Abstract
This mini-review discusses the evolution of fluorescence as a tool to study living cells and tissues in vitro and the present role of fluorescent protein biosensors (FPBs) in microphysiological systems (MPSs). FPBs allow the measurement of temporal and spatial dynamics of targeted cellular events involved in normal and perturbed cellular assay systems and MPSs in real time. FPBs evolved from fluorescent analog cytochemistry (FAC) that permitted the measurement of the dynamics of purified proteins covalently labeled with environmentally insensitive fluorescent dyes and then incorporated into living cells, as well as a large list of diffusible fluorescent probes engineered to measure environmental changes in living cells. In parallel, a wide range of fluorescence microscopy methods were developed to measure the chemical and molecular activities of the labeled cells, including ratio imaging, fluorescence lifetime, total internal reflection, 3D imaging, including super-resolution, as well as high-content screening. FPBs evolved from FAC by combining environmentally sensitive fluorescent dyes with proteins in order to monitor specific physiological events such as post-translational modifications, production of metabolites, changes in various ion concentrations, and the dynamic interaction of proteins with defined macromolecules in time and space within cells. Original FPBs involved the engineering of fluorescent dyes to sense specific activities when covalently attached to particular domains of the targeted protein. The subsequent development of fluorescent proteins (FPs), such as the green fluorescent protein, dramatically accelerated the adoption of studying living cells, since the genetic “labeling” of proteins became a relatively simple method that permitted the analysis of temporal–spatial dynamics of a wide range of proteins. Investigators subsequently engineered the fluorescence properties of the FPs for environmental sensitivity that, when combined with targeted proteins/peptides, created a new generation of FPBs. Examples of FPBs that are useful in MPS are presented, including the design, testing, and application in a liver MPS.
Keywords: Microphysiology systems, fluorescent protein biosensors, high-content screening, fluorescent probes, fluorescent proteins, fluorescence microscopy
Introduction
This mini-review describes the development and use of fluorescent protein biosensors (FPBs) from a historical perspective and then discusses the use of these tools in microphysiology systems (MPS). MPS aim to reproduce significant human and/or animal organ physiology on a small scale, typically from the milli-human to the micro-human scale, usually based on mass or volume of the organs.1–3 MPS integrates multiple cell types representing key organ functions engineered to reflect maximal, native, 3D structure and function in microfluidic devices.1,2,4,5 This review will discuss FPBs in the context of MPS, an area of active investigation by the authors. A summary of FPBs is presented in the context of demonstrations of the design, testing, and use of these tools. It is expected that FPBs will have a major impact on the use of MPS to study normal organ physiology, drug toxicity, and disease models in single organ MPS, as well as in multiple, integrated organ MPS to investigate more complex physiological processes involved in organ interactions.
Fluorescence-based reagents have been used extensively to elucidate and quantify fundamental biological processes within and between cells both in vitro6–9 and in vivo.10,11 Our focus in this mini-review is on in vitro applications. The use of one category of fluorescence-based reagents, FPBs, to define and quantify the temporal–spatial dynamics of protein functions has been well established in the literature.7 FPBs can be defined as sensors containing two component systems: a sensing domain that recognizes a specific molecular modification or binding partner that is linked to a reporter module that generates the fluorescence signal. Sensing domains can detect specific ligand(s), post-translational modifications, protein–protein interactions, conformational changes, reflect the cellular microenvironment (e.g. pH), and other relevant molecular/cellular processes. The detection of events occurs via altered fluorescence spectroscopic property(s). FPBs can exhibit a change in fluorescence excitation or emission wavelengths, fluorescence intensity, fluorescence lifetime of the excited state, or a change from a non-fluorescent to fluorescent state upon activation or vice versa.8
Despite major challenges, the relatively new field of MPS is exhibiting rapid progress.1 An important goal for the MPS field is to refine, reduce, and ultimately replace the current “gold standard” of animal-based toxicity and disease models that are not fully concordant with human toxic liabilities and disease processes.12 A major goal is to create a “human or partial human on a chip” that links multiple human organ modules to model key functions such as drug absorption, metabolism, and toxicity. The authors are focused on the implementation of a human liver on a chip and, as part of a broad effort with collaborators, the coupling of the liver with gut and kidney organs on chips. Historically, drug-induced liver injury (DILI) was the most common cause for postmarket pharmaceutical drug withdrawal and continues to be a leading cause of drug attrition.13 The potential exists to improve the early identification of DILI that arises from the exposure to toxic substances and intermediates, using MPS models and real-time monitoring of multiple mechanisms of toxicity (MOT), such as alterations in intracellular calcium flux, the generation of reactive oxygen species, and apoptosis.14 We have developed a human, 3D, microfluidic, four-cell, sequentially layered, self-assembly liver model (SQL-SAL) for studying liver toxicology and disease.15 Fundamental components of the SQL-SAL include the use of FPBs for real-time analyses of mechanisms of toxicity and disease via high-content screening (HCS) and the integration of a microphysiological system database to capture, analyze, and model data generated within the MPS, in the context of reference data available from external databases).*16
FPBs: A historical perspective
FPBs evolved from an early technology called fluorescent analog cytochemistry (FAC), originally named molecular cytochemistry.17–20 This technology involved the purification of a target protein, the covalent labeling with an environmentally insensitive fluorescent dye, the demonstration of native functions in vitro, incorporation of the analogs into living cells through microinjection or bulk loading methods, and then microscopic analysis. In addition, a large number of fluorescent probes engineered through organic chemistry have been developed to measure intracellular physiological parameters including membrane potential, pH, pCa++, and a growing list of metabolites.6,7,21
A variety of fluorescence microscopy methods have been developed to quantify the temporal–spatial dynamics of the fluorescent analogs and fluorescent probes.18,22–32 Ratio imaging enabled quantitation without 3D reconstruction of the signals33 and permitted the quantification of cellular pH, pCa++ and other environmental factors, as well as the activation of a variety of biosensors.6,7,34–36 Recent advances in light microscopy, such as super-resolution microscopy, go beyond the diffraction barrier to image at greater resolution.37 HCS was developed to create an automated platform to acquire image data and then analyze, display, database, and report on the data from a large number of cells/tissues or even small experimental organisms.38–42 Significant statistical analyses on large datasets from HCS have demonstrated the critical role of heterogeneity in biological processes and the importance of measuring it in experimental studies.43,44 Measuring and interpreting the temporal and spatial heterogeneity in the responses of the MPS disease and toxicity models will be a critical component of investigations using MPS.
A natural progression from labeling proteins with environmentally insensitive fluorescent dyes to quantifying protein dynamics in living cells was to label targeted proteins and/or protein fragments with environmentally sensitive fluorescent dyes6,7,44 to measure dynamic chemical and molecular changes in living cells.6,34,35,45–48 The reagents, originally named “optical biosensors” have been termed FPBs, involved: the purification of the wild type or site-specific modified protein; site-specific covalent labeling with an environmentally sensitive dye;6,48 the demonstration of native functions in vitro, including sensing the specific event such as binding ions, metabolites, proteins, and other macromolecules; and then incorporation into living cells through microinjection or bulk loading methods. These early reagents, FAC and FPBs, were difficult to design, construct, and to deliver to cells, but played a critical role in defining a variety of mechanisms of cellular functions and paved the way for the development of the next-generation molecular-based reagents.49,50
The advent of using molecular biology to “label” proteins by fusing the DNA sequence of the protein with the DNA sequence of a fluorescent protein such as green fluorescent protein (GFP) accelerated the use of fluorescence-based reagents in living cells and will be the focus of the remainder of the mini-review.
Genetically encoded FPBs
The discovery and implementation of intrinsically fluorescent proteins, termed fluorescent proteins (FPs), from aquatic species has had a broad impact on the use of fluorescence and FPBs in biological experiments.51 The GFP, originally obtained from the jellyfish (Aequorea victoria) and later derivations through mutagenesis, as well as the discovery of novel target proteins, provided a wide range of fluorescence excitation and emission options for analyzing fusion proteins of interest.52 The use of FPs made the creation of fluorescent analogs and FPBs simpler and more powerful. Subsequent protein engineering resulted in the emergence of genetically encoded FPBs that could be used to transfect or transduce cells and detect protein–protein interactions based on fluorescence resonance energy transfer (FRET).31 Protein engineering of GFP and related FPs has produced distinct FPs with a variety of excitation and emission properties.53 Table 1 presents the major modes of genetically encoded FPBs available and lists specific examples and key reviews related to the class of FPBs. Intrinsic fluorescence modes of FPBs are based on the inherent fluorescence of FPs, while extrinsic fluorescence modes of FPBs are based on the genetic incorporation of the sensing portion of the biosensor and the diffusion of the reporter module into the cells after expression of the targeting portion in the cells. Table 2 lists some commercial sources of standard FPB constructs that can be used to build specific FPBs and provides a starting point for researchers to begin evaluation of suitable candidate FPBs. The broad spectrum of FPs available allow researchers to fuse de novo specific protein tags or use pre-existing subcellular tagged FPs to monitor subcellular structures (Evrogen, Addgene, Systems BioSciences Inc.). PAmCherry (Clontech) is an example of a photo-inducible sensor whereby activation with ultraviolet light induces an alteration in the conjugation of the chromophore such that 560 nm light is absorbed and fluorescence emitted at 590 nm.54 Various FRET-based FPBs are available to detect post-translational modifications of proteins (Addgene, Clontech, Evrogen). Translocation and receptor modulation sensors can monitor cell membrane dynamics, as has been established for GPCR oligomerization on COS7 cells55 (Promega). Various cell cycle and protein–protein interaction biosensors are available, largely based on detection by split GFP (BiFC) or monitoring the production of fluorescent fusion proteins attached to cyclins56 (Invitrogen, Clontech). The characteristics of optimal protein–protein interaction FPBs have been discussed in detail elsewhere.50 It is also possible to use FPs to create biosensors of gene expression.57 However, we limit our discussion in this mini-review to the use of functional protein biosensors.
