Summary
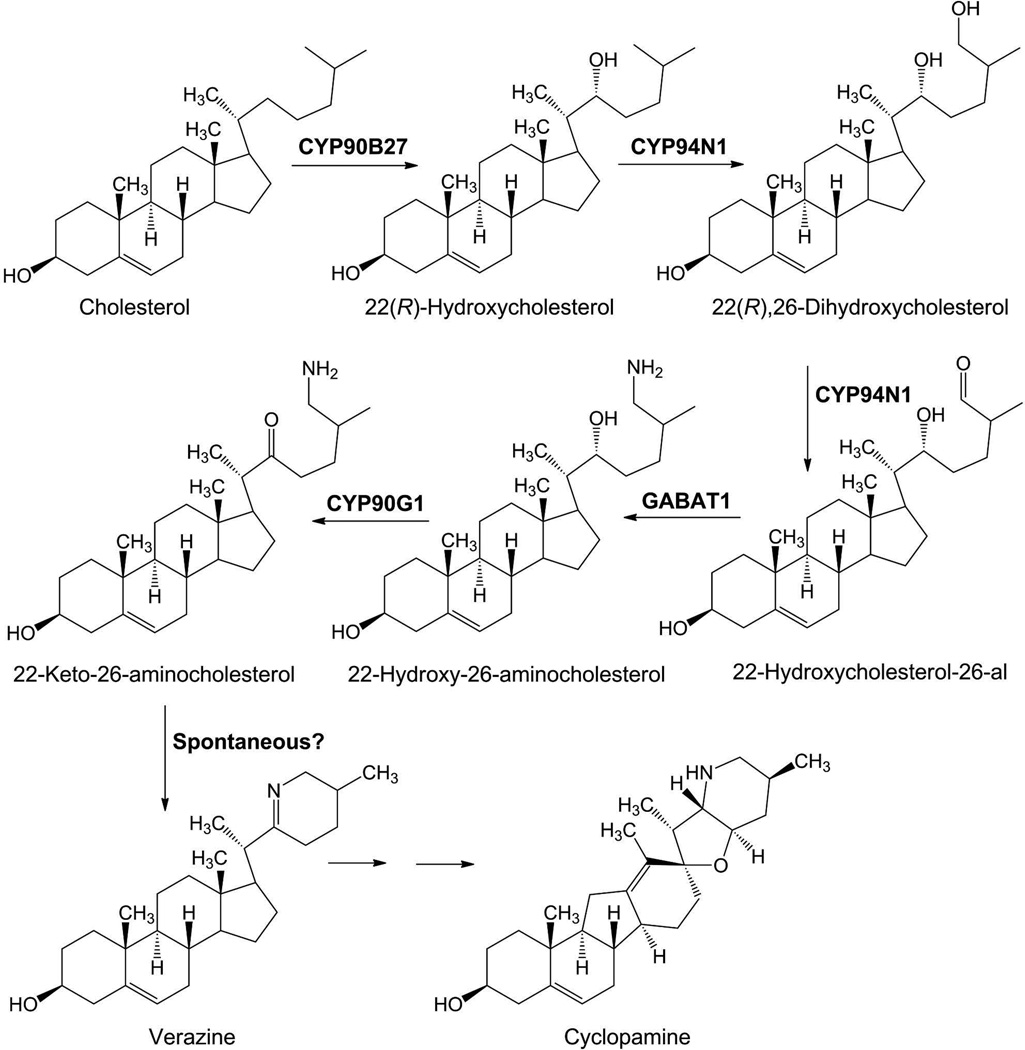
Steroid alkaloids have been shown to elicit a wide range of pharmacological effects that include anticancer and antifungal activities. Understanding the biosynthesis of these molecules is essential to bioengineering for sustainable production. Herein, we investigate the biosynthetic pathway to cyclopamine, a steroid alkaloid that shows promising antineoplastic activities. Supply of cyclopamine is limited, as the current source is solely derived from wild collection of the plant Veratrum californicum. To elucidate the early stages of the pathway to cyclopamine, we interrogated a V. californicum RNA-seq dataset using the cyclopamine accumulation profile as a predefined model for gene expression with the pattern-matching algorithm Haystack. Refactoring candidate genes in Sf9 insect cells led to discovery of four enzymes that catalyze the first six steps in steroid alkaloid biosynthesis to produce verazine, a predicted precursor to cyclopamine. Three of the enzymes are cytochromes P450 while the fourth is a γ-aminobutyrate transaminase; together they produce verazine from cholesterol.
Keywords: Verazine, Veratrum californicum, California corn lily, Haystack, steroid alkaloids, cyclopamine, KJ869252, KJ869253, KJ869254, KJ869255, KJ869256, KJ869257, KJ869262, KJ869263, KJ869264, KJ869258, KJ869261, KJ869260, KJ869259
Introduction
Plants synthesize numerous specialized metabolites that are used in medicines today (Mishra and Tiwari 2011). Their endogenous function is thought to play a role in communication of the plant with its environment, as these compounds possess an array of biological activities (Mithofer and Boland 2012, Mizutani 2012). An understanding of how these molecules are formed serves a dual role: enabling study of the in planta function, as well as development of synthetic biology production platforms for source plants that are not amenable to cultivation or genetic manipulation.
In seeking medicines from natural sources, plants synthesizing the nitrogen containing alkaloids have proven to be a vital source. Of particular pharmaceutical interest are the steroid alkaloids. Members of the families Liliaceae, Apocynaceae, Buxaceae, and Solanaceae are a rich source of these alkaloids. The structural similarity of steroid alkaloids to mammalian steroid hormones may be responsible for their therapeutic effects (Cordell 1998, Jiang et al. 2005, Zhou 2003). Our study focuses on the steroid alkaloid cyclopamine from the Liliaceae family. Members of Liliaceae contain three sub-classes of steroidal alkaloids: the jerveratrum-type, the cerveratrum-type, and the solanidine-type (Cordell 1998). Cyclopamine, also known as 11-deoxojervine, is a jerveratrum-type alkaloid that exhibits potent pharmacological properties.
Cyclopamine was originally discovered due to its teratogenic effect that resulted in craniofacial anomalies (cyclopia) in lambs born at high elevation in the northwestern United States in the middle of the 20th century (Keeler 1970a, Keeler 1970b, Keeler and Binns 1966). Studies supported by the United States Department of Agriculture found that the California corn lily Veratrum californicum was the source of the teratogen in the natural diet of the pregnant ewes (Keeler 1968, Keeler 1969, Keeler 1970a). Cyclopamine was discovered to inhibit the Hedgehog signaling pathway by direct binding to the G protein-coupled receptor Smoothened (Chen et al. 2002). As such, cyclopamine has shown promising antineoplastic activities against several cancers in which Hedgehog signaling malfunction is implicated, including pancreatic cancer, renal cell carcinoma, medulloblastoma, basal cell carcinoma, and leukemia (Bahra et al. 2012, Behnsawy et al. 2013, Berman et al. 2002, Gailani et al. 1996, Lin et al. 2010, Olive et al. 2009, Taipale et al. 2000). A semi-synthetic analog of cyclopamine, IPI-926, has been in clinical trials for treatment of several cancers including pancreatic cancer and leukemia (Lin et al. 2010, Olive et al. 2009, Tremblay et al. 2009). Due to complicated total synthesis, wild-collected V. californicum is the current commercial source of cyclopamine. Commercial cultivation of the plant has not yet been successful. Cyclopamine is thereby an attractive target for biotechnological production. The first obstacle to this approach is, however, obtaining knowledge of the underlying biosynthetic genes.
Cyclopamine biosynthesis is believed to begin with cholesterol, a common precursor to most steroidal alkaloids (Cordell 1998, Ko Kaneko et al. 1970b). Early studies on species of the Liliaceae and Solanaceae families have proposed transformation of cholesterol to metabolites identified in plant extracts that include dormantinol, dormantinone, verazine, and etioline. All of these molecules are proposed intermediates in the biosynthesis of cyclopamine (Chandler and McDougal 2014, Ko Kaneko et al. 1977). Previous feeding studies shed limited light on pathway order and suggested that L-arginine is the origin of the nitrogen atom (Ko Kaneko et al. 1976, Ko Kaneko et al. 1977, Tschesche 1980). A more recent report on the biosynthesis of Solanaceous alkaloids utilizing gene co-expression analysis and RNAi knock-out transgenic plants in Solanum tuberosum and Solanum lycopersicum outlines a biosynthetic pathway leading to the synthesis of the spirosolane-type steroid alkaloid α-tomatine. The enzyme classes identified in the Solanum pathway include several cytochrome P450 enzymes, a transaminase, and a 2-oxoglutarate-dependent dioxygenase (Itkin et al. 2013). Questions remain, however, concerning the formation of steroid alkaloids in Veratrum as no genes or enzymes have been reported.
Biochemical pathway elucidation in non-model systems has historically taken decades to complete, but bioinformatic technologies are revolutionizing the approach. A prominent example of former methods used in pathway discovery is morphine biosynthesis in opium poppy, Papaver somniferum. Though morphine was discovered in the early 1800’s, the biosynthetic pathway is still incomplete at the gene level. Genes encoding only 6 of the 8 enzymes committed to the biosynthesis of morphine have been isolated and characterized from the 1990’s to the present – several decades of work to uncover fewer than 8 genes (Gesell et al. 2009, Grothe et al. 2001, Hagel and Facchini 2010, Unterlinner et al. 1999, Ziegler et al. 2006). Now, high-throughput sequencing technology enables new approaches to biochemical pathway discovery in the non-model system by providing nucleotide sequence data acquisition at a previously unparalleled rate. A combination of bioinformatics and high-throughput sequencing has the potential to shorten natural product pathway discovery if the challenges of interrogating such large datasets from non-model systems is overcome. We present herein the discovery of four V. californicum enzymes that catalyze the first six steps from cholesterol to verazine, a predicted precursor to the steroid alkaloid cyclopamine. We utilize a biosynthetic gene discovery method founded on correlating metabolite accumulation with RNA-seq generated gene expression data. By stepwise refactoring of the pathway in Spodoptera frugiperda Sf9 cells, we eliminate the need to synthesize biosynthetic intermediates for validation of pathway enzyme activity.
