SUMMARY
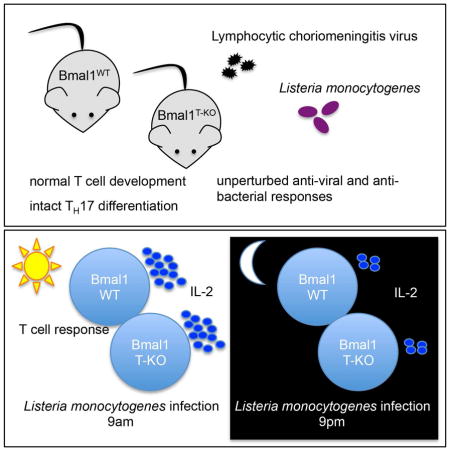
Circadian rhythms regulate many aspects of physiology ranging from sleep-wake cycles and metabolic parameters to susceptibility to infection. The molecular clock, with transcription factor BMAL1 at its core, controls both central and cell-intrinsic circadian rhythms. Using a circadian reporter we observed dynamic regulation of clock activity in lymphocytes. However, its disruption upon conditional Bmal1 ablation did not alter T or B cell differentiation or function. Although the magnitude of IL-2 production was affected by the time of bacterial infection, it was independent of cell-intrinsic expression of BMAL1. The circadian gating of the IL-2 response was preserved in Bmal1-deficient T cells despite a slight reduction in cytokine production in a competitive setting. Our results suggest that, contrary to the prevailing view, the adaptive immune response is not affected by the cell-intrinsic clock, but is likely influenced by cell-extrinsic circadian cues operating across multiple cell types.
Graphical Abstract
INTRODUCTION
Circadian clocks emerged during evolution to enable uni- and multicellular organisms to efficiently anticipate and adapt to environmental changes during day-night cycles (Bass, 2012; Bass and Takahashi, 2010). Circadian rhythms affect almost all aspects of physiology including daily activity patterns (Bunger et al., 2000), mood regulation (Chung et al., 2014), cell cycle control (Matsuo et al., 2003), metabolism (Turek et al., 2005), and immune responses (Curtis et al., 2014; Scheiermann et al., 2013). In mammals, the central clock is located in the suprachiasmatic nucleus of the brain. This central pacemaker integrates light cues and synchronizes peripheral clocks by various means including hormone secretion and neurotransmitter release by the sympathetic nervous system (Bell-Pedersen et al., 2005).
On the molecular level, the circadian clock consists of a network of interlocking transcriptional/translational feedback loops, which operate in a cell autonomous manner and are shared by the central and peripheral clocks. The transcription factors BMAL1 and CLOCK form the core of the central oscillatory loop. The BMAL1:CLOCK heterodimer binds E-box sites and drives expression of its own repressors Period (Per1-3), Cryptochrome (Cry1-2), and Rev-Erb (α and β) establishing 24h periodicity (Dibner et al., 2010; Koike et al., 2012).
The central role of BMAL1 in circadian biology is underscored by the complete loss of all rhythmic behavior in mice with germ-line Bmal1 deletion (Bunger et al., 2000). The importance of peripheral clocks operating in a given cell type has been established by cell-type specific deletion of a conditional Bmal1 allele (Storch et al., 2007). For example, circadian control of gene expression in epidermal stem cells has been implicated in the maintenance of a pool of dormant stem cells and prevention of premature epidermal ageing (Janich et al., 2011). Furthermore, genetic ablation of Bmal1 in pulmonary epithelial cells results in an altered inflammatory response to bacterial challenge, whereas adipocyte-specific deletion of Bmal1 has a strong impact on feeding behavior and body weight (Gibbs et al., 2014; Paschos et al., 2012).
Circadian effects on the immune system have been extensively documented (Curtis et al., 2014; Haus and Smolensky, 1999; Scheiermann et al., 2013). Early studies described a strong circadian effect on survival in animal models of sepsis (Halberg et al., 1960). In addition, the host response to pathogens like Salmonella and Listeria is affected by the time of infection. The circadian clock affects both the magnitude of the inflammatory response as well as the clearance of pathogenic bacteria (Bellet et al., 2013; Nguyen et al., 2013).
Functional circadian clocks have been described in cells of both the innate and adaptive immune system (Curtis et al., 2014; Scheiermann et al., 2013). Both B and T cells express the core components of the molecular clock machinery (Bollinger et al., 2011; Silver et al., 2012a). T cell proliferation and cytokine production show diurnal variation (Bollinger et al., 2011; Fortier et al., 2011). Furthermore, the adaptive response to vaccination has been described to be under circadian control. In this regard, in mice immunized with a TLR9 ligand-based vaccine, the magnitude of the adaptive immune response was dependent on the time of immunization (Silver et al., 2012b). Recently, a model for cell-intrinsic circadian regulation of TH17 differentiation has been proposed. In this model, the transcription factor NFIL3 was suggested to act as a repressor of a key driver of TH17 differentiation nuclear receptor RORγt. The diurnal expression of NFIL3 is regulated by the circadian network through direct repression by the BMAL1:CLOCK target REV-ERBα and could, therefore, result in circadian oscillation of RORγt-dependent TH17 cell generation (Yu et al., 2013).
