylation. MDC1 then sets in motion a series of signaling modifications that recruit repair factors such as RNF8, RNF168, 53BP1 and the BRCA1 A, B and C complexes (Ciccia and Elledge, 2010).
PCNA and the FANCI-FANCD2 (ID) complex respond to events following replication stress (Moldovan and D’Andrea, 2009; Moldovan et al., 2007). In response to replication blocks, PCNA undergoes monoubiquitination that recruits translesion polymerases and stimulates direct replication bypass. Additionally, PCNA can become polyubiquitinated with K63-linked chains to promote recombination-dependent DNA synthesis across DNA lesions by template switching mechanisms and recruits ZRANB3 to sites of replication stress (Ciccia et al., 2012). FANCI and FANCD2 respond to DNA interstrand crosslinks that arrest replication forks and are recruited to damaged DNA where they are ubiquitinated and promote repair.
PARP1, 2 and 3 enzymes recognize and are activated by a variety of DNA lesions, including single-strand (SSBs), DNA crosslinks, or DSBs (Beck et al., 2014). They catalyze the rapid and transient synthesis of poly-(ADP-ribose) (PAR) structures and are responsible for the recruitment of a number of factors to sites of DNA damage, including ALC1, RBMX, and components of MRN and NuRD complexes among others (Adamson et al., 2012; Ahel et al., 2009; Chou et al., 2010; Haince et al., 2008; Polo et al., 2010). Additionally, the scaffold protein XRCC1 is recruited to SSBs in a PARP1-dependent manner where it mediates the accumulation of a cohort of SSB repair factors (PNKP, APTX, and Pol β) (Caldecott, 2008). These recruited proteins then process ssDNA ends to facilitate repair.
Here we describe a focused analysis to identify new proteins recruited to DNA damage. We evaluated spatial changes to candidate protein localization after DNA damage by UV laser microirradation of BrdU prelabeled cell nuclei. We identified at least 120 novel factors that show evidence for recruitment to sites of DNA damage. Among these is the candidate amyotrophic lateral sclerosis (ALS) factor TAF15. We found that TAF15 has independent domains that allow it to localize to sites of DNA damage in a PARP1-dependent manner. Additionally, we identified 5 components of SWI/SNF chromatin remodeling complexes that localize to sites of damage. We discovered several steroid hormone receptors and transcription factors that localize to DNA damage, and we show that several of these are recruited in a manner dependent upon PARP and/or their DNA-binding domains (DBDs). We propose that chromatin remodeling may play a role in making DNA accessible to many proteins at sites of DNA damage.
RESULTS
A focused screen to identify proteins that localize to damaged chromatin
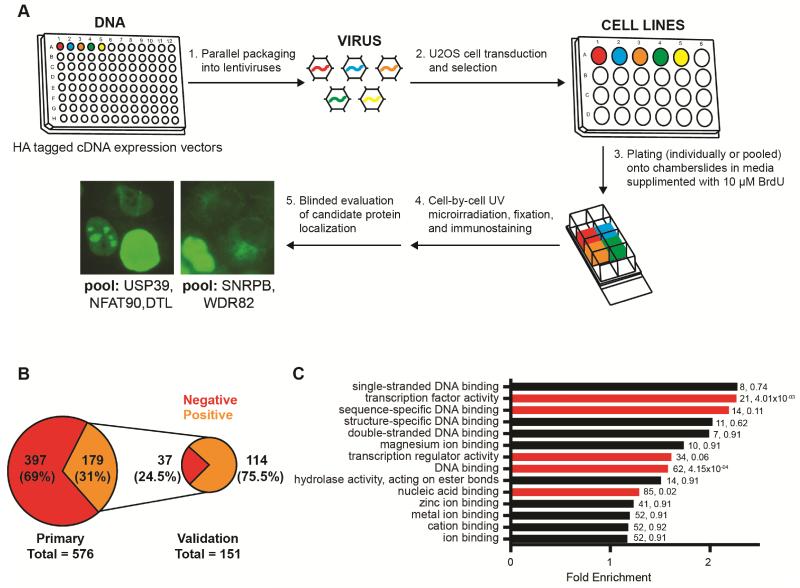
To better understand the complement of proteins that respond to DNA lesions and control localized DNA repair, we conducted a focused screen for proteins that are recruited to DNA breaks induced by UV laser microirradiation (Figure 1A). We manually curated a list of 726 candidate genes (851 ORFs) using published and unpublished datasets relevant to the DDR, including a set of genes that encode proteins experimentally identified to be chromatin-enirched after exposure to DNA damage (Tables S1-3) (Adamson et al., 2012; Chou et al., 2010; Cotta-Ramusino et al., 2011; Hurov et al., 2010; Matsuoka et al., 2007; O’Connell et al., 2010; Paulsen et al., 2009; Slabicki et al., 2010; Smogorzewska et al., 2010). Candidate ORFs were selected from sequence-verified human ORFeome collections and were individually transduced into U2OS cells for expression as N- or C-terminal HA fusion proteins using lentiviruses (Figure 1A). Selected candidate cell lines were then microirradiated (individually or as sub-pools) over a period of 5 minutes and evaluated for tagged-candidate recruitment to laser-induced DNA damage tracks (marked by γH2AX co-staining) immediately and/or after 25 min. of recovery. Pooled candidates with at least one HA-stained laser track were deconvolved and reevaluated individually at one or both time points.
Figure 1.
A spatial DNA damage localization screen. (A) Schematic of the screen. (B) Graphical representation of results from primary analysis and validation. Numbers indicate genes associated with evaluated ORFs. (C) GO molecular function (MF) terms identified by DAVID among the positive genes from primary screening (fold enrichment compared to the candidate genes evaluated). The numbers of genes classified by each GO term and FDR corrected P-values (Benjamini) are indicated. Enriched GO terms with an FDR < 0.25 are red.
We eventually evaluated, 576 unique ORFs for post-damage localization (Table S4). At various stages, candidates were eliminated from the screen for both technical and biological reasons. Technical reasons include unavailability of bacterial DNA stocks, lack of bacterial growth, and poor growth or selection of transduced cell lines. Biological reasons included selective elimination of some ORFs corresponding to genes already evaluated or encoding proteins observed to have non-nuclear localization in U2OS cells without mircoirradiation. Of candidates evaluated by Sanger sequencing at various stages of analysis, 15 ORFs were found to be incorrect (i.e. candidates not intentionally included in our original list) (Table S4). Our final candidate list was enriched for genes with known roles in DNA repair and other nuclear processes, including transcription, RNA splicing, and chromatin organization (Figure S1). Of those, 178 (179 ORFs including 2 distinct ORFs assigned to C18orf25) encoded proteins that we observed to accumulate at sites of DNA damage (scored by colocalization with least one γH2AX stained mircoirradiation track), yielding a positive identification rate of 31% (Figure 1B). These “hit” genes include ones that encode proteins with previously known localization to DNA breaks. Interestingly, we observed a higher incidence of candidate recruitment at the earlier (0-5 minute) time point (Table S4). Specifically, of 139 positive ORFs that we evaluated at both 0-5 and 25-30 minutes post microirradiation, proteins encoded by 42 of these demonstrated recruitment at both time points, 2 (ZNF384 and RHNO1) were recruited only at 25-30 minutes, and 95 (68% of these positives) demonstrated tracks at only the early (0-5 minute) time point. Additionally, of those that scored positive at both time points, 81% were observed to have a higher percentage of damaged cells with colocalizing HA and γH2AX tracks in the earlier sample (Table S4). Notable exceptions to this were CtIP, TOPBP1 and RAD18, which demonstrated increased percentages of damaged cells with stripes over time.