Results
RNA-seq and de novo transcriptome assembly
Multiplex paired-end sequencing of V. californicum cDNA produced from bulb, flower, leaf, fall rhizome, spring rhizome, fall root, green shoot, white shoot, and tissue culture samples on two 2 × 50 bp Hi-Seq channels produced over 201 million raw reads which were analyzed and filtered for artifacts/contaminants. The de novo short read assembly generated 56,994 contigs with a median length of 323 bp and an N50 of 1,471 bp (refer to Table S1 and Table S2 for Illumina reads generated per file and transcriptome statistics, respectively). Information on accessing the raw reads and assembled transcriptome can be found in Data S1. Raw mapped read-counts were utilized as a metric of relative gene expression. The average contig sequence length indicates high quality assembly and was sufficient for downstream sequence alignment and phylogenetic gene tree estimation.
Transcriptome dataset interrogation
To provide annotation, predicted peptide sequences were queried against the Pfam and Superfamily databases in addition to BLAST searches at NCBI using the non-redundant protein sequence database. Expression data for each contig was normalized using total reads per organ type to serve as the dataset for Haystack (http://haystack.mocklerlab.org/)(Mockler et al. 2007).
LC-MS/MS determination of the steroid alkaloid profile in the same V. californicum tissues used for RNA-seq identified a pronounced accumulation of cyclopamine in rhizome, followed by root and bulb (Figure S1), suggesting that biosynthesis occurs in underground organs of the plant. Since secondary metabolites are most often synthesized at or near their site of accumulation (Huang and Kutchan 2000, Nims et al. 2006, Onrubia et al. 2011, Weid et al. 2004), we initially hypothesized that underground tissues (rhizome, root, and bulb) in Veratrum are biosynthetic for cyclopamine.
Since ca. 20 times more cyclopamine accumulates in subterranean organs, these tissues were given a value of 20 for the Haystack input while the above ground organs were designated a value of 1 in order to create a generalized model for cyclopamine biosynthesis. In our approach, the LC-MS/MS alkaloid data for Veratrum is the input model used to search the deep transcriptome experimental dataset of Veratrum, correlating metabolite accumulation and gene expression data as previously reviewed (Saito et al. 2008). We obtained 3,219 contigs that fit the 20:1 subterranean organ:aerial organ cyclopamine accumulation model.
In addition to identification of genes exhibiting correlating expression patterns with cyclopamine accumulation, the protein-coding gene sequences in the Veratrum RNA-seq transcriptome dataset were classified into putative gene families using PlantTribes 2.0 (http://fgp.bio.psu.edu/tribedb/10_genomes/index.pl) (Wall et al. 2008). This dataset was used to better define and cluster the Veratrum transcripts that are of most interest to the cyclopamine pathway. Cytochromes P450 were chosen from the resulting dataset due to the hypothesized oxidative transformations necessary to convert cholesterol into cyclopamine. The alternative enzyme class would be 2-oxoglutarate-dependent dioxygenases, if the cytochrome P450 dataset did not yield positive results.
Top candidate genes from the Haystack and tribe clustering analysis with the highest homology to cytochrome P450 enzymes involved in triterpene and fatty acid hydroxylation/ biosynthesis were selected for downstream functional characterization (Table S3). Incorporation of nitrogen into the steroid skeleton is required; therefore, aminotransferases fitting the Haystack model were also included in the candidate gene list (Table S4). Full-length candidate cDNAs were expressed in S. frugiperda Sf9 cells using a baculovirus-based expression system allowing for the accommodation of multi-virus infections. S. frugiperda Sf9 cells provide a facile synthetic biology platform for the systematic refactoring of plant biosynthetic pathways (Diaz Chavez et al. 2011).
Cholesterol 22-hydroxylase
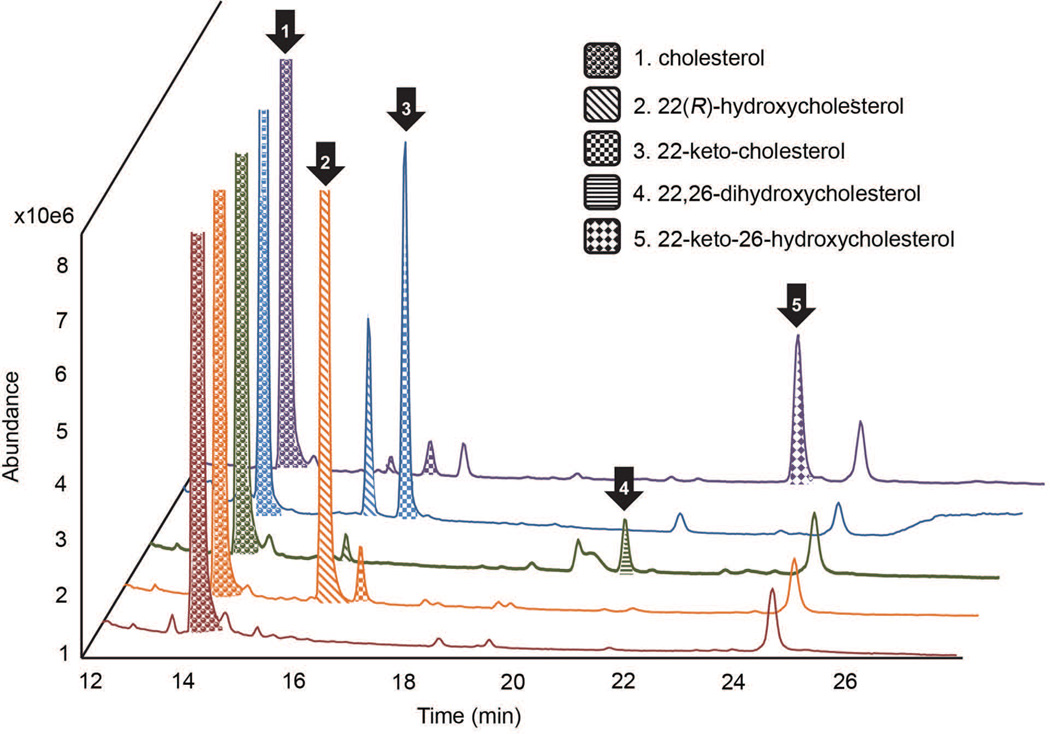
The top-scoring candidate cDNAs resulting from interrogation of the V. californicum transcriptome dataset were systematically and individually introduced, together with Eschscholzia californica cytochrome P450 reductase (CPR), into S. frugiperda Sf9 insect cells, which were harvested as previously described and used in enzyme assays with cholesterol as substrate (Diaz Chavez et al. 2011, Rosco et al. 1997). Most cytochrome P450 enzymes require the addition of a reductase for heterologous expression. In our hands, differences in enzyme activity were not dependent on the plant species from which the CPR was derived (Gesell et al. 2009). Cholesterol was chosen as the initial precursor for study based upon existing knowledge of steroid alkaloid biosynthesis (Cordell 1998, Ko Kaneko et al. 1970b). Related compounds were also tested to determine enzyme specificity (Table S5). The contig designated 2646, which is annotated as a steroid C-22 hydroxylase, added a hydroxyl group to the 22-position of cholesterol exclusively in the R orientation as determined by GC-MS (Figure 1; Figure S2 a; Figure S3 a). The hydroxylation demonstrated in Figure 1 used extracts of Sf9 cells co-expressing contig 2646 and E. californica CPR as we have discovered their ability to utilize endogenous Sf9 cholesterol as substrate. One variant of 2646 was identified by RT-PCR having 99.8% identity and catalyzing the same enzymatic reaction. Cytochrome P450 (CYP) assignments for both homologs are CYP90B27v1 and CYP90B27v2, where version CYP90B27v1, accession KJ869252, was chosen for subsequent analysis. We have designated this enzyme cholesterol 22-hydroxylase. CYP90B27 also hydroxylated 26-hydroxycholesterol (27(25R)-hydroxycholesterol) and 7β-hydroxycholesterol in the 22-position as determined by enzyme assay, but was unable to hydroxylate the 24-methylsterol campesterol, a metabolite leading into brassinosteroid biosynthesis (Figure S2 e; Figure S4; Figure S5). CYP90B27 oxidizes the hydroxyl group at the 22-position to a ketone, but only to a low degree (Figure 1). The identity of the enzymatic product of CYP90B27 acting on cholesterol was confirmed by comparison to 22(R)-hydroxycholesterol standard. The diastereomers 22(R)- and 22(S)-hydroxycholesterol are chromatographically resolved by the GC-MS method used (Figure S6).
Figure 1. GC-MS Overlay of S. frugiperda Sf9 extracts expressing Veratrum californicum genes.
