Abstract
Background
Simultaneous inactivation of pig GGTA1 and CMAH genes eliminates carbohydrate xenoantigens recognized by human antibodies. The β4GalNT2 glycosyltransferase may also synthesize xenoantigens. To further characterize glycan-based species incompatibilities, we examined human and non-human primate antibody binding to cells derived from genetically modified pigs lacking these carbohydrate-modifying genes.
Methods
The Cas9 endonuclease and gRNA were used to create pigs lacking GGTA1, GGTA1/CMAH, or GGTA1/CMAH/β4GalNT2 genes. Peripheral blood mononuclear cells were isolated from these animals and examined for binding to IgM and IgG from humans, rhesus macaques, and baboons.
Results
Cells from GGTA1/CMAH/β4GalNT2 deficient pigs exhibited reduced human IgM and IgG binding compared to cells lacking both GGTA1 and CMAH. Nonhuman primate antibody reactivity with cells from the various pigs exhibited a slightly different pattern of reactivity than that seen in humans. Simultaneous inactivation of the GGTA1 and CMAH genes increased nonhuman primate antibody binding compared to cells lacking either GGTA1 only or to those deficient in GGTA1/CMAH/β4GalNT2.
Conclusions
Inactivation of the β4GalNT2 gene reduces human and nonhuman primate antibody binding resulting in diminished porcine xenoantigenicity. The increased humoral immunity of nonhuman primates towards GGTA1/CMAH-deficient cells compared to pigs lacking either GGTA1 or GGTA1/CMAH/β4GalNT2 highlights the complexities of carbohydrate xenoantigens and suggests potential limitations of the nonhuman primate model for examining some genetic modifications. The progressive reduction of swine xenoantigens recognized by human immunoglobulin through inactivation of pig GGTA1/CMAH/β4GalNT2 genes demonstrates that the antibody barrier to xenotransplantation can be minimized by genetic engineering.
Keywords: β4GalNT2, xenoantigen, antibody, CRISPR, Cas9, primate, swine, genetic engineering
Introduction
Clinical transplantation continues to be limited by the shortage of suitable human donor organs. The UNOS waitlist continues to grow and now more than 120,000 people await transplantation in the United States [1]. Xenotransplantation using pig organs is a possible solution to the organ shortage, but has not been applied clinically because antigens on the surface of pig cells lead to antibody mediated rejection (AMR) in pig-to-nonhuman primate models [2].
Two well-characterized xenoantigens on pig cells are the carbohydrates Galactose α1-3 galactose (αGal) and N-glycolylneuraminic acid (Neu5Gc) [3, 4]. Though disrupting the swine GGTA1 gene eliminated expression of αGal and increased the survival of pig-to-primate transplants, AMR remained as a significant problem [5]. Nuclease-based genome editing has simplified the manipulation of mammalian genomes enabling the disruption of additional pig genes and the production of GGTA1/CMAH KO pigs [6-14]. Inactivation of both GGTA1 and CMAH created pigs with reduced human antibody binding to their red blood cells and peripheral blood mononuclear cells (PBMC) when compared to GGTA1 knockout (KO) pigs [7, 15]. In addition, there was less human antibody binding to PBMCs from these double KO pigs than to chimpanzee PBMCs [16]. This finding was significant since chimpanzee kidneys were not hyperacutely rejected by humans in Reemtsma's series in 1963 despite using very crude immunosuppression available at that time [17]. It may be possible to consider transplanting GGTA/1CMAH KO pig organs into humans taking advantage of new desensitization protocols, but xenoreactive antibodies are still a concern for clinical xenotransplantation [16].
Byrne and McGregor identified additional candidate xenoantigens using cDNA expression libraries from GGTA1 KO pigs and screened them with serum from baboons that had rejected GGTA1 KO pig hearts [5, 18]. Glycans produced by β1,4 N-acetylgalactosaminyl transferase (β4GalNT2) were of interest because the baboons had preformed antibodies to this glycan [19]. We describe the production of a GGTA1/CMAH/β4GalNT2 KO pig using Cas9 and gRNA technology, and evaluate human, rhesus macaque, and baboon antibody binding to PBMCs from this pig. Our data suggest that glycans produced by β4GalNT2 are xenoantigens for many humans.
Methods
Generation and selection of porcine liver derived cells deficient in GGTA1, CMAH and β4GalNT2
Oligo annealing and cloning into the PX330 plasmid to drive gRNA expression was performed as described previously [9] using Addgene plasmid 42230 [http://www.addgene.org/42230/, and reference 20]. Oligo pairs for the targeted genes are: GGTA1 (NCBI Accession: XM_005660398.1) 5’- CACCGAGAAAATAATGAATGTCAA-3’ (forward), 5’-AAACTTGACATTCATTATTTTCTC-3’ (reverse); CMAH (NCBI Accession: NM_001113015.1) 5’-CACCGAGTAAGGTACGTGATCTGT-3’ (forward), 5’-AAACACAGATCACGTACCTTACTC-3’ (reverse); β4GalNT2 (NCBI Accession: NM_001244330.1) 5’-CACCGTGTATCGAGGAACACGCTT-3’ (forward), 5’-AAACAAGCGTGTTCCTCGATACAC-3’ (reverse).
Liver derived cells [21] were co-transfected with all three gRNA/Cas9 plasmids. After 48 hours, the treated cells were passed over an IB4 lectin column to isolate αGal null cells [6]. Two million α-Gal negative cells were further stained with fluorescein labeled Dolichos biflorus Agglutinin (DBA)-FITC (Vector Laboratories, Burlingame, CA, USA) at 2 ug/ml in 500ul HBSS with 0.5% BSA and flow sorted for DBA negative cells using a BD FACSAria sorter (BD Bioscience, San Jose, CA). The presence of Neu5Gc, an indicator of CMAH gene function, was not analyzed prior to somatic cell nuclear transfer (SCNT).
