Summary
Most chronic and recurrent bacterial infections involve a biofilm component, the foundation of which is the extracellular polymeric substance (EPS). Extracellular DNA (eDNA) is a conserved and key component of the EPS of pathogenic biofilms. The DNABII protein family includes integration host factor (IHF) and Histone-like protein (HU); both are present in the extracellular milieu. We have shown previously that the DNABII proteins are often found in association with eDNA and are critical for the structural integrity of bacterial communities that utilize eDNA as a matrix component. Here, we demonstrated that Uropathogenic E. coli (UPEC) strain UTI89 incorporates eDNA within its biofilm matrix and that the DNABII proteins are not only important for biofilm growth, but are limiting; exogenous addition of these proteins promotes biofilm formation that is dependent on eDNA. In addition, we show that both subunits of IHF, yet only one subunit of HU (HupB), are critical for UPEC biofilm development. We discuss the roles of these proteins in context of the UPEC EPS.
Keywords: EPS, Biofilm, UPEC, UTI89, IHF and HU
Introduction
Urinary tract infections (UTIs) are the most common bacterial infection of the developed world, with nearly 50% of all women and approximately 12% of men experiencing at least one urinary tract infection (UTI) during their lifetime (Foxman and Brown, 2003). Most importantly, UTIs are recurrent infections, with at least 25% of women enduring a second UTI within 6–12 months (Foxman et al., 2000). Also, the use of indwelling urinary catheters in hospitals leads to a high incidence of catheter-associated urinary tract infections (CAUTI) that cause significant morbidity and mortality (Jaggi and Sissodia, 2012). According to the most recent national hospital discharge survey in 2007, UTIs account for approximately one half million emergency room visits annually in the US alone (Hall et al., 2010) and this leads to a substantial financial and health burden.
UTIs can be caused by a number of bacteria including E. coli, Klebsiella, Proteus, Enterococcus and Staphylococcus spp (Zhang and Foxman, 2003). However a diverse group of E. coli strains [collectively termed Uropathogenic E. coli (UPEC)] are the causative agents of more than 80% of UTIs (Zhang and Foxman, 2003). Despite the flow of urine in the urinary tract, UPEC adhere to and invade the urinary tract epithelium, wherein they replicate rapidly to form intracellular bacterial communities (IBCs) (Mulvey et al., 1998; Anderson et al., 2003). IBCs are a biofilm-like state that is observed during the course of UPEC infection in animal models and human infections (Mulvey et al., 1998; Anderson et al., 2003; Trautner and Darouiche, 2004). Also, biofilm formation on urinary catheters plays a primary role in the pathogenesis of CAUTI and creates a reservoir for chronic UTIs (Saint and Chenoweth, 2003; Trautner and Darouiche, 2004). Antibiotics are the first-line of treatment for UTIs (Kodner and Thomas Gupton, 2010). According to the Antimicrobial Resistance Epidemiological Survey on cystitis, approximately 48% of E. coli isolates that cause cystitis are resistant to ampicillin, 30% are resistant to trimethoprim/sulfamethoxazole, 19% are resistant to nalidixic acid and more than 10% are resistant to at least three different classes of antimicrobial agents (Schito et al., 2009). Also the high incidence of recurrence of UTIs results in repeated antibiotic treatment that negatively impacts the commensal microbiota and, in turn, can result in secondary infections in the gastrointestinal tract and vagina (MacDonald et al., 1993; Dethlefsen et al., 2008). Therefore, there is a critical need for the development of novel ways to undermine UPEC pathogenesis mediated by biofilms.
According to the Centre of Disease Control and Prevention, two-thirds of bacterial infectious processes involve biofilms (Davies, 2003). Growth in a biofilm lifestyle results in a highly organized, sessile but metabolically active, multicellular microbial community enclosed in Extracellular Polymeric Substance (EPS) that is attached to an abiotic or biotic surface. The composition of biofilm EPS is dictated by the microbe and its surrounding environment and is primarily comprised of water, exopolysaccharides, extracellular proteins and extracellular DNA (eDNA) (Flemming and Wingender, 2010). The EPS plays several functional roles including aggregation of bacterial cells, adhesion to biotic and/or abiotic surfaces, cohesion of biofilms, nutrient source, retention of water and providing a protective barrier (Flemming and Wingender, 2010). By acting as a protective barrier, the EPS renders the biofilm recalcitrant to removal by antimicrobial agents and by effectors of both the innate and adaptive immune systems (Slinger et al., 2006; Starner et al., 2008). Since disruption of the biofilm matrix and the release of bacteria resident within the biofilm is sufficient to increase susceptibility to the action of both the immune system and antimicrobials (Flemming and Wingender, 2010; Goodman et al., 2011; Cavaliere et al., 2014), it is essential to understand and elucidate the molecular interactions that contribute to the stability of the biofilm in order to define the key components of the matrix that promote resistance.
Extracellular DNA (eDNA) is common to most bacterial biofilms and serves as a structural component of these biofilms (Steinberger and Holden, 2005; Qin et al., 2007; Izano et al., 2008; Guiton et al., 2009; Flemming and Wingender, 2010; Nur et al., 2013). We have shown, that in biofilms formed in vivo by non-typeable Haemophilus influenzae (NTHI), eDNA is organized in an interwoven mesh-like structure (Jurcisek and Bakaletz, 2007). A similar organization of eDNA is also apparent in sputum samples recovered from cystic fibrosis (CF) patients that were culture positive for Burkholderia cenocepacia (Novotny et al., 2013), Pseudomonas aeruginosa, and Staphylococci (Gustave et al., 2013). This organization of eDNA in the biofilm is similar in apparent architecture to the preferred substrates of the DNABII family of proteins (Kamashev and Rouviere-Yaniv, 2000).
The DNABII proteins are a family of proteins that are ubiquitously expressed by all eubacteria and play an integral role in shaping the intracellular bacterial nucleoid structure and function (Browning et al., 2010). DNABII proteins include IHF (integration host factor) and HU (histone-like protein), which bind to DNA as dimers and condense DNA upon binding (Rice et al., 1996). The DNABII proteins have a high affinity (nM, Kd) to pre-bent secondary structures of DNA (Kamashev and Rouviere-Yaniv, 2000). Although both IHF and HU bind to and bend DNA, IHF exhibits sequence specificity (WATCAANNNNTTR where W is A or T and R is a purine) while HU binds to DNA in a sequence independent manner (Rice et al., 1996). The DNABII family of proteins is also found in great abundance in the extra-bacterial milieu of Gram-positive bacteria (Winters et al., 1993; Lunsford et al., 1996; Gao, 2000; Boleij et al., 2009; Nur et al., 2013). We have previously shown that not only are the DNABII proteins extra-bacterial, but that they also bind to and stabilize the interwoven mesh-like structure of eDNA in biofilms formed in vivo by NTHI (Goodman et al., 2011), Pseudomonas aeruginosa (Gustave et al., 2013), Staphylococci (Gustave et al., 2013) and Burkholderia cenocepacia (Novotny et al., 2013). Nur et al., have also demonstrated that in S. intermedius, Histone-like protein (HLP) is found co-localized with eDNA in the extra-bacterial milieu and suggested that this eDNA-HLP complex may play a role in biofilm formation (Nur et al., 2013).
In UPEC strain UTI89, IHF is also found in the matrix of IBCs formed in vivo in a female mouse bladder during experimental UTI (Justice et al., 2012). Further, we have shown that in UPEC strain UTI89, both IHFA and IHFB are required for both colonization of the mouse bladder and type I piliation (Justice et al., 2012). We have also demonstrated that antiserum directed against E. coli IHF destabilizes the biofilm architecture and results in disruption of biofilms formed by multiple human pathogens in vitro (Goodman et al., 2011). Also, immunological targeting of extracellular IHF results in clearance of NTHI biofilms from the middle ear in a chinchilla model of otitis media (Goodman et al., 2011), and dispersal of polymicrobial sputum aggregates recovered from cystic fibrosis patients (Gustave et al., 2013; Novotny et al., 2013). Collectively, these observations suggest that sequestration of DNABII proteins from the biofilm matrix facilitates biofilm disruption in vitro (Goodman et al., 2011; Brandstetter et al., 2013; Brockson et al., 2014) and in vivo (Goodman et al., 2011; Gustave et al., 2013; Novotny et al., 2013) with subsequent immune mediated clearance that serves to eradicate pre-formed biofilms in vivo (Goodman et al., 2011). While our studies to date have demonstrated that DNABII proteins are important in maintaining the structural integrity of bacterial biofilms, how the individual components of the biofilm matrix contribute to overall biofilm architecture has remained unclear.
To determine the contribution of DNABII proteins to the biofilm architecture, we supplemented UPEC strain UTI89 biofilms via the exogenous addition of recombinant DNABII proteins and also analyzed the ability of DNABII deficient mutants to form biofilms in vitro. We found that exogenous addition of DNABII proteins dramatically increased the size of biofilms formed in vitro by UPEC strain UTI89. We also found that while both subunits of IHF were critical for UPEC biofilm formation, only one subunit of HU (HupB) was required for in vitro biofilm formation. These data suggested that DNABII proteins were limiting and reinforced our hypothesis that they play a critical role in maintaining the structural integrity of UPEC biofilms.
