Abstract
Chromosomal deletions associated with human diseases, such as cancer are common, but synteny issues complicate modeling of these deletions in mice. We use cellular reprogramming and genome engineering to functionally dissect the loss of chromosome 7q [del(7q)], a somatic cytogenetic abnormality present in myelodysplastic syndromes (MDS). We derive del(7q)- and isogenic karyotypically normal induced pluripotent stem cells (iPSCs) from hematopoietic cells of MDS patients and show that the del(7q) iPSCs recapitulate disease-associated phenotypes, including impaired hematopoietic differentiation. These disease phenotypes are rescued by spontaneous dosage correction and can be reproduced in karyotypically normal cells by engineering hemizygosity of defined chr7q segments, in a 20 Mb region. We use a phenotype-rescue screen to identify candidate haploinsufficient genes that might mediate the del(7q)- hematopoietic defect. Our approach highlights the utility of human iPSCs both for functional mapping of disease-associated large-scale chromosomal deletions and for discovery of haploinsufficient genes.
Introduction
Large hemizygous deletions are found in most tumors and might be both hallmarks and drivers of cancer1. Hemizygous segmental chromosomal deletions are also frequent in normal genomes2. Apart from rare prototypic deletion syndromes (e.g. Smith-Magenis, Williams-Beuren, 22q11 deletion syndromes), genome wide association studies (GWAS) have implicated genomic deletions in neurodevelopmental diseases like schizophrenia and autism3, prompting the hypothesis that deletions might account for an important source of the “missing heritability” of complex diseases3, 4.
Unlike translocations or point mutations, chromosomal deletions are difficult to study with existing tools because primary patient material is often scarce and incomplete conservation of synteny (homologous genetic loci can be present on different chromosomes or in different physical locations relative to each other within a chromosome across species) complicate modeling in mice. Dissecting the role of specific chromosomal deletions in specific cancers entails, first, determining if a deletion has phenotypic consequences; second, determining if the mechanism fits a “classic” recessive (satisfying Knudson’s “two-hit” hypothesis) or a haploinsufficiency model and finally identifying the specific genetic elements critically lost. Classic tumor suppressor genes (TSGs) were discovered through physical mapping of homozygous deletions5. More recent data suggest that sporadic tumor suppressor genes are more likely to be monoallelically lost and to function through haploinsufficiency (wherein a single functional copy of a gene is insufficient to maintain normal function)6,7.
MDS are clonal hematologic disorders characterized by ineffective hematopoiesis and a propensity for progression to acute myeloid leukemia (AML)8. Somatic loss of one copy of the long arm of chromosome 7 [del(7q)] is a characteristic cytogenetic abnormality in MDS, well-recognized for decades as a marker of unfavorable prognosis. However, the role of del(7q) in the pathogenesis of MDS remains elusive. The deletions are typically large and dispersed along the entire long arm of chr7ref9. Homology for human chr7q maps to 4 different mouse chromosomes.
Genetic engineering of human pluripotent stem cells (hPSCs) has been used to model point mutations causing monogenic diseases in an isogenic setting10, 11, but not disease-associated genomic deletions. We used reprogramming and chromosome engineering to model del(7q) in an isogenic setting in hPSCs. Using different isogenic pairs of hPSCs harboring one or two copies of chr7q, we characterized hematopoietic defects mediated by del(7q). We used spontaneous rescue and genome editing experiments to show that these phenotypes are mediated by a haploid dose of chr7q material, consistent with haploinsufficiency of one or more genes. We functionally map a 20 Mb fragment spanning cytobands 7q32.3 – 7q36.1 as the crucial region and identify candidate disease-specific haploinsufficient genes using a phenotype-rescue screen. Finally, we show that the hematopoietic defect is mediated by the combined haploinsufficiency of EZH2, in cooperation with one or more out of three additional candidate genes residing in this region. The strategy reported here could be applied to study the phenotypic consequences of segmental chromosomal deletions and for haploinsufficient gene discovery in a variety of human cancers, neurological and developmental diseases.
RESULTS
Generation of del(7q)- and normal isogenic iPSCs
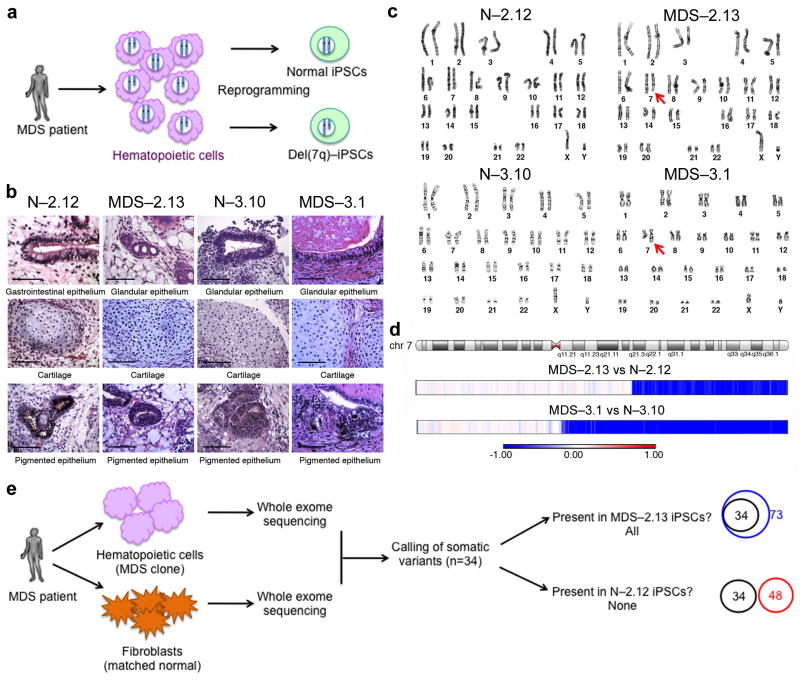
We derived iPSC lines from hematopoietic cells of two patients (patient no. 2 and no. 3) with del(7q)-MDS (Fig. 1 and Supplementary Tables 1 and 2). We reasoned that residual normal hematopoietic cells are likely to co-exist with the MDS clone in the bone marrow (BM) of MDS patients and that we could exploit this to reprogram both del(7q)- and isogenic karyotypically normal cells in parallel. We were able to derive del(7q)- and karyotypically normal (N-) iPSC lines at the same reprogramming round from both patients (Supplementary Table 3). We used an excisable lentiviral vector expressing OCT4 (also known as POU5F1), KLF4, c-MYC (also known as MYCBP) and SOX2 for reprogramming12, 13 and performed vector integration analysis to exclude iPSC lines derived from the same starting cell from being considered independent lines and thus obtain true biological replicate lines from each patient (Supplementary Fig. 1a, b). Karyotyping showed that the iPSC lines harbored identical deletions to those present in the starting patient cells (Fig. 1c), which we mapped by array-based comparative genomic hybridization (aCGH) (Fig. 1d). These iPSC lines met all standard criteria of pluripotency, before and after excision of the reprogramming vector, including expression of pluripotency markers, demethylation of the OCT4 promoter and formation of trilineage teratomas after injection into immunodeficient mice (Fig. 1b and Supplementary Fig. 1c–f). We selected from patients no.2 and no.3 respectively, two and three del(7q)-iPSC lines (MDS-2.13, MDS-2.A3, MDS-3.1, MDS-3.4, MDS-3.5), as well as four and one karyotypically normal iPSC lines (N-2.8, N-2.12, N-2.A2, N-2.A11, N-3.10) before or after vector excision, for further studies (Supplementary Table 1).
Figure 1. Generation of del(7q)- and isogenic karyotypically normal iPSCs from patients with MDS.
(a) Scheme of strategy for the generation of del(7q)- and karyotypically normal iPSCs from patients with MDS.
(b) Histology of representative teratomas derived from one normal (N-) and one del(7q)-MDS-iPSC line derived from each of the two patients (no. 2 and no. 3), showing trilineage differentiation (upper panels: endoderm, middle panels: mesoderm, lower panels: ectoderm). Scale bars, 100 μm.
(c) Representative karyotypes of one normal (N-) and one del(7q)- iPSC line derived from each patient.
(d) aCGH analysis of one representative iPSC line from each patient with a corresponding isogenic normal iPSC line as diploid control. The blue probe color indicates deletion (1 copy) and the white color normal diploid dosage. Both patients harbor large terminal chromosome 7q deletions starting at position 92,781,474 (patient no. 2) and 62,024,527 (patient no. 3).
(e) Whole exome genetic characterization of del(7q)- and karyotypically normal iPSCs from MDS patient no. 2. Venn diagrams on the right: the black circle represents the somatic variants (n=34) identified in the patient BM; the blue and red circles represent the variants found in the MDS-2.13 del(7q)-iPSC line and the N-2.12 isogenic normal iPSC line, respectively. The former completely overlap with the variants of the MDS clone, indicating that this iPSC line captures the entire genetic repertoire of the MDS clone. There is no overlap between the MDS clone variants and the variants found in the normal isogenic line, demonstrating that the latter is derived from a normal residual hematopoietic cell that is unrelated to the cell that gave rise to the MDS clone.