Somatic cell nuclear transfer
SCNT was performed as described [22] using in vitro matured oocytes (DeSoto Biosciences Inc., St. Seymour, TN). Cumulus cells were removed from the oocytes by pipetting in 0.1% hyaluronidase. Only oocytes with normal morphology and a visible polar body were selected for SCNT. Oocytes were incubated in manipulation media (Ca-free NCSU-23 with 5% FBS) containing 5 μg/mL bisbenzimide and 7.5 μg/mL cytochalasin B for 15 min. Oocytes were enucleated by removing the first polar body plus metaphase II plate, and one cell was injected into each enucleated oocyte. Couples were fused and activated simultaneously by two DC pulses of 180 V for 50 μsec (BTX cell electroporator, Harvard Apparatus, Hollison, MA, USA) in 280mM Mannitol, 0. 1mM CaCl2, and 0.05mM MgCl2. Activated embryos were placed back in NCSU-23 medium with 0.4% bovine serum albumin (BSA) and cultured at 38.5 °C, 5% CO2 in a humidified atmosphere for less than 1 h, before being transferred into the recipient. Recipients were synchronized occidental gilts on their first day of estrus. Swine used in this study followed protocols approved by the Institutional Biosafety and Institutional Animal Care and Use Committees of IUPUI.
DNA sequencing of cloned pig
DNA sequencing analysis of the gRNA/Cas9 targeted GGTA1, CMAH, and β4GalNT2 regions in the cloned pig Genomic DNA from the cloned pig was extracted using GenElute Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich, St. Louis, MO). PCR was performed with GGTA1, CMAH, and β4GalNT2-specific primer pairs, respectively. The primers were designed to flank the gRNA/Cas9 target sites and amplified 428 bases of GGTA1, 5’-CCTTAGTATCCTTCCCAACCCAGAC-3’ (forward), 5’-GCTTTCTTTACGGTGTCAGTGAATCC-3’ (reverse); 485 bases of CMAH, 5’-CTTGGAGGTGATTTGAGTTGGG-3’ (forward), 5’-CATTTTCTTCGGAGTTGAGGGC-3’ (reverse); 530 bases of β4GalNT2, 5’-CGCAAGTGACCAGACATCGTTC 3’ (forward), 5’ AAAGCCACAGGAGGAGCCAG-3’ (reverse). Pwo SuperYield DNA polymerase, dNTPack (Roche Applied Science, Indianapolis, IN) was used and PCR conditions for GGTA1 were as follows: 94°C, 2 min; 94°C, 15 sec, 54°C, 30 sec, and 72°C, 45 sec for 15 cycles; 94°C, 15 sec, 54°C, 30 sec, 72°C, 45 sec with additional 5 sec each cycle for 25 cycles; and a final extension step of 72°C for 5 min. For CMAH, 94°C, 2 min; 94°C, 15 sec, 56°C, 30 sec, and 72°C, 45 sec for 15 cycles; 94°C, 15 sec, 56°C, 30 sec, 72°C, 45 sec with additional 5 sec each cycle for 25 cycles; and a final extension step of 72°C for 5 min. For B4GALNT2, 94°C, 2 min; 94°C, 15 sec, 62°C, 30 sec, 72°C, 40 sec for 15 cycles, 94°C 15 sec, 62°C, 30 sec, 72°C 40 sec with additional 5 sec each cycle for 25 cycles; and a final extension step of 72°C for 5 min. The PCR products were separated on 1% agarose gel, purified by GenElute Gel Extraction Kit (Sigma-Aldrich, St. Louis, MO) and sequenced by the Sanger method (DNA Sequencing Core Facility, Indiana University School of Medicine) with the specific sequencing primer, 5’-CCTTAGTATCCTTCCCAACCCAGAC-3’ for GGTA1; 5’-CATTTTCTTCGGAGTTGAGGGC-3’ for CMAH; 5’-AAAGCCACAGGAGGAGCCAG-3’ for β4GalNT2. PCR products were inserted into pCR4blunt-TOPO vector, transformed into E. Coli, and individual clones were sequenced to analyze individual alleles of each targeted gene.
Flow cytometric phenotyping of PBMCs
Whole blood was obtained in Acid-Citrate-Dextrose (ACD) and prepared using Ficoll-Paque Plus (GE) to isolate PBMCs. Cells were stained with isolectin Griffonia simplicifolia GS-IB4 (IB4 lectin), Alexa Fluor 488 (Molecular Probes, Grand Island, NY, USA) and compared to cells alone. Neu5Gc phenotype was determined with chicken anti-Neu5Gc antibody, donkey anti-chicken Dylight 649, and negative control isotype from Neu5Gc kit (BioLegend, San Diego, CA, USA). β4GalNT2 phenotype determined with Dolichos biflorus agglutinin (DBA lectin), labeled with fluorescein (Vector Laboratories, Burlingame, CA, USA). All cells were suspended at 2×106/mL in HBSS with Neu5Gc free blocking buffer for 15 minutes and then incubated for 30min at room temperature with appropriate lectin or antibody. Cells were washed with blocking buffer. Flow cytometric analysis was completed on BD Accuri C6 flow cytometer with the C6 Software (BD Biosciences, San Jose, CA, USA).