Results
DNABII proteins are limiting in UTI89 biofilms
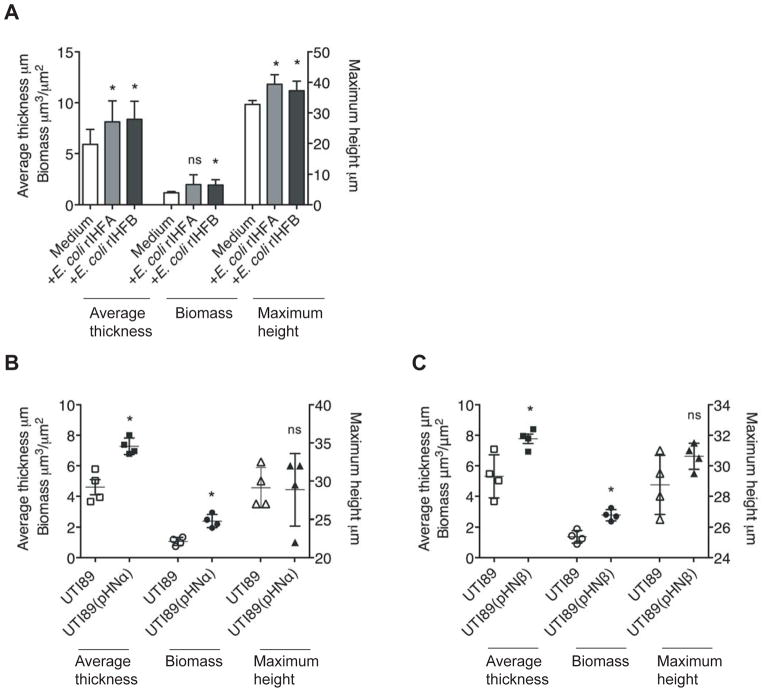
To determine the contribution of IHF to biofilm stability, we grew biofilms in vitro with recombinant E coli IHF (rIHF; to 0.5 μM) added exogenously at the time of seeding and maintained throughout the 40 hours of biofilm growth. In preliminary experiments, E. coli rIHF was titrated from 5 nM to 1.0 μM and we found that 0.5 μM E. coli rIHF was optimal for in vitro biofilm analysis. When compared to the biofilm formed in the absence of exogenous E. coli rIHF (Figure 1A), there was a dramatic increase in the size of the biofilm formed when E. coli rIHF was added exogenously (Figure 1B). Through COMSTAT analysis of images generated in multiple replicate assays, the measured parameters of biofilm maximum height, average thickness and biomass increased by 53%, 88% and 61%, respectively upon addition of E. coli rIHF (Figure 1C, D). Similar increases in biofilm architectural parameters could also be achieved by the addition of 0.5 μM recombinant E. coli HU (rHU) as well as recombinant HU from the Gram-positive microbe Streptococcus mutans (UA159), another member of the DNABII protein family, to UTI89 biofilms at seeding (Figure S1). Importantly, the exogenous addition of DNABII proteins (E. coli rIHF or E. coli rHU; to 0.5 μM) to three additional clinical isolates of UPEC at seeding resulted in an increase in the size of the biofilm formed as evidenced by a significant increase in the measured parameters of maximum biofilm height, average thickness, and biomass (Figure S2). Collectively, our results suggested that DNABII proteins are generally limiting in biofilm formation by UPEC and further, that other matrix components are likely present at sufficient levels to support additional biofilm growth in the presence of an excess of extra-bacterial recombinant DNABII proteins.
Figure 1. Effect of exogenous addition of E. coli rIHF on UPEC strain UTI89 biofilm formation.
(A, B) Biofilm growth was initiated then maintained in the absence (A) or presence (B) of exogenous E.coli rIHF (0.5 μM) for 40 hours. (C, D) Images were analyzed by COMSTAT to calculate maximum biofilm height, average thickness and biomass. Asterisks indicate statistical significance (p < 0.05) compared to control. All images were captured with a 63X objective.
Extracellular DNA (eDNA) is critical for DNABII protein mediated biofilm formation by UPEC strain UTI89
To our knowledge the presence of eDNA in the matrix of biofilms formed by UPEC strain UTI89 has not been documented. To determine whether UTI89 incorporated eDNA within its biofilm matrix, we labeled unfixed in vitro preformed UTI89 biofilms with a monoclonal antibody against double stranded DNA (dsDNA) (red). The biofilm biomass was labeled with FilmTracer™ FM® 1–43 (green). The in vitro biofilm formed by UPEC strain UTI89 incorporated dsDNA that was uniformly distributed throughout the biofilm matrix as evident from the distribution of red fluorescence observed (Figure 2A). The addition of 1 unit of DNase to the biofilms at seeding dramatically decreased the observed red fluorescence (Figure 2B) and served as a control. Further, in the presence of DNase, the relative abundance of dsDNA as determined by the ratio of the dsDNA (anti-dsDNA labeling) to the bacteria (FilmTracer™) revealed a statistically significant decrease in the amount of eDNA incorporated within the biofilm matrix compared to the control (Figure S3). To determine the contribution of eDNA to biofilm formation, we grew biofilms in vitro with DNase added exogenously at the time of seeding and maintained throughout the 40 hours of biofilm growth. When compared to the control, there was a dramatic decrease in the size of the biofilm formed in the presence of DNase as evidenced by the significant decrease in maximum height (Figure 2C), average thickness (Figure 2D), and biomass (Figure 2D). Biofilm formation in the presence of both DNase and E. coli rIHF was significantly decreased compared to the control but was similar to that of biofilms formed in the presence of DNase (Figure 2C, D) suggesting that eDNA is required for DNABII protein-mediated biofilm formation. Moreover, biofilm formation in the presence of calf thymus DNA was not significantly different compared to the control and also the biofilm formation in the presence of both calf thymus DNA and E. coli rIHF was not significantly different compared to the biofilms formed in the presence of just E. coli rIHF (data not shown) suggesting that eDNA is not limiting in biofilm formation in UPEC strain UTI89. Collectively, these data demonstrated that UTI89 biofilms formed in vitro incorporated dsDNA within the biofilm matrix, and that eDNA is not limiting but is nonetheless critical for DNABII protein-mediated biofilm formation.
Figure 2. Distribution of DNA in a UTI89 biofilm and the effect of addition of DNase on biofilms formed by UPEC strain UTI89.
(A, B) Biofilm growth was initiated then maintained in the absence (A) or presence (B) of DNase (1 unit) for 40 hours. Unfixed 40 hour biofilm was labeled with anti-dsDNA monoclonal antibody (DNA - red) and FilmTracer™ FM® 1–43 (biomass – green). All images were captured with a 63X objective. (C, D) Biofilm growth was initiated then maintained in the absence (control) or presence of DNase (+DNase) or DNase and exogenous E.coli rIHF (+E. coli rIHF + DNase) for 40 hours. Biofilms were stained with LIVE/DEAD® stain and the images were analyzed by COMSTAT to calculate maximum biofilm height (C), average thickness (D), and biomass (D). A p ≤ 0.05 was represented by * and a p ≤ 0.01 was represented by **.
Overproduction of E. coli rIHF increases biofilm formation by UTI89
The above results demonstrated that exogenous addition of E. coli rIHF (or other DNABII family members) can increase biofilm formation and that IHF is limiting in UTI89 biofilms. To confirm this result, we repeated the in vitro biofilm formation assay using a merodiploid UPEC strain that overproduced E. coli IHF via generation of a mutant wherein one copy of the IHF genes was placed under control of an inducible promoter. When compared to the biofilm formed without induction of E. coli rIHF (‘−IPTG’ lanes in Figure 3A, B), biofilms formed under conditions of overproduction of rIHF (‘+IPTG’ lanes), showed a significant increase in maximum height, average thickness, and biomass. Since overproduction of E. coli rIHF in the intra-bacterial milieu resulted in increased biofilm formation, we reasoned that there was an increase in the accumulation of IHF in the extracellular milieu that mediated the observed biofilm increase. To directly test this, we purified proteins from spent medium of either wild type UPEC strain UTI89 or UPEC that was induced to overproduce E. coli rIHF when grown statically (for 6h or 16h, respectively) to compare the extra-bacterial steady state levels of IHF. We could not detect IHF in the spent medium of the wild type (wt) strain at either of the time points tested. However there was an accumulation of HU at 16h (Figure 3C). This outcome suggested that IHF was either limiting or at the detection limit of the assay under the growth conditions employed. However, IHF and relatively more HU accumulated in the spent medium in the strain that overproduced E coli rIHF in comparison to the wild type in each of the conditions tested. These results suggested that there is a coordination of the extra-bacterial and intra-bacterial concentrations of IHF. Overproduction of rIHF also resulted in a significant increase in the abundance of eDNA within in vitro preformed biofilms (Figure S3). Taken together with data shown in Figure 1, these findings are supportive of our hypothesis that IHF is part of the extracellular matrix that contributes to the structural integrity of the UPEC strain UTI89 biofilm.
Figure 3. Effect of overproduction of IHF on UPEC strain UTI89 biofilm formation.
(A, B) Ectopic expression of E. coli rIHF was accomplished by IPTG induction of plasmid pHNαβ. Biofilm growth was initiated in the presence or absence of IPTG for 40 hours. Images were analyzed by COMSTAT to calculate biofilm maximum height (A), average thickness and biomass (B). Left Y-axis in panel B corresponds to average thickness and right Y-axis corresponds to biomass. (C) Western blot analysis of the spent medium from wild type or wild type overproducing E. coli rIHF [wt(pHNαβ)] using rabbit anti-IHF antibody directed against E. coli rIHF. Anti-IHF antibody specifically recognizes the two DNABII proteins IHF and HU. Asterisks indicate statistical significance (p < 0.05) compared to −IPTG lane.