Recent large-scale sequencing studies have shown that MDS undergoes genetic evolution through multiple cycles of mutation acquisition14. To determine whether the karyotypically normal iPSC lines originate from a completely normal hematopoietic cell or from a potential “founding” clone harboring mutations that might have arisen prior to the del(7q), we performed whole exome sequencing of BM mononuclear cells (BMMCs) and fibroblasts (as paired normal sample) from MDS patient no. 2, as well as of one del(7q)- and one normal iPSC line derived from BMMCs of this patient (MDS-2.13 and N-2.12, respectively) and identified the somatic variants. 34 single nucleotide variants (SNVs) were identified with confidence in the BMMCs by comparison to the fibroblast sample, after filtering out intronic variants, variants with less than 30 reads in BMMCs and fibroblasts and variants with variant allele frequency (VAF) higher than 1% in fibroblasts or less than 10% in BMMCs. (Fig. 1e and Supplementary Table 4). Similarly, 73 and 48 somatic SNVs were identified in the del(7q)- and isogenic normal iPSC line, respectively. The del(7q)-iPSC line contained all 34 variants of the MDS clone, whereas the isogenic normal line harbored none. These variants included two genes found recurrently mutated in MDS, SRSF2 and PHF6. Re-sequencing of the rest of the iPSC lines derived from this patient (Supplementary Table 2) showed that all of the del(7q)- (6 out of 6) and none of the karyotypically normal iPSC lines (0 out of 15) harbored these mutations. These results unequivocally demonstrate that the isogenic iPSCs represent normal cells that contain no premalignant lesions and that the del(7q)-iPSCs accurately capture the genetic composition of the MDS clone.
Decreased hematopoietic differentiation of del(7q)-MDS iPSCs
To assess the hematopoietic differentiation potential of the MDS-iPSCs we optimized an embryoid body-based differentiation protocol and monitored the course of differentiation using CD34, a hemato-endothelial marker, and CD45, the key marker of definitive hematopoietic cells15. In this differentiation protocol, normal hESC and iPSC lines typically express CD34 by day 6. Co-expression of CD45 appears by day 10, peaks at day 14, followed by loss of CD34, as the cells become more differentiated along hematopoietic lineages. By day 18, typically more than 90% of cells are CD45+ (Supplementary Fig. 2a). We assessed the intra- (Supplementary Fig. 2a, b, panels I, II and c) and inter- (Supplementary Fig. 2a, b, panels III, IV) line variation in this differentiation process using normal iPSC lines derived from BM cells of patients no. 2 and no. 3, as well as one additional iPSC line derived previously from umbilical cord blood CD34+ cells, CB-3.1-Creref13. On day 10 we observed some inter-experimental variation in expression of CD34 and CD45 markers, whereas little variation was present on day 14 and even less on day 18. Notably, increasing passage number did not alter the differentiation potential (Supplementary Fig. 2a, b, panel II) and distinct iPSC lines derived from the same individual (in the same or different reprogramming experiments) exhibited variation similar to the intra-line variation (Supplementary Fig. 2a,b, compare panels II and III) and substantially lower than that seen in iPSC lines derived from different genetic backgrounds on day 14 (Supplementary Fig. 2a,b, compare panels III and IV). These results show that different genetic background is the main source of variation and that variation among different lines from the same individual is no greater than the intra-line variation.
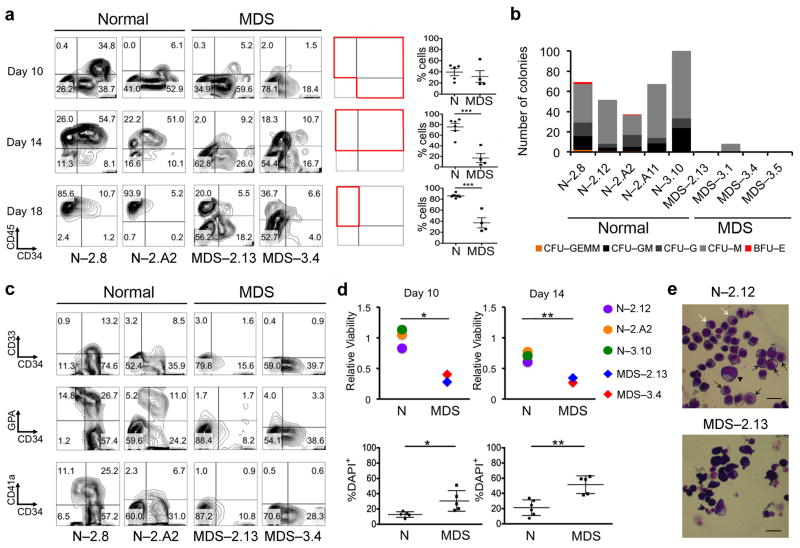
Having established the reproducibility and range of normal variation in the hematopoietic differentiation of normal iPSCs, we then tested the hematopoietic potential of MDS-iPSCs. All MDS-iPSC lines exhibited greatly reduced hematopoietic differentiation potential (Fig. 2a) and clonogenic capacity (Fig. 2b and Supplementary Fig. 3a) affecting all myeloid hematopoietic lineages (Fig. 2c). The MDS-iPSC lines also showed increased cell death during differentiation compared to their isogenic and non-isogenic normal iPSC lines (Fig. 2d and Supplementary Fig. 3b). In addition to an absolute decrease in the efficiency of differentiation, MDS-iPSC lines differentiated with slower kinetics (Supplementary Fig. 3c, f). This observation was corroborated by assessing the kinetics of the emergence and disappearance of a transient wave or primitive-like hematopoiesis, marked by co-expression of CD41a and CD235 (glycophorin A; GPA)16 (Supplementary Fig. 3d, e). Furthermore, upon differentiation along the erythroid lineage, MDS-iPSC lines, unlike their normal isogenic lines, failed to acquire cell-surface markers or morphological features of erythroid cells (Fig. 2e and Supplementary Fig. 3g, h). These phenotypes of ineffective hematopoiesis, reduced or absent clonogenicity and increased cell death are consistent with the phenotype of ex vivo cultured primary MDS cells from BM or peripheral blood17, 18.
Figure 2. MDS-iPSCs have diminished hematopoietic differentiation potential.
(a) Left panels: CD34 and CD45 expression at days 10, 14 and 18 of hematopoietic differentiation in representative normal and del(7q)-MDS-iPSC lines. Right panels: CD34 and CD45 expression and co-expression at days 10, 14 and 18 of hematopoietic differentiation, as indicated, in all iPSC lines tested. Each graph shows the percentage of cells within the quadrants included in the corresponding red box. Mean and SEM are shown. Each line was tested in 1–4 independent differentiation experiments. For those lines that were differentiated more than once, the mean value is shown. ***P<0.001.
(b) Hematopoietic colony assays in methylcellulose at day 14 of hematopoietic differentiation. The number of colonies from 5,000 seeded cells is shown. (CFU-GEMM: colony-forming unit-granulocyte, erythrocyte, monocyte, megakaryocyte; CFU-GM: colony-forming unit-granulocyte, monocyte; CFU-G: colony-forming unit-granulocyte; CFU-M: colony-forming unit-monocyte; BFU-E: burst-forming unit-erythrocyte)
(c) Assessment of lineage markers CD33 (myeloid), GPA (Glycophorin A) or CD235 (erythroid) and CD41a (megakaryocytic) at day 10 of hematopoietic differentiation.
(d) Cell viability measured by a luminescence assay based on ATP quantitation (upper panels) and by DAPI staining (lower panels) on days 10 and 14 of hematopoietic differentiation, as indicated. Viability in the upper panels is given relative to viability on day 1 of hematopoietic differentiation. Mean and SEM are shown. Each line was tested in 1–4 independent differentiation experiments. For those lines that were differentiated more than once, the mean value is shown. *P<0.05, **P<0.01.
(e) May-Giemsa staining of cells cultured for an additional 12 days in erythroid differentiation media. In normal cells we can morphologically identify cells at several stages of maturation from proerythroblast (arrowhead) to basophilic, polychromatophilic (black arrows) and orthochromatic (white arrows) erythroblasts. No morphological changes of progression to maturation are seen in MDS cells. Scale bars, 10 μm.