Human antibody binding to Swine PBMCs
Human sera were obtained from volunteers or de-identified remnant clinical lab samples with IRB approval. Swine PBMCs were isolated using Ficoll-Paque Plus and suspended in EX-CELL 610-HSF Serum-Free Medium (Sigma, St. Louis, MO, USA). A 25% mixture of heat-inactivated serum and 2×105 cells in EX-CELL with 0.1% sodium azide were incubated for 30 min at 4 °C. Cells were washed three times with EX-CELL + azide and stained with Goat anti-human IgG Alexa Fluor 488 and Donkey anti-human IgM Alexa Fluor 488 (Jackson ImmunoResearch Laboratories Inc.) for 30 min at 4 °C. Cells were washed using EX-CELL medium as above and flow cytometric analysis completed on BD Accuri C6 flow cytometer.
Rhesus and baboon antibody binding to triple β4GALNT2 -KO PBMCs
Sera samples were obtained from thirty-four Rhesus Macaques following IACUC-approved protocols (Yerkes Primate Center, Atlanta, GA, USA) and heat inactivated at 57 °C for 30 min. Baboon sera were obtained from ten animals (Texas Biomedical Research Institute, San Antonio, TX, USA) and heat inactivated. Multiple anti-immunoglobulin products were tested for cross reactivity with Baboon and Rhesus immunoglobulin using serial dilution. Goat anti-human IgG Alexa Fluor 488 and Donkey anti-human IgM Alexa Fluor 488 (Jackson ImmunoResearch Laboratories Inc.) were selected. A 25% mixture of serum was incubated with 2×105 cells for 30min at 4°C, washed three times with EX-CELL plus azide, and stained with anti-IgG or IgM secondary. Cells were washed again and analyzed flow cytometrically.
Statistical Analysis
Antibody binding results were reported as median fluorescence intensity (MFI) of the FL1 channel. Graph and data analyses were completed using Prism 6 for windows (GraphPad Software Inc. La Jolla, CA, USA). Human serum antibody binding assays were analyzed using two-tailed paired t-test comparing single MFI results for each animal. The primate serum antibody binding was completed in duplicate, and averaged values used in the graphs and analyzed using one-way ANOVA, comparing column means with Tukey correction for multiple comparison.
Results
The β4GalNT2 pig was generated as described in Figure 1. Primary liver-derived cells were simultaneously transfected with three plasmids encoding the Cas9 endonuclease and gRNA specific for the GGTA1, CMAH, and β4GalNT2 genes. Treated cells were passed over an IB4 lectin column, which binds αGal. Cells having disrupted GGTA1 pass through the column because they lack this carbohydrate xenoantigen. αGal deficient cells were then subjected to fluorescence-activated cell sorting to isolate cells that failed to bind the DBA lectin, which targets carbohydrate structures produced by the β4GalNT2 enzyme. Cells determined to lack GGTA1 and β4GalNT2 function by phenotypic selection were used in somatic cell nuclear transfer. Expression of the Neu5Gc isoform of sialic acid, a marker of CMAH gene activity, was not examined prior to SCNT as this analysis is challenging because CMAH deficient cells acquire Neu5Gc from cell culture components [9, 23]. Pig cloning statistics are shown in Table 1.
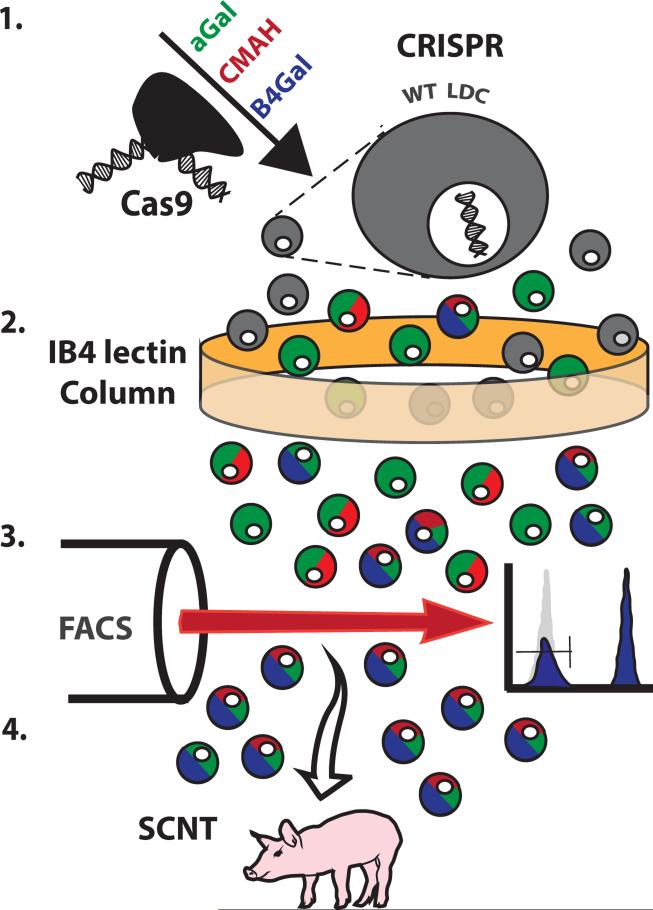
Figure 1. Genetic modification and cell selection.