Both subunits of IHF contribute to in vitro biofilm formation in UTI89
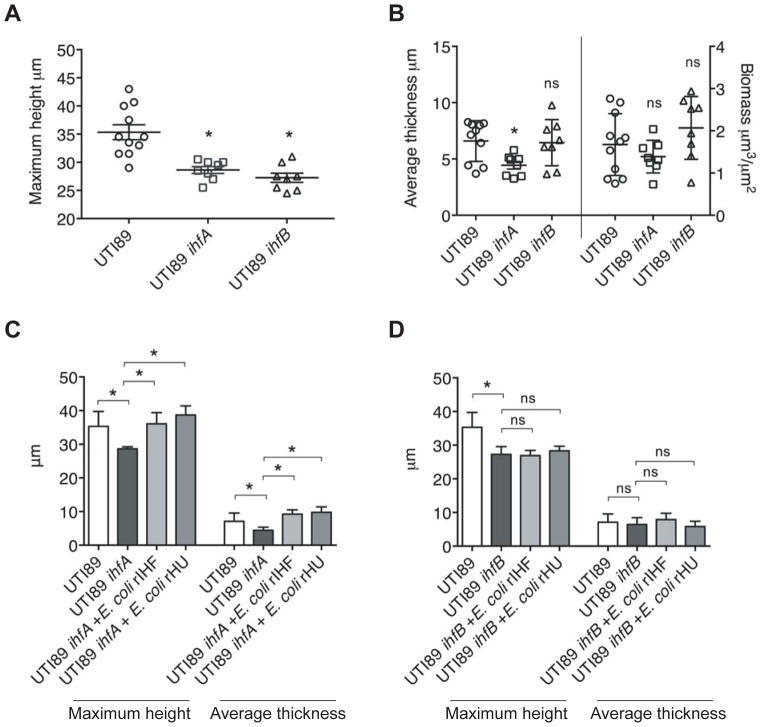
To determine the contribution of each of the subunits of IHF to biofilm formation, we employed the wild type (WT) UPEC strain UTI89 or isogenic IHFA deficient strain UTI89 ihfA11::Tn10 (Table S1) or isogenic IHFB deficient strain UTI89 ΔihfB (Table S1) in our in vitro biofilm formation assay. As shown in Figure 4A, B, whereas the WT strain produced a robust biofilm in vitro, both UTI89 ihfA11::Tn10 and UTI89 ΔihfB were less robust as judged by the significant defect in maximum height. UTI89 biofilm deficient in IHFA had a significant defect in average thickness, while the average thickness of UTI89 ΔihfB biofilm was comparable to the WT strain (Figure 4B). The biofilm biomass of UTI89 strains deficient in IHFA and IHFB was comparable to the WT (Figure 4B). These results demonstrated that IHFA and IHFB contribute to in vitro biofilm formation in UPEC strain UTI89 and also suggested that the structure of the biofilm is differentially affected as evident from average thickness (Figure 4B) in the absence of either subunit of IHF. This result is in agreement with previous findings (Justice et al., 2012) that there are significant differences in the overall architecture of the biofilm formed by UTI89 ihfA11::Tn10 and UTI89 ΔihfB in vivo in a female mouse bladder. Notably, in the presence of exogenous recombinant E. coli DNABII proteins, the maximum height and average thickness of the biofilms formed by UTI89 ihfA11::Tn10 increased significantly (Figure 4C) and were comparable to that of the WT. However, as shown in Figure 4D, there was no significant increase in the maximum height or average thickness of UTI89 ΔihfB biofilms in the presence of any of the exogenously added recombinant DNABII proteins. These data suggested that IHFA played a major role in the extracellular milieu while IHFB may have additional intracellular functions that cannot be complemented with exogenously added recombinant protein. To determine the contribution of each of the subunits of IHF to the incorporation of eDNA within the biofilm matrix, we determined the distribution and abundance of eDNA in in vitro preformed biofilms of UTI89 ihfA11::Tn10 and UTI89 ΔihfB. We observed no significant difference in the distribution of eDNA by visual inspection (data not shown), and abundance of eDNA compared to the control (Figure S3) within the limits of this assay as presented. These data suggested that the observed defect in biofilm formation by UTI89 strains deficient in IHFA or IHFB could be attributed solely to the absence of each respective subunit of IHF from the extracellular matrix.
Figure 4. Relative ability of UPEC strain UT189 IHF mutants to form biofilms in vitro.
UTI89, UTI89 ihfA11::Tn10 or UTI89 ΔihfB biofilms were grown for 40 hours in vitro in a chamber slide. Biofilms were stained with Live/Dead stain and visualized by a confocal microscope to acquire 3-dimensional datasets. Images were analyzed by COMSTAT to calculate biofilm height (A) and average thickness and biomass (B). (C) Biofilm growth of UTI89 ihfA11:Tn10 was initiated in the presence or absence of exogenous E. coli rIHF (0.5 μM) for 40 hours and the images were analyzed by COMSTAT to measure maximum height and average thickness. (D) Biofilm growth of UTI89 ΔihfB was initiated in the presence or absence of exogenous E. coli rIHF (0.5 μM) for 40 hours and the images were analyzed by COMSTAT to measure maximum height and average thickness. A p-value ≤ 0.05 was considered significant.
Individual IHF subunits increase in vitro biofilm formation by UPEC strain UTI89
The above results demonstrated that both subunits of IHF were involved in biofilm formation. To more thoroughly examine the roles of the individual IHF subunits, we exogenously supplemented in vitro formed UTI89 biofilms with 1.0 μM recombinant E coli IHFA or recombinant E coli IHFB at the time of seeding and biofilms were grown for 40 hours. When compared to the biofilm formed in the absence of exogenous rIHFA or rIHFB, as shown in Figure 5A, there was a significant increase in biofilm formation in the presence of either rIHFA or rIHFB as judged by maximum height and average thickness. We also observed a significant increase in average thickness and biomass of the in vitro formed biofilms when either of these subunits was overproduced (Figure 5B, C). These data reinforced the idea that both subunits of IHF play an integral role in UTI89 biofilm formation and also suggested that individual IHF subunits retain the ability to bind to eDNA.
Figure 5. Effect of the addition of individual recombinant IHF subunits on UPEC strain UTI89 biofilm formation.
(A) Biofilm growth of UTI89 was initiated in the presence (+E. coli rIHFA or +rIHFB) or absence (medium) of exogenous E. coli rIHFA or rIHFB (~ 1.0 μM) for 40 hours. Images were analyzed by COMSTAT to calculate biofilm maximum height, average thickness and biomass. (B) Ectopic expression of E. coli rIHFA was accomplished by IPTG induction of plasmid pHNα. Biofilm growth was initiated in the presence or absence of IPTG for 40 hours. Images were analyzed by COMSTAT to calculate maximum biofilm height, average thickness and biomass. (C) Ectopic expression of E. coli rIHFB was accomplished by IPTG induction of plasmid pHNβ. Biofilm growth was initiated in the presence or absence of IPTG for 40 hours. Images were analyzed by COMSTAT to calculate biofilm maximum height, average thickness and biomass. Left Y-axis in panels A, B and C correspond to average thickness and biomass and right Y-axis corresponds to maximum height. A p-value ≤ 0.05 was considered significant.
Effect of disruption of HU expression on UTI89 biofilm formation
Since exogenous addition of recombinant HU from E. coli or S. mutans increased in vitro biofilm formation (Figure S1) and HU accumulated in the spent medium of UTI89 (Figure 3C), we were interested in the role of the two subunits of HU in biofilm formation. To this end, we employed the wild type (WT) UPEC strain UTI89 or the isogenic strain deficient in HupA (UTI89 hupA::cam) or the isogenic strain deficient in HupB (UTI89 hupB::kan) in our in vitro biofilm formation assay. As shown in Figure 6A, B the UTI89 strain deficient in HupA did not exhibit a defect in biofilm formation in comparison to the WT strain when grown under the same conditions. However, as shown in Figure 6C, the biofilms formed by the HupB deficient strain exhibited a significant defect in maximum height and average thickness in comparison to the WT strain. The biomass of the biofilm formed by UTI89 hupB::kan strain was comparable to that of the WT strain (Figure 6D). Remarkably, the height and average thickness of the biofilms formed by UTI89 hupA::cam and UTI89 hupB::kan increased significantly in the presence of exogenous DNABII proteins (Figure 6A, C). We have also demonstrated that the qualitative distribution (data not shown) and abundance of eDNA in the biofilms formed by UPEC strains deficient in HupA or HupB was similar to that of the control (Figure S3) within the limits of this assay as presented, suggesting that the observed defect in biofilm formation in UTI89 hupB::kan was solely due to the lack of HupB. Taken together, these results suggested that HupB was directly involved in UTI89 biofilm formation.
Figure 6. Effect of HU deficiency on biofilm formation by UPEC strain UTI89.
(A, B) Biofilm growth of UTI89 hupA::cam was initiated in the presence or absence of exogenous E. coli rIHF or E. coli rHU (0.5 μM) for 40 hours and the images were analyzed by COMSTAT to measure maximum height (A), average thickness (A) and biomass (B). (C, D) Biofilm growth of UTI89 hupB::kan was initiated in the presence or absence of exogenous E. coli rIHF or E. coli rHU (0.5 μM) for 40 hours and the images were analyzed by COMSTAT to measure maximum height (C), average thickness (C) and biomass (D). A p-value ≤ 0.05 was considered significant.
UPEC strain UTI89 partitions towards the biofilm state in the presence of E. coli rIHF
As COMSTAT analysis demonstrated that there was a significant increase in biofilm biomass upon incubation with E. coli rIHF (Figure 1B, D), and since the growth rate of UTI89 is not affected by exogenous addition of E. coli rIHF (data not shown), we reasoned that the presence of added rIHF was inducing partitioning of bacteria to the biofilm/adherent state from the planktonic state. To determine whether there was a relative increase in adherent (i.e. biofilm) bacteria upon incubation with E. coli rIHF, we enumerated the total number of bacteria present within the chamber slide, both free (planktonic, in the spent medium) and adherent (part of the biofilm). As shown in Figure 7, there was a significant increase in the adherent bacteria and a corresponding significant decrease in the planktonic bacteria in the presence of E. coli rIHF. These results agree well with the COMSTAT analysis (Figure 1D) and reinforce our hypothesis that in the presence of E. coli rIHF, more bacteria partition towards the biofilm state.
Figure 7. Partitioning of UPEC strain UTI89 in a biofilm in the presence of E. coli rIHF.