To validate our primary analysis, positive candidate expression vectors were clonally isolated and the encoded ORFs were sequenced to verify gene identity. These constructs were then used to reconstruct individual expression lines in U2OS cells, and these cells were used reevaluate candidate recruitment to mircoirradiation tracks. Of the 179 positive ORFs identified by primary analysis, we reevaluated 151 (150 genes) at one or both time points. Most candidates not selected for reevaluation were known to have established roles in DNA repair or maintaining genomic stability and many were shown previously to localize at DNA lesions. Of these, 114 (113 genes) demonstrated recruitment to one or more γH2AX stained mircoirradiation tracks (a validation rate of 75.5%) (Figure 1B). Possible factors that may have influenced this validation rate include primary analysis false positives caused by well-to-well candidate contamination during parallel DNA preparation and validation false negatives caused by rare localization events. Indeed, candidates that did not score in reevaluation (37) had, on average, fewer damaged cells with visible colocalization tracks in the primary screen than those that validated (~8.0% compared to ~17% at 0-5 minutes and ~1.6% compared to ~3.4% at 25-30 minutes).
Proteins that localize to sites of damage include ALS-associated TAF15 and EWSR1
Surprisingly, we observed that our validated positive genes included many encoding transcription regulators. DAVID functional annotation of hits in our primary screen revealed that “transcription factor activity” and “transcription regulator activity” molecular function terms were enriched over our set of evaluated genes (FDR < 0.25), a curated list that is itself enriched for genes involved in nuclear processes (Figure 1C and S1). Additionally, these positives were enriched for “DNA binding,” “sequence-specific DNA binding,” and “nucleic acid binding” terms (Figure 1C). Specific transcription-associated genes among our validated positives included NFIA, MYBL2, YY1, ZSCAN5A, ZNF829, ZBTB10, ZNF281, ZNF384, ATF2, ATF7, BATF3, POU2F2, and PBX2 (Figure 2A). These 13 genes were also among the 62 identified by DAVID analysis as “DNA binding” (Figure 1C), and interestingly, the DNA binding regions of these genes are varied. In our analysis, ORFs encoding YY1, ZSCAN5A, ZNF829, ZBTB10, ZNF281, and ZNF384 all had intact zinc finger domains, those encoding ATF7 and BATF3 contained leucine zippers, POU2F2 and PBX2 were homeobox-containing, and both NFIA and MYBL2 contained other DBDs. Of note, ATF2 was previously shown to be an ATM phosphorylation substrate that accumulates at DNA breaks in its phosphorylated form after exposure to ionizing radiation (Bhoumik et al., 2005).
Figure 2.
Transcription factors and chromatin remodeling factors localize to sites of DNA damage. (A) Validation images of cells expressing indicated HA fusion proteins from ORFs encoded by transcription-associated genes. Each image was individually adjusted during exposure and processing to best demonstrate protein localization. Scale bars, 10 μm. Inset numbers represent the percentages of damaged cells with colocalizing HA and γH2AX damage tracks. (B) Cells expressing DPF2, BCL7A, BCL7C or PHF10 HA fusion proteins or no exogenous fusion protein were mircoirradiated and immunostained with antibodies against HA or SMARCC1 and γH2AX (0-5 minutes after microirradiation) as in (A). Table lists the percentages of cells with γH2AX microirradiation tracks observed that have visible accumulation of the indicated protein or HA fusion colocalizing with γH2AX; these data are from screen primary and validation analyses found in Tables S4 and S5 or independent experiments (indicated with asterisks). N/A indicates data not collected
Our list also contains ORFs assigned to chromatin-associated proteins, with known and as yet unknown links to DNA repair. These include components of the Polycomb (BMI1) and nucleosome remodelling deacetylase (NuRD) (RBBP4) repressive complexes, subunits of which were previously shown to localize to DNA breaks (Chou et al., 2010; Ismail et al., 2010; Polo et al., 2010), as well as one isoform (α) of the chromodomain and chromoshadow domain-containing heterochromatin protein 1 (HP1, encoded by CBX5) and the HP1-binding protein 3 (HP1BP3). The three human isoforms of HP1, including β and γ (not included in our library), have emerging roles in DNA repair tied to damage-induced changes in protein mobility (Dinant and Luijsterburg, 2009). Additionally, of 5 ORFs encoding subunits of the human SWI/SNF complexes (BAF or PBAF) included in our library, 4 were positive for damage tracks (Figure 2B). These ORFs all encode components of the BAF complex: DPF2, BCL7A, BCL7C, and PHF10 (Kadoch et al., 2013). Using antibodies against a fifth BAF protein, we showed that SMARCC1 also accumulates at microirradiation tracks with similar kinetics (Figure 2B).
Our validated list of ORFs also includes a few that encode canonical components of the DDR, including RFC3, RFC4 and RAD18, known mediators of BER (POLB) and TC-NER (TCEA1), and one that encodes a protein recently characterized in the DDR and described at DNA breaks (PPM1G) (Beli et al., 2012; Khoronenkova et al., 2012). These serve as positive controls. CHTF18, a component of an alternative replication factor C (RFC) complex, which along with DCC1, CTF8 and RFC1-4 loads PCNA onto DNA (Bermudez et al., 2003), is present among our validated positives. Finally, 3 RING domain-containing proteins (RNF138, RNF113A, UHRF2) and the SCF-like ubiquitin ligase specificity factor, CDT2 (DTL), which degrades CDT1 in response to DNA damage to control replication firing (Jin et al., 2006), are also present on this list.