S. frugiperda Sf9 cells infected with varying combinations of baculovirus containing genes from V. californicum were extracted and analyzed by gas chromatography mass spectrometry. Each colored chromatograph corresponds to the following: Red-CYP719A14 (control cytochrome P450) and CPR, Orange-CYP90B27 (cholesterol 22-hydroxylase) and CPR, Green-CYP90B27, CYP94N1 (22-hydroxycholesterol 26-hydroxylase/oxidase), and CPR, Blue-CYP90B27, CYP90G1 (22-hydroxy-26-aminocholesterol 22-oxidase), and CPR, Purple-CYP90B27, CYP94N1, CYP90G1, and CPR. Metabolites are numbered according to the legend and shaded for clarity. CPR refers to the cytochrome P450 reductase from Eschscholzia californica and control P450 refers to CYP719A14 cheilanthifoline synthase from Argemone mexicana.
22-Hydroxycholesterol 26-hydroxylase/oxidase
To identify the second enzyme in the pathway, a series of triple infections of S. frugiperda Sf9 cells were carried out that contained CYP90B27 and E. californica CPR, but varied the second enzyme. Candidates for the second enzyme were the remaining top-scoring candidate cDNAs resulting from interrogation of the V. californicum transcriptome dataset. Contig 12709, which annotated as a fatty acid hydroxylase, was found to hydroxylate 22(R)-hydroxycholesterol at the C-26 position forming 22,26-dihydroxycholesterol by GC-MS (Figure 1; Figure S4). This enzyme also oxidizes the hydroxyl group at the 26 position creating a highly reactive 22-hydroxycholesterol-26-al as detected by LC-MS/MS (Figure S7 a). Four variants were generated by RT-PCR with identities ranging from 93–99%; all catalyze the same reaction. Hydroxylation of cholesterol by 12709 was not detected (Figure S3); this enzyme was therefore designated 22-hydroxycholesterol 26-hydroxylase/oxidase. CYP assignments are CYP94N1v1, CYP94N1v2, CYP94N2v1, and CYP94N2v2. In figures and subsequent text, CYP94N1 refers to accession KJ869255 which was chosen as the representative variant for this study. The identity of 22,26-dihydroxycholesterol produced by action of 12709 on 22(R)-hydroxycholesterol was ultimately determined using the 22-hydroxylating activity of CYP90B27 to produce 22,26–dihydroxycholesterol from pure 26-hydroxycholesterol and comparing the mass spectra of the two enzymatic products (Figure S4).
22-Hydroxycholesterol-26-al transaminase
To identify the third enzyme in the pathway, a series of quadruple infections of S. frugiperda Sf9 cells were carried out that contained CYP90B27, CYP94N1, and E. californica CPR, but varied the third enzyme. Candidates for the third enzyme were the remaining top-scoring candidate cDNAs. A γ-aminobutyric acid (GABA) transaminase (contig 12084) was shown to incorporate nitrogen into the 26-position of 22-hydroxycholesterol-26-al using GABA as an amino group donor to produce 22-hydroxy-26-aminocholesterol as detected by LC-MS/MS (Figure S8 a). Three variants were found by RT-PCR, each with over 99% identity and all able to catalyze the same reaction. Evidence for the structure of 22-hydroxy-26-aminocholesterol was obtained by high resolution MS (Figure S9). 22-Hydroxy-26-aminocholesterol was not produced in Sf9 enzyme assays that included CYP90B27, CYP90G1 (see below), GABAT1, and CPR but lacked CYP94N1 (Figure S7 b; orange), indicating that addition of nitrogen to position 26 is likely. This enzyme was designated 22-hydroxycholesterol-26-al transaminase. In figures and subsequent text, GABAT1 refers exclusively to accession KJ89263, the variant chosen for subsequent study.GABAT1 was expressed and purified from E. coli and used in enzyme assays to test potential co-substrates GABA, L-arginine, and L-glutamine (Figure S10). GABA was the only accepted amino group donor. Minor peaks in the L-arginine and L-glutamine assays are likely due to GABA present in E. coli (Dhakal et al. 2012).
22-Hydroxy-26-aminocholesterol 22-oxidase
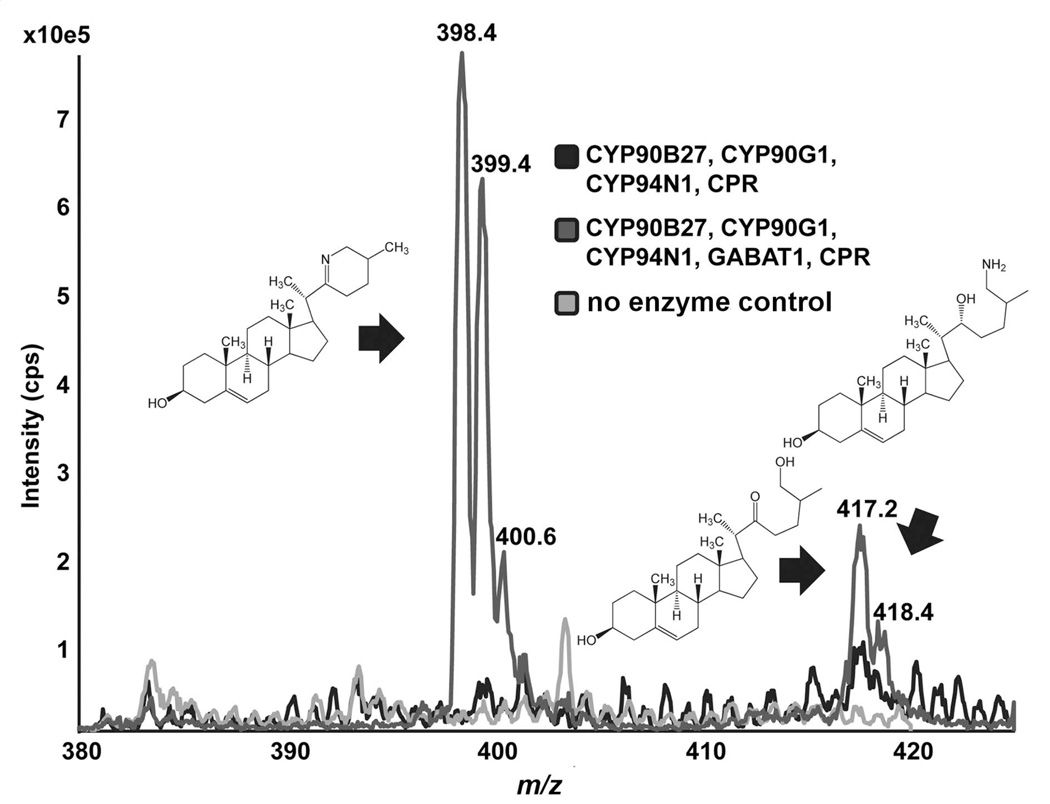
To identify the fourth enzyme in the pathway, a series of quintuple infections of S. frugiperda Sf9 cells were carried out that contained CYP90B27, CYP94N1, GABAT1, and E. californica CPR, but varied the fourth enzyme. Candidates for the fourth enzyme were the remaining top-scoring candidate cDNAs. Contig 13284 was also annotated as a steroid C-22 hydroxylase, but hydroxylation at the 22-position of cholesterol was not readily detected. It was, however, able to oxidize an existing hydroxyl group at position 22 to a much greater degree than CYP90B27 as shown by GC-MS (Figure 1; Figure S2 b). 13284 oxidizes the 22-hydroxy position of 22(R)-hydroxycholesterol to form 22-keto-cholesterol (Figure 1), 22,26-dihydroxycholesterol to form 22-keto-26-hydroxycholesterol (Figure 1; Figure S8 b), and 22-hydroxy-26-aminocholesterol to form a short lived intermediate that cyclizes to verazine (Figure 2; Figure S8 c). Four variants were isolated using RT-PCR, each having more than 97% identity and all able to perform the same reaction. CYP designations for each sequence are: CYP90G1v1, CYP90G1v2, CYP90G1v3, and CYP90G2. In figures and subsequent text, CYP90G1 refers to accession KJ869260 which was selected for further analysis. The structures of the enzymatic products 22-keto-cholesterol and 22-keto-26-hydroxycholesterol were confirmed by NMR spectroscopy (Data S2). The structure of verazine was supported by high resolution MS (Figure S9). The enzyme was subsequently designated 22-hydroxy-26-aminocholesterol 22-oxidase. Accession numbers and designations for all characterized enzymes can be found in Table S6.
Figure 2. Production of verazine by heterologous expression of Veratrum californicum genes in S. frugiperda Sf9 cells.
Select genes were introduced into S. frugiperda Sf9 cells using a baculovirus expression system. Metabolites were extracted and analyzed by LC-MS/MS in the full scan Enhanced MS mode detecting 380 – 425 m/z. Each chromatogram represents the combination of genes as follows: Black-CYP90B27 (cholesterol 22-hydroxylase), CYP94N1 (22-hydroxycholesterol 26-hydroxylase/oxidase), CYP90G1 (22-hydroxy-26-aminocholesterol 22-oxidase), and CPR; Dark Grey-CYP90B27, CYP94N1, CYP90G1, GABAT1 (22-hydroxycholesterol-26-al transaminase), and CPR; Light Grey-no enzyme control. CPR refers to the cytochrome P450 reductase from Eschscholzia californica, and GABAT1 refers to the γ-aminobutyric acid transaminase 1 from V. californicum. Peak at 398.4 is verazine, peak at 417.2 is 22-keto-26-hydroxycholesterol, and peak at 418 .4 is for 22-hydroxy-26-aminocholesterol.
Biosynthetic pathway to verazine
The substrate specificities determined for these enzymes suggested a potential metabolic grid in the metabolism of cholesterol. CYP90B27 catalyzes the 22-hydroxylation of cholesterol; this is most likely the first step in the biosynthesis of steroid alkaloids in V. californicum, confirmed by the inability of CYP94N1 and CYP90G1 to accept cholesterol as substrate (Figure S3).