The majority of previous studies, that have implicated the cell-intrinsic clock in different aspects of immune cell function, relied on the analysis of immune cell subsets isolated from mice with germ-line deficiency of a circadian oscillator. Thus, it remains unknown whether the observed circadian regulation of the adaptive immune response is affected by a cell-intrinsic clock or is due to indirect effects of clocks operating elsewhere.
We addressed this major outstanding question by employing T- and B cell- specific deletion of Bmal1 to characterize the cell-intrinsic requirement of the circadian clock in cells of the adaptive immune system. Although our analysis of a circadian reporter showed dynamic regulation of the molecular clock in T and B cells, it did not affect their differentiation and all the observed circadian effects on adaptive immunity were independent of cell-intrinsic expression of BMAL1. These results challenge the notion that lymphocyte-intrinsic clocks are required for adaptive immune responses and imply that circadian regulation of adaptive immunity is likely due to circadian molecular signals emanating from the central clock or peripheral clocks operating in other cell types.
RESULTS
Expression of a Circadian Reporter is Dynamically Regulated in Lymphocytes
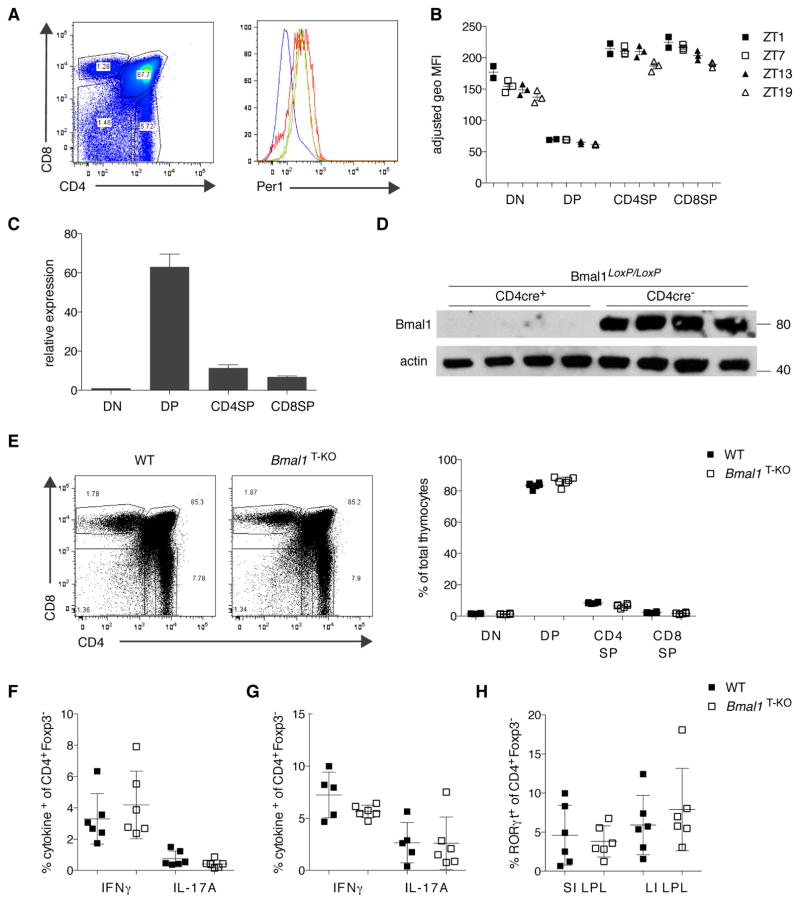
To explore the expression of the circadian clock in lymphocytes, we utilized a fluorescence-based circadian reporter, the Per1Venus mouse (Cheng et al., 2009; Janich et al., 2011). Per1 is a direct transcriptional target of the BMAL1:CLOCK heterodimer and serves by itself as a repressor of Bmal1. We analyzed reporter expression in lymphocyte subsets over a 24h period, sampling mice every six hours, starting at Zeitgeber (ZT) 1 (where ZT0 refers to lights on and ZT12 refers to lights off). We detected minor circadian regulation of Per1Venus expression in thymocytes, splenic T cells, and B cells, with levels peaking at ZT1 and dropping to their lowest point at ZT19 (Figure 1B and S1C, and data not shown). We also observed a pronounced variation in reporter expression, which coincided with progression between thymic developmental stages. We detected a sharp decline of PER1Venus expression at the DN to DP transition and a rebound as thymocytes progressed to the CD4SP and CD8SP stages, respectively (Figure 1A and 1B). To confirm that this dynamic expression of Per1 during thymic development was related to the circadian machinery, we FACS purified thymocyte subsets and assessed the expression of Bmal1 mRNA by quantitative PCR analysis (Figure 1C and Figure S3). As expected, Bmal1 expression peaked at the DP stage when PER1 levels were the lowest, since BMAL1 represents the positive limb of the clock and PER1 the negative limb. When we analyzed thymic development in even greater detail, we observed that the increase in PER1Venus levels coincided with positive selection, one of the major checkpoints in T cell development (Figure S1A).