Table 1.
Major modes of genetically encoded fluorescent protein biosensors (FPBs) with specific examples
| Fluorescence mode | FPB type | Specific examples | Reference(s) |
|---|---|---|---|
| Intrinsic fluorescence | Engineered fluorescent proteins | ||
| Fluorescent proteins | GFP | 118 | |
| Spectral variants | Cerulean & mCherry | 119 | |
| Ca++ | Pericam, Cameleons | 53, 73, 120 | |
| pH | pHluorins, EGFP | 121, 122 | |
| Fluorescent protein fusions | |||
| Complementation | BiFC* | 123 | |
| Modified fluorescent proteins | cpFP† | 123 | |
| FRET photoconvertible | CFP → YFP FRET | 52 | |
| Intracellular localization | PAFPs‡, FRAP, P-PIB§ | 8, 31, 50, 124 | |
| Intracellular chloride | Clomeleon | 125 | |
| Optogenetic biosensors | |||
| Action potential modulation | Halorhodopsins | 65 | |
| Channel rhodopsins | 63, 126 | ||
| Archaerhodopsins | 65 | ||
| Voltage sensitive | Mermaid | 69 | |
| Calcium indicators | GECI | 66, 127 | |
| Extrinsic fluorescence | Antibody-based systems | FAP** | 128 |
| Quench bodies | 129 | ||
| Covalent labels | SNAP-tag, HaloTag | 130,131 | |
| FlAsH | 9 | ||
| Inteins | 132 | ||
| Biotinylation | 132,133 | ||
Bimolecular fluorescence complementation.
Circularly permuted fluorescence protein.
Photoactivatable fluorescence proteins.
Protein–protein interaction biosensor.
Fluorogen-activating protein.
Table 2.
Some commercial sources of FPBs
| Company | Types of FPBs offered |
|---|---|
| Evrogen | Ion detection, photoinducible, photoactivatable, subcellular localization, FRET, cell death http://www.evrogen.com/ |
| Invitrogen | Subcellular tracking, cell cycle http://www.lifetechnologies.com/us/en/home.html |
| Systems BioSciences | Subcellular localization http://www.systembio.com/ |
| Promega | Protein dynamics, subcellular tracking, protein–protein interaction, protein translocation assays http://www.promega.com/ |
| Addgene | Subcellular tracking, various ions and small molecules, protein–protein (FRET) pairs, protein modification https://www.addgene.org/ |
| Clontech | Cell cycle, photoactivatable, photoconvertible, subcellular labeling, protein–protein (FRET) pairs, destabilized proteins http://www.clontech.com/ |
Subsequent derivations of FPs led to novel photodynamic properties and biosensors. For example, the covalent linkage of two FPs with distinct excitation and emission properties produced FPBs with FRET capabilities, known as FRET-based sensors.58 These constructs contain a donor and acceptor FRET pair, such that the donor FP when in close physical proximity to the acceptor FP allows for energy transfer from donor to acceptor FP.9 This configuration has since been altered for biosensors to monitor conformational changes induced by small molecule ligands/ions (e.g. Ca++), protein cleavage sites (e.g. caspases), and post-translation modifications (e.g. phosphorylation).59 Further experimentation produced hybrid FPs consisting of artificially cleaved and ligated protein configurations. For example, the circularly permutated FPs create new amino and carboxyl termini by cleavage which results in altered fluorescence spectra. The circularly permutated fluorescent proteins have been used to create additional biosensors for Ca++.60
Specialty FBPs
A number of fast kinetic, genetically encoded light sensitive proteins have been developed that can reversibly activate, silence, or report on molecular processes of the action potential and other cell functions in neuronal cells and circuits. Halorhodopsins (chloride channel), channel rhodopsins (light gated ion channel), and archaerhodopsins (proton pump) are examples of photoactivatable proteins that are inserted into the genome to control neuromodulatory events in vitro or in vivo.61–65 Other FBPs have been engineered to monitor the rapid molecular processes associated with chloride and calcium ion modulation, as well as voltage-sensing membrane proteins.65–69 These FPBs allow long-term, non-invasive imaging of neuronal and other cell types by converting physiological signals into measureable changes of intrinsic fluorescence on the order of milliseconds to seconds. Furthermore, the FBPs developed to modulate or monitor neuronal activity can be encoded into the genome by a variety of viral and non-viral methods, and hence, can be successfully used in mammalian and non-mammalian cells for targeting specific cell populations.61,66,68
Intrinsic engineered FPs include spectral variants of GFP, FP fusions used to monitor protein dynamics and post-translational modifications as well as calcium and pH biosensors. Engineered fluorescent fusion proteins include photoactivatable (PAFPs) or photobleachable Fluorescence recovery after photobleaching (FRAP) protein fusions that undergo spectral changes when exposed to a particular wavelength of light and are used to track protein dynamics.70,71 Potentially interacting proteins can be investigated with bimolecular protein complementation (BiFC) linking proteins under investigation to split domains of FPs.72 Intracellular calcium quantitation includes the ratiometric, FRET-based cameleon biosensor utilizing calmodulin and an M13 calcium sensing domain.53 The calcium sensor pericam also provides reversible calcium sensing using a protein scaffold that is based on circularly permutated GFP.73 The EGFP derivative of GFP, and subsequently the pHluorins, allow for pH measurements in the cytosol and other organelles with spectral shifts occurring at different pH ranges.74,75
Extrinsic FPBs are covalent or non-covalent protein labels that are not inherently fluorescent but selectively bind fluorescent molecules. The FlAsH and HaloTag, for example, are peptide fusion tags that bind different fluorescent reagents and may be advantageous when larger FP fusions perturb protein function or localization. Fluorogen activating proteins (FAPs) permit protein localization, dynamics, and pH studies through fusions of proteins of interest to single-chain antibodies that bind fluorophores with low background fluorescence such that the fluorophores exhibit a large increase in fluorescence signal only when bound to the FAP.76 FAPs also do not require washout of excess reagent prior to imaging due to the low fluorescence background of the fluorophore when not bound to the FAP protein fusion.77
FPBs versus the use of diffusible fluorescent probes in MPS
An exciting next frontier for FPBs is their application in MPS. There are several advantages to the use of stable FPBs in MPS compared to widely used diffusible fluorescent probes.21 Most fluorescent probes are not targetable to specific cells and therefore cannot be directed to a certain cell type, or specific subcellular compartments in only those cells, within a complex, multicellular system such as that found in an MPS. The lack of cell type targeting may lead to non-specific interference from neighboring cells when performing experiments. A further limitation of diffusible fluorescent probes is the relatively short half-life in cells. The fluorescent probes are pumped out or sequestered and must be re-added to the cells when studying the MPS over extended times. Another consequence of using diffusible fluorescent probes is the relatively high cytotoxicity induced upon excitation. Phototoxicity can occur within cells due to the generation of reactive oxygen radicals.78 Most of the fluorescent probes exhibit greater phototoxicity when compared to FPs exposed to the same dose of light (Unpublished observation).Light-induced cell damage is of particular concern in MPSs that aim to function for several weeks in order to monitor chronic toxicities or disease states. The critical role of minimizing the dose of illumination in live cell studies has been discussed elsewhere.79
In contrast to diffusing fluorescent probes into cells, FPBs that are genetically encoded can be targeted to specific cells and subcellular compartments before assembling multiple cell types into the MPS. Integration of the FPBs into the cellular genome allows the expression of the biosensors over extended times. Inducible systems or gene-specific promoter elements can also be used to “turn on” the expression of the selected biosensor, increasing the control of the measurements. Some examples of inducible gene expression include the Tet-On system (Clontech) that allows for inducible gene induction or repression in response to doxycycline in the culture media.80 This system has been recently expanded upon in an effort to create lentiviral delivery of the reverse tetracycline-controlled transactivator to ease development of the transactivator cell line.81 Gene-specific promoter elements can be placed upstream of the FPB encoding region for cell-type specific temporal and spatial expression.82 Such molecular approaches can then be targeted into a subpopulation of cells within the overall MPS device. The subpopulation of cells in an MPS that have been transduced with FPBs has been termed “sentinel cells.” The sentinel cells allow for real-time monitoring of key physiological events and the development of specific molecular signatures.16 The variety of different excitation and emission properties of FPB and FP allows a great deal of flexibility in terms of multiplexing different sentinel cells within an MPS.