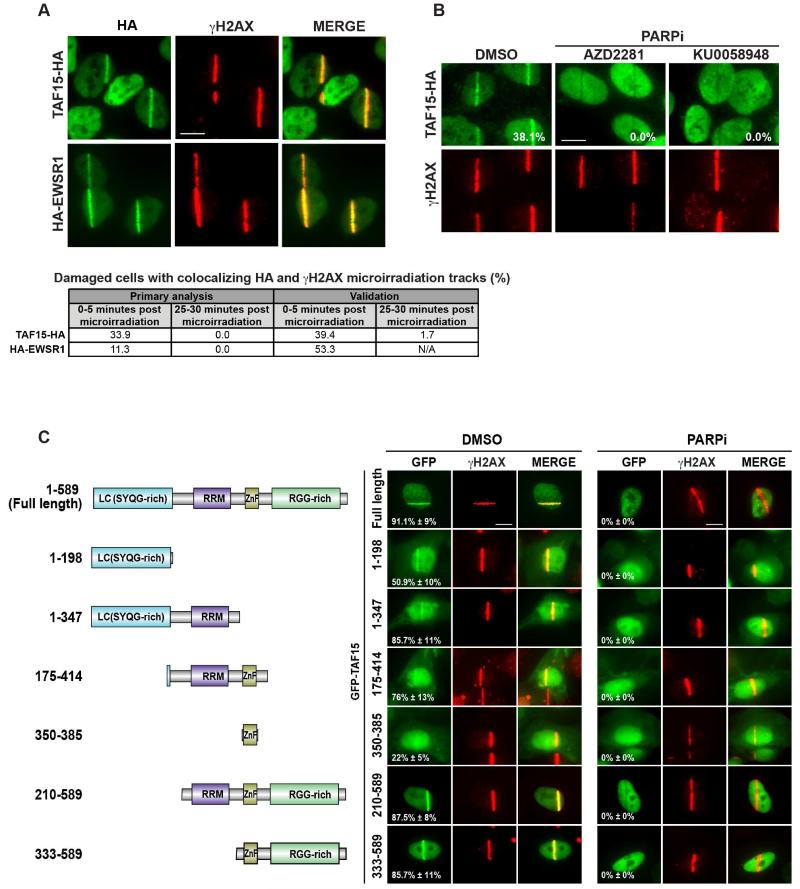
Among our strongest candidates were TAF15 and EWSR1, two members of the FET (FUS, EWSR1, TAF15) family of proteins (formerly called TET). These proteins have similar domain structures, each with a low complexity (LC)/transcriptional activation domain at the N-terminus, a zinc finger domain, an RNA recognition motif (RRM), and RGG-rich regions that could also mediate RNA-binding (Kovar, 2011). FET proteins are ubiquitously expressed and notorious for their presence at the break points of chromosomal translocations associated with human cancers, including Ewing’s sarcomas, liposarcoma, chondrosarcoma, and various leukemias (Kovar, 2011). In our validation analysis, TAF15 and EWSR1 yielded damage tracks colocalizing with γH2AX in 39.4% and 53.3% of damage cells, respectively (Figure 3A and Table S5). Endogenous TAF15 localization was confirmed using antibodies against TAF15 protein (Figure S4). The third member of the FET protein family (FUS) was not included in our library; however, consistent with our results, both FUS and EWSR1 were recently observed to accumulate at sites of DNA damage (Mastrocola et al., 2013; Qiu et al., 2014; Wang et al., 2013). Interestingly, TAF15 and FUS were also among the top 10 proteins recently identified by proteome-wide mass-spectrometry to be PARylated after exposure to genotoxic stress (Jungmichel et al., 2013), a list which also included the RNA-binding protein RBMX and transcription factor ZNF384. We previously found RBMX to be recruited to sites of DNA damage in a transient and PARP-dependent manner (Adamson et al., 2012), and ZNF384 scored for recruitment to DNA lesions in our validation analysis (Figure 2A and Table S5). Of note, ZNF384 has also been observed in recurrent rearrangements with TAF15 or EWSR1 in acute leukemia (Martini et al., 2002). Similar to RBMX, TAF15 damage tracks observed in our primary and validation analyses were diminished by 25-30 minutes post microirradiation (Figure 3A), and additional work revealed that TAF15 recruitment is also PARP-dependent (Figure 3B). Furthermore, in the absence of PARP, microirradiation of cells expressing HA-TAF15 produced antistripes as was previously reported for a number of RNA-proteins (Chou et al., 2010; Adamson et al., 2012; Beli et al., 2012).
Figure 3.
Analysis of TAF15 localization to chromatin in response to DNA damage. (A) Cells expressing TAF15 or EWSR1 HA fusion proteins were microirradiated and immunostained with antibodies against HA and γH2AX (0-5 minutes after microirradiation). Each image was individually adjusted during exposure and processing to best demonstrate protein localization. Scale bars, 10 μm. Table (below) lists the percentages of damaged cells with γH2AX microirradiation tracks observed to colocalize with accumulation of the indicated HA fusion protein; these data are from screen primary and validation analyses found in Tables S4 and S5. For TAF15, images were collected from an independent experiment. N/A indicates data not collected. (B) Cells expressing TAF15-HA were treated with PARP inhibitors (5 μM AZD2281 or 1 μM KU0058948) or DMSO for 1 hour, microirradiated for 5 minutes, and then fixed immediately for immunostaining with antibodies against HA and γH2AX. Scale bars, 10 μm. (C) Structure-function analysis of TAF15 localization to damaged chromatin. Cells expressing GFP-TAF15 (full length or the indicated truncations) were treated with DMSO (right panel) or PARP inhibitor (5 μM AZD2281, left panel) for 1 hour, microirradiated for 5 minutes, and then fixed immediately for immunostaining with antibodies against γH2AX. Scale bars, 10 μm. Data represent the mean percentages of damaged cells with GFP and γH2AX colocalizing damage tracks ± Standard deviation (N=2).
To gain further insight into the mechanism by which TAF15 is recruited to sites of DNA damage, we examined a series of GFP-tagged TAF15 truncations (Figure 3C). Interestingly, we observed that two non-overlapping truncation mutants of the N- and C-terminus localize to DNA regions damaged by microirradiation (Figure 3C). One fragment contains the entire LC domain while the second contains an RGG-rich domain. A third fragment that contains just 24 overlapping residues with the N-terminal fragment and includes the RRM and ZnF motifs also localizes to damaged regions; however, a fragment with the ZnF motif but lacking the RRM demonstrated impaired localization. Preincubation of cells expressing full-length GFP-TAF15 or these mutants with the PARP inhibitor AZD2281, completely abolished localization of the full-length fusion protein and all of the different truncated versions to microirradiation tracks (Figure 3C). Thus, two and possibly three, independent regions of TAF15 are capable of localizing to sites of DNA damage in a PARP-dependent manner.
Nuclear receptors localize to sites of DNA breaks in a PARP-dependent manner
One factor examined was ESRRA, a member of a subfamily of orphan nuclear receptor transcription factors. ESRRA shares significant homology with the estrogen receptor (ESR1) and thus is referred to estrogen receptor-related (ESRR). The other subfamily members are ESRRB and ESRRG. These proteins have established roles as regulators of gene networks and are involved in energy homeostasis (Lanvin et al., 2008). To date, there is no evidence implicating any subfamily members in the DNA damage response. We found an ESRRA-HA fusion protein was recruited to DNA damage tracks colocalizing with γH2AX in 61% of the damaged cells (Figures 4A and 4C). Colocalization tracks were visible at 0-5 minutes. These diminished by 25-30 min. post microirradiation (data not shown). ESRRB-HA and ESRRG-HA also localize DNA damage (Figures 4A and 4C). Both the frequency of visible ESRRB-HA and ESRRG-HA accumulation (48% and 46%, respectively) and its transient nature (Figure 4C) were similar to ESRRA recruitment (data not shown).