Step 1: Wild type liver-derived cells were treated with the Cas9 endonuclease and gRNA targeting the GGTA1, CMAH, and β4GalNT2 genes. Step 2: This heterogenous population was subjected to an IB4 lectin column to enrich for GGTA1 mutant cell types. Step 3: GGTA1 mutant cells were incubated with DBA lectin and sorted to isolate DBA-negative cells, which do not express β4GalNT2-derived carbohydrates. Step 4: SCNT was performed with cells determined to lack GGTA1 and β4GalNT2 gene function by the phenotypic selection. Phenotypic analysis of CMAH gene activity is difficult to perform on cells in culture and was not attempted prior to SCNT.
Table 1.
Somatic cell nuclear transfer with B4GALCMAH genetically modified cells.
| Recipient | Transferred embryos | Pregnancy | Fetuses collected | Piglets | KO | Cloning efficiency** |
|---|---|---|---|---|---|---|
| 1 | 82 | No | ||||
| 2 | 102 | Yes | Aborted | |||
| 3 | 69 | No | ||||
| 4 | 65 | Yes | Aborted | |||
| 5 | 69 | No | ||||
| 6 | 96 | Yes | 1 | 1 | 1.04 | |
| Total | 483 | 50%* | 0 | 1 | 1 | 0.2 (1.04%***) |
Pregnant animals/ total recipients
Fetuses and piglets/ total embryos transferred
Fetuses and piglets/ embryos transferred to pregnant recipients
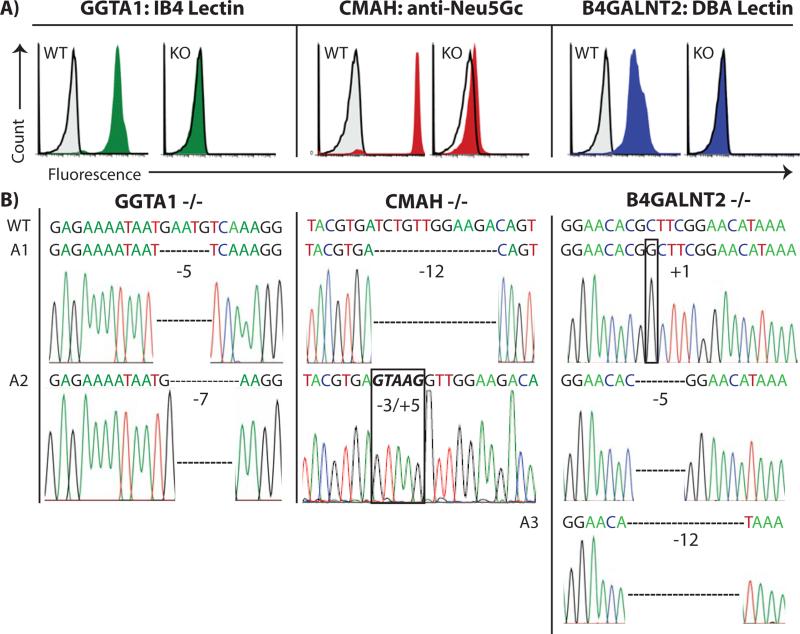
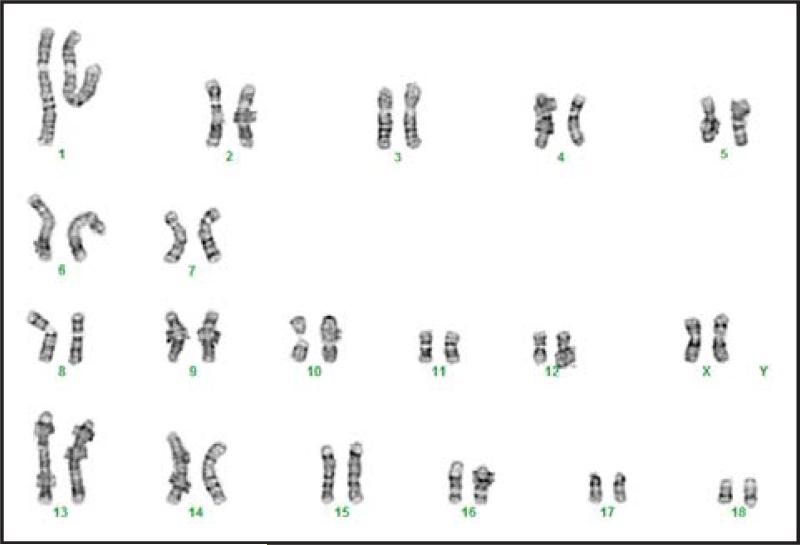
PBMC collected from a single engineered pig were subjected to phenotypic analysis, with PBMC from WT pigs serving as positive controls (Figure 2A). The individual alleles from all three targeted genes were also subjected to DNA sequence analyses (Figure 2B). Deletions of 5 and 7 bases from exon 3 of the GGTA1 gene (Figure 2B) prevented expression of the αGal carbohydrate (Figure 2A) by creating frameshifts immediately after the start codon in both alleles. One copy of the CMAH gene had a 3 base deletion combined with a 5 base insertion, and the second allele contained a 12 base deletion (Figure 2B). Each of these modifications may have disrupted exon 6 of CMAH by shifting its reading destroying the immediate upstream canonical splice acceptor site. Either defective mRNA splicing or frame shift mutations would cause an enzyme deficiency eliminating Neu5Gc synthesis (Figure 2A). β4GalNT2 exhibited three distinct, and apparently mutually exclusive, mutant alleles consisting of either a 1 base insertion, or a 5 base deletion, or a 12 base deletion. These modifications all occur in exon 2 and precede the coding region for the catalytic domain of the β4GalNT2 enzyme. The 1- and 5- base deletions lead to frameshifts in the translational reading frame. The 12-base deletion likely disrupts targeting of the β4GalNT2 enzyme to the secretory pathway by eliminating four amino acids from the signal sequence that targets the enzyme to the secretory pathway. The presence of more than two alleles may represent gene duplication rather than aneuploidy because the animal has a normal karyotype and only carries two copies of chromosome 12 (Figure 3), which is the location of the β4GalNT2 gene (Ensembl database, β4GALNT2 ENSSSCG00000030269).