Figure 1. Dynamic Regulation of Circadian Reporter Expression During Lymphocyte Development but T Cell-Specific Deletion of Bmal1 has no Effect on T Cell Development.
(A) PER1Venus expression in thymocytes was analyzed by flow cytometry. The gated populations in the left plot correspond to the histogram overlay in the right plot (DN: red line; DP: blue line; CD4SP: green line; CD8SP: orange line).
(B) Data as in (A) plotted for individual mice (n= 2–3 animals per time point) analyzed at four time points (ZT1, ZT7, ZT13, ZT19).
(C) Bmal1 expression analysis in FACS purified thymocyte populations from three individual mice. Expression is shown relative to the DN population.
(D) BMAL1 protein levels were assessed by Western blotting in lysates from total thymus of WT or Bmal1T-KO mice. Equal loading was confirmed by β-actin expression.
(E) Thymocyte development is unperturbed in Bmal1T-KO mice.
(F, G) IFNγ and IL-17A production in CD4+ T cells after αCD3/αCD28 stimulation in spleen (F) and large intestine LPL (G) is not affected by BMAL1-deficiency.
(H) Frequencies of RORγt expressing CD4+ T cells in small and large intestine LPL are unchanged in Bmal1T-KO mice.
(B) Error bars represent mean ± SEM. (C, E–H) Error bars represent mean ± SD. All data are representative of at least three individual experiments. See also Figure S1, 3.
Next we assessed whether this dynamic regulation of the circadian clock was observable during B cell development in the bone marrow. Analogous to thymocyte development, we detected dynamic regulation of PER1Venus levels during B cell development, with a sharp increase of reporter expression at the Hardy fraction C to C′ transition, which coincides with the pre-BCR checkpoint (Figure S1B)(Hardy et al., 1991). These data suggested a potential role for the molecular circadian clock in lymphocyte development. Alternatively, it was possible that the cell-intrinsic circadian clock, while operating in cells of the adaptive immune system, was inconsequential in functional terms.
Bmal1B-KO Mice Have Normal B Cell Development and Function
To assess if the circadian clock machinery is required for B cell development and function, we generated mice with B cell-specific deletion of Bmal1 (mb1CRE/Bmal1LoxP/LoxP, hereafter referred to as Bmal1B-KO). Deletion of BMAL1 in splenic B cells was confirmed by immunoblotting and quantitative PCR (Figure S2A and data not shown). Bmal1B-KO mice had normal cell numbers in the bone marrow, spleen and lymph nodes (Figure S2B). B cell development in the bone marrow was unperturbed (Figure S2C). When we compared the distribution of various B cell subsets in spleen, lymph nodes and peritoneal cavity, we did not observe any differences between wild type (WT) and Bmal1B-KO mice (Figure S2D and S2E, and data not shown). Bmal1-deficient B cells matured into germinal center B cells and plasma cells (Figure S2F) and underwent class-switch recombination and affinity maturation upon immunization with sheep red blood cells as efficiently as their wild-type counterparts (data not shown). Thus, our results suggest that B cell differentiation or function is unaffected by a cell-intrinsic circadian clock.
T Cell Differentiation and Function is Unperturbed in the Absence of BMAL1
Since we observed dynamic expression of components of the molecular clock during thymic differentiation, we generated mice with a T cell-specific deletion of Bmal1 (Cd4CRE/Bmal1LoxP/LoxP, hereafter referred to as Bmal1T-KO). Efficient deletion of Bmal1 was confirmed by quantitative PCR and immunoblotting for BMAL1 in total thymocytes (Figure S3A and 1D). We also assessed the effect of Bmal1 deletion on the expression of other core clock genes in developing thymocytes, which revealed disrupted expression of Rev-Erbβ and affected Clock, Per2 and Dbp mRNA expression (Figure S3). We analyzed T cell development in the thymus and did not observe a difference in frequency or numbers of developing thymocytes (Figure 1E and data not shown).