Figure 4.
Localization of steroid hormone receptor family members to damaged chromatin. (A) Accumulation of ESRR family proteins at UV laser-induced DNA damage tracks. Cells expressing ESRRA-, ESRRB-, or ESRRG-HA (C-terminal) fusion proteins were microiradiated for 5 minutes and fixed immediately for immunostaining with antibodies against HA and γH2AX. Each image was individually adjusted during exposure and processing to best demonstrate protein localization. Scale bars, 10 μm. (B) Cells treated with PARP inhibitor (5 μM AZD2281) for 1 hour prior to microiradiation were processed and analyzed similar to those in (A). (C) Quantification of microirradiated cells with ESRRA-, ESRRB- or ESRRG-HA accumulation at γH2AX damage tracks. Data represent the mean percentages of damaged cells with colocalizing tracks ± standard deviation (N=3). (D) Localization of seven additional nuclear receptors from 3 different subfamilies after UV laser-induced DNA damage. Cells and images processed as described in (A).
Three members of another subfamily of orphan nuclear receptors (NR4A) were recently shown to be recruited to IR- and UV-induced DNA damage foci in a PARP-dependent manner (Jagirdar et al., 2013; Malewicz et al., 2011). To test whether PARP activity is similarly required for ESRRA, ESRRB, and ESRRG, we evaluated their damage-induced recruitment after pretreatment with a PARP inhibitor (AZD2281) and found the frequency and intensity of colocalization tracks were greatly diminished compared to untreated controls (Figures 4A-4C).
Given that six nuclear receptors from two different subfamilies accumulate at DNA damage sites, we decided to test several other nuclear receptors for their ability to localize to DNA damaged by UV microirradiation. In total, we tested seven more nuclear receptors (RARB, RORC, THRB, NR2C2, NR5A2, RXRG, NCOA5). Surprisingly, 6 of these (with the exception of NCOA5) demonstrated recruitment to DNA damage as HA-fusion proteins (Figure 4D).
Multiple classes of transcription factors localize to damaged chromatin in a manner dependent on intact DNA-binding domains
The observation that many nuclear receptors localize to sites of DNA damage, along with the fact that our list of validated hits is enriched for transcription factors, prompted us to speculate that localization to DNA damage in these cases may be in part linked to a common property shared by proteins with transcription factor activity. To test this possibility we compiled a list of 35 additional transcription factors (Table 1). These transcription factors were selected based on the sole criterion that they have no known involvement in the cellular response to DNA damage. Otherwise, the list is composed of transcription factors of various types, some with tissue specific expression patterns and/or defined developmental roles, and others with wide expression patterns and functions. As most transcription factors from the primary screen maximally localized to UV-microirradiation sites at the 0-5 min. time point, we chose to test their localization at 0-5 minutes post microirradiation. Strikingly, 71% (25/35) accumulated at UV laser-induced micro-irradiation tracks (Table 1, Figures 5F and S2). The positive scoring group could be divided into 3 categories based on the intensity of the fluorescent signal. Of these, 28% (7/25) formed high intensity stripes, 28% (7/25) medium, and 44% (11/25) low intensity (Table 1 and Figure S2). Among this group, the percentage of damaged cells with candidate accumulation at γH2AX tracks varied between 17% and 80% (Figure 5F). Together, these results demonstrate that localization of transcription factors to site of DNA damage is a wide-ranging phenomenon.
Table 1. A set of 35 transcription factors evaluated for localization to damaged chromatin.
Proteins were selected from 836 transcription factors present in the ORFeome collection (hORFeome V8.1), excluding ORFs with a previous association to DNA damage responses, DNA repair, and related GO terms. Selected ORFs were stably expressed as N-terminal HA-fusions in U2OS cells. These cells were microirradiated for 5 minutes, fixed immediately, and immunostained with antibodies against HA and γH2AX. For each transcription factor, the intensity of the fluorescent signal from the colocalizing tracks were evaluated by eye and assigned to one of four intensity categories (Strong, Medium, Weak, or Negative) based on the researcher’s discretion.
| Gene symbol |
ORF ID |
Accession Version |
Localization to DNA damage |
Intensity of colocalization with γH2AX |
DNA-binding domains identified using Pfam or blastp |
|---|---|---|---|---|---|
| CTCF | 6173 | BC014267.2 | Yes | Strong | Zinc-finger double domain (7); C2H2-type zinc finger (2) |
| HOXC10 | 1844 | BC001293.1 | Yes | Homeobox (1) | |
| ZNF625 | 5197 | BC007868.2 | Yes | Zinc-finger double domain (7) | |
| HOXA9 | 5870 | BC006537.2 | Yes | Homeobox (1) | |
| ZNF366 | 56668 | BC121053.1 | Yes | Zinc-finger double domain (2); C2H2-type zinc finger (8) | |
| SATB2 | 52671 | BC098136.1 | Yes | CUT domain (2); Homeobox (1) | |
| HOXC9 | 11938 | BC053894.1 | Yes | Homeobox (1) | |
|
| |||||
| NEUROG1 | 2872 | BC008687.1 | Yes | Medium | Helix-loop-helix (1) |
| ZNF219 | 2058 | BC036105.1 | Yes | Zinc-finger double domain (1); C2H2-type zinc fingers (7) | |
| HOXD10 | 13989 | BC069619.1 | Yes | Homeobox (1) | |
| SOX5 | 10885 | BC060773.1 | Yes | HMG box (1) | |
| ZNF324** | 4991 | BC007717.2 | Yes | ||
| PITX2 | 6983 | BC013998.2 | Yes | Homeobox (1); OAR domain (1) | |
| NR1H4 | 54905 | BC130573.1 | Yes | C4-type zinc finger (2) | |
|
| |||||
| MYOG | 11922 | BC053899.1 | Yes | Weak | Myogenic Basic domain (1); Helix-loop-helix (1) |
| TSHZ3 | 56855 | BC127095.1 | Yes | C2H2-type zinc finger (4) | |
| STAT5A | 8201 | BC027036.1 | Yes | STAT_bind (1) | |
| HOXA10 | 14955 | BC071843.1 | Yes | Homeobox (1) | |
| ZNF786 | 55871 | BC128392.1 | Yes | Zinc-finger double domain (9); C2H2-type zinc finger (1) | |
| ZIM3 | 56365 | BC114503.1 | Yes | Zinc-finger double domain (9) | |
| MYCL1** | 7111 | BC011864.2 | Yes | ||
| TCF7L2 | 9999 | BC032656.1 | Yes | HMG box (1) | |
| ZNF184 | 8172 | BC022992.1 | Yes | Zinc-finger double domain (17); C2H2-type zinc finger (1) | |
| ESR2 | 1485 | BC024181.2 | Yes | C4-type zinc finger (2) | |
| C17orf49 | 11390 | BC040036.1 | Yes | SANT domain (1) | |
|
| |||||
| CTCFL | 54781 | BC130486.1 | No | Negative | Zinc-finger double domain (7); C2H2-type zinc finger (2) |
| ZNF174** | 2748 | BC000876.1 | No | ||
| WT1 | 1923 | BC032861.2 | No | Zinc-finger double domain (3) | |
| ZNF434** | 4197 | BC002859.2 | No | ||
| HOXB1** | 55783 | BC096192.1 | No | ||
| MYPOP | 72064 | BC044311.1 | No | Myb/SANT-like DNA-binding domain (1) | |
| MYB | 12304 | BC064955.1 | No | Myb/SANT-like DNA-binding domain (2) | |
| PPARG | 2704 | BC006811.1 | No | C4-type zinc finger (2) | |
| MIER2 | 9505 | BC028203.1 | No | Myb/SANT-like DNA-binding domain (1); ELM2 domain (1) | |
| TADA2A | 3358 | BC001172.1 | No | Myb/SANT-like DNA-binding domain (1) | |
cDNAs lack DNA-binding domains annotated by Pfam and blastp
Figure 5.