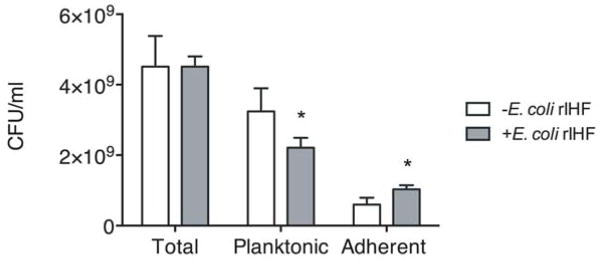
Biofilm formation was initiated and then maintained in the presence or absence of E. coli rIHF (0.5 μM) in a chamber slide for 40 hours. The contents of a well were re-suspended in the spent medium and plated on LB agar to calculate total CFU/ml. To calculate planktonic CFU/ml, the spent medium was carefully removed from the biofilm and the wells were washed twice with sterile PBS. The washes were combined with the spent medium, diluted and plated on LB agar. To calculate adherent CFU/ml, the biofilm (after two washes) was resuspended in sterile PBS, diluted and plated on LB agar. A p-value ≤ 0.05 was considered significant.
UPEC strain UTI89 biofilm formed in the presence of E. coli rIHF is functionally similar to that formed with no added rIHF
Since the addition of recombinant DNABII proteins resulted in a bigger biofilm as judged by maximum height, biomass and average thickness, it was imperative to determine if the structures formed in the presence of exogenously added recombinant DNABII proteins were similar to native biofilms in terms of several selected biological phenotypes. To address this, first we determined the Minimum Inhibitory Concentration (MIC100) of ciprofloxacin (CIP) for UT189 grown either planktonically or as a biofilm in either the presence or absence of exogenously added E. coli rIHF. As shown in Table 1, the MIC of CIP for UTI89 biofilm in the presence of rIHF was indistinguishable from that of the native UTI89 biofilm. Consistent with previous observations of increased resistance of bacterial biofilms to antimicrobial agents (Slinger et al., 2006; Kaji et al., 2008; Starner et al., 2008), the MIC of CIP was more that 2500 times greater than that of UT189 in the planktonic state. Second, we exposed the planktonic and biofilm state UTI89 that had been grown in the presence or absence of exogenous E. coli rIHF to hydrogen peroxide for 30 min and then enumerated the bacteria that survived the challenge. As shown in Figure 8A, while none of the UTI89 grown in the planktonic state (with or without added exogenous E. coli rIHF) survived exposure to 0.2% hydrogen peroxide, the biofilm state (irrespective of the presence or absence of exogenous rIHF) was resistant to incubation with 0.8% hydrogen peroxide as demonstrated by ~107 viable CFU/ml. Lastly, we measured the relative expression of Antigen 43 (Ag43), a UPEC protein that is specifically expressed in the biofilm state when cultured in minimal medium (Schembri et al., 2003; Biscola et al., 2011) by RTPCR analysis and Western blot. As shown in Figure 8B, and C, while there was no expression of Ag43 by UPEC when grown in the planktonic state, there was similar expression of this bacterial protein when grown in the biofilm state either with or without added exogenous E. coli rIHF. Collectively, these results demonstrated that the biofilm formed with exogenous E. coli rIHF retains several functional attributes of a native biofilm. Together, our data suggested that the presence of E. coli DNABII proteins during biofilm development was limiting and that additional protein promoted the formation of the biofilms that were indistinguishable from native biofilms in specific biological functions tested here.
Table 1.
Minimum Inhibitory Concentration (MIC) for Ciprofloxacin MIC (μg ml−1)
| UTI89 | No added E. coli IHF | +E. coli rIHF 0.5 μM |
|---|---|---|
| Planktonic (~108 CFU/ml) | 0.8 | 0.8 |
| 24h Biofilm (~108 CFU/ml) | >2000 | >2000 |
Figure 8. E. coli rIHF treated UTI89 biofilms maintain sensitivity to hydrogen peroxide and express Ag43.

(A) Effect of hydrogen peroxide on UPEC strain UT189 biofilm. Biofilm formation was initiated then maintained in the presence or absence of E. coli rIHF (0.5 μM) in a chamber slide for 24 hours. The biofilms were then treated with the indicated concentration of hydrogen peroxide for 30 min. The plot represents the CFU/ml of UPEC that survived the hydrogen peroxide challenge for 30 min. (B) Representative RT-PCR analysis for identification of antigen43 (Ag43) from cDNA of planktonic (P) and biofilm (in the presence or absence of 0.5 μM E. coli rIHF) state UTI89.
Discussion
In this study, we have demonstrated that DNABII proteins are critical components of the UPEC strain UTI89 biofilm matrix and further, that these proteins effectively directly or indirectly facilitate partitioning of bacteria to the biofilm state when added exogenously. The observed increase in biofilm maximum height, average thickness and biomass in response to the exogenous addition of recombinant DNABII proteins (Figure 1, S1, S2) suggested that these proteins are limiting in biofilms formed by multiple clinical isolates of UPEC. In this regard, it is interesting that such a critical component of the matrix is limiting, given that biofilms are the preferred state of growth for most bacteria. We speculate that there is a delicate balance between IHF bound to eDNA within the biofilm matrix and free IHF in the extracellular milieu such that there is sufficient IHF in the EPS in order to maintain the structural integrity of the biofilm yet an insufficient excess to elicit an adaptive immune response. Indeed, it is evident from the resolved structure of IHF bound to DNA, as elucidated by x-ray crystallography, that a large portion of the protein is occluded by bound DNA, namely specific regions of the N and C terminus and β-ribbons that contain the contact points for DNA (Rice et al., 1996). We have also shown that while immunization of chinchillas with E. coli rIHF resulted in a protective adaptive immune response, immunization with E. coli rIHF pre-bound to an excess of DNA failed to induce an effective immune response in a chinchilla model of experimental otitis media (Goodman et al., 2011).
The DNABII family is a group within a class of proteins called Nucleoid Associated Proteins (NAPs) where members, while not related in either primary or secondary structure, all affect the global conformation of bacterial chromatin. In E. coli the NAP protein family includes at least 11 other proteins in addition to IHF and HU (Ali Azam et al., 1999). Interestingly, two NAPs Fis and H-NS that are highly expressed in the exponential phase (Ali Azam et al., 1999), have a predominant role in DNA structure intra-bacterially (Shiraishi et al., 2007; Cendra Mdel et al., 2013; Flatten and Skarstad, 2013; Ohniwa et al., 2013; Sato et al., 2013), yet have no effect on biofilm formation when added exogenously within the limits of the assays as presented here (Figure S4). This outcome suggested that the two tested NAPs do not have a dominant extracellular role in biofilm formation in this UPEC strain, under these growth conditions.
Extracellular DNA (eDNA) has been identified as a major component of the biofilm matrix of several Gram-positive and Gram-negative bacteria (Steinberger and Holden, 2005; Qin et al., 2007; Izano et al., 2008; Guiton et al., 2009; Flemming and Wingender, 2010). In this study, we have demonstrated that UPEC strain UTI89 incorporated dsDNA within its biofilm matrix and that the eDNA was distributed throughout the biofilm matrix (Figure 2A, B). We have previously shown that the biofilm formed by NTHI during experimental otitis media and sputum samples recovered from cystic fibrosis patients that were culture positive for Burkholderia cenocepacia included an abundant amount of double stranded, extra-bacterial DNA that assembled into an intricate lattice-like network that resembled cruciform DNA structures (Goodman et al., 2011). E. coli HU has been shown to bind to DNA with bent architectures and to branch-like DNA structures (Kamashev and Rouviere-Yaniv, 2000). It has also been shown that E. coli HU has a high affinity to cruciform DNA structures (Pontiggia et al., 1993). Although the nature of eDNA has yet to be determined, if the interwoven mesh-like structures that have been observed is a derivative of the cruciform structure, these data suggest that the DNABII family of proteins likely play a predominant role in maintaining the structural integrity of eDNA in a biofilm through high affinity binding and DNA secondary structure stabilization (Tjokro et al., 2014). In support of this hypothesis, we have shown that the exogenous addition of DNase or of DNase and E. coli rIHF in combination to UTI89 biofilms resulted in a significant decrease in the size of the biofilm compared to the control (Figure 2C, D), suggesting that DNABII proteins mediate biofilm formation likely by stabilizing the secondary structure of eDNA.
It is not well understood as to how the DNABII proteins that do not contain a signal sequence for export exit the bacterial cell into the extracellular milieu. In this study we have shown that overproduction of E. coli rIHF in UTI89 resulted in accumulation of IHF in the spent medium (Figure 3) and an increased amount of eDNA in the biofilm matrix (Figure S3), suggesting that the release of IHF and DNA is likely a stochastic process. It has recently been demonstrated in Burkholderia cenocepacia strain K56-2, that the Type VI secretion system (T6SS) contains a binding site for IHF directly upstream of the coding sequence and that the export of IHF into in vitro formed biofilms was dependent on an active T6SS suggesting that IHF could possibly regulate its own export via the T6SS in B. cenocepacia (K56-2) (Novotny et al., 2013). Such an active mechanism of export of DNABII proteins has not been described in UPEC strain UTI89. We envision that these proteins and DNA gain access to the extracellular milieu by passive or active cell lysis or autolysis (Claverys and Havarstein, 2007; Rice and Bayles, 2008). It is intriguing that HU is indispensable for the signal recognition particle complex (SRP) in Gram-positive bacteria (Nakamura et al., 1999); recently it was shown that HU may even associate with the 4.5s RNA of the SRP of E. coli (Macvanin et al., 2012). It is possible that HU could exit the bacterial cell via this system. Each of these mechanisms is being further investigated.