To establish the pathway order after 22-hydroxylation of cholesterol, a series of enzyme assays were carried out using S. frugiperda Sf9 cell extracts containing each individual candidate enzyme. The order of addition for each enzyme was varied, and products were analyzed by GC-MS or LC-MS/MS. The flow chart for both sets of experiments is presented in Figure S11. As seen in Figure S12 c and e, 22-keto-26-hydroxycholesterol was only produced at detectable levels by CYP90G1 from 22,26-dihydroxycholesterol. CYP94N1 was unable to hydroxylate 22-keto-cholesterol at levels detected by GC-MS. The ability of CYP90G1 to accept 22,26-dihydroxycholesterol as substrate and produce 22-keto-26-hydroxycholesterol, along with the lack of product detection for CYP94N1 incubated with 22-keto-cholesterol, provided evidence that CYP94N1 acted directly after CYP90B27. This evidence was substantiated with another set of enzyme assays, beginning with CYP90B27, showing that CYP94N1 produced little product when provided with 22-keto-cholesterol (using the increased sensitivity of LC-MS/MS for detection) as seen in Figure S7 b; blue, as compared to the large amount of product produced when 22,26-dihydroxycholesterol is acted upon by CYP90G1 (Figure S7 a; blue).
22-Hydroxy-26-aminocholesterol was produced in Sf9 cells by CYP90B27, CYP94N1, GABAT1, and CPR in the presence or absence of CYP90G1 (Figure S8 a; c). GABAT1, therefore, did not require a 22-ketone moiety on the substrate. When CYP90B27 acted in the presence of CYP94N1, several side products were detected in addition to 22,26-dihydroxycholesterol (Figure S7 a; red). These compounds included 22-keto-26-hydroxycholesterol, 22-hydroxycholesterol-26-al and two unidentified products. Since an amino group was not added to the 22-ketone moiety of 22-keto-26-hydroxycholesterol, 22-keto-26-hydroxycholesterol most likely does not participate in this steroid alkaloid pathway. The short lived and highly reactive 22-hydroxycholesterol-26-al must therefore be the substrate of GABAT1. After an amino group is transferred to the C-26 aldehyde, CYP90G1 oxidizes the C-22-hydroxyl moiety to a ketone, and cyclization to verazine occurs (Figure 2; Figure S8 c).
Evidence for the short-lived intermediate 22-hydroxycholesterol-26-al was obtained by dimedone aldehyde trapping (Figure S13). The amino group is added prior to oxidation of the C-22 hydroxyl group; therefore, the amino group must be transferred to a C-26 aldehyde. The structure of the predicted cyclic imine verazine was supported by borohydride reduction of the double bond (Figure S14) and exact mass analysis as demonstrated by high resolution MS (Figure S9).
The biosynthetic pathway proposed herein (Figure 3) is similar to a hypothesized pathway presented in earlier studies of steroid alkaloids in the genus Veratrum. We found, however, that 22-keto-26-hydroxycholesterol (dormantinone) was not readily accepted as substrate for GABAT1, as verazine was not formed when supplied this compound (Figure S7 a; purple). In contrast, verazine was formed when 22-hydroxy-26-amino-cholesterol was supplied to CYP90G1 (Figure S7 a; green). In further support of our proposed pathway, the biosynthetic intermediates 22-hydroxy-26-aminocholesterol, 22-keto-26-hydroxycholesterol, and verazine were detected in V. californicum extracts by LC-MS/MS (Figure S15) and the distribution of these intermediates followed the same accumulation pattern as cyclopamine (Figure S1). 22,26-Dihydroxycholesterol (dormantinol) and verazine have also been previously detected in steroid alkaloid producing Veratrum species and hypothesized to be intermediates in steroid alkaloid biosynthesis (Adam et al. 1967, Ko Kaneko et al. 1977).
Figure 3. Proposed Veratrum californicum cyclopamine biosynthetic pathway leading from cholesterol.
Cholesterol is first hydroxylated at position C-22 in the R-orientation by CYP90B27 (cholesterol 22-hydroxylase), followed by hydroxylation/oxidation at position C-26 by CYP94N1 (22-hydroxycholesterol 26-hydroxylase/oxidase). Next, a transamination reaction by GABAT1 (22-hydroxycholesterol-26-al transaminase) transfers an amino group from γ-aminobutyric acid to the C-26-aldehyde, forming 22-hydroxy-26-aminocholesterol. The C-22-hydroxy group is then oxidized to a ketone by CYP90G1 (22-hydroxy-26-aminocholesterol 22-oxidase) to form 22-keto-26-aminocholesterol, a reactive intermediate that cyclizes to verazine. GABAT1 refers to the γ-aminobutyric acid transaminase 1 from V. californicum.
Site of steroid alkaloid biosynthesis in V. californicum
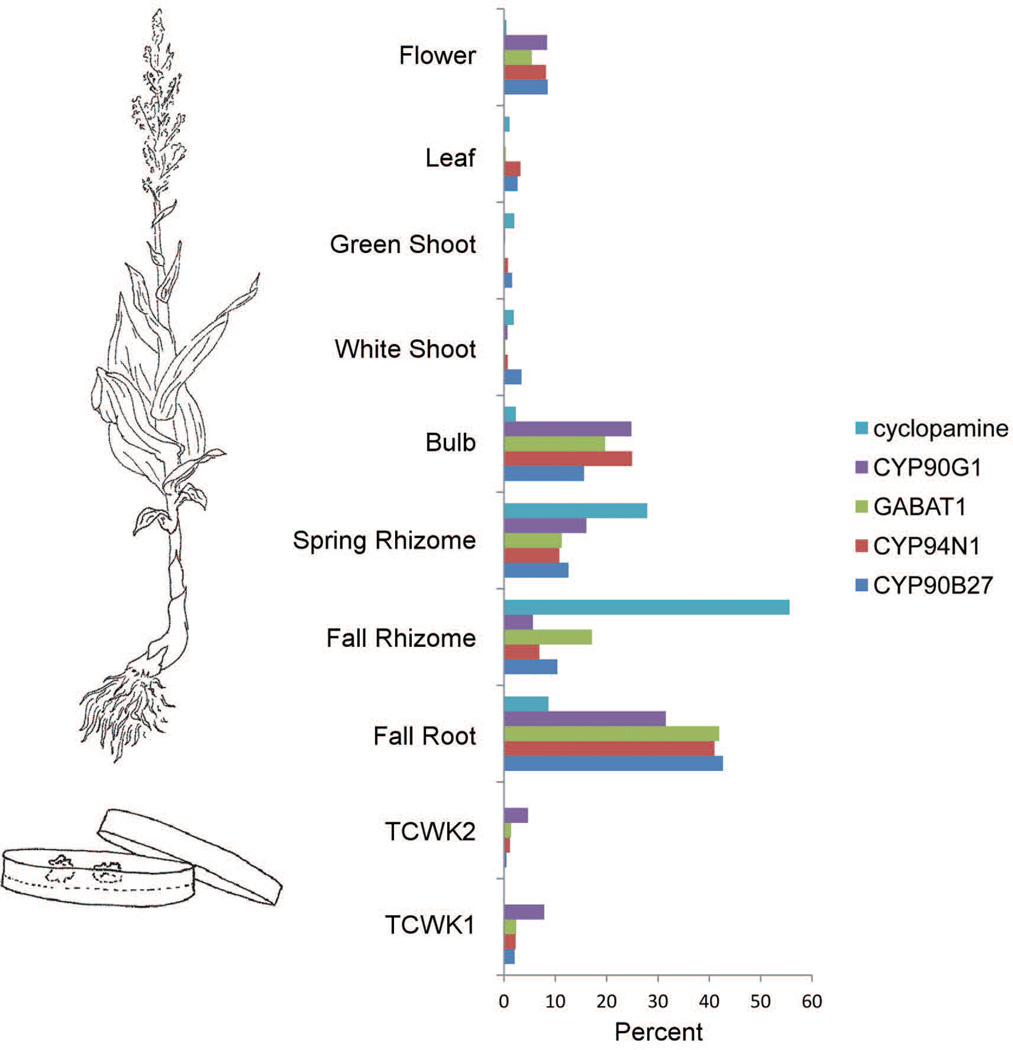
A comparison was made between biosynthetic gene expression profiles and cyclopamine accumulation in V. californicum (Figure 4). A pattern emerged that indicates biosynthetic genes are most highly expressed in root, bulb, and spring rhizome, while cyclopamine (Figure 4; Figure S1) is highest in spring- and fall rhizome. The hypothesized intermediates 22-hydroxy-26-aminocholesterol, 22-keto-26-hydroxycholesterol, and verazine follow a similar trend with highest accumulation in root, spring- and fall rhizome (Figure S15). Moreover, the higher level of cyclopamine in fall rhizome compared to spring rhizome suggests an accumulation of the steroid alkaloid during rhizome growth in summer (Figure S1). Interestingly, fall rhizome has significantly more cyclopamine relative to transcript level; the opposite is true for bulb (Figure 4).
Figure 4. Cyclopamine accumulation vs gene expression of steroid alkaloid biosynthetic genes.