MDS-iPSC hematopoiesis rescue by chr7q dosage correction
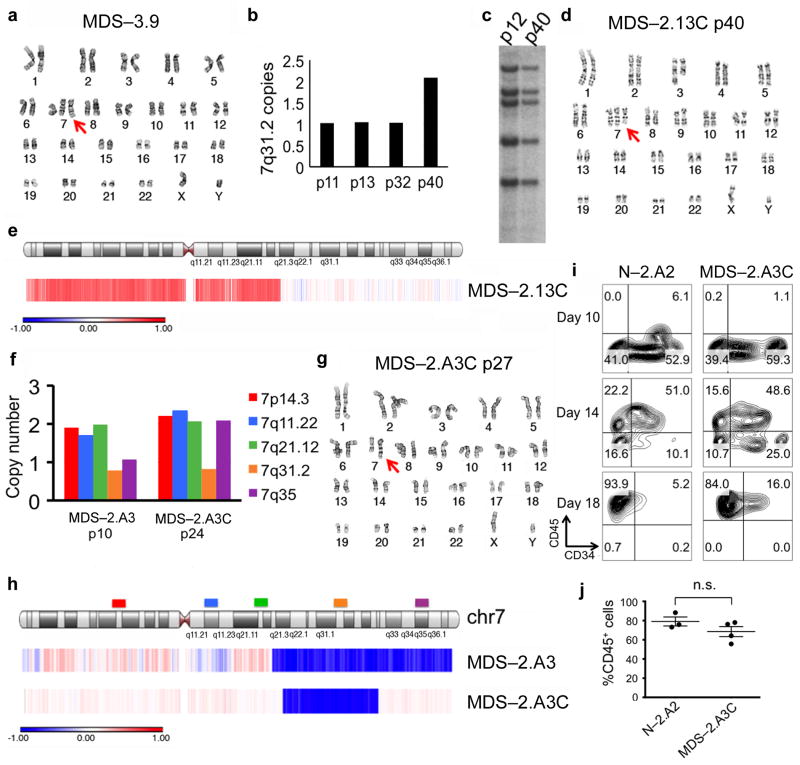
Apart from the profound hematopoietic defects, at least some of the MDS-iPSC lines exhibited slower growth in culture at the undifferentiated state compared to their isogenic normal iPSCs, as observed by smaller colony size (Supplementary Fig. 4a) and confirmed by growth competition assays against a GFP-marked normal iPSC line derived from N-2.12 (Supplementary Fig. 4b, c). Because hPSCs can spontaneously acquire chromosomal abnormalities and those that provide a growth advantage can be selected over time in culture (as the clone that harbors them outgrows the rest of the population), we reasoned that we might be able to isolate clones that spontaneously acquired a second copy of chr7q material. Indeed, by screening additional iPSC lines derived from patient no. 3 for chr7q dosage using a qPCR assay, we identified one line, MDS-3.9, that had acquired an additional chromosome 7, an event not detectable in the starting BM cells (Fig. 3a). Additionally, by monitoring chr7q dosage in the del(7q)-MDS-iPSC lines over time, we were able to detect spontaneous dosage correction in one del(7q)-MDS-iPSC line derived from patient no. 2, MDS-2.13, at passage 40 (Fig. 3b). We excluded the possibility of contamination of the culture with an unrelated line by reprogramming vector integration analysis (Fig. 3c). Karyotyping and aCGH showed that dosage correction had again occurred through duplication of the normal intact chromosome 7 (Fig. 3d, e). As expected, the derivative line MDS-2.13C, grew faster than its parental line in subsequent culture, albeit not as fast as its isogenic karyotypically normal iPSCs (Supplementary Fig. 4d). The same phenomenon of spontaneous acquisition of an extra chromosome 7 occurred in two additional independent clones of the same iPSC line, obtained after single-cell subcloning of early passage cells (before or after reprogramming vector excision), selection of a chr7q haploid clone and subsequent expansion for more than 10 passages. Notably, a large chromosome 3q deletion also initially present in this iPSC line (and in the patient BM cells it was derived from) remained unchanged (Fig. 1c and Fig. 3d). Moreover, we never observed acquisition of extra chr7 copies in normal iPSC lines by close qPCR monitoring over more than 50 passages, in line with reports from systematic studies of chromosomal aberrations commonly observed in hPSCs19,20.
Figure 3. Spontaneous compensation for chromosome 7q dosage imbalance rescues the hematopoietic defect of MDS-iPSCs.
(a) Karyotype of line MDS-3.9, derived from patient no. 3, harboring a derivative chromosome from a 1;7 chromosomal translocation [der(1;7)(q10;p10), the entire long arm of one copy of chromosome 7q is missing and part of chromosome 1q is translocated in its place, identical to the translocation seen in all MDS-iPSC lines from patient no. 3, see also Fig. 1c, MDS-3.1], in addition to two normal chromosomes 7.
(b) qPCR measurement of copy number of a region on 7q31.2 in the del(7q)-iPSC line MDS-2.13 at increasing passage numbers, as indicated.
(c) Southern blot probing integration sites of the vector used for reprogramming of the MDS-2.13 line at passage number 12 (haploid for 7q) and 40 (diploid for 7q).
(d) Karyotyping of MDS-2.13 (see Fig. 1c) at passage number 40 (MDS-2.13C) showing duplication of the normal chromosome 7 without additional karyotypic changes.
(e) aCGH analysis confirming the karyotypic finding. The red color indicates amplification (3 copies) and the white color normal diploid dosage.
(f) qPCR measurement of copy number with different probes along the length of chromosome 7, as indicated, in the del(7q)-iPSC line MDS-2.A3 at passage 10 and 24 (MDS-2.A3C).
(g) Karyotype of line MDS-2.A3C.
(h) aCGH analysis of the del(7q)-iPSC line MDS-2.A3 at passage 10 (MDS-2.A3) and passage 40 (MDS-2.A3C). The blue color indicates deletion (1 copy) and the white color normal diploid dosage.
(i) CD34 and CD45 expression at days 10, 14 and 18 of hematopoietic differentiation. Representative panels of the normal isogenic line N-2.A2 and the dosage corrected MDS line MDS-2.A3C.
(j) CD45 expression at day 14 of hematopoietic differentiation. Mean and SEM are shown. n.s.: not significant.
These events of spontaneous compensation of chr7q dosage imbalance provided evidence for strong in vitro selection for these events. We therefore screened additional del(7q)-MDS-iPSC lines using more probes along the length of chr7 to potentially find clones with duplications of only part of chr7q. We were able to identify an informative clone derived from a second del(7q)-iPSC line from patient 2, MDS-2.A3, which over time acquired a second copy of a telomeric part of chr7q without duplication of the entire chr7 (Fig. 3f, g). aCGH analysis showed that the duplicated region was approximately 30 Mb of size and spanned bands 7q32.3 – 7qter (Fig. 3h, and Supplementary Table 5). The hematopoietic differentiation potential of this “corrected” line, MDS-2.A3C, was fully restored to levels comparable to those of normal iPSCs (Fig. 3i, j). The “corrected” line maintained the SRSF2 and PHF6 mutations. This data strongly suggest that reduced chr7q dosage is responsible for the hematopoietic defect of MDS-iPSCs, as the latter is fully rescued by acquisition of a second copy of chr7q material derived from duplication of the existing copy, and pinpoint a ~30Mb 7q32.3 – 7qter fragment (nucleotides approximately 131,706,336 – 159,128,530) as the critical region.
Engineering heterozygous chr7q loss in normal hPSCs
To further determine the impact of chr7q hemizygosity on the cellular phenotype, we generated perfectly isogenic pairs of hPSCs harboring one or two copies of chr7q by engineering chr7q deletions in normal hPSCs. To this end, we combined a recently reported strategy for deleting supernumerary chromosomes using adeno-associated vector (AAV)-mediated gene targeting of an HSV-tk transgene21 with a previously described strategy using Cre-mediated recombination of inverted loxP sites22,23 (Fig. 4a). We constructed an AAV vector containing two loxP sites in inverted orientation and a positive (puro) and negative (HSV-tk) selection marker. The vector was targeted to a near-telomeric region of chromosome 7 (7q36.3) by ATG-trap of the DNAJB6 gene, highly expressed in hPSCs (Supplementary Fig. 5a). The hESC line H1, as well as the normal iPSC line N-2.12, were transduced with the vector. 14 and 5 puromycin-resistant clones from line N-2.12 and H1, respectively, were picked, initially screened by PCR specific for the targeted allele and subsequently by Southern Blot analysis (Supplementary Fig. 5b). 13 of the 14 N-2.12 clones were found to be correctly targeted and one to have a random vector integration. 5 out of the 5 H1 clones were correctly targeted, one of which harbored an additional random integration. One targeted clone from each line (H1-D-1-1 and N-2.12-D-1-1, respectively, hereafter referred to as H1-D and N-2.12-D) were selected on the basis of lowest background resistance to ganciclovir (GCV) and transduced with an integrase-deficient lentiviral vector (IDLV) transiently expressing Cre re-combinase12, 13. After single-cell subcloning and selection with ganciclovir, clones were first screened by qPCR probing different regions along the length of chromosome 7. Three H1-derived and seven N-2.12-derived clones (H1-D-Cre1, H1-D-2Cre6, H1-D-Cre7, N-2.12-D-Cre10, N-2.12-D-Cre32, N-2.12-D-2Cre4, N-2.12-D-8Cre21, N-2.12-D-8Cre23, N-2.12-D-Cre44 and N-2.12-D-6Cre6) were selected after screening 23 and 46 clones, respectively, and after excluding clones with additional chromosomal abnormalities by karyotyping (Supplementary Fig. 5c). aCGH showed that these clones harbored different deletions spanning variable lengths along the entire chromosome 7q (Fig. 4b and Supplementary Table 6). Clones N-2.12-D-Cre-44 and N-2.12-D-6Cre6 were among the selected ganciclovir-resistant clones, although the chr7q deletion in these clones does not include the tk gene. Ganciclovir resistance may have been conferred by a mutation or other mechanisms. Clones N-2.12-D-Cre10 and N-2.12-D-Cre32 shared the exact same deletion and hence were presumably derived from the same clone, but were treated as separate clones.