Figure 2. Phenotype and genotype of GGTA1/CMAH/β4GalNT2 knockout pig.
A) PBMCs from wild type (WT) or KO pigs were incubated with fluorescent probes: IB4 lectin to detect αGal carbohydrates (green histograms); chicken antibodies specific for the Neu5Gc carbohydrate (red histograms); DBA lectin specific for carbohydrates produced by the β4GalNT2 gene (blue histograms). Gray histograms outlined in black represent negative controls that were unstained cells for lectin experiments and an irrelevant isotype matched antibody for Neu5Gc staining. The appearance of a single histogram in KO samples indicates overlap with negative control. B) DNA sequence analysis was performed on isolated alleles of GGTA1, CMAH and β4GalNT2.
Electropherograms revealed mutations in all alleles of each gene. WT sequence (WT) is shown for comparison to the modified alleles.
Figure 3. Normal Karyotype of the GGTA1/CMAH//β4GalNT2 knockout pig.
Chromosome 12 contains the β4GalNT2 gene.
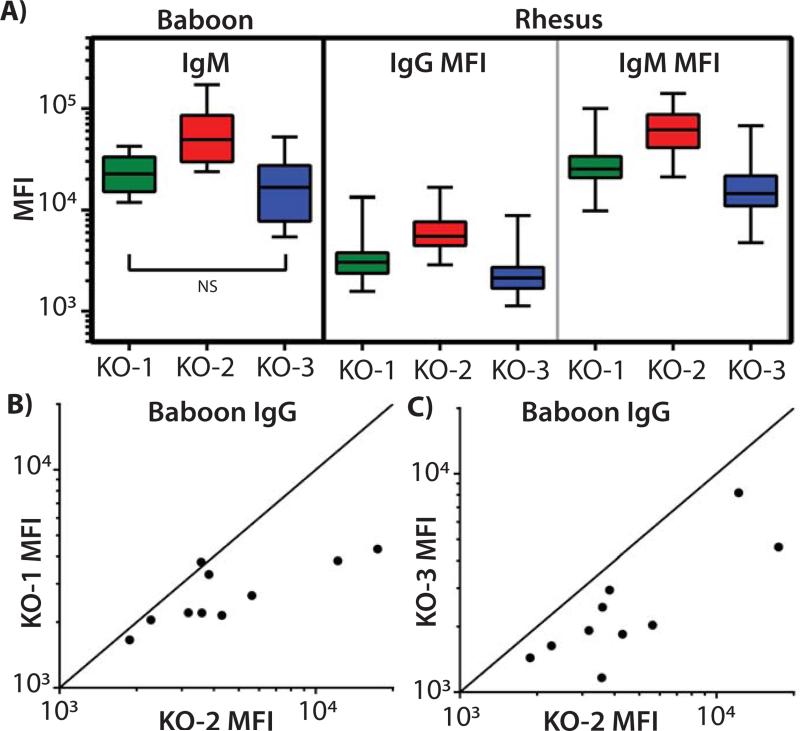
Next we examined antibody binding to cells from the triple knockout animal because Byrne and McGregor have demonstrated that baboon antibodies recognize xenoantigens produced by the β4GalNT2 gene. We compared IgM and IgG binding of NHP antibodies to PBMC isolated from swine deficient in GGTA1 only (KO-1); GGTA1 and CMAH (KO-2); or GGTA1, CMAH, and β4GalNT2 (KO-3).
Swine PBMC, deficient in GGTA1 and CMAH, bound more baboon IgM than did cells lacking only GGTA1 (Figure 4A; KO-1 vs. KO-2, p=0.041). Disrupting β4GalNT2 in addition to CMAH and GGTA1 reduced pig xenoantigen expression as shown by diminished baboon IgM fluorescence (Figure 4A; KO-2 vs. KO-3, p<0.010). The grouped analyses did not reflect significant differences in the binding of baboon IgG to the various swine PBMC (KO-1 vs. KO2, p=0.132; KO-2 vs. KO-3, p=0.073). This may have resulted from analyzing too few samples. When examined individually, the IgG from 9 of 10 baboon sera recognized more antigens on KO-2 swine PBMC than on KO-1cells (Figure 4B). Disruption of swine β4GalNT2 in the KO-3 cells reduced binding of IgG from all 10 baboons compared to KO-2 cells (Figure 4C). These patterns of IgG reactivity, increased binding in the absence of CMAH activity and diminished binding as a result of β4GalNT2 inactivation, mirrored the results with baboon IgM.