Tissues from Veratrum californicum were extracted and analyzed by liquid chromatography mass spectrometry for cyclopamine quantitation. Transcript abundance was analyzed by alignment of individual reads to the assembled transcriptome for gene expression. Both gene expression and cyclopamine accumulation are shown as a percent of the total for comparison. The abbreviations TCWK1 and TCWK2 stand for tissue culture one- and two weeks after transfer to fresh media (respectively). CYP90G1 refers to 22-hydroxy-26-aminocholesterol 22-oxidase, GABAT1 refers to by 22-hydroxycholesterol-26-al transaminase (γ-aminobutyric acid transaminase 1 from V. californicum), CYP94N1 refers to 22-hydroxycholesterol 26-hydroxylase/oxidase, and CYP90B27 refers to cholesterol 22-hydroxylase.
Discussion
The evolutionary origin of specialized metabolism like that in the Veratrum genus is of particular interest not only for understanding species divergence but also for the recognition of additional species containing related compounds with potential therapeutic value. Biochemical analysis of the Veratrum steroid alkaloid pathway herein alongside a recently proposed biosynthetic pathway for steroid glycoalkaloids in Solanum lycopersicum (Itkin et al. 2013) suggests independent origins of the Solanum steroid alkaloids and the Veratrum steroid alkaloids. In S. lycopersicum, initial transformations of cholesterol include C-22 hydroxylation followed by C-26 hydroxylation and closure of the E-ring. Oxidation at C-26 and subsequent transamination at that position occur without the formation of the intermediate verazine. In conjunction with our results, previous work on steroid alkaloid formation in Veratrum does not support E-ring closure prior to aldehyde formation and transamination (Ko Kaneko et al. 1975, Ko Kaneko et al. 1970a, Ko Kaneko et al. 1976, Ko Kaneko et al. 1977). Verazine production requires formation of the F-ring following transamination prior to E-ring closure. If the pathway was identical to that proposed in S. lycopersicum, E-ring closure prior to amination would not allow for the formation of verazine. Oxidation at C-26 by the multifunctional CYP94N1 may be the gateway reaction when comparing the point of divergence between the two pathways. This oxidation in Veratrum allows for the immediate addition of nitrogen, leading to F-ring closure and verazine formation. In contrast, the action of GAME11, the 2-oxoglutarate-dependent dioxygenase in the Solanum pathway, in conjunction with other unidentified enzymes after C-26 hydroxylation, leads to closure of the E-ring prior to C-26 oxidation and nitrogen addition. The possibility exists that multiple steroid alkaloid biosynthetic pathways occur in one or both families and those respective genes are yet to be discovered, as solanidine-type and verazine-like compounds have been identified in both members of the Liliaceae and Solanaceae (Cordell 1998).
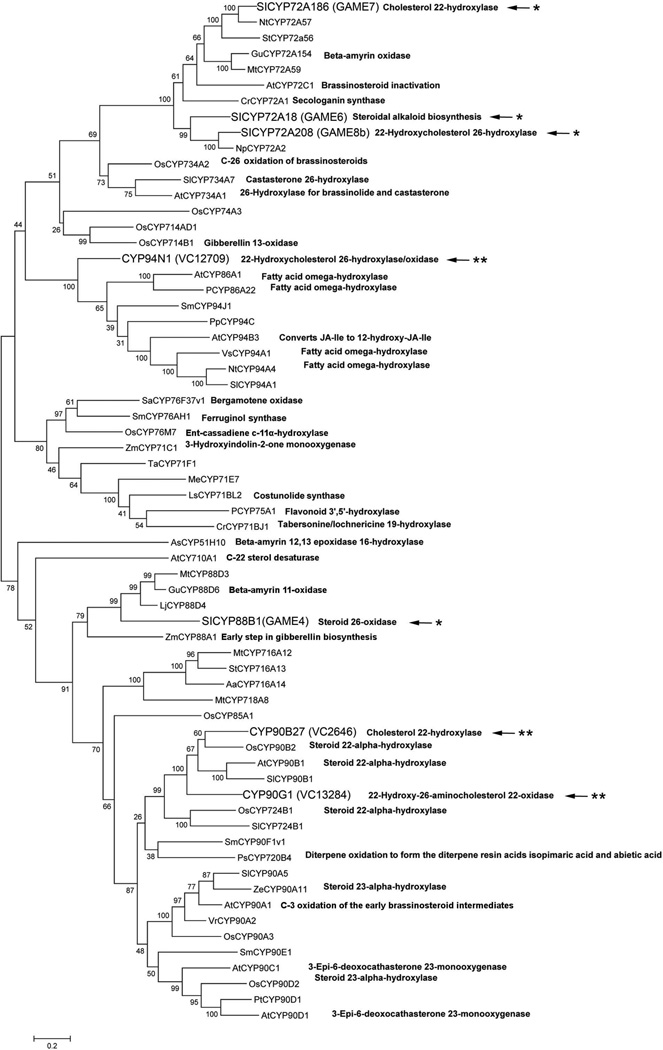
The independent origin of the two pathways is further corroborated by the phylogenetic relationship of the enzymes involved. A phylogenetic analysis of select cytochrome P450 enzymes, including several involved in steroid metabolism (Figure 5, Table S7), demonstrates this distinction. CYP90B27 in V. californicum and CYP72A186 (GAME7), the proposed cholesterol 22-hydroxylase in S. lycopersicum, are in distinct pan-angiosperm clades suggesting recruitment from separate P450 subfamilies; likewise for CYP72A208 (26-hydroxylase in S. lycopersicum, GAME8) and V. californicum CYP94N1. Similarly, CYP88B1 (GAME4), the S. lycopersicum enzyme that performs oxidation at position 26, falls into a well-supported clade distinct from the clade containing CYP94N1. The phylogenetic placement of these enzymes alludes to potential convergent evolution of these genes. The similarity of V. californicum enzymes CYP90B27 and CYP90G1 with the CYP90B1s, enzymes known to participate in primary brassinosteroid metabolism, may be indicative of steroid alkaloid biosynthesis evolution, in part, deriving from the brassinosteroid pathway.
Figure 5. Phylogenetic tree of select plant cytochrome P450 enzymes.
Nucleotide sequences obtained from Genbank, Uniprot, and the Sol Genomics Network of selected cytochrome P450 enzymes were aligned by codon with the Muscle algorithm. Cytochrome P450 designations, species, and their corresponding function can be found in Table S7. Only experimentally determined functions are designated in the figure. Veratrum californicum enzymes involved in cyclopamine biosynthesis are represented by two stars; Solanum lycopersicum enzymes involved in steroid alkaloid metabolism are represented with one star. Phylogenetic reconstruction was performed using the Maximum Likelihood statistical method with bootstrapping in MEGA version 6.06 with default parameters.
An in-depth phylogenetic analysis (Figure S16, Table S8) of the steroid modifying genes from V. californicum and S. lycopersicum alongside homologous cytochrome P450 genes taken from transcriptome assemblies generated by the 1KP and MonAToL sequencing projects (Data S1) substantiate the hypothesis of independently derived pathways. Three major cytochrome P450 clades emerge. Both V. californicum CYP90B27 and CYP90G1 are localized in Clade 1. Clade 1 contains a number of ancient gene duplications that show evidence of paralog retention with expression of multiple copies within species, especially in monocots. In agreement with Figure 5, SlCYP90B1 falls into the same well-supported subclade as these two Veratrum genes with CYP90B27 being more closely related to SlCYP90B1. Clade 2 contains the S. lycopersicum genes GAME6, GAME7, and GAME8, but no sampled Veratrum genes. Interestingly, this clade shows several eudicot gene duplications, most likely genome-specific tandem duplications that are not observed in the monocot species included in this study. V. californicum CYP94N1 is found in Clade 3. Clade 3 appears to be the least diverse of the three P450 clades containing multiple copies of homologs only in a few taxa, potentially related to whole genome duplication events.
We functionally investigated the relatedness of the V. californicum GABAT1 that incorporates nitrogen into 22-hydroxycholesterol-26-al and the S. lycopersicum GABA transaminase isozyme 2 involved in steroid alkaloid biosynthesis. These enzymes share 64% identity at the amino acid level. Due to the similarities of the two pathways, we tested whether S. lycopersicum GABA transaminase isozyme 2 can incorporate nitrogen into 22-hydroxycholesterol-26-al. We demonstrated that S. lycopersicum GABA transaminase isozyme 2 was able to convert 22-hydroxycholesterol-26-al to 22-hydroxy-26-aminocholesterol and, in conjunction with CYP90G1, resulting in subsequent cyclization to verazine (Figure S8 d). S. lycopersicum GABA transaminase isozyme 2 was used as query to BLAST the V. californicum transcriptome; the best hit was another transaminase, contig 674. 674, designated GABAT2, has 68% identity to S. lycopersicum GABA transaminase isozyme 2 and 69% identity to V. californicum GABAT1 at the amino acid level. Despite the sequence homology to both V. californicum GABAT1 and S. lycopersicum GABA transaminase isozyme 2, GABAT2 was unable to convert 22-hydroxycholesterol-26-al to 22-hydroxy-26-aminocholesterol at detectable levels (Figure S17).
Overall, our data indicates that CYP90B27, CYP94N1, CYP90G1, and GABAT1 catalyze the first six steps of steroid alkaloid biosynthesis in V. californicum to form verazine, a proposed intermediate of the antineoplastic cyclopamine. We refactored the pathway to verazine in S. frugiperda Sf9 cells by infections with viruses containing each gene, thereby demonstrating gene-product function. Verazine exhibits strong antifungal properties and can serve as a synthon for the synthesis of bioactive molecules with additional pharmaceutical properties including liver protection and anti-tumor activities (Gan et al. 1993, Jiang et al. 2005, Kusano et al. 1987, Zhou 2003). We also demonstrated that the precursor in this pathway is 22(R)-hydroxycholesterol rather than 22(S)-hydroxycholesterol, as previously proposed (Ko Kaneko et al. 1977). Our data suggests that dormantinone is not an intermediate in the formation of verazine, and the amino donor for GABAT1 is GABA rather than L-arginine or L-glutamine (Ko Kaneko et al. 1976).