Methods to deliver FPBs to cells
A key step in biosensor implementation is determining the optimal method of delivery into target cells to maximize efficiency and minimize toxicity or other side effects. Several examples of these methods are summarized in Table 3. For initial studies designed to test the newly created FPBs, easily cultured and transfected cells can be transiently transfected.83 However, not all cell lines are amenable to chemical transfection nor will the delivery gene be integrated into the host cell genome in the absence of some of selection procedures.84 Viral delivery methods, such as adenovirus transduction, are often used for primary or otherwise difficult cell lines.85 Lentiviral particles can be produced with the FPB construct of interest whereby the FPB is randomly inserted into a target cell genome. The advantage of using the lentiviral approach is that most cell lines, primary cells, and induced-pluripotent stem (iPS) iPS-derived cells can be transduced for evaluation of new FPBs.86 The use of site-specific gene targeting has been a topic of great interest recently and is discussed in detail below in regards to iPSC.87,88,89
Table 3.
Methods for the incorporation of genetically encoded FPBs into cells
| Technique | Advantage | Limitation | Review(s) | |
|---|---|---|---|---|
| Transient transfection | Rapid | Temporary expression Low efficiency Cytotoxicity | 83 | |
| Virally mediated transfection | Primary cell compatible Transient or stable | Random integration Cytotoxicity | 85 | |
| Electroporation-based methods | High efficiency Primary cell compatible | Cytotoxicity Expensive | 84 | |
| Knock-in | Targeted and stable gene insertion Replaces native gene | Time consuming Requires significant expertise | 89 | |
| Gene editing | Targeted and stable gene insertion Replaces native gene | Time consuming Technically challenging Expensive | 87, 88, 95 | |
Case study: Implementation of FPBs in the liver MPS (SQL-SAL)
Strategy of testing functions of FPBs in cells
The successful application of FPBs involves a phased approach of implementation and validation (Figure 1). This approach must balance practical as well as functional parameters of the chosen FPBs for testing and eventual use in MPS. The phased approach for this purpose begins by testing the function of FPBs in relevant cell lines grown in static cultures in microplates starting with the transfection of the cells with the FPB construct. The best candidate FPBs suited for a particular MPS are then chosen including any further molecular engineering of the constructs. The FPBs are then tested and validated in human primary cells in static culture using the lentiviral delivery system, since this system can transduce many types of cells. As a final step, induced-pluripotent stem cells matured to the selected cell types and transduced with the lentiviral system are tested. The use of iPSC is advantageous for two reasons: (a) provides unlimited source of otherwise expensive, difficult to obtain, and/or variable quality human primary cells; and (b) allows for the use of patient- or disease-specific cells for study. iPSC can also be modified by targeted genetic manipulations.90,91
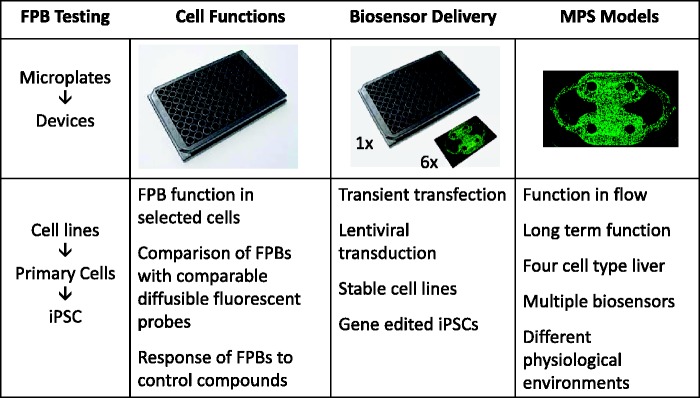
Figure 1.
Diagram of the implementation of fluorescent protein biosensors (FPBs) in microphysiological systems (MPS). The first phase involves testing of the function of FPBs in immortalized cell lines, primary cells, and induced pluripotent stem (iPSC) cells in static plate cultures that utilizes transient or stable gene delivery methods. The second phase involves optimal delivery to primary cells and iPSC in both microplates and microfluidic devices (microfluidic device is enlarged 6 x relative to the microplate for visualization purposes). The final stage involves the incorporation of the FPBs in MPS models that include multiple cell types, media perfusion, and multiplexing of different FPBs to model distinct physiological environments and disease states for long-term function.
The development of MPS systems can take advantage of cell lines that are easy to grow and are less expensive to test the engineering, biocompatibility of materials, robustness, and reproducibility in the context of the requirements for high-quality imaging. Cell lines are also useful for testing and characterizing the quantitation of FPB readouts as dictated by the particular application of the MPS. Initial work performed in our lab used immortalized cell lines, such as HepG2, starting in static culture to determine the optimal strategy to validate the FPBs for HCS. The rationale for this approach was to test specific FPBs, in a cost-effective and less time-consuming way, for use as live-cell sensors (“sentinel cells”) that can potentially give predictive data of compromised liver health and function. Investigations can then be performed in static plate cultures of primary human cells, in our case primary human hepatocytes, to confirm the organ-specific utility of a particular FPB. Subsequent validation in the relevant primary cells can be performed with drugs and biological modulators with known mechanisms of action.
Finally, the use of FPBs in MPS allows for closed loop testing. Closed loop testing is the process by which the readouts from the system are used to modulate the experimental protocol. For example, the real-time readout of ROS can be used to maintain a low level of ROS by adjusting the test compound concentration, allowing a better determination of the chronic effects of ROS generation. Although a follow-up study can be initiated using a concentration optimized from an earlier study, the advantage of live readout FBPs allows for real-time dosing adjustment during the ongoing study for a specific measurement such as defining the no observed adverse effect level, the maximum tolerated dose, or the optimal concentration for drug efficacy or disease treatment.
Optimizing delivery of FPBs into cells
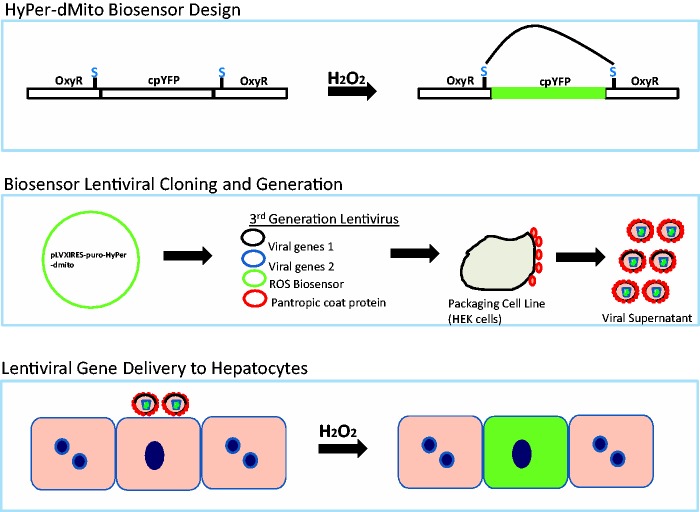
The second phase of implementation is optimizing the delivery of FPBs. As a case study for using a stable lentiviral gene delivery system for FPB incorporation into MPS, primary human hepatocytes were investigated. The use of primary human hepatocytes in the SQL-SAL is preferred over cell lines due to their retention of liver metabolic and clearance functions in vitro. Figure 2 shows an example of steps required to incorporate a commercially available FPB construct, in this case the HyPer FPB, into a lentiviral expression vector. After testing and initial studies in HepG2 liver carcinoma cells confirmed the ROS sensitivity, the lentiviral supernatant was incubated with primary human hepatocytes. Transduced hepatocytes were then used for validation studies and subsequently incorporation into MPS models. The efficiency of transduction ranged from ca. 30 to 60% in human primary hepatocytes and was higher in HepG2 cells.
Figure 2.
Schematic representing steps required for generation of lentiviral packaged FPBs. The top panel shows the concept of the biosensor design of the hydrogen peroxide sensing FBP (Hyper-dMito, Evrogen) with the cpYFP inserted into the oxidation sensitive bacterial transcription factor, OxyR. In the presence of hydrogen peroxide a disulfide bridge is formed in OxyR, resulting in fluorescence signal from the cpYFP. The middle panel describes the subcloning of the HyPer-dMito into a lentiviral vector and lentiviral packaging. The lower panel indicates lentiviral supernatant incubated with primary human hepatocytes to produce a subpopulation of hepatocytes stably expressing the Hyper-dMito. The subset of cells containing a FBF to provide live cell readouts is referred to as sentinel cells.