Figure 4. Engineering chr7q deletions in normal hPSCs.
(a) Overview of strategy for engineering chromosome 7q deletions. An AAV carrying a positive (puro) and a negative (HSV-tk) selection gene is targeted into chr7q and correctly targeted clones are positively selected in a first step. In a second step, Cre recombinase is transiently expressed and clones that have lost the targeted copy of chr7q are selected with GCV. IDLV, integrase-deficient lentiviral vector.
(b) aCGH specific for chromosome 7 in all engineered del(7q)-hPSC clones analyzed, as indicated. The blue color indicates deletion (1 copy), the red color amplification (3 copies) and the white color normal diploid dosage. Lower panel: chromosome 7 ideogram. The purple box indicates the ~40 Mb region (approximately 112,920,418 – 152,127,281) functionally mapped in the panel of clones with engineered chromosome 7q deletions. Its 5′ and 3′ borders are defined by the 3′ border of the deletion in clone N-2.12.D-6Cre6 and the 3′ end of the deletion in clone H1-D-2Cre6, respectively.
(c, d) CD45 expression at day 14 of hematopoietic differentiation of del(7q)- engineered clones derived from the H1 hESC (c, upper panel and d, left panel) and the N-2.12 iPSC (c, middle and lower panels and d, right panel) line. Mean and SEM are shown. *P < 0.05, **P < 0.01, ***P < 0.001, n.s.: not significant.
(e) Methylcellulose assays at day 14 of hematopoietic differentiation. The number of colonies from 5,000 seeded cells is shown. (CFU-GEMM: colony-forming unit-granulocyte, erythrocyte, monocyte, megakaryocyte; CFU-GM: colony-forming unit-granulocyte, monocyte; CFU-G: colony-forming unit-granulocyte; CFU-M: colony-forming unit-monocyte; BFU-E: burst-forming unit-erythrocyte)
(f) Schematic summary of the approach to defining the chr7q critical region. The purple box indicates the ~40 Mb region functionally mapped in the panel of clones with engineered chromosome 7q deletions from b. The orange box denotes the ~30 Mb region duplicated in the rescued clone MDS-2.A3C, shown in Fig. 3h. Their overlap defines a critical region of ~20 Mb spanning cytobands q32.3-q36.1 (nucleotides approximately 131,706,336- 152,127,281).
Based on our previous phenotypic characterization data (shown in Fig. 2 and Supplementary Fig. 2), we selected the percentage of CD45+ cells at day 14 of hematopoietic differentiation as the most appropriate readout. Eight of the ten clones showed markedly diminished hematopoietic differentiation potential and clonogenic capacity (Fig. 4c–e), similarly to our del(7q)-MDS-iPSCs. Notably, two of the seven clones (N-2-12-D-Cre-44 and N-2.12-D-6Cre6), harboring a smaller deletion spanning 7q11.21 – 7q.31.1 retained comparable differentiation ability to that of normal isogenic and non-isogenic hPSCs (Fig. 4c–e). The chr7q region that these two clones, but none of the other 8 clones, maintained in a diploid dosage was a ~40 Mb region spanning 7q31.1 - q36.1 (Fig. 4f), which shared ~20 Mb of overlap with the region acquired by the “rescued” MDS-2.A3C clone (Fig. 3h). The phenotypic analysis of our collection of engineered del(7q)- hESC and iPSC clones, in combination with our findings in the spontaneously corrected line MDS-2.A3C, functionally delineate a ~20 Mb region, spanning 7q32.3 – 7q36.1 (nucleotides approximately 131,706,336 – 152,127,281), as the critical region in del(7q)-MDS (Fig. 4f).
Phenotype-rescue screen of chr7q genes
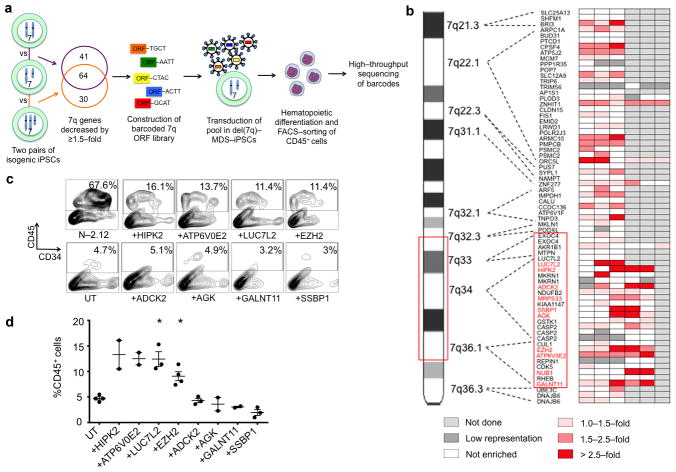
Our data provide functional evidence that chr7q hemizygosity mediates MDS phenotypes through a dosage effect, since these phenotypes are rescued by acquisition of a duplicated copy of chr7q material and recapitulated by the artificial engineering of hemizygosity in normal cells. Indeed, gene expression analyses in our isogenic iPSC pairs show that hemizygosity of chr7q results in reduced expression levels of a large number of genes in the chr7q deleted region, which are restored upon chr7q dosage correction (Supplementary Fig. 6). There is increasing evidence that cancer-associated deletions may produce collective phenotypes through the cumulative haploinsufficiencies of multiple genes that opportunistically co-localize in the deleted clusters24, 25,7. It is therefore likely that the del(7q) mediates multiple gene haploinsufficiencies that co-operate in producing the hematopoietic phenotype, similarly to what was shown in del(5q)-MDS26, 27. To identify candidate haploinsufficient genes in chr7q (whose reduction in dosage from two copies to one is expected to result in a substantial reduction in expression and function), we selected genes with significantly reduced expression at the haploid state by analyzing two pairs of isogenic iPSCs harboring one or two copies of chr7q, derived through reprogramming (MDS-2.13 vs N-2.12) and through spontaneous correction (MDS-2.13 vs MDS-2.13C). 105 and 94 genes, respectively, were found to be reduced by at least 1.5-fold (log2(FC) ≥ 0.585), 64 of which were found in both comparisons (Fig. 5a and Supplementary Table 7). Prioritizing these 64 genes, as well as genes with the highest differences in expression, and after excluding genes with very low expression in human HSPCs28, we selected 62 genes and constructed a lentiviral open reading frame (ORF) library with each ORF tagged to a unique 4-nucleotide barcode sequence (Supplementary Fig. 7a–c and Supplementary Table 8). 76 vectors encoding a total of 75 ORFs (62 genes, including 13 alternative transcripts) and one control reporter gene (mCherry), tagged to 70 different barcodes, were constructed. These were packaged as pools in 3 different batches (Supplementary Table 8) and transduced into two different del(7q)-iPSC lines, MDS-2.13 and MDS-2.A3, in 12 independent transduction experiments (Supplementary Table 9). To maximize the chance that a gene will present as a hit, we performed multiple independent transductions at a high Multiplicity of Infection (MOI). To assess representation of all vectors/barcodes and test for potential biases introduced by sample preparation, we performed high-throughput sequencing of the barcodes in two samples derived from the same transduced cell pool (transduction #4) processed independently from collection to sequencing. This analysis showed representation of all the barcodes included in the transduced library batch (batch #1), which was practically identical between the two separately processed samples (Supplementary Table 10 and Supplementary Fig. 7d). The transduced cell pools were monitored weekly for vector silencing (by flow cytometry for GFP) and for spontaneous chr7q duplication by chr7q copy number qPCR. Experiments where GFP expression became undetectable soon after transduction or where chr7q copy number became >1.5 were discontinued (transductions #1, #4 and #5, Supplementary Table 9). For each screening experiment, transduced del(7q)-iPSCs were differentiated along the hematopoietic lineage, CD45+ cells were sorted on day 15 of differentiation and paired samples of the sorted CD45+ cells and the starting undifferentiated cells were subjected to high-throughput sequencing of the barcodes (Supplementary Fig. 7a). A total of 8 screening experiments in 6 independently transduced cell pools from the two del(7q)-iPSC lines were done and enrichment in CD45+ hematopoietic progenitors relative to undifferentiated cells was calculated for each ORF/barcode (Fig. 5b and Supplementary Table 11). Focusing on hits residing in the critical region 7q32.3 – 7q36.1, identified above, ten genes were found enriched (by >1.5-fold) recurrently (i.e. in at least two independent experiments) (Supplementary Fig. 7e and Supplementary Table 11). These were LUC7L2, HIPK2, MRPS33, ADCK2, SSBP1, AGK, EZH2, ATP6V0E2, NUB1 and GALNT11. Six of them (LUC7L2, HIPK2, ADCK2, EZH2, ATP6V0E2 and GALNT11) presented as hits in more than half of the experiments they were tested in and were considered as “stronger” hits. These six genes, as well as two of the four remaining hits (SSBP1 and AGK) were individually tested for rescue of hematopoiesis in a different del(7q)-iPSC line than the ones used for the primary screen (Fig. 5c, d and Supplementary Fig. 8a, b). Four genes, all included among the “stronger” hits, could be confirmed to partially rescue the emergence of CD45+ hematopoietic progenitors when overexpressed in del(7q)-iPSCs (Fig. 5c, d). These were HIPK2, ATP6V0E2, LUC7L2 and EZH2.