IHF is crucial for regulation of a number of genes that contribute to virulence including production of elastase in Vibrio vulnificus (Jeong et al., 2010), cholera toxin in Vibrio cholera (Stonehouse et al., 2008), and type III secretion effectors in E. coli (Li et al., 2004). Regulation of type I pilus and P- pilus in UTI89 (Justice et al., 2012), adhesins and capsule in Neisseria (Hill et al., 2002) and E. coli (Rowe et al., 2000; Corcoran and Dorman, 2009) are also dependent on expression of IHF. In this study we have shown that both subunits of IHF play a role in UTI89 biofilm formation. However, exogenous E. coli rIHF could only restore biofilm formation in the absence of IHFA but not IHFB, suggesting that IHFB likely plays a more critical role in the intracellular milieu. It is conceivable that IHFB indirectly affects biofilm formation through the Type I pilus. In support of this hypothesis, we have previously shown that while both subunits of IHF are required for type I piliation in UPEC strain UTI89, only IHFB maintains the type I pilus promoter in the ‘ON’ state. Further, the ectopic production of Type I pilus in UTI89 ΔihfB fully restored virulence, which suggests that the major defect in the absence of IHFB is the lack of expression of Type I pilus (Justice et al., 2012). Since the Type I pilus of UPEC has been linked to virulence and biofilm formation (Hultgren et al., 1985; Pratt and Kolter, 1998; Martinez et al., 2000; Bahrani-Mougeot et al., 2002; Justice et al., 2006; Snyder et al., 2006; Ulett et al., 2007; Wright et al., 2007; Crepin et al., 2012; Hung et al., 2013), we envision that IHFB affects biofilm formation via the expression of Type I pilus. Also, the ability of other components of the EPS to support biofilm growth in the absence of IHFB remains to be evaluated.
The characterization of biofilms formed by UTI89 ihfA11::Tn10 and UTI89 ΔihfB suggested that the two subunits of IHF have a differential effect on the structure of the biofilm as judged by the average thickness (Figure 4B). This may indicate that individual homodimers of IHF have independent and unique functions, which is in sharp contrast to the null phenotype observed in laboratory E. coli strains that lack either subunit of IHF (Werner et al., 1994; Zulianello et al., 1994). To support our data, we have previously demonstrated that while both IHFA and IHFB are required for colonization of the mouse bladder, the community architecture of UTI89 that could not express IHFA was strikingly different from that of the UTI89 mutant that could not express IHFB. In the absence of IHFA, the IBCs formed by UTI89 exhibited a more globular architecture while in the absence of IHFB the IBCs were more open and elongated suggesting that the homodimers of IHF have unique functions (Justice et al., 2012). Alas, we were previously unable to construct a mutant that contained a deletion of both ihf alleles in UTI89 to test if there would be, as we might expect, an even stronger biofilm phenotype in a ihfA ihfB double mutant (Justice et al., 2012). Other studies have shown that individual IHF subunits form homodimers in vitro in complex with DNA and that either subunit can support Lambda site-specific recombination (Werner et al., 1994; Zulianello et al., 1994) in vitro, suggesting that the homodimers of IHF are functional. Together, these data reinforced our hypothesis that individual IHF homodimers have independent functions and it is likely that homodimers and heterodimers of IHF play unique roles in biofilm formation by UTI89.
We have yet to determine if the roles of IHF and HU in eDNA dependent biofilm EPS are different, similar or redundant. IHF and HU are both highly expressed in the early and late stationary phases (Ali Azam et al., 1999) and an increased accumulation of these proteins in the spent medium is evident as shown in Figure 3C. IHF and HU have some redundant functions in terms of DNA binding and it is possible that these proteins act independently, compensate or substitute for the lack of the other in order to maintain the structural integrity of eDNA. It is also conceivable that these proteins coordinate with each other to support biofilm formation by UTI89 and likely other strains. In support of this hypothesis, Bonnefoy et al., have shown that homo and hetero dimers of HU modulate the DNA binding activity of IHF to random linear dsDNA sequences (Bonnefoy and Rouviere-Yaniv, 1991). One could also imagine possible interaction of IHF monomers with HU monomers to form mixed heterodimers that modulate the structural integrity of eDNA. To further investigate this, we determined the distribution of eDNA within in vitro preformed UPEC biofilms and showed that in the presence of homodimers of IHF (strains UTI89 ihfA11::Tn10 and UTI89 ΔihfB) or homodimers of HU (strains UTI89 hupA::cam and UTI89 hupB::kan), the qualitative distribution (data not shown) and abundance of eDNA (Figure S3) within the biofilm matrix was similar to that of the wild type UPEC biofilm within the limits of the assay as presented here which suggested that these proteins could compensate or substitute for the lack of the other in order to maintain the distribution and likely structural integrity of eDNA. The fine structural analysis of eDNA in the presence of homodimers of IHF or HU remains to be further evaluated. Also, the biofilms formed in the presence of homodimers of IHF or HU (same strains as above) were not more susceptible to hydrogen peroxide compared to the wild type UPEC biofilm (data not shown). The susceptibility of these biofilms to antimicrobial peptides remains to be evaluated. Hence it remains an open question as to how redundant each DNABII protein is with respect to one another in terms of biological functions performed within the extracellular matrix.
Most intriguing is that HU from the Gram-positive bacterium S. mutans, when added exogenously to UPEC strain UTI89, resulted in a dramatic increase in the size of the biofilm (Figure S1). This result suggested that the DNABII proteins are likely interchangeable between various bacterial genera and, in turn, implies a similar eDNA structure that is compatible with a consortia of heterologous bacteria within a biofilm. In support of this hypothesis, we have previously shown that antiserum directed against E. coli rIHF destabilizes the biofilm architecture and notably disrupts biofilms formed by multiple Gram-positive and Gram-negative human pathogens in vitro (Goodman et al., 2011). We envision that the structural arrangement of eDNA in a biofilm and the proteins that stabilize the secondary structure of the eDNA have evolved to support a compatible EPS such that most of the constituent bacterial species within a given ecological niche could co-exist and intermingle.
This study identifies key constituents of the EPS that contribute to the integrity of the biofilm matrix formed by UPEC strain UTI89. Given that IHF affects biofilm formation in vitro and community architecture of UTI89 in vivo and also regulates expression of a number of genes associated with virulence such as Type I pilus, P pilus and capsule (Rowe et al., 2000; Justice et al., 2012) suggests that a therapeutic approach that targets DNABII proteins would affect both intracellular and extracellular targets of UPEC pathogenesis and thereby would severely impair the virulence of UTI89. Although this study is focused on UTI89 biofilms, these components of the EPS (i.e. DNA, DNABII proteins) are common to many pathogenic bacteria (Flemming and Wingender, 2010; Goodman et al., 2011). Hence, these observations will likely aid in the development of novel therapies to treat and/or prevent acute and chronic bacterial infection during the course of UTI that could likely be applicable to a multitude of biofilm-mediated diseases.
Experimental Procedures
Bacteria strains and plasmids
The strains and plasmids used in this study are shown in supplementary Table 1. The hupA::cam and hupB::kan mutations were independently introduced into UTI89. The hupA::cam and hupB::kan mutations were transduced from MG1655 hupA::cam and MG1655 hupB::kan into UTI89 respectively using standard P1 transduction (Slihavy, 1984) and transductants were selected on LB agar (Fisher Scientific) containing 25 μg/ml cholramphenicol (Fisher Scientific) and 25 μg/ml kanamycin sulfate (Fisher Scientific), respectively. Ectopic production of either or both IHF subunits was accomplished using pHNα (Granston and Nash, 1993), pHNβ (kindly provided by Howard Nash), or pHNαβ (Lee et al., 1992). Ectopic production of E. coli HU was accomplished using pRLM118 (kindly provided by Roger McMacken).
Purification of DNABII proteins
IHF was purified from E. coli strain HN880 (Nash et al., 1987). The cells were grown at 30°C to an Optical Density (OD) of 0.4–0.6 at 600 nm. At mid-log phase, the growth temperature was shifted to 42°C for 4h to induce expression of IHF. Bacterial cells were pelleted at 15000g for 5 min and resuspended in lysis buffer (20 mM Tris pH 7.4, 1 mM EDTA, 10% glycerol, 20 mM NaCl, 1 mM PMSF, 5 mM MgCl2, 5 mM CaCl2). The suspension was treated with DNase (100 μg/ml) for 30 min on ice followed by cell lysis by three passes in a French press at 20,000 psi. The cell lysate was clarified at 25,000g for 30 min at 4°C and passed through a 0.45 μm filter. The proteins were precipitated with ammonium sulphate (50% saturation) and the cell lysate was clarified further at 25,000g for 15 min at 4°C. Ammonium sulphate was added to 80% saturation; the proteins were pelleted and resuspended in binding buffer (10 mM Phosphate buffer pH 7, 500 mM NaCl). This IHF enriched protein fraction was dialyzed (3000 MWCO) in binding buffer overnight at 4°C. The dialyzed fraction was bound to heparin-Sepharose column equilibrated in binding buffer. The column was washed and the bound IHF was eluted with 10 column volumes of linear gradient of elution buffer (10 mM Phosphate buffer pH 7, 2M NaCl). The collected fractions were analyzed by SDS PAGE, pooled and dialyzed in 10 mM Tris pH 7.4, 50 mM KCl. The fraction was further purified by a Co-Talon column (GE Healthcare) and pure IHF was eluted with 10 mM Tris pH 7.4, 300 mM KCl. The protein was concentrated in a centrifugal filter (3000 MWCO) and dialyzed in storage buffer (50 mM Tris pH 7.4, 600 mM KCl, 1 mM EDTA, 10% glycerol) and stored at −80°C.
HU was purified from E. coli strain N99 himAΔsma himΔ::cat made by D. Friedman that lacks both subunits of IHF (Granston et al., 1988) and overproduces HU from an inducible plasmid. HU was purified using the same procedure as IHF except that the binding buffer with 200 mM NaCl and the elution buffer for Co-Talon column with 200 mM KCl was used.
IHFA and IHFB were purified from E. coli strains containing the plasmid pHNα and pHNβ, respectively. Both proteins were purified using the same procedure as HU except that the proteins were eluted with 10 column volumes of linear gradient of elution buffer (10 mM Phosphate buffer pH 7, 800 mM NaCl) from the heparin-Sepharose column.