The reproducibility of delivery of FPBs into cells would improve with targeted insertion rather than random FPB integration into the genome. Considering the technical challenges of targeting the insertion site on the host genome in primary cells, it is likely such methods are best deployed with iPSC planned for the final design of an MPS. The latter would provide a renewable resource of cells with an FPB of interest that could be differentiated into the required adult cell type. Knock-in strategies may be used to perform specific gene targeted insertions, using Cre/lox methods for gene targeting.89 More recently, gene editing methods of zinc-finger nucleases,92 TALENs,93 and CRISPR-based editing94 have all been used to modify iPSCs. These methods are evolving rapidly and allow for targeted insertion events that do not leave extensive non-gene-associated flanking DNA sequences, thereby decreasing the size of the insertion and reducing any potential interference from insertion of non-gene DNA.
When comparing CRISPR-Cas9 gene editing to lentiviral gene delivery, lentiviral-based methods are more straightforward but with a higher potential for confounding artifacts due to random insertions. Gene editing techniques, while growing in popularity, require careful controls to be performed to screen for off-target effects of FPB insertion.95 This has been of particular concern with CRISPR-Cas9, due to the small base pair nuclease recognition site that has the potential for off-target cutting.96 Good experimental design can reduce the potential for off-target effects. For example, multiple CRISPR-Cas9 nucleases can be designed for a single target cut site and two or more separate cell lines can be carried forward into development and phenotype screening,97 or double-nicking nucleases can be used to increase the cleavage recognition site.98 These efforts may vary in their success rate and thus delay progress of FPB incorporation into iPSCs, making these efforts worthwhile only after demonstration and validation of physiological responses. Taken together, and until iPSC knock in lines are more readily available, lentiviral gene delivery is a rapid, stable, and reliable method to deliver FPBs across many cell types. The lentiviral delivery also allows experiments to be performed when time and cost are at a premium.
Validation of FPBs for liver functions
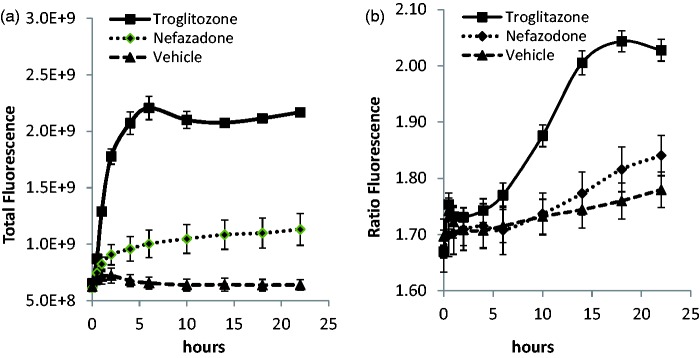
Figure 3 depicts the validation of the HyPer-dMito ROS biosensor (Evrogen) transduction of HepG2 cells. Drugs known to induce ROS production were used to test both a standard diffusible fluorescence probe for ROS, as a positive control, and the FPB. Hepatocellular damage is known to occur in response to the generation of mitochondrial reactive oxygen species.99 Mitochondrial ROS detection in both parenchymal and non-parenchymal cells is therefore an important molecular signature for early indication of liver damage. Dihydroethidium (DHE) reacts with superoxide radicals and the resulting product is a red-fluorescent, DNA binding 2-hydroxyethidium100 that intercalates into DNA.101 Either DHE was added to the media and diffused into the HepG2 cells, or the cells were transduced with a lentiviral HyPer-dMito FPB, and the cells were treated with either vehicle or a liver toxic drug (nefazodone or troglitazone). HCS was used to quantify the generation of ROS. Troglitazone and nefazodone, but not vehicle, produced ROS species as detected by increased DHE fluorescence (Figure 3(a)). The attenuated nefazodone response, compared to troglitazone, is due to both DHE substrate depletion,102 as well as a reduction in nefazodone-generated ROS in metabolism-deficient HepG2 cells. The HyPer-dMito FPB exhibited ROS-induced fluorescence when challenged with troglitazone and nefazodone comparable to the response observed with DHE (Figure 3(b)). The HyPer-dMito FPB can be measured over at least 28 days and is reversible with drug washout (data not shown), while DHE diffuses out of cells over time and is not reversible. We found that analysis by ratiometric or single wavelength emission exhibited increased fluorescence signal versus vehicle control (Figure 3(b)). See supplemental materials for details.
Figure 3.
Determination of mitochondrial ROS induction by hepatotoxic drugs. (a) Monitoring ROS production with a standard diffusible probe, dihydroethidium (DHE) fluorescence, in HepG2 cells in static culture treated with 60 µM troglitazone, a validated drug that induces ROS, 60 µM nefazodone that also induces ROS production, and 1% DMSO in media (vehicle). Error bars are ± standard error of the mean on duplicate wells and fields. All curves are normalized to the initial time 0 value. (b) Mitochondrial ROS production (hydrogen peroxide measured with the HyPer-dMito biosensor fluorescence) in lentiviral transduced HepG2 cells treated with 60 µM nefazodone, 60 µM troglitazone, and vehicle. Data are presented as total fluorescence or ratiometric values of 488/405 nm. Error bars are ± standard error of the mean on duplicate wells and fields.
The HyPer-dMito sensor can be used in live cell experiments to monitor generation of mitochondrial hydrogen peroxide.36 Importantly, this FPB is sensitive to low concentration ranges of liver toxic drugs, including troglitazone, similar to what has been previously described. Ratiometric imaging can be used to control for heterogeneous biosensor expression within a population of cells and the potential for pH effects. Use of only the 488 nm excitation, however, does indicate overall generation of hydrogen peroxide. The use of the single wavelength excitation might be useful to limit phototoxicity within a MPS that must be viable for several weeks and a relative measure of ROS change is sufficient.
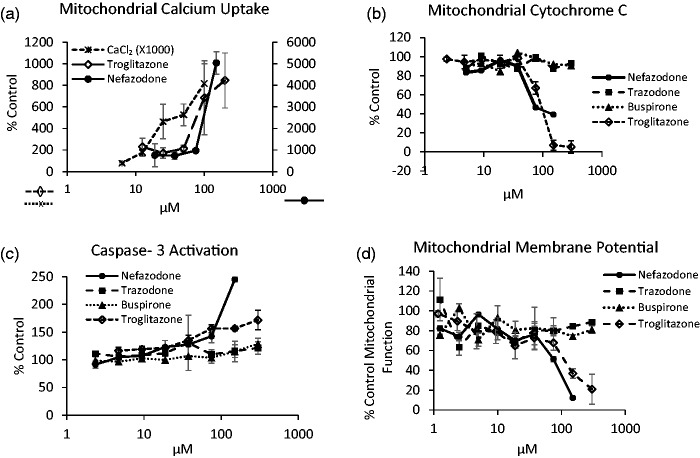
Apoptosis is an important MOT that can result in hepatic injury. Loss of hepatocytes, as well as the programmed cell death of liver non-parenchymal cells such as Kupffer cells has been associated with DILI.103 Early determination of hepatocellular damage and apoptosis would provide better insight into the relevant MOT of drug treatment under investigation.104 Mitochondria are a key organelle during the induction of apoptosis, as the loss of the mitochondria membrane potential and integrity can result in the release of pro-apoptotic factors such as cytochrome c.105 Additionally, mitochondrial calcium fluxes are associated with the onset of apoptosis, as well as additional liver cell pathology.106 Three FPBs provide three different indications of the induction of apoptosis: mitochondrial calcium uptake, cytochrome c release, and caspase-3 activation. These three FPBs were evaluated in HepG2 cells and compared to the potentiometric mitochondrial probe tetramethylrhodamine ethyl ester (TMRE) or titrations of calcium into calcium free media (Figure 4).
Figure 4.
Steps in the induction of intrinsic apoptosis established through the use of three genetically encoded biosensors and the diffusible probe TMRE in HepG2 cells in static culture. (a) Apoptosis induction resulting from mitochondrial calcium uptake measured by the biosensor Case12-mito (Evrogen). Mitochondrial free calcium is increased at 16 h following addition of CaCl2 in microplates containing Case12-mito transduced HepG2 cells or by addition of the hepatotoxins nefazodone (2nd axis) and troglitazone. (b) Apoptosis arising from mitochondrial damage is monitored at 16 h in HepG2 cell transduced with the cytochrome c-GFP biosensor in response to nefazodone and troglitazone but not the non-hepatotoxins trazodone or buspirone. (c) Apoptosis induction monitored at 16 h by the activation of caspase-3 in HepG2 cells transduced with the Casper-3BG (Evrogen) FPB by nefazodone and troglitazone but not trazodone or buspirone. (d) Mitochondrial membrane potential in HepG2 cells with TMRE (200 nM) for 1 h demonstrates the decrease in mitochondrial membrane potential as a loss of 605 nm fluorescence under increasing nefazodone and troglitazone dosing. Values represent mean and SD of triplicate measurements.