Figure 5. A phenotype-rescue screen identifies candidate haploinsufficient genes in del(7q)-MDS.
(a) Scheme of genetic screen. Expression microarray analyses of two isogenic pairs of iPSCs with one or two copies of chr7q were used to select candidate haploinsufficient genes, which were then each tagged to a unique barcode and cloned to construct a lentiviral ORF library. The library was transduced as a pool into del(7q)-MDS-iPSCs and genes that rescued hematopoiesis were identified by means of enrichment in sorted CD45+ progenitors differentiated from the cells.
(b) Heat map of gene enrichment from 3–6 independent experiments. Fold enrichment was calculated from the sequencing reads as %representation in CD45+ cells over % representation in undifferentiated cells. Values between 0–1.0 are depicted as “not enriched”. Barcodes with read counts lower than 100 (marked as “low representation”) were not included in the analysis. Genes within the critical region (red box) that were recurrent (i.e. in at least 2 independent experiments) hits (i.e. had enrichment >1.5-fold) are shown in red. See also Supplementary Table 11.
(c) Day 14 of hematopoietic differentiation of a del(7q)-iPSC line (MDS-2.12-D-8Cre23) transduced with lentiviral vectors expressing HIPK2, ATP6V0E2, LUC7L2, EZH2, ADCK2, AGK, GALNT11 or SSBP1, as indicated, compared to the untransduced (UT) line. One representative experiment is shown for each condition.
(d) CD45 expression at day 14 of hematopoietic differentiation of the groups shown in (c). Mean and SEM are shown. *P<0.05.
Validation of chr7q haploinsufficient genes
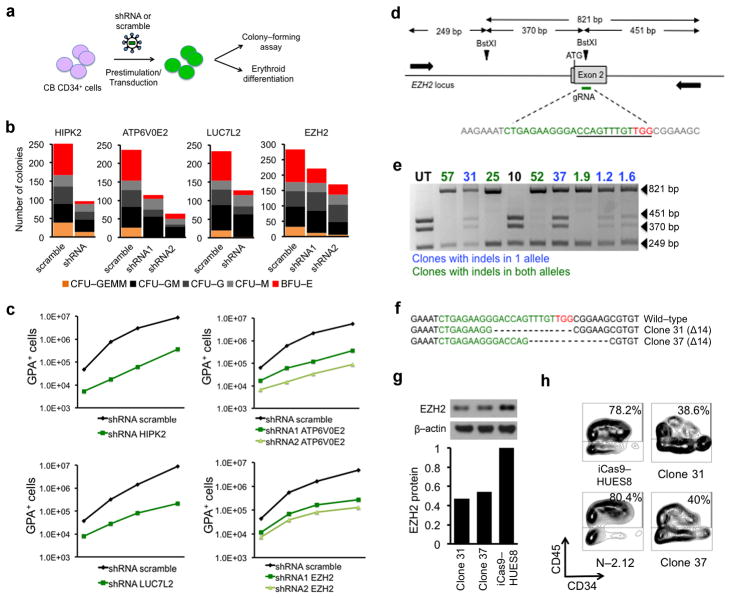
These four hits were further tested in short hairpin RNA (shRNA) knockdown experiments in normal cord blood (CB) CD34+ hematopoietic progenitors. Different shRNAs were tested and those that conferred a reduction in expression of approximately 50% on average upon transduction of nearly 100% of the cells were selected to approximate haploinsufficiency (Fig. 6a and Supplementary Fig. 9a, b). Knockdown of each of the four genes reduced colony formation, affecting primarily multipotent and erythroid progenitors rather than more mature myeloid precursors (Fig. 6b). In vitro expansion and differentiation of erythroblasts were also diminished (Fig. 6c and Supplementary Fig. 9c). Thus, all four confirmed hits could be validated by knockdown assays to diminish hematopoietic potential when knocked down to approximately half of their normal levels.
Figure 6. Validation of chr7q haploinsufficient genes.
(a) Experimental scheme. CB CD34+ cells were prestimulated for 48 h and transduced with a lentiviral vector encoding shRNA or scramble. 48 h later the cells were plated in colony-forming assays or in liquid erythroid induction culture.
(b) Methylcellulose assays of CB CD34+ cells expressing shRNAs against HIPK2, ATP6V0E2, LUC7L2, EZH2, or scramble, as indicated. Number of colonies from 1000 seeded cells is shown. The mean of duplicate experiments (from two independent transductions) is shown. (CFU-GEMM: colony-forming unit-granulocyte, erythrocyte, monocyte, megakaryocyte; CFU-GM: colony-forming unit-granulocyte, monocyte; CFU-G: colony-forming unit-granulocyte; CFU-M: colony-forming unit-monocyte; BFU-E: burst-forming unit-erythrocyte)
(c) Absolute number of GPA+ cells in vitro generated from CB CD34+ cells expressing shRNAs against HIPK2, ATP6V0E2, LUC7L2, EZH2 or scramble in erythroid media. Absolute numbers of GPA+ cells were calculated from the total cell counts and the percentage of GPA+ cells by flow cytometry. The mean values of duplicate experiments from two independent transductions are shown.
(d) Schematic representation of the EZH2 locus with the position of the gRNA target sequence, the BstXI restriction sites and the primers used for screening of CRISPR/Cas9-targeted alleles indicated. The gRNA sequence is shown in green and the PAM motif in red. The sequence of the BstXI restriction site is underlined. The beginning of the gRNA sequence is 23 nt downstream of the ATG.
(e) Representative image of RFLP analysis for screening of single cell clones. The 821 band (resistant to BstXI digest) indicates the presence of indels. Clones with indels in only one allele are shown in blue and clones with indels in both alleles are shown in green. Clones with both alleles intact are shown in black.
(f) Sequences of the wild-type EZH2 locus and the two clones (clone 37 and clone 31) harboring monoallelic indels, as indicated. Sequences shown were obtained from the 821 bp band and therefore correspond to the disrupted allele. The other allele was confirmed to be intact by sequencing the 370 and 451 bp bands (not shown). Each clone harbors a different 14-nucleotide deletion introducing a frame-shift.
(g) Western blot analysis (upper panels) and quantification (lower panels) of EZH2 expression in the two clones harboring monoallelic inactivation of EZH2, compared to that in the parental hESC line HUES8.
(h) Day 14 of hematopoietic differentiation of the two clones with monoallelic inactivation of EZH2, compared to that of the parental hESC line HUES8 and of normal iPSCs. One of two replicate experiments is shown.