In vitro biofilm analysis
For in vitro biofilm analysis, all strains were cultured on LB agar for 18 - 20 h at 37°C, in a humidified atmosphere containing 5% CO2, and then suspended in LB broth to an OD of 0.65 at 490 nm. Cultures were diluted (1:6) in LB broth, then incubated statically at 37°C, 5% CO2 until reaching an OD490nm of 0.65. The cultures were diluted 1:2500 in LB, and 200 μl of this suspension was inoculated into each well of an eight-well chamber slide (Fisher Scientific). After 16h of incubation at 37°C, 5% CO2, the medium was replaced with fresh medium. After an additional 8h incubation period, the medium was replaced again as described above and the chamber slides were incubated for an additional 16 h as before. For exogenous addition of DNABII proteins or DNase, the appropriate protein was added at 0.5 μM and RNase-free DNase (Promega) was added at 1unit at the time of seeding. Also, proteins and RNase-free DNase were added at 0.5 μM every time the medium was replaced. For all bacterial isolates, after the final incubation (40 h total), the medium was removed and the biofilms were washed twice with saline (0.9% NaCl) and stained with LIVE/DEAD® stain (Molecular probes, Eugene, OR) according to manufacturer’s instructions. Biofilms were then washed twice in saline and fixed with 1.6% paraformaldehyde, 0.025% glutaraldehyde and 4% acetic acid in 0.1M phosphate buffer at pH 7.4. The biofilms were imaged using a x63 objective on a Zeiss 510 Meta-laser scanning confocal microscope (Carl Zeiss, Thornwood, NY). Three-dimensional images were reconstructed with AxioVision Rel. 4.8 (Carl Zeiss) and maximum biofilm height, average thickness and biomass were determined via COMSTAT analysis (Heydorn et al., 2000). For overexpression of DNABII proteins, IPTG was used at 1 mM to induce the expression of IHFA, IHFB or IHFAB at the time of seeding and throughout biofilm growth for 40h. All in vitro biofilm assays were repeated a minimum of three times on separate days, and all individual biofilm assays were done in duplicates on each assay day. Data are presented as mean values ± SEM.
Distribution of DNA within a UTI89 biofilm
The distribution of dsDNA within a UTI89 biofilm was determined as previously described (Novotny et al., 2013). Briefly, unfixed 40 hour UTI89 biofilms were labeled with mouse anti-dsDNA monoclonal antibody (Abcam, Inc.), which was revealed with goat anti-mouse IgG conjugated to AlexaFluor 647 (Molecular Probes). Bacterial cells were labeled with FilmTracer™ FM® 1–43 (Molecular Probes) according to manufacturer’s instructions. The biofilms were imaged using a x63 objective on a Zeiss 510 Meta-laser scanning confocal microscope (Carl Zeiss, Thornwood, NY). Three-dimensional images were reconstructed with AxioVision Rel. 4.8 (Carl Zeiss). Zeiss image acquisition software was employed to determine the relative fluorescence of anti-dsDNA labeling to FilmTracer™. The relative abundance of dsDNA was determined by the ratio of the dsDNA (anti-dsDNA labeling) to the bacteria (FilmTracer™).
Number of bacteria in the biofilm or planktonic state
Biofilm formation was initiated in the presence or absence of E. coli rIHF at 0.5 μM in a chamber slide for 40 h at 37°C in a humidified atmosphere containing 5% CO2. The biofilms were cultured in either medium (LB) alone or in medium containing E. coli rIHF (0.5 μM) at 16 h and 24 h to maintain viability. At the end of 40 h, the contents of the well was resuspended, serially diluted and plated on LB agar to calculate the total number of bacteria. For planktonic bacteria, the spent medium from chamber slides was carefully removed. For the remaining adhered bacteria, biofilms were washed twice with sterile PBS and the washes were combined with the bacteria from the spent medium as the source of bacteria in the planktonic state. After the washes, the biofilm was resuspended in 200 μl of sterile PBS. Both planktonic and biofilm bacteria were serially diluted and plated on LB to calculate the number of bacteria (colony forming units). The assay was repeated a minimum of three times on separate days and all individual assays were done in duplicates on each assay day. Statistical significance was assessed by unpaired t-test (GraphPad Prism version 6.0). A p value ≤ 0.05 was considered significant.
Purification of proteins from spent medium
UTI89 was cultured in M9 medium (modified M9) supplemented with 0.1 mg/ml thiamine, 4 mM cysteine and vitamins (Desai et al., 2012) at 37°C, 200rpm for 16h. The cells were removed by spinning at 15000g for 5 min at 4°C and then by passing the spent medium through a 0.45 μm filter. PMSF (1 mM) was added to the spent medium to prevent protein degradation. Proteins in the spent medium were precipitated with ammonium sulphate (80% saturation) at 4°C for 3h. The spent medium was centrifuged at 15000g, 4°C for 15 min to pellet the proteins. The proteins were resuspended in 10 mM Tris pH 7.4, 200 mM KCl; dialyzed (3 KDa MWCO) in the same buffer overnight at 4°C and then concentrated with a centrifugal filter (3 KDa MWCO). The proteins were resolved by SDS-PAGE and probed with polyclonal antiserum from rabbits directed against E. coli rIHF in a western blot analysis. Briefly, 2 μl protein suspension was separated on a 16% resolving, 5% stacking gel in Tris/Glycine/SDS buffer, then transferred on to nitrocellulose membrane (Invitrogen). Membrane was blocked overnight at 4°C in 5% Blotto in Tris-buffered saline with 0.5% Tween 20 (Fisher Scientific) prior to incubation with rabbit anti-IHF followed by anti-rabbit IgG-HRP (Invitrogen). Blots were developed with ECL plus substrate kit (Pierce) and images were captured with Typhoon scanner (GE Healthcare).
Hydrogen peroxide survival assay
Biofilm formation was initiated in the presence or absence of E. coli rIHF at 0.5 μM in a chamber slide for 24 h at 37°C in a humidified atmosphere containing 5% CO2. The biofilms were cultured in either medium (LB) alone or in medium containing E. coli rIHF (0.5 μM) at 16 h to maintain viability. At the end of 24 h, the biofilms were incubated with 0 – 0.8% hydrogen peroxide for 30 min at 37°C. Then hydrogen peroxide was removed and the biofilms were resuspended in sterile PBS, diluted and plated on LB agar to calculate the number of bacteria that survived the challenge. Planktonic state cells were obtained by suspending UPEC strain UTI89 in LB broth to an OD of 0.65 at 490 nm. Cultures were diluted (1:6) in LB broth, then incubated statically at 37°C, 5% CO2 until an OD490nm of 0.65. 100 μl of this culture was used for the hydrogen peroxide challenge. All assays were repeated a minimum of three times on separate days. Data are presented as mean values ± SEM.
Determination of Minimum Inhibitory Concentration (MIC) of ciprofloxacin (CIP) for UPEC strain UTI89
Biofilm formation was initiated in the presence or absence of E. coli rIHF at 0.5 μM in a chamber slide for 24 h at 37°C in a humidified atmosphere containing 5% CO2. The biofilms were cultured in either medium (LB) alone or in medium containing E. coli rIHF (0.5 μM) at 16 h to maintain viability. At the end of 24 h, the biofilms were treated with 0 – 2000 μg ml−1 CIP for 16 h at 37°C. The CIP was removed and the biofilms were washed twice with sterile PBS, resuspended in sterile PBS, diluted and spotted on LB agar. MIC was defined as the lowest concentration of CIP at which no growth was observed (Andrews, 2001). Planktonic state cells were obtained by suspending UTI89 in LB broth to an OD of 0.65 at 490 nm. Cultures were diluted (1:6) in LB broth, then incubated statically at 37°C, 5% CO2 until an OD490nm of 0.65. 100 μl of this culture was used for the CIP treatment. All assays were repeated a minimum of three times on separate days. Data are presented as mean values ± SEM.
Detection of autotransporter protein antigen43 (Ag43) by RT-PCR
Biofilm formation was initiated in the presence or absence of E. coli rIHF at 0.5 μM in a chamber slide for 16 h at 37°C in a humidified atmosphere containing 5% CO2 in modified M9 medium. At the end of 16 h, biofilms were resuspended and centrifuged at 5000g for 10 min. RNA extraction was performed with the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. RNase-free DNase (Qiagen) was used according to the manufacturer’s specifications to remove DNA. As a planktonic control, exponentially growing UTI89 in the same medium was used. To obtain cDNA, Superscript III (Life technologies) was used according to the manufacturer’s specifications. Primers for 16S ribosomal RNA (Leverton and Kaper, 2005) were used as an internal control for the PCR, and the assay was then carried out using specific primers for Ag43 5′-atgacaaccagattgcctcggtac-3′ and 5′-ggcatctccgtagacggtct- 3′.
Statistical evaluation
Significance in maximum height, mean thickness and biomass was assessed by unpaired t-test (GraphPad Prism version 6.0). A p ≤ 0.05 was represented as * and a p ≤ 0.01 was represented by **.
Supplementary Material
Figure S1. Effect of exogenous addition of DNABII Proteins on UPEC strain UTI89 biofilm formation.
Biofilm growth was initiated then maintained in the presence or absence of the indicated recombinant exogenous proteins (0.5 μM) from E. coli and S. mutans (Sm) for 40 hours. Images were analyzed by COMSTAT to calculate biofilm maximum height (A), average thickness (B) and biomass (C). Asterisks indicate statistical significance (p ≤ 0.05) compared to control.
Figure S2. Effect of exogenous addition of DNABII Proteins on biofilm formation by clinical isolates of UPEC
Biofilm growth was initiated then maintained in the presence or absence of the indicated exogenously added recombinant proteins (0.5 μM) from E. coli for 40 hours. Biofilms were stained with LIVE/DEAD® stain and the images were analyzed by COMSTAT to calculate biofilm maximum height (A, C, E), average thickness, and biomass (B, D, F). Panels A and B represent biofilms formed by UPEC #925. Panels C and D represent biofilms formed by UPEC #932. Panels E and F represent biofilms formed by UPEC #933. Asterisks indicate statistical significance (p ≤ 0.05) compared to control.