Transcription factor localization to damaged chromatin is pervasive and DNA-binding domain-dependent. (A) Schematic of deletions in the DNA-binding region of the homeodomain of HOXC10. (B) Structure-function analysis of HOXC10. Cells expressing the indicated HA-HOXC10 fusion proteins (illustrated in A) were microiradiated for 5 minutes and fixed immediately for immunostaining with antibodies against HA and γH2AX. Each image was individually adjusted during exposure and processing to best demonstrate protein localization. Scale bars, 10 μm. (C) Schematic of HA-PITX2 DNA-binding domain mutants and corresponding structure-function analysis. Cells expressing the indicated deletion mutants were treated and analyzed as in (B). (D) Schematic of HA-NR1H4 DNA-binding domain mutants and corresponding structure-function analysis. Cells expressing the indicated deletion mutants were treated and analyzed as in (B). (E) Quantification of damaged cells with colocalizing HA and γH2AX tracks from the experiments depicted in (B-D). Data represent the mean percentages ± standard deviation of damaged cells with colocalizing tracks (N=3). (F) Cells expressing HA-fusion proteins of the indicated transcription factors were microiradiated for 5 minutes, fixed immediately for immunostaining with antibodies against HA and γH2AX, and quantified. Scale bars, 10 μm. Data represent the percentages of damaged cells with colocalizing HA and γH2AX tracks.
The ability to bind DNA is a defining feature of transcription factors. Although different types of DNA-binding domains (DBDs) can dramatically vary in structure, in general, they all convey high affinity toward DNA and could, therefore, potentially mediate the localization of transcription factors to DNA damage sites. To determine if DBDs are indeed the common trait that facilitates recruitment, we generated mutated versions of three transcription factors (HOXC10, PITX2, NR1H4) with disrupted DNA-binding domains and tested their ability to localize to microirradiation tracks.
HOXC10 is a member of the Hox family of transcription factors, which play an important role in morphogenesis in all multi-cellular organisms (Philippidou and Dasen, 2013). HOXC10 contains a homeodomain, a DNA-binding domain that is highly conserved among members of the Hox family. The HOXC10 homeodomain has two residue clusters that make direct contact with DNA (LaRonde-LeBlanc and Wolberger, 2003). We generated a series of three N-terminal HA-tagged HOXC10 deletion mutants: (1) HA-HOXC10 del269-275 that disrupts the N terminal cluster and lacks seven residues, (2) HA-HOXC10 del311-324 that disrupts the C-terminal cluster and lacks 14 residues, and (3) HA-HOXC10 del269-324 that disrupts the entire homoeodomain and lacks 55 residues (Figure 5A). Each deletion impairs the accumulation at microirradiation-induced DNA damage (Figures 5A-5B and 5E). We observed that the fluorescence intensity of the HA-HOXC10 del269-275 signal at damage sites was much weaker and the fraction of cells with visible tracks colocalizing with γH2AX was also reduced by 47% (Figures 5A-B and 5E). Deleting the C-terminal cluster had an even more dramatic effect with 67% reduction of colocolizing stripes compared to the full-length protein. Moreover, the HA-HOXC10 del269-324 colocalization signal was barely visible and the fraction of cells with visible colocalization tracks was reduced by 96% compared to the wild-type protein (Figures 5A-B and 5E).
We also evaluated PITX2, another homoeodomain-containing transcription factor, using a similar structure-function analysis. PITX2 plays an important role in the establishment of the left to right axis of various organs during embryogenesis (Varon and Havsteen, 1990). We generated a single mutant version of PITX2 (HA-PITX2_del126-184) that lacks 59 residues corresponding to the entire homeodomain of the protein. In the absence of this DNA-binding domain, localization of the protein to microirradiation tracks was nearly abolished: we observed tracks of colocalizing HA-PITX2_del126-184 and γH2AX in only 15% of irradiated cells compared to 82% for the full-length protein, and for these, the fluorescent signal was extremely faint (compared to the relatively strong signal of the wild-type protein) (Figures 5C and 5E).
We next examined the localization of NR1H4 (the nuclear receptor family subfamily 1, group H, member 4). This protein responds to bile acid ligands by binding its cognate DNA-binding sites through a pair of Zn-fingers and activating transcription (Staudinger et al., 2013). Similar to HOXC10 and PITX2, deletion of the first Zn-finger of NR1H4 (amino acids 129-152) significantly diminished its ability to localize to damaged chromatin (Figures 5D and 5E). HOXC10 and all of its derivatives are dependent upon PARP for localization (Figure 6E). Thus, the ability to bind DNA is critical for the localization of these transcription factors.
Figure 6.
PARP-dependent localization is frequent for proteins identified by our screening analysis. (A) Cells expressing the indicated HA fusion proteins were treated with a PARP inhibitor (5 μM AZD2281) or DMSO for 1 hour, microirradiated for 5 minutes, and fixed immediately for immunostaining with antibodies against HA and γH2AX. Each image was individually adjusted during exposure and processing to best demonstrate protein localization. The top 16 images are of validated candidates identified by screening. The lower panel displays a set of six transcription factors described in Table 1. Scale bars, 10 μm. (B) Cells expressing the indicated HA fusion proteins were processed as in (A) except that PARG was depleted by siRNA in order to strengthen candidate localization tracks (Adamson et al., 2012). Four of these 13 HA-fusion proteins (RPS27A, DTL, CCDC82, and GATAD1) were also tested without PARG depletion and are shown in (A). Images were processed as in (A) (C) Quantification of cells depicted in (A). Data represent the percentages of damaged cells with colocalizing HA and γH2AX tracks (D) Quantification of cells depicted in (B). (E) Localization of HA-HOXC10 fusion proteins, and two of its DNA-binding domain mutants depicted in Figure 5A, in the presence or absence of PARP inhibitor. Cells and images were processed as in (A).