One of the four hits, EZH2, is a gene encoding a histone methyltransferase that constitutes the catalytic unit of the polycomb repressive complex 2 (PRC2) and is found recurrently mutated in MDS29–32. EZH2 mutations found in MDS are commonly loss-of-function heterozygous mutations, consistent with a haploinsufficiency mechanism8, 33. EZH2 expression was reduced but not abolished in our del(7q)-MDS-iPSCs (Supplementary Fig. 8d) and we found no mutations in the second allele. We thus selected EZH2 to validate further. First, we confirmed that restoring EZH2 expression in two additional del(7q)-iPSC lines (MDS-2.13 and N-2.12-D-2Cre4) by lentiviral transduction again partially rescued their hematopoietic differentiation potential (Supplementary Fig. 8c–i). To further characterize the effect of EZH2 haploinsufficiency in the hematopoietic phenotype, we designed a CRISPR/Cas9-based strategy to inactivate the EZH2 gene in hESCs, by mediating a double-strand break downstream of the ATG and exploiting the error-prone non-homologous end joining repair mechanism to introduce frame-shifting insertions or deletions of a few nucleotides (indels) (Fig. 6d). The hESC line, iCas9-HUES8, carrying an inducible Cas9 transgene inserted into the AAVS1 locus34, was transduced with a lentiviral vector expressing a guide RNA (gRNA) targeting EZH2 and clones with mono-allelic indels were isolated (Fig. 6e and Supplementary Fig. 10a, b). Two clones (clones 31 and 37), each harboring a different frame-shifting deletion (Fig. 6f), found to express EZH2 protein at approximately 50% of the normal level (Fig. 6g) and to be clonal (Supplementary Fig. 10c) were selected. The hematopoietic differentiation and colony formation potential of both EZH2 haploinsufficient clones was substantially diminished at levels intermediate between those of normal and del(7q)-iPSCs (Fig. 6h, Supplementary Fig. 10d, e and compare to Fig. 4c–e). This is consistent with the partial phenotypic rescue mediated by restoration of EZH2 expression in del(7q)-iPSCs (Fig. 5c, d and Supplementary Fig. 8) and supports a model of cooperation between haploinsufficiencies of more than one genes to generate the del(7q) hematopoietic phenotype.
In summary, these data show that haploinsufficiency of EZH2 decreases the hematopoietic potential of cells, and that the hematopoietic defects caused by chr7q hemizygosity are mediated through the haploinsufficiency of EZH2, in combination with one or more additional genes, which might include LUC7L2, HIPK2 and/or ATP6V0E2 and possibly additional genes.
DISCUSSION
We used cellular reprogramming for the first time, to our knowledge, to model a somatically acquired genetic lesion in an isogenic human setting, by exploiting the somatic mosaicism in the hematopoietic compartment of MDS patients. Our detailed characterization of a panel of normal isogenic and non-isogenic iPSC lines (Supplementary Fig. 2) showing that the genetic background is the major source of line-to-line variation in differentiation potential, highlights the advantage of using isogenic controls. The efficiency of reprogramming of MDS hematopoietic cells was highly variable between the two patient samples compared to that of isogenic normal cells (Supplementary Table 2). It is likely that clonal or subclonal gene mutations increase or diminish reprogramming “fitness”35. As reprogramming is an inherently clonal process and can “capture” genetic lesions present at low frequency in the tissue of origin36, reprogramming MDS cells could provide a powerful tool to dissect the clonal architecture and evolution of this disease and to link genotypes to cellular phenotypes14, 37. Further dissecting the MDS-iPSC phenotypes at the molecular level could provide insights into the pathogenesis of myelodysplasia and leukemic transformation. Moreover, the robustness of our cellular MDS phenotypes could provide a platform for phenotype-driven drug screens to identify small molecules that ameliorate these phenotypes. Our 20 Mb region coincides with some proposed commonly deleted regions from patient studies, but not with others38, 9, 39. Among other candidate haploinsufficient genes on chr7q, MLL3ref40 is located inside our critical region, but it was not found differentially expressed between del(7q)- and isogenic normal iPSCs in our analysis and therefore not included in our library screen (Supplementary Table 7). Clearly, additional work is needed for a more complete understanding of the chr7q genetic elements critically involved in del(7q)-MDS and to this end our model can provide a powerful platform for gene complementation assays, as we describe here.
We combined and extended previous strategies to develop a method for introducing large-scale chromosomal deletions (of Mb range) in normal human cells. Previous studies utilizing recombination between inverted loxP sites obtained loss of entire chromosomes rather than segmental deletions22,23. In contrast, we did not find clones with complete monosomy 7. As previous studies attempted to eliminate only supernumerary or sex chromosomes, the different outcome in our study likely reflects incompatibility of the complete loss of an autosome by normal diploid iPSCs with growth in culture. Our methods for engineering deletions can enable studies of the functional consequences of alterations in gene copy number, so far feasible mostly in yeast, for the first time in human cells41 and may unveil species-specific dosage sensitivities that may not manifest in mice or other model organisms3. Furthermore, hPSCs with chromosomal deletions can provide a haploid background for engineering complete gene knock-outs (by inactivating the remaining allele) or for high-throughput classical forward genetics mutagenesis screens42.
Our ‘functional mapping approach’ could complement genetic mapping and physical mapping approaches, providing a framework to study commonly deleted chromosomal regions of interest. Any cancer or other disease in which a deletion is suspected to be pathogenetic could be modeled with this approach. Somatic mosaicism, if present, as in all cancers and for deletions arising de novo during late stages of development, provides the opportunity to derive isogenic diseased and normal iPSCs directly through reprogramming from the same individual, as we show here. For germline deletions, isogenic controls can only be derived through chromosome engineering approaches.
Gene haploinsufficiency is increasingly appreciated as an important mechanism of phenotypic variation and disease24, 43, 44. Genes whose function is dependent on dosage cannot be revealed by genomic approaches and require functional assays. Among the latter, gain-of-function (rescue) assays, as we present here, provide substantial advantages over knockdown/knockout screens. We thus envision a generalizeable approach to the discovery of haploinsufficient genes starting with genomic data pointing to commonly deleted regions from copy number variant (CNV) databases or cancer genomes (as indicators of mono-allelic loss of function), following a workflow similar to the one we propose here: generate isogenic hPSCs harboring the specific deletion, determine a phenotype in a disease-relevant cell type derived from hPSCs, identify candidate haploinsufficient genes based on their differential expression in the diploid vs haploid state and filter them through phenotype-rescue screens to finally select one or few candidates for follow-up studies. Because a gene can be mono-allelically inactivated in different ways, our approach can be integrated with data on different classes of mutation (CNVs, single nucleotide polymorphisms (SNPs)) from cancer genome databases to inform gene prioritization. Indeed, two of the haploinsufficient genes identified in our study, EZH2 and LUC7L2, also harbor recurrent inactivating mutations in MDS/AML genomes8, 33,45, 46.
In summary, we present a strategy for functional cancer genetics that should prove generally applicable to the study of disease-associated chromosomal deletions.
METHODS
iPSC generation
Cryopreserved BM or peripheral blood mononuclear cells were thawed and cultured in X-VIVO 15 media with 1% nonessential amino acids (NEAA), 1 mM L-glutamine and 0.1 mM β-mercaptoethanol (2ME) and supplemented with 100 ng/ml stem cell factor (SCF), 100 ng/ml Flt3 ligand (Flt3L), 100 ng/ml thrombopoietin (TPO) and 20 ng/ml IL-3 for 1–4 days. The excisable lentiviral vector CMV-fSV2A expressing OCT4, KLF4, c-MYC and SOX2 (derived from pLM-fSV2A12 after replacing CMV for hPGK) was used and produced as described13. For induction of reprogramming, 10,000–250,000 cells were plated on retronectin-coated 24-well dishes and transduced at an MOI of 30 or higher for 16h. Two days later, the cells were harvested and plated on mitotically inactivated MEFs (GlobalStem) in 6-well plates and the plates were centrifuged at 500g for 30 min at RT. The next day and every day thereafter, half of the medium was changed to hESC medium with 0.5 mM valproic acid (VPA). Colonies with hPSC morphology were manually picked and expanded, as described13.
Culture of hPSCs on MEFs or in feeder-free conditions was performed as previously described13. Characterization of pluripotency (flow cytometry, OCT4 promoter methylation analysis, teratoma formation assays) and Cre-mediated vector excision were done as previously described12, 13, 47. Patient samples were obtained with informed consent under protocols approved by an Institutional Review Board at Memorial Sloan-Kettering Cancer Center and the Fred Hutchinson Cancer Research Center.
Whole exome sequencing and calling of somatic variants
Exome sequencing was performed at the University of Washington. Library construction, exome capture, sequencing, and mapping were performed as previously described48. Briefly, 1 ug of genomic DNA was subjected to a series of shotgun library construction steps, including fragmentation through acoustic sonication (Covaris), end-polishing and A-tailing, ligation of sequencing adaptors and PCR amplification with 8 bp barcodes for multiplexing. Libraries underwent exome capture using the Roche/Nimblegen SeqCap EZ v3.0 (~62 MB target). Prior to sequencing, the library concentration was determined by triplicate qPCR and molecular weight distributions verified on the Agilent Bioanalyzer (consistently 250 ± 25 bp). Sequencing reads were processed through a pipeline consisting of a combined suite of Illumina software and other “industry standard” software packages (i.e., Genome Analysis ToolKit [GATK], Picard, BWA, SAMTools, and in-house custom scripts) that included base calling, alignment, local realignment, duplicate removal, quality recalibration, data merging, variant detection, genotyping and annotation. Somatic variant calling was performed using the MuTect method49.