Figure S3. Quantification of eDNA within the biofilm matrix
Biofilm growth by the indicated strains was initiated then maintained for 40 hours. Lanes indicated as UTI89+DNase and UTI89+E. coli rIHF were incubated with DNase (1 unit) or E. coli rIHF (0.5 μM) at seeding, respectively. Unfixed 40 hour biofilms were then labeled with anti-dsDNA monoclonal antibody and FilmTracer™ FM® 1–43. The relative abundance of eDNA was determined by the ratio of dsDNA (anti-dsDNA labeling) to biomass (FilmTracer™).
Figure S4. Effect of exogenous addition of E. coli rHNS or E. coli rFis (Nucleoid Associated Proteins) on UPEC strain UTI89 biofilm formation.
Biofilm growth was initiated in the presence or absence of the indicated recombinant E. coli exogenous proteins (0.5 μM) for 40 hours. Images were analyzed by COMSTAT to calculate biofilm maximum height (A), average thickness (B) and biomass (C).
Table S1. Strains and plasmids
Acknowledgments
This work was supported by NIH grant R01DC011818 to SDG and LOB. We thank Dr. Reid Johnson for his generous gift of purified E. coli Fis protein, Linda Kenney for purified E. coli H-NS protein, and Dr. Mark Schembri for anti-Ag43 antibody. We also thank John Buzzo for purification of the following E. coli proteins: IHF, HU, IHFA and IHFB. We thank Jason B. Navarro for help with figures. We thank the anonymous reviewers for suggesting additional experiments that generated highly supportive data.
Footnotes
The authors declare no conflict of interest.
References
- Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- Bahrani-Mougeot FK, Buckles EL, Lockatell CV, Hebel JR, Johnson DE, Tang CM, Donnenberg MS. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol Microbiol. 2002;45:1079–1093. doi: 10.1046/j.1365-2958.2002.03078.x. [DOI] [PubMed] [Google Scholar]
- Biscola FT, Abe CM, Guth BEC. Determination of Adhesin Gene Sequences in, and Biofilm Formation by, O157 and Non-O157 Shiga Toxin-Producing Escherichia coli Strains Isolated from Different Sources. Appl Environ Microbiol. 2011;77:2201–2208. doi: 10.1128/AEM.01920-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij A, Schaeps RM, de Kleijn S, Hermans PW, Glaser P, Pancholi V, et al. Surface-exposed histone-like protein a modulates adherence of Streptococcus gallolyticus to colon adenocarcinoma cells. Infect Immun. 2009;77:5519–5527. doi: 10.1128/IAI.00384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy E, Rouviere-Yaniv J. HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein-DNA complexes with short DNA fragments. EMBO J. 1991;10:687–696. doi: 10.1002/j.1460-2075.1991.tb07998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstetter KA, Jurcisek JA, Goodman SD, Bakaletz LO, Das S. Antibodies directed against integration host factor mediate biofilm clearance from Nasopore. Laryngoscope. 2013;123:2626–2632. doi: 10.1002/lary.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockson ME, Novotny LA, Mokrzan EM, Malhotra S, Jurcisek JA, Akbar R, et al. Evaluation of the kinetics and mechanism of action of anti-integration host factor mediated disruption of bacterial biofilms. Mol Microbiol. 2014 doi: 10.1111/mmi.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Grainger DC, Busby SJ. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Cavaliere R, Ball JL, Turnbull L, Whitchurch CB. The biofilm matrix destabilizers, EDTA and DNaseI, enhance the susceptibility of nontypeable Hemophilus influenzae biofilms to treatment with ampicillin and ciprofloxacin. Microbiologyopen. 2014 doi: 10.1002/mbo3.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Cendra MM, Juarez A, Madrid C, Torrents E. H-NS is a novel transcriptional modulator of the ribonucleotide reductase genes in Escherichia coli. J Bacteriol. 2013;195:4255–4263. doi: 10.1128/JB.00490-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d’etre. Nat Rev Microbiol. 2007;5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- Corcoran CP, Dorman CJ. DNA relaxation-dependent phase biasing of the fim genetic switch in Escherichia coli depends on the interplay of H-NS, IHF and LRP. Mol Microbiol. 2009;74:1071–1082. doi: 10.1111/j.1365-2958.2009.06919.x. [DOI] [PubMed] [Google Scholar]
- Crepin S, Houle S, Charbonneau ME, Mourez M, Harel J, Dozois CM. Decreased expression of type 1 fimbriae by a pst mutant of uropathogenic Escherichia coli reduces urinary tract infection. Infect Immun. 2012;80:2802–2815. doi: 10.1128/IAI.00162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J Bacteriol. 2012;194:3774–3780. doi: 10.1128/JB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatten I, Skarstad K. The Fis protein has a stimulating role in initiation of replication in Escherichia coli in vivo. PLos One. 2013;8:e83562. doi: 10.1371/journal.pone.0083562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Foxman B, Brown P. Epidemiology of urinary tract infections: transmission and risk factors, incidence, and costs. Infect Dis Clin North Am. 2003;17:227–241. doi: 10.1016/s0891-5520(03)00005-9. [DOI] [PubMed] [Google Scholar]
- Foxman B, Gillespie B, Koopman J, Zhang L, Palin K, Tallman P, et al. Risk factors for second urinary tract infection among college women. Am J Epidemiol. 2000;151:1194–1205. doi: 10.1093/oxfordjournals.aje.a010170. [DOI] [PubMed] [Google Scholar]
- Gao QS. S mutans GtfB and GtfC gene expression. Los Angeles: University of Southern Calfornia; 2000. [Google Scholar]
- Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, et al. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- Granston AE, Nash HA. Characterization of a set of integration host factor mutants deficient for DNA binding. J Mol Biol. 1993;234:45–59. doi: 10.1006/jmbi.1993.1562. [DOI] [PubMed] [Google Scholar]
- Granston AE, Alessi DM, Eades LJ, Friedman DI. A point mutation in the Nul gene of bacteriophage lambda facilitates phage growth in Escherichia coli with himA and gyrB mutations. Mol Gen Genet. 1988;212:149–156. doi: 10.1007/BF00322458. [DOI] [PubMed] [Google Scholar]
- Guiton PS, Hung CS, Kline KA, Roth R, Kau AL, Hayes E, et al. Contribution of autolysin and Sortase a during Enterococcus faecalis DNA-dependent biofilm development. Infect Immun. 2009;77:3626–3638. doi: 10.1128/IAI.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustave JE, Jurcisek JA, McCoy KS, Goodman SD, Bakaletz LO. Targeting bacterial integration host factor to disrupt biofilms associated with cystic fibrosis. J Cyst Fibros. 2013;12:384–389. doi: 10.1016/j.jcf.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MJ, DeFrances CJ, Williams SN, Golosinskiy A, Schwartzman A. National Hospital Discharge Survey: 2007 summary. Natl Health Stat Report. 2010:1–20. 24. [PubMed] [Google Scholar]
- Heydorn A, Ersboll BK, Hentzer M, Parsek MR, Givskov M, Molin S. Experimental reproducibility in flow-chamber biofilms. Microbiology. 2000;146 (Pt 10):2409–2415. doi: 10.1099/00221287-146-10-2409. [DOI] [PubMed] [Google Scholar]
- Hill SA, Samuels DS, Nielsen C, Knight SW, Pagotto F, Dillon JA. Integration host factor interactions with Neisseria gene sequences: correlation between predicted binding sites and in vitro binding of Neisseria -derived IHF protein. Mol Cell Probes. 2002;16:153–158. doi: 10.1006/mcpr.2001.0403. [DOI] [PubMed] [Google Scholar]
- Hultgren SJ, Porter TN, Schaeffer AJ, Duncan JL. Role of type 1 pili and effects of phase variation on lower urinary tract infections produced by Escherichia coli. Infect Immun. 1985;50:370–377. doi: 10.1128/iai.50.2.370-377.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C, Zhou Y, Pinkner JS, Dodson KW, Crowley JR, Heuser J, et al. Escherichia coli biofilms have an organized and complex extracellular matrix structure. MBio. 2013;4:e00645–00613. doi: 10.1128/mBio.00645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggi N, Sissodia P. Multimodal supervision programme to reduce catheter associated urinary tract infections and its analysis to enable focus on labour and cost effective infection control measures in a tertiary care hospital in India. J Clin Diagn Res. 2012;6:1372–1376. doi: 10.7860/JCDR/2012/4229.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HS, Kim SM, Lim MS, Kim KS, Choi SH. Direct interaction between quorum-sensing regulator SmcR and RNA polymerase is mediated by integration host factor to activate vvpE encoding elastase in Vibrio vulnificus. J Biol Chem. 2010;285:9357–9366. doi: 10.1074/jbc.M109.089987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–3875. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Lauer SR, Hultgren SJ, Hunstad DA. Maturation of intracellular Escherichia coli communities requires SurA. Infect Immun. 2006;74:4793–4800. doi: 10.1128/IAI.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Li B, Downey JS, Dabdoub SM, Brockson ME, Probst GD, et al. Aberrant community architecture and attenuated persistence of uropathogenic Escherichia coli in the absence of individual IHF subunits. PLoS One. 2012;7:e48349. doi: 10.1371/journal.pone.0048349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji C, Watanabe K, Apicella MA, Watanabe H. Antimicrobial effect of fluoroquinolones for the eradication of nontypeable Haemophilus influenzae isolates within biofilms. Tohoku J Exp Med. 2008;214:121–128. doi: 10.1620/tjem.214.121. [DOI] [PubMed] [Google Scholar]
- Kamashev D, Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodner CM, Thomas Gupton EK. Recurrent urinary tract infections in women: diagnosis and management. Am Fam Physician. 2010;82:638–643. [PubMed] [Google Scholar]