The pattern of early recruitment among positives correlates with PARP dependency
To get a sense of the prevalence of PARP-dependent accumulation at sites of DNA damage among our set of proteins that localize to microirradiation-induced damage tracks, we reevaluated 31 in the presence and absence of the PARP inhibitor AZD2281 (Figures 6A-D). Of these, 8 showed complete or partial PARP-independence (RAD18, RPL27A, DTL, CCDC82, UHRF2, CTCF, RFC4 and POU2F2). However, the remaining 23 factors demonstrated nearly complete PARP-dependency for recruitment (74%). Proteins with PARP-dependent localization to damaged chromatin are quickly and transiently recruited (Li and Yu, 2014), and intriguingly, data from our primary screen suggests that many of our positives were similarly transiently recruited. In particular, nearly two-thirds of the positives that were evaluated at both 0-5 and 25-30 minutes demonstrated tracks at only the early time point. Together, these conclusions suggest that many of our positives overall may be strongly PARP-dependent for localization. Therefore, we hypothesize that any factor that similarly accumulates transiently at microirradiation tracks are also likely to be PARP-dependent.
PARP1 induces chromatin changes at sites of UV laser microirradiation
A potential explanation for how DNA binding proteins relocalize to laser induced UV damage is that the DNA becomes accessible by means of relaxing the chromatin structure at the site of the damage. To test this notion, we examined the intensity of DNA staining with Hoechst upon laser striping. UV laser microirration resulted in a dark stripe pattern due to reduced staining along the path of the beam, which is indicative of chromatin decompaction (Mazumder et al., 2008) (Figure S3). Importantly this “antistripe” pattern was mediated by PARP1, as inhibition of PARP activity or depletion of PARP1 using siRNA prevented the formation of this pattern. One possible mechanism for chromatin decompaction would be the recruitment of chromatin remodeling factors. We have attempted to identify factors that might mediate this chromatin expansion and have depleted them using RNAi and examined localization of DNA binding proteins. We examined ALC1, CHD4, INO80, TIP60, KAT2A, SMARCC1, SMARCC2, BAZ1B, EZH2, SUZ12 and p300 and none of these appeared to affect localization after UV laser irradiation (data not shown). While the effects could be due to as yet unidentified chromatin remodelers or a combination of factors we tested that act redundantly, an alternative hypothesis is that PARP1 modification of chromatin can directly affect its structure. While the precise nature of the chromatin alteration is not known, there is clearly a physical change in the DNA and the possible mechanisms allowing that are discussed below.
DISCUSSION
The localization of proteins to sites of DNA damage is thought to be a hallmark of involvement in DNA damage sensing and repair processes. Here, we present a focused screen of a large set of proteins, enriched for factors with potential roles in DNA repair and other nuclear processes, that identified a set of proteins that localize to sites of UV laser microirrradiation-induced DNA damage at either 0-5 minutes or 25-30 minutes post irradiation. Of 576 genes evaluated in our primary screen from our focused library, 179 appeared positive (31%), and of these, we retested 151 to identify a set of 113 genes encoding proteins that localize to damaged chromatin. Interestingly, approximately two-thirds of candidates evaluated were only detected at damage sites in the early time point. Additionally, DAVID analysis using molecular function GO terms implicated transcriptional control in the DNA damage response. Another 9 nuclear receptors, 25 additional transcription factors, 1 additional component of the BAF complex, were also found to localize to sites of damaged chromatin, bringing the number of genes encoding these damage localizing factors to 148. The majority of these had not previously been reported to localize to sites of DNA damage.
A number of chromatin remodeling proteins are known to localize to sites of DNA damage, for example components of the NuRD and Polycomb complexes (Chou et al., 2010; Polo et al., 2010; Smeenk et al., 2010). Here we identified one isoform of the chromodomain and chromoshadow domain-containing protein HP1 and the HP1-binding protein 3 (HP1BP3), and 5 components of mammalian SWI/SNF complexes (DPF2, BCL7A, BCL7C, SMARCC1 and PHF10) (Kadoch et al., 2013). Consistent with these results, previous reports demonstrated that BRG1, a component of SWI/SNF complexes, colocalizes with γH2AX (Park et al., 2006) and binds both γH2AX nucleosomes (Lee et al., 2010) and BRCA1 (Bochar et al., 2000). Mammalian SWI/SNF complexes are involved in chromatin remodeling and contain known tumor suppressors. Together their subunits have been reported to be mutated in nearly 20% of cancers (Kadoch et al., 2013). Additionally, the PBAF-specific SWI/SNF subunit BRD7 has been shown to regulate the function of p53, a key mediator of DNA damage-induced cell cycle arrest, and both BRD7 and an additional subunit (BAF180) are required for oncogene-induced and/or replicative senescence (Burrows et al., 2010). A role in responding to DNA damage could also be important for this complex.
Among transcription factors strongly recruited to sites of damage is TAF15. TAF15 is a component of TFIID (Bertolotti et al., 1996), a multiprotein general transcription factor complex composed of 13 evolutionarily conserved TBP-associated factors (TAF proteins) and the TATA-binding protein (TBP) (Bhaumik, 2011). TFIID is a component of the RNA polymerase II (RNAPII) preinitiation complex, which positions RNA polymerase for transcriptional start (Thomas and Chiang, 2006). TAF15 is also a member of the FET family of DNA/RNA binding proteins, the other members of which are FUS and EWSR1 (Kovar, 2011). These proteins possess a number of conserved structural features, including an RNA-binding domain and a characteristic low complexity (LC) domain. The LC domains bind the CTD region of RNAPII and act as transcriptional activation domains (Kwon et al., 2013). They are also partners in translocations with a variety of different DNA-binding domains in cancer (Kovar, 2011). Interestingly, LC domains can form hydrogels (composed of polymeric, amyloid-like fibers) that self-assemble in a concentration-dependent manner and may heterotypically trap other LC-containing proteins (Kato et al., 2012). Importantly, accumulation of FET proteins within neuronal and glial cytoplasmic inclusions is a pathological characteristic of amyotrophic lateral sclerosis (ALS) associated with mutations in FUS and in subtypes of frontotemporal lobar degeneration (FTLD), with FUS accumulation observed in ALS and all FET proteins observed in FTLD inclusions (Mackenzie and Neumann, 2012; Neumann et al., 2011). TAF15 has also been described as a candidate ALS protein (Couthouis et al., 2011).