The candidate gene selection criteria with emphasis on clades that contained only Veratrum genes fitting the Haystack model increased our rate of gene identification. Of the 3,219 genes fitting the Haystack model, 9 cytochrome P450 genes were assigned high priority for biochemical characterization. Seven were tested; 3 catalyzed the first reactions in the verazine biosynthesis pathway. Likewise, 3 GABATs were prioritized for functional characterization; 1 GABAT was found to participate in the verazine pathway. Based upon the success of this methodology, we expect continued, facile elucidation of this and other biochemical pathways from non-model systems.
Experimental Procedures
Plant Material and RNA extraction
Liquid Chromatography Mass Spectrometry (LC-MS/MS) Method
Samples were first extracted with either ethanol (see Method S2) for plant tissue or ethyl acetate for Sf9 cell suspensions and enzymatic assays. Liquid chromatographic separation was achieved with 10 µl injections on a LC-20AD (Shimadzu) LC system coupled to a 4000 QTRAP (AB Sciex Instruments) for MS/MS analysis. Separation was achieved using a Phenomenex Gemini C-18 NX column (150 × 2.00 mm, 5 µm) with a flow rate of 0.5 ml/min and the following gradient program [solvent A (0.05% formic acid/0.04% ammonium hydroxide (25%) v/v in H2O; solvent B (0.05% formic acid/0.04% ammonium hydroxide (25%) v/v in 90% acetonitrile]: Solvent B was held at 20% for 2 min, then 2–11 min 20–30% B, 11 – 18 min 30–100% B, 18–22 min 100% B, 22–23 min 100–20% B, and held at 20% B for an additional 5 minutes. Program parameters included a TurboIonSpray ionization source temperature of 500°C and low resolution for Q1 and Q3 done with MRM (Multiple Reaction Monitoring) scans in the positive ion mode. Specific ion fragments and parameters can be found in Table S10. In conjunction, EMS (Enhanced MS) scan with a mass range of 380 to 425 m/z, and EPI (Enhanced Product Ion) scans for 398, 417, and 418 m/z were included. Compound identification was determined by comparison of retention time and fragmentation pattern to the authentic standard cyclopamine (Infinity Pharmaceuticals) (where applicable). Quantitation was performed by plotting peak area versus pmol of standard using Analyst 1.5 (Applied Biosystems).
Gas Chromatography Mass Spectrometry (GC-MS) Method
Samples were first extracted with either hexane:isopropanol 3:2 followed by hexane only or ethyl acetate. Dried extracts were derivatized with 40 µl Sylon HTP (Sigma) for 1 hour at 90°C prior to injection with a 7683B autosampler onto a 7890A gas chromatograph coupled to a 5975C mass spectrometer inert XL MSD with triple-axis detector (Agilent Technologies). Both full scan and SIM methods were run in the splitless mode with 1 µl injection volume and a flow rate of 1 ml/min with helium as the carrier gas. Separation was performed on a Zebron ZB-5MSi column (Phenomenex) with guardian 5M (30 m × 0.25 mm × 0.25 µm) with 5% Polysilarylene - 95% Polydimethylsiloxane copolymer composition and 106 relative voltage. The initial temperature of 240°C was held for 5 minutes and increased to 300°C at a rate of 10°C/min and held for 25 minutes. The full scan method measured mass from 50 to 800 amu and ions detected in the SIM mode included: 99.1, 129, 165, 171, 173.1, 187, 261, 314.1, 329.3, 330, 370, 382.3, 417.4, 456.4, 458, 460, 470, 472.3, 486, 546, 560, and 634.
Veratrum californicum metabolite extraction for quantitation by LC-MS/MS
See Method S2.
Transcriptome assembly and determination of relative contig expression
cDNA library construction, Illumina paired-end sequencing, and de novo transcriptome assembly were performed at the National Center for Genome Resources (Santa Fe, New Mexico). Please refer Method S3 for transcriptome assembly details.
Transcriptome dataset interrogation using Haystack and Plant Tribes
Identification of genes whose expression pattern correlated with accumulation of cyclopamine was determined using the Haystack program (Michael et al. 2008, Mockler et al. 2007). The LC-MS/MS cyclopamine quantitation data for the different V. californicum tissues was used to formulate a model based upon the ratio of biosynthetic tissues. 95 % of the total cyclopamine was found in the subterranean tissues (root, bulb, and rhizome) whereas 5 % was found above ground (leaf, shoot, and flower). For the input model, each subterranean tissue was given a value of 20 and all above ground tissues including the tissue culture samples was designated 1. Parameters for Haystack were as follows: correlation cut off = 0.7 fold change = 2, p-value = 0.05 and background = 1. Due to the large data input, Haystack analysis was performed on a UNIX server in-house as opposed to the version available online. Annotation data was then merged with the gene outputs from each of the models. Subsequent alignments and phylogenetic analysis were performed using Muscle algorithm (Edgar 2004) and Mega v6.06 (Tamura et al. 2011). See Method S4 for dataset interrogation using PlantTribes and a detailed description of methods for contig prioritization.
Construction of viral expression vectors
Candidate contigs obtained from Haystack and phylogenetic analysis were subjected to BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and global alignments to homologous, experimentally characterized gene sequences with the CLC Main Workbench 6.8, for prediction of the open reading frame. Where the reading frame appeared incomplete, Rapid Amplification of cDNA Ends (RACE) was used to obtain the complete coding sequence. V. californicum cDNA was prepared from root RNA extracts using M-MLV Reverse Transcriptase (Invitrogen) according to manufacturer’s instructions. All primer sequences and PCR programs can be found in Table S11 and S12, respectively. Please refer to Method S5 for specific cloning details.
Virus co-transfection, amplification, and protein production
Extraction of multiple infections for Sf9 in vivo product production
See Method S7.
Enzyme assays
Each cytochrome P450 co-expressed with CPR in S. frugiperda Sf9 cells and GABAT1 was subjected to individual enzyme assays with the compounds designated in Table S5 including cholesterol (Sigma Aldrich), 22(R)-hydroxycholesterol (Sigma Aldrich), 26-hydroxycholesterol [27(25R)-hydroxycholesterol] (Avanti Polar Lipids Inc.), 22(S)-hydroxycholesterol (American Radiolabeled Chemicals), 24(S)-hydroxycholesterol (American Radiolabeled Chemicals), 4β-hydroxycholesterol (Research Plus Inc.), 7β-hydroxycholesterol (Sigma Aldrich), campesterol (Avanti Polar Lipids Inc.), β-sitosterol (Sigma Aldrich), stigmasterol (Sigma Aldrich), GABA (Sigma Aldrich), L-arginine (Sigma Aldrich), and L-glutamine (Sigma Aldrich) to determine substrate specificity. Compounds were prepared to 1 mM stock solutions of 100% DMSO and diluted with H2O, except for the amino acids, which were prepared as 200 mM stocks in pure H2O. For GC-MS analysis, 5 individual assays per substrate were pooled after incubation at 30°C for 2 hours; one assay produced sufficient product for analysis by LC-MS/MS. Assay conditions were as follows: 80 µl S. frugiperda Sf9 cell suspension (obtained by re-suspension of 50 ml viral infected culture pellet in 3.5 ml of 100 mM tricine pH 7.4/ 5 mM thioglycolic acid), 60 mM potassium phosphate buffer pH 8, 1.25 mM NADPH, 7.5 µM substrate, and H2O in a total volume of 200 µl. Controls were performed with no enzyme and S. frugiperda Sf9 cells expressing an unrelated cytochrome P450 (CYP719A14 cheilanthifoline synthase from Argemone mexicana), or CPR-only, for each assay.
The initial GABAT1 enzyme assay contained 55 µl S. frugiperda Sf9 cell suspension infected with CYP90B27, CYP94N1, CYP90G1, and CPR modified baculoviruses (to provide 22-hydroxycholesterol-26-al substrate), 40 µl S. frugiperda Sf9 cells expressing GABAT1, 60 mM potassium phosphate buffer pH 8, 1.5 mM DTT, 100 µM pyridoxal-5-phosphate (PLP), 16 mM GABA, 500 µM NADPH, and H2O to a total volume of 200 µl. Assay mixes lacking either enzyme or GABA, and control cytochrome P450 assays were run in parallel and each was allowed to proceed for 2 hours at 30°C. Samples were extracted twice with 400 µl ethyl acetate. For determination of the amino group donor, assays were prepared as above for GABAT1 with the following exceptions. The GABAT1 gene was cloned into the EcoRI/NdeI sites of pET28a and expressed in E.coli PlusE cells. The GABAT1 protein was purified in the presence of 2 µg/ml PLP using TALON metal affinity resin (Clontech) and PD-10 desalting columns (GE Healthcare) according to manufacturer’s instructions to obtain purified enzyme. The substrate 22-hydroxycholesterol-26-al was obtained by extracting 2 ml of Sf9 cells expressing CYP90B27, CYP94N1, and CPR with 2 volumes of ethyl acetate and dried with N2. The dried extract was resuspended in 400 µl of 10% DMSO. Assays were performed in duplicate and each run with 22-hydroxycholesterol-26-al and either GABA, L-arginine, or L-glutamine alongside CPR only controls and controls using GABAT1 expressed in Sf9 cells. Samples were then dried under N2, re-suspended in 50–100 µl 80% methanol, and injected onto LC-MS/MS with conditions described above. All cytochrome P450 enzyme assays utilized crude S. frugiperda Sf9 protein extracts that contain endogenous metabolites, including cholesterol.