Case12 is a FPB that indicates cytosolic or mitochondrial calcium concentrations depending on the targeting peptide engineered into the FPB and can be applied as an apoptosis FPB.107 The reversible FP provides sensitivity to nanomolar concentrations of intracellular or mitochondrial calcium. It is based on a circularly permutated yellow fluorescent protein modified with the calcium-binding domain of calmodulin and an M13 peptide. The biosensor is not fluorescent until calcium ions bind and induce a conformational change that restores the fluorescence emission of the FP chromophore.108 Case12-transduced HepG2 cells responded to titrations of calcium into media as well as the hepatotoxins troglitazone and nefazodone (Figure 4(a)), compounds known to increase cytosolic calcium.109
Another apoptosis FPB uses a mitochondrial targeting sequence derived from cytochrome c oxidase VIII linked to copGFP (Systems Biosciences Inc.) that is a surrogate for cytochrome c release from mitochondria. HepG2 cells transduced with the biosensor packaged in a lentiviral delivery system (Figure 2) indicate a decrease in the mitochondrial fluorescence following treatment with nefazodone and troglitazone but not buspirone or trazodone (Figure 4(b)).
Finally, the FRET-based casper-3BG (Evrogen) FPB is specific for caspase-3 cleavage, with improved efficiency.110,111 The biosensor construct is comprised of two covalently linked FPs, TagBFP and TagGFP2, containing the caspase-3 amino acid cleavage recognition sequence DEVD in the linker (Evrogen). An increase in the expression and activation of caspase-3 results in a loss of FRET in the casper-3BG biosensor and an increase in 450 nm fluorescence. Nefazodone and troglitazone are compounds reported to activate caspase-3 in human liver cells112,113 but not buspirone or trazodone in agreement with our results (Figure 4(c)). The results can be compared to the loss of the mitochondrial potential, as indicated by TMRE fluorescence, with nefazodone and troglitazone, but not buspirone or trazodone (Figure 4(d)).
Table 4 lists some of the commercially available, genetically encoded FPBs constructs tested in our liver MPS program and Table 5 demonstrates the validation of the biosensors, using standard screening statistics. Each of these probes in combination with real-time, live cell imaging provides data for determining MOT for toxicological assessment.40,114
Table 4.
Selected commercially available, genetically encoded FPBs used in liver MPS
| FPB | Source | What it indicates | References |
|---|---|---|---|
| HyPer-dMito | Evrogen | H2O2 concentration | 36 |
| Case12-Mito | Evrogen | Mitochondrial calcium uptake | 107 |
| Casper3-BG | Evrogen | Caspase-3 activity | 110 |
| pCT-Mito-GFP | Systems BioSciences | Mitochondrial membrane integrity | 134 |
| pCT-H2B-GFP | Systems BioSciences | Cell localization | 134 |
Table 5.
Validation of FPBs in primary hepatocytes
| FPB | FPB colors | Control compound | Conc (µM) | SSMD |
|---|---|---|---|---|
| Nuclear/cell position (Histone H2B-FP) | Blue Green Red | Menadione | 100 | >2 (at 5 h) |
| Apoptosis (cytochrome C oxidase VIII subunit tagged-FP) | Green Red | Menadione | 100 | >2 (at 5 h) |
| Mitochondrial reactive oxygen species (HyPer-dMito) | Green | Menadione | 100 | >2 (at 2.5 h) |
| Mitochondrial calcium uptake (Case12-mito) | Green | Menadione | 50 | >2 (at 4.5 h) |
The early success of monitoring liver-specific MOTs has encouraged ongoing efforts in our lab to develop new biosensors to monitor additional crucial liver cell pathologies, such as general oxidative stress fluctuations and non-alcoholic fatty liver disease, to name two.
Example of FPBs used in a liver MPS device
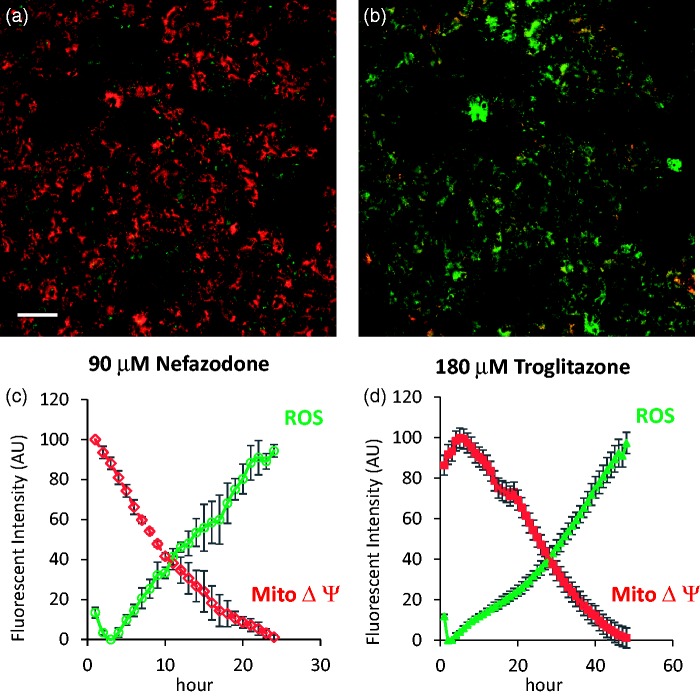
The final phase in the development and application of FPBs is the incorporation of the “sentinel” cells that contain the FPBs into an MPS organ model. Figure 5 demonstrates the functioning of the ROS FPB in a two-cell type (hepatocyte and endothelial cell) human liver MPS treated over a 24 h period with an acute dose of nefazodone or troglitazone, showing the loss of the membrane potential followed closely by the increase of the ROS production. The FPBs remained functional over at least 28 days.15
Figure 5.
Demonstration of HyPer-dMito FPB activity in a human two cell liver MPS model. (a) Image of TMRE fluorescence and HyPer-dmito fluorescence at time 0 in human primary hepatocytes in a microfluidic device also containing endothelial cells. Image is dominated by TMRE (red) from active mitochondria (white bar = 50 µm). (b) Image of TMRE fluorescence and HyPer-dMito fluorescence in the same field after 20 h exposure to nefazodone (90 µM). Image is dominated by HyPer-dMito (green) following loss of mitochondrial membrane potential and the generation of ROS. (c) Time course of induction of ROS and loss of mitochondrial membrane potential resulting from exposure to nefazodone (90 µM). (d) Time course of induction of ROS and loss of mitochondrial membrane potential resulting from exposure to troglitazone (180 µM). Values are linearly scaled from minimum to maximum and represent the mean ± SD of nefazodone (n=3) or troglitazone (n=4) images.
Pitfalls/precautions in using FPBs
FPBs are powerful tools for real-time monitoring of physiology in vitro. However, care must be taken to ensure proper controls are performed such that the interpretation of the data is not influenced by artifacts. Many of the FPs are well documented to be sensitive to perturbations in pH, thereby producing changes in their fluorescence spectra. The HyPer-dMito (ROS) FPB, for example, has been a subject of debate in the literature with regards to increases in the fluorescence measured at a single excitation wavelength that may be attributable to changes in pH.115 Controls for pH changes can include the use of diffusible probes or genetically encoded pH sensing FPBs with different spectral properties from the functional biosensor of interest.116 For example, we compare performance of FPBs in static microplate cultures with pH sensitive, diffusible probes (e.g. SNARF-1AM, Molecular Probes, data unpublished). Useful discussion related to proper controls and imaging of diffusible probes has been published.117 Many drugs are fluorescent and the fluorescence level of the drug in the cells without a biosensor can be used to correct for the background, provided it is identified before the experiment. It is advisable to perform compound or drug dosing studies with the cells of interest in microplates first and to optimize imaging conditions to identify drug fluorescence before investing in the use of MPS organ models.
Future of FBPs in MPS
It is clear that real-time readouts of key cell/tissue/organ functions will be an important component of the MPS models. There are a number of valuable FPBs that are already available from commercial suppliers (Table 2) and this is an important starting place for investigators. New FPBs developed for specific organ physiological parameters as well as combining different types of FPBs within an MPS (Table 1) will be explored to increase the ability to measure and to manipulate individual cells within the MPS. For example, monitoring one type of FPB in one cell type while activating another to modulate the cell physiology will permit real-time experimentation and testing.
ACKNOWLEDGEMENTS
We thank the University of Pittsburgh Cancer Institute (UPCI) Vector Core Facility and Dr Robert W. Sobol for assistance with lentiviral materials and constructs. Support for the UPCI Vector Core Facility was provided by the Cancer Center Support Grant from the National Institutes of Health P30 CA047904. This work was supported in part by the National Institutes of Health Common Fund and the National Center for Advancing Translational Sciences (#5UH2TR000503-02, #4UH3TR000503–03, 3UH2TR000503-02S1) and National Institutes of Health P30 CA047904 (University of Pittsburgh Cancer Institute). Research reported in this publication was supported in part by the Office Of The Director, National Institutes Of Health of the National Institutes Of Health under Award Number S10OD012269. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes Of Health. Authors declare no competing interests.
Footnotes
Gough A et al. in preparation.
Authors’ notes
The materials and methods for the experimental data presented here are included in the supplementary material.