Hematopoietic differentiation
iPSC colonies were collected with dispase and plated in low-attachment dishes in DMEM with 20% Fetal Bovine Serum (FBS), 1% NEAA, 1 mM L-glutamine and 0.1 mM 2ME, supplemented with 30 ng/ml bone morphogenetic protein 4 (BMP4) and 50 ug/ml ascorbic acid. One day later, the medium was switched to StemPro-34 SFM (Gibco) with 1% NEAA, 1 mM L-glutamine and 0.1 mM 2ME supplemented with 30 ng/ml BMP4, 50 ng/ml FGF2 and 50 ug/ml ascorbic acid. At day 4, the cytokine cocktail was changed to: 20 ng/ml vascular endothelial growth factor (VEGF), 10 ng/ml FGF2, 100 ng/ml stem cell factor (SCF), 20 ng/ml Flt3 ligand (Flt3L), 20 ng/ml thrombopoietin (TPO), 40 ng/ml IL-3 and 50 ug/ml ascorbic acid and replaced at day 6. At day 8 the cytokine cocktail was changed to: 100 ng/ml stem cell factor (SCF), 20 ng/ml Flt3 ligand (Flt3L), 20 ng/ml thrombopoietin (TPO), 40 ng/ml IL-3 and 50 ug/ml ascorbic acid and replaced every two days. At the end of the embryoid body differentiation culture (day 10, 14 or 18) cells were dissociated with accutase into single cells. For flow cytometry, the following antibodies were used: CD34-PE (clone 563, BD Pharmingen) or CD34-Brilliant Violet 421 (clone 581, BD Pharmingen), CD45-PerCP (clone HI30, Biolegend), CD45-APC (clone HI30, BD Pharmingen), CD43-APC (clone 1G10, BD Pharmingen), CD235a-PE-Cy5 (clone GA-R2, BD Pharmingen) or CD235a-RPE (clone JC159, Dako), CD71-APC (clone M-A712, BD Pharmingen), CD41a-APC-H7 (clone HIP8, BD Pharmingen) and CD33-PE-CF594 (clone WM53, BD Pharmingen). For methylcellulose assays, the cells were resuspended in StemPro-34 SFM medium at a concentration of 3 × 104/ml. 500μl of cell suspension were mixed with 2.5 ml ColonyGel Human Enriched Complete Medium (ReachBio) and 1 ml was plated in duplicate 35-mm dishes. Colonies were scored after 14 days and averaged between the duplicate dishes. Erythroid differentiation was performed as previously described12.
aCGH
aCGH was performed on a chromosome 7-specific array platform (NimbleGen Human CGH 385K Chromosome 7 Tiling Array) with a mean spacing of 365 bp or on a custom array manufactured by Agilent Technologies, consisting of 162,863 unique probes (159,157 on chr7) with a median spacing of 795 bp. Labeling and hybridization were performed according to the manufacturer’s instructions. The dataset was assessed for quality and median background subtracted signal intensities were normalized using the Bioconductor package limma. The data was further normalized by mean centering the logFC values, where the mean was calculated from all autosomes, except chromosome 7. Aberrations were detected by applying the Genomic Segmentation algorithm (minimum genomic markers: 20; p-value threshold: 0.0001; signal to noise: 1) within Partek software, version 6.6 (Partek Inc., St. Louis MO, USA).
Growth assays
For growth curves, 4 × 104 cells per well were seeded in triplicate Matrigel-coated 6-well-plates, harvested at different time points and counted on a hemocytometer. Cell counts were calculated relative to the cell count 48h hours after plating. For growth competition assays, clone N-2.12-GFP was generated by transducing N-2.12 cells with a lentiviral vector expressing eGFP, manually picking a few colonies homogeneously expressing GFP under a fluorescent microscope, expanding them and selecting one with a narrow distribution of GFP expression. iPSCs were mixed 1:1 with the N-2.12-GFP clone and 1 × 105 cells/well were plated in triplicate wells of a 6-well plate. One well was harvested the next day (day 1) and the population size of GFP− cells was measured by flow cytometry. Tra-1-81 staining was used to exclude MEFs. The remaining duplicate wells were passaged every 4 days for 16 days, one third was replated and the rest was used for flow cytometry. The relative population size of GFP− cells at each time point was calculated relative to the population size at day 1.
Cell viability assay
An equal number of cells from each iPSC line were seeded on MEFs in one well of a 6-well plate per time point. 5 days later, the cells from each well were harvested separately and induced towards hematopoietic differentiation as described above. On different days, embryoid bodies were collected and treated with accutase to obtain single cells. Cell viability was determined using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega) following the manufacturer’s instructions. Luminescence was measured on a Perkin Elmer Envision Multilabel Detector Plate Reader.
AAV-mediated gene targeting and selection of clones with deletions
The 5′ and 3′ homology arms of the AAV vector consisted of chromosome 7 nucleotides 157,150,063 – 157,151,258 and 157,151,259 – 157,152,447 (hg19 human genome assembly), respectively. The entire sequence was amplified from H1 gDNA in two independent PCR reactions, cloned and sequenced, and one allele was chosen for subsequent amplification and cloning. AAV vector production was performed as described21. hESCs and iPSCs were transduced in feeder-free conditions, replated on puromycin-resistant MEFs and selected with 0.5 μg/ml puromycin for 2–4 days. Southern Blot analyses were performed as previously described13. Transduction with a Cre-expressing IDLV was performed as previously described13. Ganciclovir selection was performed at a concentration of 200 μM for 10–15 days.
qPCR for chromosome 7 dosage
TaqMan qPCR was performed as previously described13 with primers and probes shown in Supplementary Table 12.
Gene expression analysis by qRT-PCR
RNA was isolated with Trizol (Life Technologies). Reverse transcription was performed with Superscript III (Life Technologies) and qPCR was performed with the SsoFast EvaGreen Supermix (Bio-Rad) using primers shown in Supplementary Table 13. Reactions were carried out in triplicate in a 7500 Fast Real-Time PCR System (Applied Bio-systems).
Microarray gene expression analysis
Total RNA integrity was tested using an Agilent 2200 TapeStation (Agilent Technologies, Inc., Santa Clara, CA) and was quantified using a Trinean DropSense96 spectrophotometer (Caliper Life Sciences, Hopkinton, MA). High quality RNA samples were converted to cDNA and biotin-labeled for microarray analysis using Ambion’s Illumina To-talPrep RNA Amplification kit (Life Technologies, Grand Island, NY). Labeled cRNAs were processed on a HumanHT-12 Expression BeadChip (Illumina, Inc., San Diego, CA) and imaged using an Illumina iScan system. Microarray data was assessed for quality and quantile normalized using the Bioconductor package lumi Initial filtering included flagging probes that were below a signal “noise floor,” which was calculated as the 75th percentile of the negative control probe signals within each array. We subsequently filtered the dataset by employing a variance filter using the “shorth” function of the Biocon-ductor package genefilter. Differential gene expression was determined using the Bio-conductor package limma, and a false discovery rate (FDR) method was used to correct for multiple testing. Significant differential gene expression was defined as |log2 (ratio) ≥ 0.585 (± 1.5-fold) with the FDR set to 5%.
Lentiviral ORF library construction and genetic screens
For construction of the 7q ORF lentiviral library, cDNAs were PCR-amplified from peripheral blood or alternative sources as indicated in Supplementary Table 8 and cloned in the mP2A lentiviral backbone (Supplementary Fig. 7b). For packaging, each vector plasmid was independently co-transfected with the two packaging plasmids and the supernatants were pooled and concentrated 400–1000 -fold. Packaging and transduction were performed as described previously13. FACS-sorted CD45+ cells and paired undifferentiated iPSCs were collected from each screening experiment, genomic DNA was isolated and a 300 bp vector sequence containing the barcode was amplified in 12 PCR cycles. The universal TruSeq adapters were tagged in a second round of 15 cycles of PCR amplification and the PCR product was purified and sequenced in the Illumina platform. Sequencing reads (36nt) were trimmed of vector sequences for barcode extraction with a Python Programming code and the representation of each barcode sequence was calculated as percentage of total number of reads of all barcodes. Fold enrichment was calculated between paired samples.
Western Blot analysis
hESCs were lysed with high salt buffer (0.3 M KCl) supplemented with protease inhibitor. Protein concentrations were determined by bicinchoninic acid assay (Pierce Biotechnology Inc) and 20 ug of protein from each extract were diluted in Laemmli sodium dodecyl sulfate sample buffer and resolved by electrophoresis on Bolt 4% to 12% Bis-Tris pre-cast gels (Invitrogen) and blotted on nitrocellulose membrane. The membranes were blocked with 5% nonfat dry milk diluted in Tris-buffered saline and incubated with primary antibody Ezh2 and β-Actin (Cell Signaling). After washing, blots were incubated with HRP-conjugated secondary antibody and developed using ECL Western Blotting Detection Reagents (Amersham GE Healthcare). Band intensity was quantified by ImageJ.
shRNA knockdown in CB CD34+ cells
CB CD34+ cells were purchased from AllCells. Two to four shRNAs against each of the four genes HIPK2, ATP6V0E2, LUC7L2 and EZH2 and one scramble sequence were inserted in the 3′UTR of the G-U6 lentiviral vector50 driven by a U6 promoter. shRNA sequences are shown in Supplementary Table 14. Vectors were packaged as described13. CB CD34+ cells were prestimulated for 48h and transduced with the vectors, as described above under “iPSC generation”. Two days after transduction, clonogenic assays in methylcellulose and erythroid differentiation were performed, as described50. Erythroid differentiation was monitored by flow cytometry for CD71 and GPA. A separate well was used for cell counts.