Assays to clarify order of enzymatic transformations
See Method S8 and Figure S11.
Enzymatic product purification for NMR and High Resolution MS for structure elucidation
Dimedone aldehyde trapping
Enzyme assays containing CYP94N1 and 22(R)-hydroxycholesterol as substrate, or CYP90B27 + CYP94N1 utilizing endogenous cholesterol in S. frugiperda Sf9 cells as substrate, or CYP90B27 + CYP94N1 and GABAT1, also utilizing endogenous cholesterol in S. frugiperda Sf9 cells as substrate with either 80 µl 10 mg/ml dimedone in 10% DMSO or 80 µl 10% DMSO were incubated overnight at 30°C. Assays were extracted twice with 2 volumes ethyl acetate and analyzed by LC-MS/MS. All cytochrome P450 enzymes were co-expressed with CPR.
Sodium borohydride reduction
2 ml S. frugiperda Sf9 cells expressing CYP90B27 + CYP94N1 + GABAT1 + CYP90G1 + CPR were extracted twice with equal volume ethyl acetate. Extracts were divided equally, dried under N2, and re-suspended in 50 µl 80% methanol each. One sample was treated with 50 µl 1 M NaBH4 in 1 M NaOH for 15 minutes. 100 µl H2O were added to both samples, and each extracted twice with equal volumes of chloroform. Samples were dried under N2, re-suspended in 50 µl 80% methanol and analyzed by LC-MS/MS as described above. S. frugiperda Sf9 cells expressing CPR only were run in parallel as control.
Phylogenetic analysis of cytochrome P450 enzymes across species using deep transcriptome sequence data from 1KP and MonAToL projects
See Method S10 and Table S8.
Supplementary Material
Acknowledgments
We greatly thank Infinity Pharmaceuticals for providing authentic cyclopamine. This work was supported by funds from Infinity Pharmaceuticals to T.M.K., NSF DEB-1442071 to E.A. Kellogg, NSF DBI-0521250 to PMSF, an Indo-US (IUSSTF) Research Fellowship to A.K.S. and NIH 1R01DA025197-02 to T.M.K. Analysis performed on the 4000 QTRAP and LTQ-Velos Pro Orbitrap was supported by The National Science Foundation under Grants DBI-0521250 and DBI-0922879, respectively. We also thank the National Science Foundation for supporting this research through Assembling the Tree of Life (DEB 0829868), and the 1000 Plants (1KP) initiative, led by GKSW, which was funded by the Alberta Ministry of Innovation and Advanced Education, Alberta Innovates Technology Futures (AITF) Innovates Centres of Research Excellence (iCORE), Musea Ventures, and BGI-Shenzhen.
Footnotes
Author contributions
D.A.M. collected V. californicum plant material. M.M.A isolated V. californicum RNA. D.R.R. analyzed transcriptome data and produced the candidate gene list. D.R.R., A.K.S, and M.M.A. cloned candidates into baculovirus expression vectors and expressed them in Sf9 cells. J.M.A. and M.M.A. developed and performed enzyme assays to determine enzymatic activity and purified metabolites for structure elucidation. C.M.S. and M.O.J. purified metabolites and determined structures by NMR. B.S.E analyzed metabolites by LTQ-Velos Pro Orbitrap. M.R.M. performed deep phylogenetic analysis of cytochromes P450. J.C.P., P.P.E., M.D.B., A.S., G.K.W., and M.K.D. contributed plant materials and RNAseq data from the 1KP project. M.M.A. and T.M.K. analyzed data and wrote the manuscript, which was reviewed by all authors. T.M.K. directed the study.
Competing financial interests
The authors declare no competing financial interests.
The following supporting information is available online:
Supporting Figures
Figure S1. Cyclopamine accumulation profile in Veratrum californicum.
Figure S2. Mass spectra of select derivatized standards and enzymatically formed products.
Figure S3. GC-MS analysis of selected Veratrum californicum cytochrome P450 enzymes with cholesterol.
Figure S4. Mass spectra of enzymatically formed 22,26-dihydroxycholesterol.
Figure S5. GC-MS analysis of Veratrum californicum CYP90B27 with 26-hydroxycholesterol and 7β-hydroxycholesterol.
Figure S6. GC-MS analysis of Veratrum californicum cytochrome P450 enzymes CYP90B27 and CYP90G1.
Figure S7. Determination of the order of the enzymatic steps to verazine.
Figure S8. LC-MS/MS of S. frugiperda Sf9 extracts expressing Veratrum californicum genes.
Figure S9. High-resolution mass spectrometric analysis of enzymatically formed 22-hydroxy-26-aminocholesterol and verazine.
Figure S10. LC-MS/MS of enzyme assays with his-tag purified GABAT1.
Figure S11. Enzyme assay workflow for clarification of cyclopamine biosynthetic pathway.
Figure S12. Enzyme assays for biosynthetic pathway order clarification in Veratrum californicum using GC-MS.
Figure S13. Trapping of enzymatically formed aldehyde intermediate.
Figure S14. Derivatization of enzymatically formed verazine.
Figure S15. Relative tissue specific accumulation of selected Veratrum californicum metabolites.
Figure S16. Phylogenetic tree of cytochromes P450 using transcriptome data from 1KP and MonAToL sequencing projects.
Figure S17. LC-MS/MS analysis of Veratrum californicum GABATs.
Supporting Tables
Table S1. Sequence reads and length generated for each RNA sample (paired end).
Table S2. Transcriptome statistics provided by the NCGR.
Table S3. Selected top-scoring cytochrome P450 candidate cDNAs for the enzymatic conversion of cholesterol to cyclopamine.
Table S4. Selected top-scoring transaminases in the steroid alkaloid biosynthetic pathway to cyclopamine.
Table S5. Substrate testing for cytochrome P450 enzymes co-expressed with CPR.
Table S6. Assigned enzyme names.
Table S7. Accession numbers and both putative and determined function for sequences in cytochrome P450 phylogenetic tree.
Table S8. Species key and source for sequences in P450 phylogenetic tree using deep transcriptome sequence data from 1KP and MonAToL projects.
Table S9. LS/MS rooting medium was prepared with the following concentrations and brought to a final pH of 5.75.
Table S10. LC-MS/MS Q-TRAP 4000 method parameters.
Table S11. Primer sequences.
Table S12. PCR Parameters.
Table S13. Viral combinations for in vivo production of metabolites in S. frugiperda Sf9 cells.
Supporting Methods
Method S1. Plant material and RNA extraction.
Method S2. Veratrum californicum metabolite extraction for quantitation by LC-MS/MS.
Method S3. Transcriptome assembly and retrieval of expression data.
Method S4. Transcriptome dataset interrogation using Haystack and Plant Tribes.
Method S5. Construction of viral expression vectors.
Method S6. Virus co-transfection, amplification, and protein production.
Method S7. Extraction of multiple infections for Sf9 in vivo product production.
Method S8. Assays to clarify order of enzymatic transformations.
Method S9. Enzymatic product purification for NMR and High Resolution MS for structure elucidation.
Method S10. Phylogenetic analysis of cytochrome P450 enzymes across species using deep transcriptome sequence data from 1KP and MonAToL projects.
Data S1. Data access for sequence information.
Data S2. NMR designations for 22-keto-cholesterol and 22-keto-26-hydroxycholesterol in MeOD.