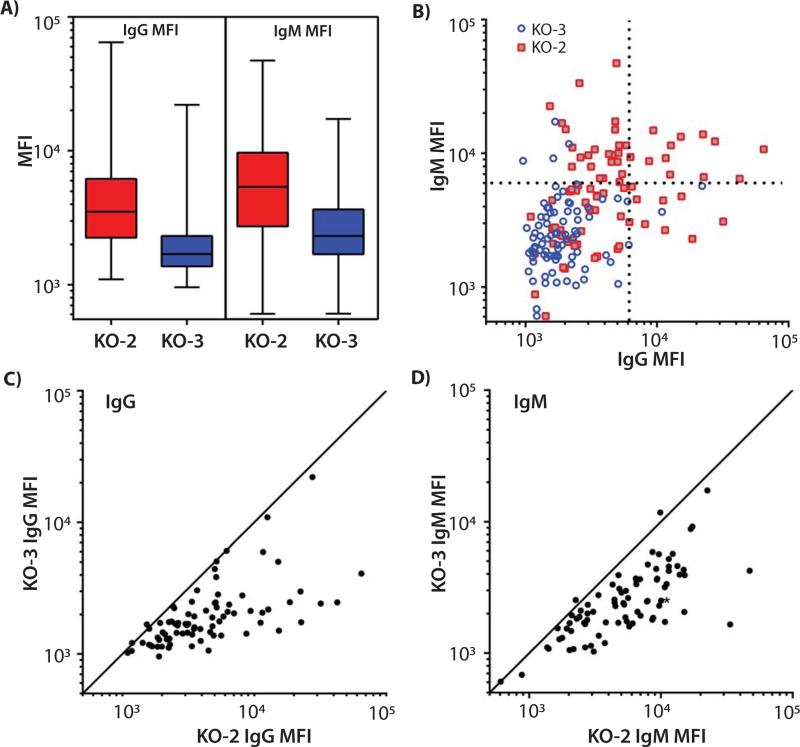
Figure 4. Plots of NHP antibody binding to single, double, and triple KO PBMCs.
NHP sera (34 rhesus macaques and 10 baboons) were incubated with PBMC from swine containing inactivated GGTA1 (KO-1), GGTA1/CMAH (KO-2), or GGTA1/CMAH/β4GALNT2 (KO-3). Secondary fluorescent antibodies were used to detect IgM and IgG binding to the cells with median fluorescence intensity being evaluated (MFI). A) Summary data are shown in box and whisker plots comparing binding of antibodies to swine PBMC of each genotype. Oneway ANOVA, and Tukey post hoc analyses were used to analyze the differences between each group. All comparisons of rhesus IgM and IgG binding yielded p values less than 0.006. GGTA1 and CMAH deficient pig cells bound the most baboon IgM (KO-1 vs. KO-2, p=0.041, and KO-2 vs. KO-3, p=0.010). Comparisons of baboon IgM binding to KO-1 versus KO-3 cells (NS, p=0.626), and baboon IgG binding comparisons did not achieve statistical significance (IgG not shown). B) MFI from baboon IgG binding to KO-2 and KO-1 PBMC were plotted individually. The diagonal line indicates equivalent binding to both KO-1 and KO-2 cells. Dots falling below the line represent samples where KO-1 cells bound fewer antibodies than KO-2 PBMC. C) Baboon IgG binding to KO-2 relative to KO-3 where data points below the line represent samples where KO-3 cells bound fewer antibodies than KO-2 PBMC.
In an effort to overcome the issues of small sample size and to extend the observations to additional NHP, we obtained serum from 34 rhesus macaques and repeated the immunoglobulin binding studies. Swine cells with disrupted GGTA1 and CMAH bound more rhesus IgM and IgG than did cells lacking GGTA1 only (Figure 4A; IgM and IgG KO-1 vs. KO-2, p<0.001). The elimination of swine β4GalNT2 activity concomitantly with CMAH and GGTA1 in KO-3 reduces antibody binding below both KO-2 and KO-1 cells (Figure 4A; IgM and IgG KO-2 vs. KO-3, and KO-1 vs. KO-3 p<0.006).
We previously showed that the majority of human donors had reduced antibody binding to PBMCs from swine deficient in both GGTA1 and CMAH gene function compared to cells devoid of GGTA1 alone [16]. Disrupting GGTA1/CMAH/β4GALNT2 further reduces swine xenoantigenicity as shown by grouped comparisons of human IgM and IgG binding (Figure 5A; KO-2 vs. KO-3, p<0.001). While three-gene disruption simultaneously minimized IgM and IgG binding in over 90% of humans (Figure 5B, lower left quadrant), a small fraction of samples retained either IgM or IgG xenoreactive antibodies (Figure 5B, upper left and lower right quadrants respectively). In contrast to results obtained for KO-2 analyses, no subject demonstrated simultaneously elevated binding levels of both IgM and IgG to cells taken from KO-3 pigs (Figure 5B, upper right quadrant). Comparing reactivity of individual sera with KO-2 and KO-3 cells (Figure 5C and 5D) confirmed that most humans have immunoglobulin specific for the products of β4GalNT2 in the background of a GGTA1 and CMAH deficient cell.
Figure 5. Plot of human IgG and IgM antibody binding to PBMCs from GGTA1/CMAH KO and GGTA1/CMAH/β4GALNT2 KO pigs.
Human sera (n=82) were incubated with PBMC from swine having inactivated GGTA1/CMAH (KO-2), or GGTA1/CMAH/β4GALNT2 (KO-3). Secondary fluorescent antibodies were used to detect IgM and IgG binding and fluorescence reported as median fluorescence intensity (MFI). A) Grouped analysis of human IgM and IgG binding to KO-2 and KO-3 swine PBMC. One-way ANOVA and Tukey post hoc analyses indicate significantly different antibody binding in ever comparison (p<0.01). B) Each data point represents a matched IgG and IgM MFI from individual human sera samples to either KO-2 or KO-3 PBMC. Dotted lines were placed at 6,000 MFI to highlight transitions from minimal to elevated antibody binding as determined by visual inspection of the data. More than 90% of samples are in left lower quadrant for KO-3. C) Individual data is plotted to allow simultaneous comparison of human IgG binding to KO-2 and KO-3 cells. The diagonal line represents equivalent binding to both sources of PBMC. Values falling below the line indicate less antibody binding to the KO-3 PBMCs relative to KO-2 cells. D) Human IgM binding to KO-2 and KO-3 was analyzed exactly as described in panel C. Dots falling below the line represent samples where KO-3 cells bound less IgM than KO-2 PBMC.