Authors’ contributions
All authors participated in the design, interpretation of the studies, and analysis of the data and review of the manuscript; NS, RB, LV, and RD conducted the experiments; NS, LV, RB, AG, and DLT wrote the manuscript. NS and LV have contributed equally to submitted work.
References
- 1.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014; 32: 760–72. [DOI] [PubMed] [Google Scholar]
- 2.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip 2013; 13: 3496–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wikswo JP. The relevance and potential roles of microphysiological systems in biology and medicine. Exp Biol Med 2014; 239: 1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz RE, Trehan K, Andrus L, Sheahan TP, Ploss A, Duncan SA, Rice CM, Bhatia SN. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci USA 2012; 109: 2544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wikswo JP (ed). Annual thematic issue: the biology and medicine of microphysiological systems [Special Issue]. Exp Biol Med 2014;239:1061–1072. [DOI] [PMC free article] [PubMed]
- 6.Giuliano KA, Post PL, Hahn KM, Taylor DL. Fluorescent protein biosensors: measurement of molecular dynamics in living cells. Annu Rev Biophys Biomol Struct 1995; 24: 405–34. [DOI] [PubMed] [Google Scholar]
- 7.Giuliano KA, Taylor DL, Waggoner AS. Reagents to measure and manipulate cell functions. High content screening. Springer, New York, NY, 2006, pp.141–63. [DOI] [PubMed]
- 8.Seward HE, Bagshaw CR. The photochemistry of fluorescent proteins: implications for their biological applications. Chem Soc Rev 2009; 38: 2842–51. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol 2002; 3: 906–18. [DOI] [PubMed] [Google Scholar]
- 10.Provenzano PP, Eliceiri KW, Keely PJ. Multiphoton microscopy and fluorescence lifetime imaging microscopy (FLIM) to monitor metastasis and the tumor microenvironment. Clin Exp Metastasis 2009; 26: 357–70. [DOI] [PubMed] [Google Scholar]
- 11.Niesner RA, Hauser AE. Recent advances in dynamic intravital multi-photon microscopy. Cytometry A 2011; 79: 789–98. [DOI] [PubMed] [Google Scholar]
- 12.Greaves P, Williams A, Eve M. First dose of potential new medicines to humans: how animals help. Nat Rev Drug Discov 2004; 3: 226–36. [DOI] [PubMed] [Google Scholar]
- 13.Holt MP, Ju C. Mechanisms of drug-induced liver injury. AAPS J 2006; 8: E48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bale SS, Vernetti L, Senutovitch N, Jindal R, Hegde M, Gough A, McCarty WJ, Bakan A, Bhushan A, Shun TY, Golberg I, DeBiasio R, Usta OB, Taylor DL, Yarmush ML. In vitro platforms for evaluating liver toxicity. Exp Biol Med 2014; 239: 1180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vernetti LA, Senutovtich N, Boltz R, DeBiasio R, Shun TY, Gough A, Taylor DL. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp Bio Med 2015; epub ahead of print. [DOI] [PMC free article] [PubMed]
- 16.Bhushan A, Senutovitch N, Bale SS, McCarty WJ, Hegde M, Jindal R, Golberg I, Berk Usta O, Yarmush ML, Vernetti L, Gough A, Bakan A, Shun TY, DeBiasio R, Lansing Taylor D. Towards a three-dimensional microfluidic liver platform for predicting drug efficacy and toxicity in humans. Stem Cell Res Ther 2013; 4: S16–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor DL, Wang YL. Molecular cytochemistry: incorporation of fluorescently labeled actin into living cells. Proc Natl Acad Sci USA 1978; 75: 857–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher GW, Conrad PA, DeBiasio RL, Taylor DL. Centripetal transport of cytoplasm, actin, and the cell surface in lamellipodia of fibroblasts. Cell Motil Cytoskeleton 1988; 11: 235–47. [DOI] [PubMed] [Google Scholar]
- 19.Taylor DL, Amato PA, Luby-Phelps K, McNeil P. Fluorescent analog cytochemistry. Trends Biochem Sci 1984; 9: 88–91. [Google Scholar]
- 20.Taylor DL, Wang YL. Fluorescently labelled molecules as probes of the structure and function of living cells. Nature 1980; 284: 405–10. [DOI] [PubMed] [Google Scholar]
- 21.Spence MT, Johnson ID. The molecular probes handbook: a guide to fluorescent probes and labeling technologies. Life Technologies Corporation, Grand Island, NY, 2010.
- 22.Giuliano K, Nederlof M, DeBiasio R, Lanni F, Waggoner A, Taylor D. Multi-mode light microscopy. In: Herman B, Jacobson K. (eds). Optical microscopy for biology, New York: Wiley-Liss, 1990, pp. 543–57. [Google Scholar]
- 23.Taylor DL, Nederlof M, Lanni F, Waggoner AS. The new vision of light microscopy. Am Scientist 1992; 80: 22–35. [Google Scholar]
- 24.Tsien R, Waggoner A. Fluorophores for confocal microscopy. In: Pawley J (ed) Handbook of biological confocal microscopy. Springer, New York, NY, US, 1995, pp.267–79.
- 25.Yengo CM, Berger CL. Fluorescence anisotropy and resonance energy transfer: powerful tools for measuring real time protein dynamics in a physiological environment. Curr Opin Pharmacol 2010; 10: 731–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gradinaru CC, Marushchak DO, Samim M, Krull UJ. Fluorescence anisotropy: from single molecules to live cells. Analyst 2010; 135: 452–9. [DOI] [PubMed] [Google Scholar]
- 27.Hoover EE, Squier JA. Advances in multiphoton microscopy technology. Nat Photon 2013; 7: 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa-Ankerhold HC, Ankerhold R, Drummen GP. Advanced fluorescence microscopy techniques—FRAP, FLIP, FLAP, FRET and FLIM. Molecules 2012; 17: 4047–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo W, He K, Xia T, Fang X. Single-molecule monitoring in living cells by use of fluorescence microscopy. Anal Bioanal Chem 2013; 405: 43–9. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda T, Nagai T. Quantitative measurement of intracellular protein dynamics using photobleaching or photoactivation of fluorescent proteins. Microscopy 2014; 63: 403–8. [DOI] [PubMed] [Google Scholar]
- 31.Miyawaki A. Development of probes for cellular functions using fluorescent proteins and fluorescence resonance energy transfer. Ann Rev Biochem 2011; 80: 357–73. [DOI] [PubMed] [Google Scholar]
- 32.Padilla-Parra S, Tramier M. FRET microscopy in the living cell: different approaches, strengths and weaknesses. Bioessays 2012; 34: 369–76. [DOI] [PubMed] [Google Scholar]
- 33.Bright GR, Fisher GW, Rogowska J, Taylor DL. Fluorescence ratio imaging microscopy: temporal and spatial measurements of cytoplasmic pH. J Cell Biol 1987; 104: 1019–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn K, DeBiasio R, Taylor DL. Patterns of elevated free calcium and calmodulin activation in living cells. Nature 1992; 359: 736–8. [DOI] [PubMed] [Google Scholar]
- 35.Taylor DL, Woo ES, Giuliano KA. Real-time molecular and cellular analysis: the new frontier of drug discovery. Curr Opin Biotechnol 2001; 12: 75–81. [DOI] [PubMed] [Google Scholar]
- 36.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods 2006; 3: 281–6. [DOI] [PubMed] [Google Scholar]
- 37.Kusumi A, Tsunoyama TA, Hirosawa KM, Kasai RS, Fujiwara TK. Tracking single molecules at work in living cells. Nat Chem Biol 2014; 10: 524–32. [DOI] [PubMed] [Google Scholar]
- 38. Taylor DL. Past, present, and future of high content screening and the field of cellomics. High content screening. Springer, New York, NY, 2006, pp.3–18. [DOI] [PubMed]
- 39.Giuliano KA, DeBiasio RL, Dunlay RT, Gough A, Volosky JM, Zock J, Pavlakis GN, Taylor DL. High-content screening: a new approach to easing key bottlenecks in the drug discovery process. J Biomol Screen 1997; 2: 249–59. [Google Scholar]
- 40.Giuliano KA, Gough AH, Taylor DL, Vernetti LA, Johnston PA. Early safety assessment using cellular systems biology yields insights into mechanisms of action. J Biomol Screen 2010; 15: 783–97. [DOI] [PubMed] [Google Scholar]
- 41.Giuliano KA, Haskins JR, Taylor DL. Advances in high content screening for drug discovery. Assay Drug Dev Technol 2003; 1: 565–77. [DOI] [PubMed] [Google Scholar]
- 42.Taylor DL. A personal perspective on high-content screening (HCS): from the beginning. J Biomol Screen 2010; 15: 720–5. [DOI] [PubMed] [Google Scholar]
- 43.Gough AH, Chen N, Shun TY, Lezon TR, Boltz RC, Reese CE, Wagner J, Vernetti LA, Grandis JR, Lee AV, Stern AM, Schurdak ME, Taylor DL. Identifying and quantifying heterogeneity in high content analysis: application of heterogeneity indices to drug discovery. PLoS One 2014; 9: e102678–e102678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gough A, Lezon T, Faeder JR, Chennubhotla C, Murphy RF, Critchley-Thorne R, Taylor DL. High content analysis with cellular and tissue systems biology: a bridge between cancer cell biology and tissue-based diagnostics. In: Mendelsohn J, Howley PM, Israel MA, Gray JW, Thompson CB. (eds). The molecular basis of cancer, 4th ed New York, NY: Elsevier, 2014. [Google Scholar]
- 45.Thompson R. Protein-based biosensors with polarization transduction. In: Geddes C, Lakowicz J (eds) Advanced concepts in fluorescence sensing. Topics in fluorescence spectroscopy. 10: Springer, New York, NY, US, 2005, pp.1–19.