Several links between FET proteins and DNA damage have been previously reported. EWSR1 is important for resistance to IR and UV (Hurov et al., 2010; Paronetto et al., 2011), and alternative–splicing mediated by EWSR1 is suppressed after exposure to UV, perhaps due to UV-induced relocalization to the nucleolus (Paronetto et al., 2011). Most relevant to our work, both EWSR1 and FUS have been shown to localize rapidly and transiently to sites of DNA damage (Mastrocola et al., 2013; Wang et al., 2013). FUS localization is PARP-dependent (Mastrocola et al., 2013), and familial ALS FUS mutants cause spontaneous DNA damage implying a role in DNA repair (Qiu et al., 2014). Furthermore, all three members of the FET protein family are PARylated under genotoxic stress conditions (Jungmichel et al., 2013).
Like FUS, we found accumulation of TAF15 to sites of DNA damage is PARP-dependent. Structural analysis of the dependency of TAF15 domains for localization revealed the presence of two separable domains that can independently localize to sites of microirradiation, one of which is the LC domain. This is reminiscent of the structure/function analysis of another RNA binding protein, RBMX, which also accumulates at microirradiated chromatin in a PARP-dependent manner and has two separable domains that can independently localize (Adamson et al., 2012). These results suggest the possibility that these proteins each contact multiple different surfaces on PAR structures, which could allow the bridging of different PAR branches, assembling them into a larger structure or perhaps even acting as a “glue” to hold distinct structures together. Such an assembly of PAR structures from both sides of a DSB could transiently hold the broken ends of DNA together to facilitate DNA repair. Indeed, while PARP is primarily thought to coordinate repair of single strand breaks, there is evidence to suggest that PARP also promotes DSB repair via a Ligase III-dependent alternative NHEJ pathway (Ciccia and Elledge, 2010). The LC domains could also serve a bridging role based on their proposed ability to engage in associations with other LC sequences (Kwon et al., 2013). The degree to which FET protein involvement with PARP-generated structures in response to DNA damage might influence neuronal survival in ALS remains to be determined; although of note, the accumulation of DNA damage in neurons is thought to play an important role in neuronal dysfunction (Ciccia and Elledge, 2010).
We identified DNA-binding proteins from multiple families that localize to damaged chromatin, including homeodomain-containing and Zn finger-containing proteins. Additionally we found that ESRRA, the nuclear receptor class orphan steroid hormone-related, is similarly recruited. Direct examination of other related factors revealed that 8 out of 9 different nuclear receptors from multiple families can localize to microirradiation tracks; these are ESRRB, ESRRG, RARB, RORC, THRB, NR2C2, NR5A2, and RXRG. This suggests that such localization may be a general property of transcription factors. A direct test of 35 additional transcription factors revealed that 71% can localize to UV laser tracts. This high incidence of positives suggests that the property of DNA-binding itself may be responsible for localization. Mutation of the DNA-binding domains of three different proteins revealed that these domains are responsible for protein localization to damaged chromatin, providing support for this hypothesis.
It is known that ectopically expressed DNA-binding proteins tend to bind regions previously established as DNAse hypersensitive sites (Wu et al., 2014), which have reduced nucleosome density established by so-called pioneer transcription factors (Zaret and Carroll, 2011). Thus, the remodeling of chromatin structure may facilitate such DNA-binding. One hypothesis for how this might be related to our localization findings rests upon our observation that several DNA-binding proteins show PARP-dependent localization (Figure 6A). If PARP were to initiate chromatin remodeling to free up DNA, DNA-binding proteins might have greater access. Several chromatin remodeling factors are recruited damaged chromatin in a PARP-dependent manner, including components of the NuRD complex (Chou et al., 2010; Polo et al., 2010) and the SNF2-related protein CHD1L (ALC1) protein (Ahel et al., 2009; Gottschalk et al., 2009). Thus, PARylation may reconfigure chromatin structure at sites of UV microbeam irradiation. This could then result in exposed DNA that could then provide accessible binding sites for transcription factors. Our experiment examining the Hoechst staining pattern of nuclear DNA in response to UV laser treatment supports a physical change in chromatin at the sites of DNA damage. In support of these findings, micro-irradiation of spots in nuclear DNA have been recently shown to expand and then shrink in response to UV laser treatment (Burgess et al., 2014). While PARP was not examined in that study, the physical changes they observe happen on a time scale consistent with PARP activity. While these effects could be due to as yet unidentified chromatin remodelers or a combination of factors we tested that act redundantly, an alternative hypothesis is that PARP1 modification of chromatin can directly affect its structure. PARP has been shown to relax chromatin structure in vitro and in Drosophila in vivo (Poirier et al., 1982; Tulin and Spradling, 2003). Furthermore, it has been shown that PARP1 can be incorporated into chromatin as a structural component that is interchangeable with the linker histone H1 (Kim et al., 2004). (ADP-ribosyl)ation of this incorporated PARP1 result in disassociation of nucleosomes and subsequent decompaction of chromatin at that site (Kim et al., 2004). Thus, PARP1’s activity on chromatin might directly relax chromatin structure leading to greater accessibility for DNA binding factors.
The large number of factors that we have found to localize to sites of damage was unexpected. In particular, the fact that 71% of independently selected transcription factors accumulated at these sites begs the question of what the physiological role of any particular factor might be. It is possible that rapid association of such factors generate a burst of transcription at the break sites, and it has been suggested that DICER-dependent siRNAs are rapidly synthesized at the sites of DSBs (d’Adda di Fagagna, 2014). However, the mechanism by which this double-stranded RNA is made is unknown. These factors could be involved in such processes. However, it is also possible that the accumulation of any particular DNA-binding factor has little or no physiological relevance but is merely associating with newly and possibly transiently accessible DNA. Nevertheless, what is clear is that the roles of these factors in DNA damage-relevant processes remains to be determined. Despite this, it is likely that many of the factors identified in this work are likely to have physiological relevance, and as such our results will be a valuable resource for future study of the DNA damage response.
MATERIALS and METHODS
Cell culture
Human HEK293T were grown in Dulbecco’s Modified Eagle Medium and U2OS cells were grown in McCoy’s 5A both media supplemented with 9% fetal bovine serum (FBS) (Clontech), 99 units / ml of penicillin, and 0.1 mg / ml streptomycin (Invitrogen). Transduced cell lines were selected and carried in media supplemented with 1 μg/ml puromycin.
Antibodies and reagents
Primary antibodies used for immunofluorescence were FITC-conjugated anti-HA (Bethyl Laboratories Inc, A190-108F), anti-phospho-Histone H2A.X (Ser139) (EMD Millipore, 05-636), anti BAF155 (SMARCC1) (Santa Cruz Biotecnology Inc, sc-10756), anti-TAF15 (Abcam, ab134916). Secondary antibodies used for immunofluorescence were: Goat anti-Mouse IgG Alexa Fluor® 594 conjugate (Life Technologies, A11005) for visualization of γH2AX and Goat anti-Rabbit IgG Alexa Fluor® 488 conjugate (Life Technologies, A11034) for visualization of SMARCC1 and TAF15. The PARP inhibitors AZD2281 (Axon Medchem) and KU0058948 (KuDOS Pharmaceuticals) were used as indicated. PARP1 was depleted from the cells using siGenome siRNA pool (Dharmacon, MU-006656-01-0002), and anti-PARP1(Cell Signaling, #9532) was used to detect PARP1 knockdown efficiency.