Discussion
Solid organ transplantation has made tremendous improvements in the past 50 years, but its success is now threatened by the shortage of suitable donor organs [1]. Xenotransplantation could eliminate the shortage of donor organs, but has never made it to the clinic because of the xenoreactive antibody barrier. In 1970, Calne coined the terms concordant and discordant to describe xenografts that were rejected more or less vigorously with discordant species combinations having higher antibody titers than concordant ones [24]. In 1989 Calne suggested that the first step toward clinical xenotransplantation using pig organs would be to convert the rejection from a discordant to a concordant form [25]. We have shown that, with respect to human antibody binding, PBMC and erythrocytes from GGTA1/CMAH deficient swine express fewer xenoantigens than chimpanzee cells suggesting the antigenic burden on pig tissues could be reduced to concordant levels [15, 16]. These results were encouraging but needed improvement with regard to further xenoantigen reduction.
Byrne and McGregor showed porcine β4GALNT2 encoded a xenoantigen using a GGTA1 deficient and CD46 transgenic pig expression library and screening for baboon antibody binding using serum from animals that had rejected porcine hearts [18, 19]. In humans and mice, β4GALNT2 catalyzes the addition of N-acetylgalactosamine to a sialic acid modified lactosamine to produce GalNAc β1-4(Neu5Ac α2-3) Gal β1-4GlcNAc β1-3Gal, the Sda blood group antigen. This gene is functional in transplantable organs (kidney, heart, liver, lung, and pancreas) and endothelial cells in the pig [19].
Mammalian immune systems typically develop humoral non-responsiveness towards molecules expressed by the host. Consequently, the lack GGTA1 and CMAH gene function in all humans prohibits their immune system from becoming tolerant to the αGal and Neu5Gc carbohydrates which likely explains their importance as xenoantigens [16]. Approximately 5% of humans possess inactive β4GaltNT2 and consequently develop antibodies against the SDa and CAD carbohydrates produced by this gene [25]. Therefore we hypothesized that a small percentage of humans would contain xeno-reactive antibodies specific for glycans produced by β4GaltNT2 in pig cells. Contrary to our hypothesis, the majority of humans demonstrated humoral immunity towards pig cells expressing the β4GalNT2 gene (Figure 5). Further studies comparing the activities of swine and human β4GalNT2 may reveal why the pig enzyme creates antigens that broadly react with human immunoglobulin.
The pig-to-nonhuman primate model has been important for the development of xenotransplantation. β4GALNT2 was identified as a candidate for genome editing using this model. Old-world NHPs have an intact CMAH gene and thus express Neu5Gc [26]. Our results show that PBMCs from GGTA1/CMAH KO pigs bind more baboon and rhesus antibodies than PBMCs from the GGTA1 KO pigs (Figure 4). It is clear that the impact of the CMAH deletion cannot be evaluated in the pig-to-NHP model [4]. Our results show that deletion of β4GALNT2 reduces baboon and rhesus antibody binding to the GGTA1/CMAH KO PBMCs. Since deletion of CMAH on the GGTA1 background increases NHP antibody binding, it is possible that a GGTA1/ β4GALNT2 KO pig will be more advantageous for testing in the pig-to-NHP xenotransplant model. Unlike what was seen in NHP, the sequential deletion of all three genes progressively reduced antigenicity of pig cells with regard to human IgM and IgG [16 and Figure 5]. The lack of antibody binding to PBMCs from the GGTA1/CMAH/β4GALNT2 KO pig suggests that organs from this animal will not face significant AMR when transplanted into humans.
Acknowledgements
This study was supported by the IU Health Transplant Institute and Indiana University Health. This study used facilities constructed with support from the National Center for Research Resources via a Research Facilities Improvement Grant C06RR10601-01. We thank the Methodist Research Institute and Laboratory Animal Research Center Staff for providing care to the animals.
Footnotes
Author Contributions:
The manuscript has been revised and approved by all authors. JLE, PL performed gene manipulations, pig cloning, and genotyping. RAS, GM, and JB performed phenotyping, and antibody binding studies. All authors participated in experimental design. ABA, KAN, MLF participated in concept and design of NHP studies. JLE, GM, PL, RAS MT, AJT drafted the article. AJT secured funding and developed study concept.
Disclosures:
A.J.T. has created Xenobridge, LLC, and has applied for patents related to this work. R.A.S. receives consulting fees relevant to xenotransplantation. The other authors report no conflicts.