- 46.Post PL, DeBiasio RL, Taylor DL. A fluorescent protein biosensor of myosin II regulatory light chain phosphorylation reports a gradient of phosphorylated myosin II in migrating cells. Mol Biol Cell 1995; 6: 1755–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeBiasio RL, LaRocca GM, Post PL, Taylor DL. Myosin II transport, organization, and phosphorylation: evidence for cortical flow/solation-contraction coupling during cytokinesis and cell locomotion. Mol Biol Cell 1996; 7: 1259–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahn KM, Waggoner AS, Taylor DL. A calcium-sensitive fluorescent analog of calmodulin based on a novel calmodulin-binding fluorophore. J Biol Chem 1990; 265: 20335–45. [PubMed] [Google Scholar]
- 49.Kurzawa L, Morris MC. Cell-cycle markers and biosensors. Chembiochem Eur J Chem Biol 2010; 11: 1037–47. [DOI] [PubMed] [Google Scholar]
- 50.Giuliano K, Premkumar D, Taylor DL. Optimal characteristics of protein-protein interaction biosensors for cellular systems biology profiling. High content screening: science, techniques and applications, Hoboken, NJ: John Wiley, 2007, pp. 371–87. [Google Scholar]
- 51.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science 1994; 263: 802–5. [DOI] [PubMed] [Google Scholar]
- 52.Frommer WB, Davidson MW, Campbell RE. Genetically encoded biosensors based on engineered fluorescent proteins. Chem Soc Rev 2009; 38: 2833–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997; 388: 882–7. [DOI] [PubMed] [Google Scholar]
- 54.Subach FV, Malashkevich VN, Zencheck WD, Xiao H, Filonov GS, Almo SC, Verkhusha VV. Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states. Proc Natl Acad Sci USA 2009; 106: 21097–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maurel D, Comps-Agrar L, Brock C, Rives ML, Bourrier E, Ayoub MA, Bazin H, Tinel N, Durroux T, Prezeau L, Trinquet E, Pin JP. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nat Methods 2008; 5: 561–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008; 132: 487–98. [DOI] [PubMed] [Google Scholar]
- 57.Soboleski MR, Oaks J, Halford WP. Green fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cells. FASEB J 2005; 19: 440–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fritz RD, Letzelter M, Reimann A, Martin K, Fusco L, Ritsma L, Ponsioen B, Fluri E, Schulte-Merker S, van Rheenen J, Pertz O. A versatile toolkit to produce sensitive FRET biosensors to visualize signaling in time and space. Science Signal 2013; 6: rs12–rs12. [DOI] [PubMed] [Google Scholar]
- 59.Morris MC. Fluorescent biosensors of intracellular targets from genetically encoded reporters to modular polypeptide probes. Cell Biochem Biophys 2010; 56: 19–37. [DOI] [PubMed] [Google Scholar]
- 60.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci USA 1999; 96: 11241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han X. In vivo application of optogenetics for neural circuit analysis. ACS Chem Neurosci 2012; 3: 577–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 2005; 8: 1263–8. [DOI] [PubMed] [Google Scholar]
- 63.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA 2003; 100: 13940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS One 2007; 2: e299–e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, Boyden ES. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 2010; 463: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderon NC, Esposti F, Borghuis BG, Sun XR, Gordus A, Orger MB, Portugues R, Engert F, Macklin JJ, Filosa A, Aggarwal A, Kerr RA, Takagi R, Kracun S, Shigetomi E, Khakh BS, Baier H, Lagnado L, Wang SS, Bargmann CI, Kimmel BE, Jayaraman V, Svoboda K, Kim DS, Schreiter ER, Looger LL. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci 2012; 32: 13819–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J 2002; 82: 509–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakai R, Repunte-Canonigo V, Raj CD, Knopfel T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur J Neurosci 2001; 13: 2314–8. [DOI] [PubMed] [Google Scholar]
- 69.Mutoh H, Perron A, Akemann W, Iwamoto Y, Knopfel T. Optogenetic monitoring of membrane potentials. Exp Physiol 2011; 96: 13–8. [DOI] [PubMed] [Google Scholar]
- 70.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 2002; 297: 1873–7. [DOI] [PubMed] [Google Scholar]
- 71.Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J. Diffusional mobility of Golgi proteins in membranes of living cells. Science 1996; 273: 797–801. [DOI] [PubMed] [Google Scholar]
- 72.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 2002; 9: 789–98. [DOI] [PubMed] [Google Scholar]
- 73.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc Natl Acad Sci USA 2001; 98: 3197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998; 394: 192–5. [DOI] [PubMed] [Google Scholar]
- 75.Mahon MJ. pHluorin2: an enhanced, ratiometric, pH-sensitive green florescent protein. Adv Biosci Biotechnol 2011; 2: 132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grover A, Schmidt BF, Salter RD, Watkins SC, Waggoner AS, Bruchez MP. Genetically encoded pH sensor for tracking surface proteins through endocytosis. Angewandte Chem 2012; 51: 4838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fisher GW, Fuhrman MH, Adler SA, Szent-Gyorgyi C, Waggoner AS, Jarvik JW. Self-checking cell-based assays for GPCR desensitization and resensitization. J Biomol Screen 2014; 19: 1220–6. [DOI] [PubMed] [Google Scholar]
- 78.Minamikawa T, Sriratana A, Williams DA, Bowser DN, Hill JS, Nagley P. Chloromethyl-X-rosamine (MitoTracker Red) photosensitises mitochondria and induces apoptosis in intact human cells. J Cell Sci 1999; 112: 2419–30. [DOI] [PubMed] [Google Scholar]
- 79.Taylor DL, Salmon ED. Basic fluorescence microscopy. In: Wang YL, Taylor DL. (eds). Methods in cell biology. Volume 29 Fluorescence microscopy of living cells in culture. Part A, San Diego, CA: Academic Press, 1989, pp. 207–37. [PubMed] [Google Scholar]
- 80.Yang T, Burrows C, Park JH. Development of a doxycycline-inducible lentiviral plasmid with an instant regulatory feature. Plasmid 2014; 72: 29–35. [DOI] [PubMed] [Google Scholar]
- 81.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA 1992; 89: 5547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng C, Baum BJ. Evaluation of promoters for use in tissue-specific gene delivery. Methods Mol Biol 2008; 434: 205–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim TK, Eberwine JH. Mammalian cell transfection: the present and the future. Anal Bioanal Chem 2010; 397: 3173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kobayashi N, Rivas-Carrillo JD, Soto-Gutierrez A, Fukazawa T, Chen Y, Navarro-Alvarez N, Tanaka N. Gene delivery to embryonic stem cells. Birth Defects Res Part C Embryo Today Rev 2005; 75: 10–8. [DOI] [PubMed] [Google Scholar]
- 85.Sakuma T, Barry MA, Ikeda Y. Lentiviral vectors: basic to translational. Biochem J 2012; 443: 603–18. [DOI] [PubMed] [Google Scholar]
- 86.Cronin J, Zhang XY, Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Curr Gene Ther 2005; 5: 387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013; 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014; 157: 1262–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Phillips MI, Tang YL. Genetic modification of stem cells for transplantation. Adv Drug Deliv Rev 2008; 60: 160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Howden SE, Thomson JA. Gene targeting of human pluripotent stem cells by homologous recombination. Methods Mol Biol 2014; 1114: 37–55. [DOI] [PubMed] [Google Scholar]
- 91.Huebsch N, Loskill P, Mandegar MA, Marks NC, Sheehan AS, Ma Z, Mathur AA, Nguyen TN, Yoo JC, Judge LM, Spencer CI, Chukka A, Russell CR, So PL, Conklin BR, Healy KE. Automated video-based analysis of contractility and calcium flux in human iPS-derived cardiomyocytes cultured over different spatial scales. Tissue Eng Part C Methods 2014. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 92.Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, Zhang L, Guschin D, Fong LK, Vu BJ, Meng X, Urnov FD, Rebar EJ, Gregory PD, Zhang HS, Jaenisch R. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 2011; 146: 318–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 2011; 29: 731–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA 2013; 110: 15644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014; 32: 347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 2014; 24: 132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014; 343: 84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013; 154: 1380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jaeschke H, Gores GJ, Cederbaum AI, Hinson JA, Pessayre D, Lemasters JJ. Mechanisms of hepatotoxicity. Toxicol Sci 2002; 65: 166–76. [DOI] [PubMed] [Google Scholar]