- Lee EC, Hales LM, Gumport RI, Gardner JF. The isolation and characterization of mutants of the integration host factor (IHF) of Escherichia coli with altered, expanded DNA-binding specificities. EMBO J. 1992;11:305–313. doi: 10.1002/j.1460-2075.1992.tb05053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverton LQ, Kaper JB. Temporal expression of enteropathogenic Escherichia coli virulence genes in an in vitro model of infection. Infect Immun. 2005;73:1034–1043. doi: 10.1128/IAI.73.2.1034-1043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Rosenshine I, Tung SL, Wang XH, Friedberg D, Hew CL, Leung KY. Comparative proteomic analysis of extracellular proteins of enterohemorrhagic and enteropathogenic Escherichia coli strains and their ihf and ler mutants. Appl Environ Microbiol. 2004;70:5274–5282. doi: 10.1128/AEM.70.9.5274-5282.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunsford RD, Nguyen N, London J. DNA-binding activities in Streptococcus gordonii: identification of a receptor-nickase and a histonelike protein. Curr Microbiol. 1996;32:95–100. doi: 10.1007/s002849900017. [DOI] [PubMed] [Google Scholar]
- MacDonald TM, Beardon PH, McGilchrist MM, Duncan ID, McKendrick AD, McDevitt DG. The risks of symptomatic vaginal candidiasis after oral antibiotic therapy. Q J Med. 1993;86:419–424. [PubMed] [Google Scholar]
- Macvanin M, Edgar R, Cui F, Trostel A, Zhurkin V, Adhya S. Noncoding RNAs binding to the nucleoid protein HU in Escherichia coli. J Bacteriol. 2012;194:6046–6055. doi: 10.1128/JB.00961-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Yahagi S, Yamazaki T, Yamane K. Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J Biol Chem. 1999;274:13569–13576. doi: 10.1074/jbc.274.19.13569. [DOI] [PubMed] [Google Scholar]
- Nash HA, Robertson CA, Flamm E, Weisberg RA, Miller HI. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J Bacteriol. 1987;169:4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny LA, Amer AO, Brockson ME, Goodman SD, Bakaletz LO. Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS One. 2013;8:e67629. doi: 10.1371/journal.pone.0067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nur A, Hirota K, Yumoto H, Hirao K, Liu D, Takahashi K, et al. Effects of extracellular DNA and DNA-binding protein on the development of a Streptococcus intermedius biofilm. J Appl Microbiol. 2013;115:260–270. doi: 10.1111/jam.12202. [DOI] [PubMed] [Google Scholar]
- Ohniwa RL, Muchaku H, Saito S, Wada C, Morikawa K. Atomic force microscopy analysis of the role of major DNA-binding proteins in organization of the nucleoid in Escherichia coli. PLos One. 2013;8:e72954. doi: 10.1371/journal.pone.0072954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontiggia A, Negri A, Beltrame M, Bianchi ME. Protein HU binds specifically to kinked DNA. Mol Microbiol. 1993;7:343–350. doi: 10.1111/j.1365-2958.1993.tb01126.x. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev. 2008;72:85–109. doi: 10.1128/MMBR.00030-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- Rowe S, Hodson N, Griffiths G, Roberts IS. Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J Bacteriol. 2000;182:2741–2745. doi: 10.1128/jb.182.10.2741-2745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint S, Chenoweth CE. Biofilms and catheter-associated urinary tract infections. Infect Dis Clin North Am. 2003;17:411–432. doi: 10.1016/s0891-5520(03)00011-4. [DOI] [PubMed] [Google Scholar]
- Sato YT, Watanabe S, Kenmotsu T, Ichikawa M, Yoshikawa Y, Teramoto J, et al. Structural change of DNA induced by nucleoid proteins: growth phase-specific Fis and stationary phase-specific Dps. Biophys J. 2013;105:1037–1044. doi: 10.1016/j.bpj.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schembri MA, Kjaergaard K, Klemm P. Global gene expression in Escherichia coli biofilms. Mol Microbiol. 2003;48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- Schito GC, Naber KG, Botto H, Palou J, Mazzei T, Gualco L, Marchese A. The ARESC study: an international survey on the antimicrobial resistance of pathogens involved in uncomplicated urinary tract infections. Int J Antimicrob Agents. 2009;34:407–413. doi: 10.1016/j.ijantimicag.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Shiraishi K, Ogata Y, Hanada K, Kano Y, Ikeda H. Roles of the DNA binding proteins H-NS and StpA in homologous recombination and repair of bleomycin-induced damage in Escherichia coli. Genes Genet Syst. 2007;82:433–439. doi: 10.1266/ggs.82.433. [DOI] [PubMed] [Google Scholar]
- Slihavy TJ, Berman ML, Engquist LW. Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- Slinger R, Chan F, Ferris W, Yeung SW, St Denis M, Gaboury I, Aaron SD. Multiple combination antibiotic susceptibility testing of nontypeable Haemophilus influenzae biofilms. Diagn Microbiol Infect Dis. 2006;56:247–253. doi: 10.1016/j.diagmicrobio.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Snyder JA, Lloyd AL, Lockatell CV, Johnson DE, Mobley HL. Role of phase variation of type 1 fimbriae in a uropathogenic Escherichia coli cystitis isolate during urinary tract infection. Infect Immun. 2006;74:1387–1393. doi: 10.1128/IAI.74.2.1387-1393.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starner TD, Shrout JD, Parsek MR, Appelbaum PC, Kim G. Subinhibitory concentrations of azithromycin decrease nontypeable Haemophilus influenzae biofilm formation and Diminish established biofilms. Antimicrob Agents Chemother. 2008;52:137–145. doi: 10.1128/AAC.00607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger RE, Holden PA. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl Environ Microbiol. 2005;71:5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonehouse E, Kovacikova G, Taylor RK, Skorupski K. Integration host factor positively regulates virulence gene expression in Vibrio cholerae. J Bacteriol. 2008;190:4736–4748. doi: 10.1128/JB.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjokro NO, Rocco CJ, Priyadarshini R, Davey ME, Goodman SD. A biochemical analysis of the interaction of Porphyromonas gingivalis HU PG0121 protein with DNA. PLoS One. 2014;9:e93266. doi: 10.1371/journal.pone.0093266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautner BW, Darouiche RO. Role of biofilm in catheter-associated urinary tract infection. Am J Infect Control. 2004;32:177–183. doi: 10.1016/j.ajic.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulett GC, Mabbett AN, Fung KC, Webb RI, Schembri MA. The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology. 2007;153:2321–2331. doi: 10.1099/mic.0.2006/004648-0. [DOI] [PubMed] [Google Scholar]
- Werner MH, Clore GM, Gronenborn AM, Nash HA. Symmetry and asymmetry in the function of Escherichia coli integration host factor: implications for target identification by DNA-binding proteins. Curr Biol. 1994;4:477–487. doi: 10.1016/s0960-9822(00)00108-1. [DOI] [PubMed] [Google Scholar]
- Winters BD, Ramasubbu N, Stinson MW. Isolation and characterization of a Streptococcus pyogenes protein that binds to basal laminae of human cardiac muscle. Infect Immun. 1993;61:3259–3264. doi: 10.1128/iai.61.8.3259-3264.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Foxman B. Molecular epidemiology of Escherichia coli mediated urinary tract infections. Front Biosci. 2003;8:e235–244. doi: 10.2741/1007. [DOI] [PubMed] [Google Scholar]
- Zulianello L, de la Gorgue de Rosny E, van Ulsen P, van de Putte P, Goosen N. The HimA and HimD subunits of integration host factor can specifically bind to DNA as homodimers. EMBO J. 1994;13:1534–1540. doi: 10.1002/j.1460-2075.1994.tb06415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Effect of exogenous addition of DNABII Proteins on UPEC strain UTI89 biofilm formation.
Biofilm growth was initiated then maintained in the presence or absence of the indicated recombinant exogenous proteins (0.5 μM) from E. coli and S. mutans (Sm) for 40 hours. Images were analyzed by COMSTAT to calculate biofilm maximum height (A), average thickness (B) and biomass (C). Asterisks indicate statistical significance (p ≤ 0.05) compared to control.
Figure S2. Effect of exogenous addition of DNABII Proteins on biofilm formation by clinical isolates of UPEC
Biofilm growth was initiated then maintained in the presence or absence of the indicated exogenously added recombinant proteins (0.5 μM) from E. coli for 40 hours. Biofilms were stained with LIVE/DEAD® stain and the images were analyzed by COMSTAT to calculate biofilm maximum height (A, C, E), average thickness, and biomass (B, D, F). Panels A and B represent biofilms formed by UPEC #925. Panels C and D represent biofilms formed by UPEC #932. Panels E and F represent biofilms formed by UPEC #933. Asterisks indicate statistical significance (p ≤ 0.05) compared to control.
Figure S3. Quantification of eDNA within the biofilm matrix
Biofilm growth by the indicated strains was initiated then maintained for 40 hours. Lanes indicated as UTI89+DNase and UTI89+E. coli rIHF were incubated with DNase (1 unit) or E. coli rIHF (0.5 μM) at seeding, respectively. Unfixed 40 hour biofilms were then labeled with anti-dsDNA monoclonal antibody and FilmTracer™ FM® 1–43. The relative abundance of eDNA was determined by the ratio of dsDNA (anti-dsDNA labeling) to biomass (FilmTracer™).
Figure S4. Effect of exogenous addition of E. coli rHNS or E. coli rFis (Nucleoid Associated Proteins) on UPEC strain UTI89 biofilm formation.
Biofilm growth was initiated in the presence or absence of the indicated recombinant E. coli exogenous proteins (0.5 μM) for 40 hours. Images were analyzed by COMSTAT to calculate biofilm maximum height (A), average thickness (B) and biomass (C).
Table S1. Strains and plasmids