References
- 1.Israni AK, Salkowski N, Gustafson S, et al. New national allocation policy for deceased donor kidneys in the United States and possible effect on patient outcomes. J Am Soc Nephrol. 2014;25(8):1842–1848. doi: 10.1681/ASN.2013070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen G, Qian H, Starzl T, et al. Acute rejection is associated with antibodies to non-Gal antigens in baboons using Gal-knockout pig kidneys. Nat Med. 2005;11(12):1295–1298. doi: 10.1038/nm1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galili U. Discovery of the natural anti-Gal antibody and its past and future relevance to medicine. Xenotransplantation. 2013;20(3):138–147. doi: 10.1111/xen.12034. [DOI] [PubMed] [Google Scholar]
- 4.Salama A, Evanno G, Harb J, Soulillou J. Potential deleterious role of anti-Neu5Gc antibodies in xenotransplantation. Xenotransplantation. 2014 doi: 10.1111/xen.12142. DOI: 10.1111/xen.12142. [DOI] [PubMed] [Google Scholar]
- 5.Byrne GW, Stalboerger PG, Davila E, et al. Proteomic identification of non-Gal antibody targets after pig-to-primate cardiac xenotransplantation. Xenotransplantation. 2008;15(4):268–276. doi: 10.1111/j.1399-3089.2008.00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li P, Estrada JL, Burlak C, Tector AJ. Biallelic knockout of the alpha-1,3 galactosyltransferase gene in porcine liver-derived cells using zinc finger nucleases. J Surg Res. 2013;181(1):e39–45. doi: 10.1016/j.jss.2012.06.035. [DOI] [PubMed] [Google Scholar]
- 7.Lutz AJ, Li P, Estrada JL, et al. Double knockout pigs deficient in N-glycolylneuraminic acid and galactose alpha-1,3-galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation. 2014;20(1):27–35. doi: 10.1111/xen.12019. [DOI] [PubMed] [Google Scholar]
- 8.Tan W, Carlson DF, Lancto CA, et al. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci U S A. 2014;110(41):16526–16531. doi: 10.1073/pnas.1310478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li P, Estrada JL, Burlak C, et al. Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single-guide RNA and carbohydrate selection. Xenotransplantation. 2014 doi: 10.1111/xen.12131. DOI: 10.1111/xen.12131. [DOI] [PubMed] [Google Scholar]
- 10.Reyes LM, Estrada JL, Wang ZY, et al. Creating class I MHC-null pigs using guide RNA and the Cas9 endonuclease. J Immunol. 2014;193(11):5751–5757. doi: 10.4049/jimmunol.1402059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato M, Miyoshi K, Nagao Y, et al. The combinational use of CRISPR/Cas9-based gene editing and targeted toxin technology enables efficient biallelic knockout of the alpha-1,3-galactosyltransferase gene in porcine embryonic fibroblasts. Xenotransplantation. 2014;21(3):291–300. doi: 10.1111/xen.12089. [DOI] [PubMed] [Google Scholar]
- 12.Whitworth KM, Lee K, Benne JA, et al. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014;91(3):78, 1–13. doi: 10.1095/biolreprod.114.121723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Xin J, Fan N, et al. Generation of CRISPR/tCas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci. 2014 doi: 10.1007/s00018-014-1744-7. DOI: 10.1007/s00018-014-1744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tector AJ, Ford ML. Literature Watch Implications for transplantation. Am J Transplant. 2015;15(1):3. [Google Scholar]
- 15.Wang ZY, Burlak C, Estrada JL, et al. Erythrocytes from GGTA1/CMAH knockout pigs: implications for xenotransfusion and testing in non-human primates. Xenotransplantation. 2014;21(4):376–384. doi: 10.1111/xen.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burlak C, Paris LL, Lutz AJ, et al. Reduced binding of human antibodies to cells from GGTA1/CMAH KO pigs. Am J Transplant. 2014;14(8):1895–1900. doi: 10.1111/ajt.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reemtsma K, McCracken BH, Schlegel JU, et al. Renal Heterotransplantation in Man. Ann Surg. 1964;160:384–410. doi: 10.1097/00000658-196409000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrne GW, Stalboerger PG, Du Z, Davis TR, McGregor CGA. Identification of new carbohydrate and membrane protein antigens in cardiac xenotransplantation. Transplantation. 2011;91(3):287–292. doi: 10.1097/TP.0b013e318203c27d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrne GW, Du Z, Stalboerger PG, Kogelberg H, McGregor CGA. Cloning and expression of porcine beta1,4 N-acetylgalactosaminyl transferase encoding a new xenoreactive antigen. Xenotransplantation. 2014;21(6):543–554. doi: 10.1111/xen.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waghmare SK, Estrada J, Reyes L, et al. Gene targeting and cloning in pigs using fetal liver derived cells. Journal of Surgical Research. 171(2)(2011):e223–e229. doi: 10.1016/j.jss.2011.07.051. [DOI] [PubMed] [Google Scholar]
- 22.Estrada J, Sommer J, Collins B, Mir B, et al. Swine generated by somatic cell nuclear transfer have increased incidence of intrauterine growth restriction (IUGR). Cloning and stem cells. 2007;9(2):229–236. doi: 10.1089/clo.2006.0079. [DOI] [PubMed] [Google Scholar]
- 23.Bardor M, Nguyen DH, Diaz S, Varki A. Mechanism of uptake and incorporation of the non-human sialic acid N-glycolylneuraminic acid into human cells. Journal of Biological Chemistry. 2005;280(6):4228–4237. doi: 10.1074/jbc.M412040200. [DOI] [PubMed] [Google Scholar]
- 24.Calne RY. Organ transplantation between widely disparate species. Transplant Proc. 1970;2(4):550–556. [PubMed] [Google Scholar]
- 25.Hardy MA. Xenograft 25 : proceedings of the International Congress, Xenograft 25, held at Arden House, Harriman, New York, 11-13 November 1988. Excerpta Medica ;Sole distributors for the USA and Canada, Elsevier Science Pub. Co.; Amsterdam ; New YorkNew York, NY, USA: [Google Scholar]
- 26.Padler-Karavani V, Varki A. Xenotransplantation. 2011;Potential impact of the non-human sialic acid N-glycolylneuraminic acid on transplant rejection risk.18(1):1–5. doi: 10.1111/j.1399-3089.2011.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]