- 100.Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, Vasquez-Vivar J, Kalyanaraman B. Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence. Proc Natl Acad Sci USA 2005; 102: 5727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Drasdo D, Bode J, Dahmen U, Dirsch O, Dooley S, Gebhardt R, Ghallab A, Godoy P, Haussinger D, Hammad S, Hoehme S, Holzhutter HG, Klingmuller U, Kuepfer L, Timmer J, Zerial M, Hengstler JG. The virtual liver: state of the art and future perspectives. Arch Toxicol 2014;88:2071–2075. [DOI] [PubMed]
- 102.Chen J, Rogers SC, Kavdia M. Analysis of kinetics of dihydroethidium fluorescence with superoxide using xanthine oxidase and hypoxanthine assay. Ann Biomed Eng 2013; 41: 327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ju C, Reilly T. Role of immune reactions in drug-induced liver injury (DILI). Drug Metab Rev 2012; 44: 107–15. [DOI] [PubMed] [Google Scholar]
- 104.Porter AG, Janicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ 1999; 6: 99–104. [DOI] [PubMed] [Google Scholar]
- 105.Meyer LH, Karawajew L, Schrappe M, Ludwig WD, Debatin KM, Stahnke K. Cytochrome c-related caspase-3 activation determines treatment response and relapse in childhood precursor B-cell ALL. Blood 2006; 107: 4524–31. [DOI] [PubMed] [Google Scholar]
- 106.Kuznetsov AV, Margreiter R, Amberger A, Saks V, Grimm M. Changes in mitochondrial redox state, membrane potential and calcium precede mitochondrial dysfunction in doxorubicin-induced cell death. Biochim Biophys Acta 2011; 1813: 1144–52. [DOI] [PubMed] [Google Scholar]
- 107.Souslova EA, Belousov VV, Lock JG, Stromblad S, Kasparov S, Bolshakov AP, Pinelis VG, Labas YA, Lukyanov S, Mayr LM, Chudakov DM. Single fluorescent protein-based Ca2+ sensors with increased dynamic range. BMC Biotechnol 2007; 7: 37–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leder L, Stark W, Freuler F, Marsh M, Meyerhofer M, Stettler T, Mayr LM, Britanova OV, Strukova LA, Chudakov DM, Souslova EA. The structure of Ca2+ sensor Case16 reveals the mechanism of reaction to low Ca2+ concentrations. Sensors 2010; 10: 8143–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim JA, Han E, Eun CJ, Tak YK, Song JM. Real-time concurrent monitoring of apoptosis, cytosolic calcium, and mitochondria permeability transition for hypermulticolor high-content screening of drug-induced mitochondrial dysfunction-mediated hepatotoxicity. Toxicol Lett 2012; 214: 175–81. [DOI] [PubMed] [Google Scholar]
- 110.Shcherbo D, Souslova EA, Goedhart J, Chepurnykh TV, Gaintzeva A, Shemiakina II, Gadella TW, Lukyanov S, Chudakov DM. Practical and reliable FRET/FLIM pair of fluorescent proteins. BMC Biotechnol 2009; 9: 24–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Gruenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Conversion of red fluorescent protein into a bright blue probe. Chem Biol 2008; 15: 1116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bova MP, Tam D, McMahon G, Mattson MN. Troglitazone induces a rapid drop of mitochondrial membrane potential in liver HepG2 cells. Toxicol Lett 2005; 155: 41–50. [DOI] [PubMed] [Google Scholar]
- 113.Szkolnicka D, Farnworth SL, Lucendo-Villarin B, Storck C, Zhou W, Iredale JP, Flint O, Hay DC. Accurate prediction of drug-induced liver injury using stem cell-derived populations. Stem Cells Translat Med 2014; 3: 141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Taylor DL, DeBiasio R, LaRocca G, Pane D, Post P, Kolega J, Giuliano K, Burton K, Gough B, Dow A, Yu J, Waggoner AS, Farkas D. Potential of machine-vision light microscopy in toxicologic pathology. Toxicol Pathol 1994; 22: 145–59. [DOI] [PubMed] [Google Scholar]
- 115.Quatresous E, Legrand C, Pouvreau S. Mitochondria-targeted cpYFP: pH or superoxide sensor? J Gen Physiol 2012; 140: 567–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wei-LaPierre L, Gong G, Gerstner BJ, Ducreux S, Yule DI, Pouvreau S, Wang X, Sheu SS, Cheng H, Dirksen RT, Wang W. Respective contribution of mitochondrial superoxide and pH to mitochondria-targeted circularly permuted yellow fluorescent protein (mt-cpYFP) flash activity. J Biol Chem 2013; 288: 10567–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Perry SW, Norman JP, Barbieri J, Brown EB, Gelbard HA. Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. BioTechniques 2011; 50: 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kremers GJ, Gilbert SG, Cranfill PJ, Davidson MW, Piston DW. Fluorescent proteins at a glance. J Cell Sci 2011; 124: 157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods 2005; 2: 905–9. [DOI] [PubMed] [Google Scholar]
- 120.Sample V, Newman RH, Zhang J. The structure and function of fluorescent proteins. Chem Soc Rev 2009; 38: 2852–64. [DOI] [PubMed] [Google Scholar]
- 121.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci USA 1998; 95: 6803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hess ST, Heikal AA, Webb WW. Fluorescence photoconversion kinetics in novel green fluorescent protein pH sensors (pHluorins). J Phys Chem B 2004; 108: 10138–48. [Google Scholar]
- 123.Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol 2006; 7: 449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Miyawaki A, Nagai T, Mizuno H. Engineering fluorescent proteins. Adv Biochem Eng/Biotechnol 2005; 95: 1–15. [DOI] [PubMed] [Google Scholar]
- 125.Watts SD, Suchland KL, Amara SG, Ingram SL. A sensitive membrane-targeted biosensor for monitoring changes in intracellular chloride in neuronal processes. PLoS One 2012; 7: e35373–e35373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Ultrafast optogenetic control. Nat Neurosci 2010; 13: 387–92. [DOI] [PubMed] [Google Scholar]
- 127.Akerboom J, Carreras Calderon N, Tian L, Wabnig S, Prigge M, Tolo J, Gordus A, Orger MB, Severi KE, Macklin JJ, Patel R, Pulver SR, Wardill TJ, Fischer E, Schuler C, Chen TW, Sarkisyan KS, Marvin JS, Bargmann CI, Kim DS, Kugler S, Lagnado L, Hegemann P, Gottschalk A, Schreiter ER, Looger LL. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci 2013; 6: 2–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fisher GW, Adler SA, Fuhrman MH, Waggoner AS, Bruchez MP, Jarvik JW. Detection and quantification of beta2AR internalization in living cells using FAP-based biosensor technology. J Biomol Screen 2010; 15: 703–9. [DOI] [PubMed] [Google Scholar]
- 129.Ueda H, Dong J. From fluorescence polarization to Quenchbody: recent progress in fluorescent reagentless biosensors based on antibody and other binding proteins. Biochim Biophys Acta 2014; 1844: 1951–9. [DOI] [PubMed] [Google Scholar]
- 130.Correa IR., Jr Live-cell reporters for fluorescence imaging. Curr Opin Chem Biol 2014; 20: 36–45. [DOI] [PubMed] [Google Scholar]
- 131.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol 2008; 3: 373–82. [DOI] [PubMed] [Google Scholar]
- 132.Jung D, Min K, Jung J, Jang W, Kwon Y. Chemical biology-based approaches on fluorescent labeling of proteins in live cells. Mol Biosyst 2013; 9: 862–72. [DOI] [PubMed] [Google Scholar]
- 133.Chen I, Ting AY. Site-specific labeling of proteins with small molecules in live cells. Curr Opin Biotechnol 2005; 16: 35–40. [DOI] [PubMed] [Google Scholar]
- 134.Fong M, Lesnik J, Li G, Antes TJ, Lu B. Cyto-Tracers™: novel lentiviral-based molecular imaging tools. BioTechniques 2010; 49: 839–40. [Google Scholar]
- 135.Tang JB, Goellner EM, Wang XH, Trivedi RN, St Croix CM, Jelezcova E, Svilar D, Brown AR, Sobol RW. Bioenergetic metabolites regulate base excision repair-dependent cell death in response to DNA damage. Mol Cancer Res 2010; 8: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dunn JC, Tompkins RG, Yarmush ML. Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol Prog 1991; 7: 237–45. [DOI] [PubMed] [Google Scholar]
- 137.Tourovskaia A, Fauver M, Kramer G, Simonson S, Neumann T. Tissue-engineered microenvironment systems for modeling human vasculature. Exp Biol Med 2014; 239: 1264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]