For more information on cell line construction and processing for primary screening, library cloning, candidate validation and other analysis, see supplemental methods.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ronald Bernardi (Baylor College of Medicine) and Michael Leichter (Rheinische Akademie, Cologne) for reagents. We thank Max A. Horlbeck for bioinformatics advice. B.A. is an HHMI Fellow and L.P.V. is a Damon Runyon Fellow. This work was supported by grants from the NIH: GM44664 to S.J.E, 1R01CA178039⍰to T.F.W., and AG011085 to J.W.H. and grants from the DOD BCRP: BC120604 to T.F.W.⍰S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Poly(ADP-ribose) polymerases in double-strand break repair: Focus on PARP1, PARP2 and PARP3. Exp Cell Res. 2014 doi: 10.1016/j.yexcr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics. 2010;9:1314–1323. doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez VP, Maniwa Y, Tappin I, Ozato K, Yokomori K, Hurwitz J. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc Natl Acad Sci U S A. 2003;100:10237–10242. doi: 10.1073/pnas.1434308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR. Distinct regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. Biochim Biophys Acta. 2011;1809:97–108. doi: 10.1016/j.bbagrm.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoumik A, Takahashi S, Breitweiser W, Shiloh Y, Jones N, Ronai Z. ATM-dependent phosphorylation of ATF2 is required for the DNA damage response. Mol Cell. 2005;18:577–587. doi: 10.1016/j.molcel.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- Burgess RC, Burman B, Kruhlak MJ, Misteli T. Activation of DNA damage response signaling by condensed chromatin. Cell Rep. 2014;9:1703–1717. doi: 10.1016/j.celrep.2014.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc Natl Acad Sci U S A. 2010;107:14280–14285. doi: 10.1073/pnas.1009559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell. 2012;47:396–409. doi: 10.1016/j.molcel.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C, McDonald ER, 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–1317. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol. 2014;24:171–178. doi: 10.1016/j.tcb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Dinant C, Luijsterburg MS. The emerging role of HP1 in the DNA damage response. Mol Cell Biol. 2009;29:6335–6340. doi: 10.1128/MCB.01048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–1950. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagirdar K, Yin K, Harrison M, Lim W, Muscat GE, Sturm RA, Smith AG. The NR4A2 nuclear receptor is recruited to novel nuclear foci in response to UV irradiation and participates in nucleotide excision repair. PLoS One. 2013;8:e78075. doi: 10.1371/journal.pone.0078075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol Cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoronenkova SV, Dianova, Ternette N, Kessler BM, Parsons JL, Dianov GL. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell. 2012;45:801–813. doi: 10.1016/j.molcel.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kovar H. Dr. Jekyll and Mr. Hyde: The Two Faces of the FUS/EWS/TAF15 Protein Family. Sarcoma. 2011;2011:837474. doi: 10.1155/2011/837474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanvin O, Bianco S, Vanacker JM. Estrogen-receptor-related receptors and hormone-dependent cancers. Adv Exp Med Biol. 2008;617:235–243. doi: 10.1007/978-0-387-69080-3_22. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc NA, Wolberger C. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 2003;17:2060–2072. doi: 10.1101/gad.1103303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yu X. The role of poly(ADP-ribosyl)ation in DNA damage response and cancer chemotherapy. Oncogene. 2014 doi: 10.1038/onc.2014.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M. FET proteins in frontotemporal dementia and amyotrophic lateral sclerosis. Brain Res. 2012;1462:40–43. doi: 10.1016/j.brainres.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Malewicz M, Kadkhodaei B, Kee N, Volakakis N, Hellman U, Viktorsson K, Leung CY, Chen B, Lewensohn R, van Gent DC, et al. Essential role for DNA-PK-mediated phosphorylation of NR4A nuclear orphan receptors in DNA double-strand break repair. Genes Dev. 2011;25:2031–2040. doi: 10.1101/gad.16872411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD, Liu S, Jimenez AE, Jin J, Zou L. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol Cell. 2014;53:235–246. doi: 10.1016/j.molcel.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko M, Vlassis A, Guialis A, Leichter M. Domains involved in TAF15 subcellular localisation: dependence on cell type and ongoing transcription. Gene. 2012;506:331–338. doi: 10.1016/j.gene.2012.06.088. [DOI] [PubMed] [Google Scholar]
- Martini A, La Starza R, Janssen H, et al. Recurrent rearrangement of the Ewing’s sarcoma gene, EWSR1, or its homologue, TAF15, with the transcription factor CIZ/NMP4 in acute leukemia. Cancer Res. 2002;62:5408–5412. [PubMed] [Google Scholar]
- Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem. 2013;288:24731–24741. doi: 10.1074/jbc.M113.497974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Mazumder A, Roopa T, Basu A, Mahadevan L, Shivashankar GV. Dynamics of chromatin decondensation reveals the structural integrity of a mechanically prestressed nucleus. Biophys J. 2008;95:3028–3035. doi: 10.1529/biophysj.108.132274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Neumann M, Bentmann E, Dormann D, Jawaid A, et al. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain. 2011;134:2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell BC, Adamson B, Lydeard JR, Sowa ME, Ciccia A, Bredemeyer AL, Schlabach M, Gygi SP, Elledge SJ, Harper JW. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol Cell. 2010;40:645–657. doi: 10.1016/j.molcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Minana B, Valcarcel J. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol Cell. 2011;43:353–368. doi: 10.1016/j.molcel.2011.05.035. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P, Dasen JS. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Blackford AN, Chapman JR, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol Cell. 2012;45:505–516. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Lee S, Shang Y, Wang WY, Au KF, Kamiya S, Barmada SJ, Finkbeiner S, Lui H, Carlton CE, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124:981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Slabicki M, Theis M, Krastev DB, Samsonov S, Mundwiller E, Junqueira M, Paszkowski-Rogacz M, Teyra J, Heninger AK, Poser I, et al. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol. 2010;8:e1000408. doi: 10.1371/journal.pbio.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Woody S, Sun M, Cui W. Nuclear-receptor-mediated regulation of drug- and bile-acid-transporter proteins in gut and liver. Drug Metab Rev. 2013;45:48–59. doi: 10.3109/03602532.2012.748793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- Varon R, Havsteen BH. Kinetics of the transient-phase and steady-state of the monocyclic enzyme cascades. J Theor Biol. 1990;144:397–413. doi: 10.1016/s0022-5193(05)80083-9. [DOI] [PubMed] [Google Scholar]
- Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci. 2013;16:1383–1391. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.