CRISPR/Cas9-mediated knockout of EZH2
A gRNA targeting the EZH2 locus 23 nt downstream of the ATG and overlapping with a BstXI restriction site (sequence shown in Fig. 6d and Supplementary Table 14) was designed and cloned in the 3′UTR of the G-U6 lentiviral vector driven by a U6 promoter50. The vector was packaged as described13. The hESC line iCas9-HUES834, harboring a doxycycline-inducible Cas9 (iCas9) and a constitutive reverse tetracycline transactivator (M2rtTA) inserted each in one allele of the AAVS1 locus, was dissociated with accutase into single cells and plated on Matrigel (day 0, Supplementary Fig. 10a). Doxycycline was added to the culture the following day (day 1) at 2 ug/ml for two days. On day 2, the cells were transduced with the gRNA lentiviral vector at varying MOIs (0.1, 1, 10 and 30 ul, Supplementary Fig. 10a). Two days later (day 4), the cells were dissociated into single cells with accutase and replated at a density of 100–500/cm2. A portion of cells was used for flow cytometry for GFP expression to evaluate transduction efficiency and Restriction Fragment Length Polymorphism (RFLP) analysis to estimate the efficiency of cleavage by Cas9. PCR with primers F: GTGGCACAAGAGGCAAAAAT and R: CGATTGCCATCCTTTCTTTG was performed. The PCR product was digested with BstXI and analysed in an agarose gel stained with EtBr. Band intensity was quantified by Image Lab (Bio-rad). After 7–10 days single colonies were picked in separate wells of a 6-well plate, allowed to grow for approximately 3–6 days and screened by PCR and RFLP analysis. Approximately 100 cells were picked directly into a 0.2 ml tube, pelleted and lysed. RFLP analysis was performed as above.
Statistical analysis
Statistical analysis was performed with GraphPad Prism software. Data are shown as the mean with standard error of the mean (SEM). Pairwise comparisons between different groups were performed using a two-sided unpaired unequal variance t-test. For all analyses, P < 0.05 was considered statistically significant. Investigators were not blinded to the different groups.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grants R00 DK087923 (EPP), R01 HL121570 (EPP), P30 CA15704 and by awards from the University of Washington Royalty Research Fund (EPP), the American Society of Hematology (EPP), the Sidney Kimmel Foundation for Cancer Research (EPP), the Aplastic Anemia & MDS International Foundation (EPP), the Ellison Medical Foundation (EPP), the Damon Runyon Cancer Research Foundation (EPP) and a John H. Tietze Stem Cell Scientist Award (EPP). We thank David Russell and Li Li for sharing their expertise in AAV-mediated gene targeting and Thalia Papayannopoulou for sharing her expertise in assessment of May-Giemsa slides and for useful discussions. We thank Chrystel Husser, Cassie Sather and Ryan Basom for excellent technical assistance and J. Overbey for statistical advice.
Footnotes
CONTRIBUTIONS
AGK, CJC and IB performed experiments and analyzed data, JJD analyzed microarray data, EKD, FP, VMK and SDN selected and procured patient samples, AN and DRH performed bioinformatics analyses, GAF and CEM performed histological analyses of teratomas, DH provided the iCas9-HUES8 cell line, TG analyzed whole exome sequencing data, TG and SDN provided critical reading of the manuscript and scientific discussions, EPP conceived, designed and supervised the study, analyzed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 4.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 6.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solimini NL, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337:104–109. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindsley RC, Ebert BL. Molecular pathophysiology of myelodysplastic syndromes. Annual review of pathology. 2013;8:21–47. doi: 10.1146/annurev-pathol-011811-132436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jerez A, et al. Loss of heterozygosity in 7q myeloid disorders: clinical associations and genomic pathogenesis. Blood. 2012;119:6109–6117. doi: 10.1182/blood-2011-12-397620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soldner F, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells--opportunities for disease modelling and drug discovery. Nat Rev Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- 12.Papapetrou EP, et al. Genomic safe harbors permit high beta-globin transgene expression in thalassemia induced pluripotent stem cells. Nat Biotechnol. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papapetrou EP, Sadelain M. Generation of transgene-free human induced pluripotent stem cells with an excisable single polycistronic vector. Nat Protoc. 2011;6:1251–1273. doi: 10.1038/nprot.2011.374. [DOI] [PubMed] [Google Scholar]
- 14.Walter MJ, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sturgeon CM, Ditadi A, Clarke RL, Keller G. Defining the path to hematopoietic stem cells. Nat Biotechnol. 2013;31:416–418. doi: 10.1038/nbt.2571. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy M, et al. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep. 2012;2:1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Flores-Figueroa E, Gutierrez-Espindola G, Guerrero-Rivera S, Pizzuto-Chavez J, Mayani H. Hematopoietic progenitor cells from patients with myelodysplastic syndromes: in vitro colony growth and long-term proliferation. Leuk Res. 1999;23:385–394. doi: 10.1016/s0145-2126(98)00176-3. [DOI] [PubMed] [Google Scholar]
- 18.Sato T, Kim S, Selleri C, Young NS, Maciejewski JP. Measurement of secondary colony formation after 5 weeks in long-term cultures in patients with myelodysplastic syndrome. Leukemia. 1998;12:1187–1194. doi: 10.1038/sj.leu.2401084. [DOI] [PubMed] [Google Scholar]
- 19.Amps K, et al. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol. 2011;29:1132–1144. doi: 10.1038/nbt.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narva E, et al. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat Biotechnol. 2010;28:371–377. doi: 10.1038/nbt.1615. [DOI] [PubMed] [Google Scholar]
- 21.Li LB, et al. Trisomy correction in down syndrome induced pluripotent stem cells. Cell Stem Cell. 2012;11:615–619. doi: 10.1016/j.stem.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewandoski M, Martin GR. Cre-mediated chromosome loss in mice. Nat Genet. 1997;17:223–225. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- 23.Matsumura H, et al. Targeted chromosome elimination from ES-somatic hybrid cells. Nat Methods. 2007;4:23–25. doi: 10.1038/nmeth973. [DOI] [PubMed] [Google Scholar]
- 24.Davoli T, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue W, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci U S A. 2012;109:8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebert BL, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starczynowski DT, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 28.Pang WW, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci U S A. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 30.Lindsley RC, Ebert BL. The biology and clinical impact of genetic lesions in myeloid malignancies. Blood. 2013 doi: 10.1182/blood-2013-06-460295. [DOI] [PubMed] [Google Scholar]
- 31.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 32.Makishima H, et al. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia. 2010;24:1799–1804. doi: 10.1038/leu.2010.167. [DOI] [PubMed] [Google Scholar]
- 33.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez F, et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young MA, et al. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abyzov A, et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Beau MM, et al. Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid diseases. Blood. 1996;88:1930–1935. [PubMed] [Google Scholar]
- 39.Dohner K, et al. Molecular cytogenetic characterization of a critical region in bands 7q35-q36 commonly deleted in malignant myeloid disorders. Blood. 1998;92:4031–4035. [PubMed] [Google Scholar]
- 40.Chen C, et al. MLL3 Is a Haploinsufficient 7q Tumor Suppressor in Acute Myeloid Leukemia. Cancer Cell. 2014;25:652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang YC, Amon A. Gene copy-number alterations: a cost-benefit analysis. Cell. 2013;152:394–405. doi: 10.1016/j.cell.2012.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 43.White JK, et al. Genome-wide Generation and Systematic Phenotyping of Knockout Mice Reveals New Roles for Many Genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolze A, et al. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science. 2013;340:976–978. doi: 10.1126/science.1234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosono N, et al. Recurrent genetic defects on chromosome 7q in myeloid neoplasms. Leukemia. 2014;28:1348–1351. doi: 10.1038/leu.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh H, et al. Putative RNA-splicing gene LUC7L2 on 7q34 represents a candidate gene in pathogenesis of myeloid malignancies. Blood cancer journal. 2013;3:e117. doi: 10.1038/bcj.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papapetrou EP, et al. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci U S A. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papapetrou EP, Korkola JE, Sadelain M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells. 2010;28:287–296. doi: 10.1002/stem.257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.