References
- Adam G, Schreiber K, Tomko J, Vassova A. Verazine, a new Veratrum alkaloid with 22, 26-imino-cholestan structure. Tetrahedron. 1967;23:167–171. doi: 10.1016/s0040-4020(01)83297-x. [DOI] [PubMed] [Google Scholar]
- Bahra M, Kamphues C, Boas-Knoop S, Lippert S, Esendik U, Schuller U, Hartmann W, Waha A, Neuhaus P, Heppner F, Pietsch T, Koch A. Combination of hedgehog signaling blockage and chemotherapy leads to tumor reduction in pancreatic adenocarcinomas. Pancreas. 2012;41:222–229. doi: 10.1097/MPA.0b013e31822896dd. [DOI] [PubMed] [Google Scholar]
- Behnsawy HM, Shigemura K, Meligy FY, Yamamichi F, Yamashita M, Haung WC, Li X, Miyake H, Tanaka K, Kawabata M, Shirakawa T, Fujisawa M. Possible role of sonic hedgehog and epithelial-mesenchymal transition in renal cell cancer progression. Korean journal of urology. 2013;54:547–554. doi: 10.4111/kju.2013.54.8.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, Beachy PA. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- Chandler CM, McDougal OM. Medicinal history of North American Veratrum. Phytochemistry reviews : proceedings of the Phytochemical Society of Europe. 2014;13:671–694. doi: 10.1007/s11101-013-9328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes & development. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell GA. The alkaloids. Chemistry and biology. San Diego, Amsterdam: Academic Press, Elsevier; 1998. [Google Scholar]
- Dhakal R, Bajpai VK, Baek KH. Production of gaba (gamma - Aminobutyric acid) by microorganisms: a review. Brazilian journal of microbiology : [publication of the Brazilian Society for Microbiology] 2012;43:1230–1241. doi: 10.1590/S1517-83822012000400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Chavez ML, Rolf M, Gesell A, Kutchan TM. Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch Biochem Biophys. 2011;507:186–193. doi: 10.1016/j.abb.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailani MR, Stahle-Backdahl M, Leffell DJ, Glynn M, Zaphiropoulos PG, Pressman C, Unden AB, Dean M, Brash DE, Bale AE, Toftgard R. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nature genetics. 1996;14:78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- Gan KH, Lin CN, Won SJ. Cytotoxic principles and their derivatives of Formosan Solanum plants. Journal of natural products. 1993;56:15–21. doi: 10.1021/np50091a003. [DOI] [PubMed] [Google Scholar]
- Gesell A, Rolf M, Ziegler J, Diaz Chavez ML, Huang FC, Kutchan TM. CYP719B1 is salutaridine synthase, the C-C phenol-coupling enzyme of morphine biosynthesis in opium poppy. The Journal of biological chemistry. 2009;284:24432–24442. doi: 10.1074/jbc.M109.033373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe T, Lenz R, Kutchan TM. Molecular characterization of the salutaridinol 7-O-acetyltransferase involved in morphine biosynthesis in opium poppy Papaver somniferum. The Journal of biological chemistry. 2001;276:30717–30723. doi: 10.1074/jbc.M102688200. [DOI] [PubMed] [Google Scholar]
- Hagel JM, Facchini PJ. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nature chemical biology. 2010;6:273–275. doi: 10.1038/nchembio.317. [DOI] [PubMed] [Google Scholar]
- Huang FC, Kutchan TM. Distribution of morphinan and benzo[c]phenanthridine alkaloid gene transcript accumulation in Papaver somninferum. Phytochemistry. 2000;53:555–564. doi: 10.1016/s0031-9422(99)00600-7. [DOI] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, Tikunov Y, Bovy A, Chikate Y, Singh P, Rogachev I, Beekwilder J, Giri AP, Aharoni A. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science. 2013;341:175–179. doi: 10.1126/science.1240230. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Li H, Li P, Cai Z, Ye W. Steroidal alkaloids from the bulbs of Fritillaria puqiensis. Journal of natural products. 2005;68:264–267. doi: 10.1021/np0497649. [DOI] [PubMed] [Google Scholar]
- Keeler RF. Teratogenic compounds of Veratrum californicum (Durand)-IV. First isolation of veratramine and alkaloid Q and a reliable method for isolation of cyclopamine. Phytochemistry. 1968;7:303–306. [Google Scholar]
- Keeler RF. Teratogenic compounds of veratrum californicum (durand) - VI: The structure of cyclopamine. Phytochemistry. 1969;8:223–225. [Google Scholar]
- Keeler RF. Teratogenic compounds in Veratrum californicum (Durand) IX. Structure-activity relation. Teratology. 1970a;3:169–173. doi: 10.1002/tera.1420030209. [DOI] [PubMed] [Google Scholar]
- Keeler RF. Teratogenic compounds of Veratrum californicum (Durand) X. Cyclopia in rabbits produced by cyclopamine. Teratology. 1970b;3:175–180. doi: 10.1002/tera.1420030210. [DOI] [PubMed] [Google Scholar]
- Keeler RF, Binns W. Teratogenic compounds of Veratrum californicum (Durand). II. Production of ovine fetal cyclopia by fractions and alkaloid preparations. Canadian journal of biochemistry. 1966;44:829–838. doi: 10.1139/o66-101. [DOI] [PubMed] [Google Scholar]
- Ko Kaneko, Seto Hideo, Motoki Chiyoko, Mitsuhashi Hiroshi. Biosynthesis of Rubijervine in Veratrum grandiflorum. Phytochemistry. 1975;14:1295–1301. [Google Scholar]
- Ko Kaneko, Mitsuhashi Hiroshi, Hirayama Koichiro, Ohmori S. 11-Deoxojervine as a precursor for jervine biosynthesis in Veratrum grandiflorum. Phytochemistry. 1970a;9:2497–2501. [Google Scholar]
- Ko Kaneko, Mitsuhashi Hiroshi, Hirayama Koichiro, Yoshida N. Biosynthesis of C-nor-D-homo-steroidal alkaloids from acetate-1-14C, cholesterol-4-14C and cholesterol-26-14C in Veratrum grandiflorum. Phytochemistry. 1970b;9:2489–2495. [Google Scholar]
- Ko Kaneko, Tanaka Mikako W, Mitsuhashi H. Origin of nitrogen in the biosynthesis of solanidine by Veratrum grandiflorum. Phytochemistry. 1976;15:1391–1393. [Google Scholar]
- Ko Kaneko, Tanaka Mikako W, Mitsuhashi H. Dormantinol, a possible precursor in solanidine biosynthesis, from budding Veratrum grandiflorum. Phytochemistry. 1977;16:1247–1251. [Google Scholar]
- Kusano G, Takahashi A, Sugiyama K, Nozoe S. Antifungal properties of solanum alkaloids. Chemical & pharmaceutical bulletin. 1987;35:4862–4867. doi: 10.1248/cpb.35.4862. [DOI] [PubMed] [Google Scholar]
- Lin TL, Wang QH, Brown P, Peacock C, Merchant AA, Brennan S, Jones E, McGovern K, Watkins DN, Sakamoto KM, Matsui W. Self-renewal of acute lymphocytic leukemia cells is limited by the Hedgehog pathway inhibitors cyclopamine and IPI-926. PLoS One. 2010;5:e15262. doi: 10.1371/journal.pone.0015262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 2008;6:e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra BB, Tiwari VK. Natural products: an evolving role in future drug discovery. European journal of medicinal chemistry. 2011;46:4769–4807. doi: 10.1016/j.ejmech.2011.07.057. [DOI] [PubMed] [Google Scholar]
- Mithofer A, Boland W. Plant defense against herbivores: chemical aspects. Annual review of plant biology. 2012;63:431–450. doi: 10.1146/annurev-arplant-042110-103854. [DOI] [PubMed] [Google Scholar]
- Mizutani M. Impacts of diversification of cytochrome P450 on plant metabolism. Biological & pharmaceutical bulletin. 2012;35:824–832. doi: 10.1248/bpb.35.824. [DOI] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- Nims E, Dubois CP, Roberts SC, Walker EL. Expression profiling of genes involved in paclitaxel biosynthesis for targeted metabolic engineering. Metabolic engineering. 2006;8:385–394. doi: 10.1016/j.ymben.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, Frese KK, Denicola G, Feig C, Combs C, Winter SP, Ireland-Zecchini H, Reichelt S, Howat WJ, Chang A, Dhara M, Wang L, Ruckert F, Grutzmann R, Pilarsky C, Izeradjene K, Hingorani SR, Huang P, Davies SE, Plunkett W, Egorin M, Hruban RH, Whitebread N, McGovern K, Adams J, Iacobuzio-Donahue C, Griffiths J, Tuveson DA. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onrubia M, Moyano E, Bonfill M, Palazon J, Goossens A, Cusido RM. The relationship between TXS, DBAT, BAPT and DBTNBT gene expression and taxane production during the development of Taxus baccata plantlets. Plant science : an international journal of experimental plant biology. 2011;181:282–287. doi: 10.1016/j.plantsci.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Rosco A, Pauli HH, Priesner W, Kutchan TM. Cloning and heterologous expression of NADPH-cytochrome P450 reductases from the Papaveraceae. Arch Biochem Biophys. 1997;348:369–377. doi: 10.1006/abbi.1997.0374. [DOI] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics - 'majority report by precogs'. Trends in plant science. 2008;13:36–43. doi: 10.1016/j.tplants.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay MR, Lescarbeau A, Grogan MJ, Tan E, Lin G, Austad BC, Yu LC, Behnke ML, Nair SJ, Hagel M, White K, Conley J, Manna JD, Alvarez-Diez TM, Hoyt J, Woodward CN, Sydor JR, Pink M, MacDougall J, Campbell MJ, Cushing J, Ferguson J, Curtis MS, McGovern K, Read MA, Palombella VJ, Adams J, Castro AC. Discovery of a potent and orally active hedgehog pathway antagonist (IPI-926) Journal of medicinal chemistry. 2009;52:4400–4418. doi: 10.1021/jm900305z. [DOI] [PubMed] [Google Scholar]
- Tschesche RBH. Side chain functionalization of cholesterol in the biosynthesis of solasodine in Solanum laciniatum. Phytochemistry. 1980;19:1449–1451. [Google Scholar]
- Unterlinner B, Lenz R, Kutchan TM. Molecular cloning and functional expression of codeinone reductase: the penultimate enzyme in morphine biosynthesis in the opium poppy Papaver somniferum. The Plant journal : for cell and molecular biology. 1999;18:465–475. doi: 10.1046/j.1365-313x.1999.00470.x. [DOI] [PubMed] [Google Scholar]
- Wall PK, Leebens-Mack J, Muller KF, Field D, Altman NS, dePamphilis CW. PlantTribes: a gene and gene family resource for comparative genomics in plants. Nucleic acids research. 2008;36:D970–D976. doi: 10.1093/nar/gkm972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weid M, Ziegler J, Kutchan TM. The roles of latex and the vascular bundle in morphine biosynthesis in the opium poppy, Papaver somniferum. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13957–13962. doi: 10.1073/pnas.0405704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CLJ, Ye W, Liu C, Tan R. Neoverataline A and B, two antifungal alkaloids with a novel carbon skeleton from Veratrum taliense. Tetrahedron. 2003;59:5743–5747. [Google Scholar]
- Ziegler J, Voigtlander S, Schmidt J, Kramell R, Miersch O, Ammer C, Gesell A, Kutchan TM. Comparative transcript and alkaloid profiling in Papaver species identifies a short chain dehydrogenase/reductase involved in morphine biosynthesis. The Plant journal : for cell and molecular biology. 2006;48:177–192. doi: 10.1111/j.1365-313X.2006.02860.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.