A recent study had proposed that the BMAL1:CLOCK heterodimer plays an important role in the differentiation of TH17 cells due to REV-ERBα-mediated repression of Nfil3, a newly identified repressor of RORγt, a key determinant of TH17 cell differentiation (Yu et al., 2013). Based on these findings, we expected to observe impaired TH1 and TH17 differentiation in Bmal1T-KO mice. However, when we analyzed the distribution of IFNγ- and IL-17A-producing T cells in the spleen and lamina propria of the large intestine, we observed similar numbers of IFNγ-producing TH1 and IL-17-producing TH17 cells in Bmal1T-KO and littermate control mice (Figure 1F and 1G). We also found comparable expression of FOXP3 and RORγt in CD4+ T cell subsets in the lamina propria of these mice (Figure 1H and data not shown).
In addition, we generated mice with a T cell-specific deletion of the Clock gene, and mice with a combined Bmal1 and Clock deficiency in T cells. Similar to the Bmal1T-KO mice, mice harboring CLOCK/BMAL1 double deficient and CLOCK single deficient T cells had no observable defects in thymocyte differentiation and generation of peripheral T cells. Moreover, the splenic and intestinal lamina propria CD4+ T cells from these mice produced comparable amounts of IFNγ and IL-17A (data not shown). These observations indicate that T cell-specific BMAL1 deficiency does not affect T cell differentiation and function and it cannot be compensated for by CLOCK expression.
Susceptibility to Experimental Autoimmune Encephalomyelitis is Unaffected by T Cell-Restricted Bmal1 Deficiency
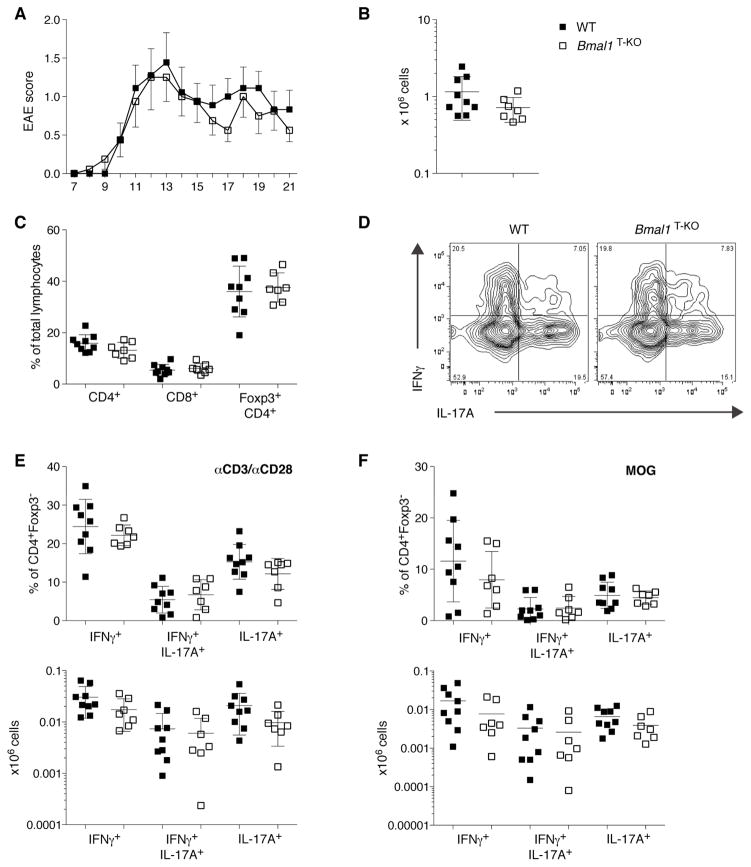
Even though we did not detect a defect in TH17 differentiation in unchallenged Bmal1T-KO mice, it remained possible that upon challenge generation of TH17 effector cells from Bmal1-deficient precursors were impaired. We tested the generation of pathogenic TH17 cells in experimental autoimmune encephalomyelitis (EAE), a mouse model of multiple sclerosis. Upon immunization with MOG peptide, both groups of mice started to show symptoms of ascending paralysis on day 8. Disease progressed comparably in Bmal1T-KO and littermate control WT mice (Figure 2A). Analysis of leukocyte infiltration in the central nervous system (CNS) at day 22 did not reveal any differences between the groups (Figure 2B and 2C). Moreover, IFNγ- and IL-17A- production by CD4+ T cells in the CNS was unaffected by T cell-specific ablation of Bmal1 (Figure 2D–F). Thus, we did not detect any defect in generation of TH1 or TH17 cells in Bmal1T-KO mice not only in the steady state, but also in an autoimmune setting.
Figure 2. T Cell-Specific BMAL1-Deficiency has no Impact on Experimental Autoimmune Encephalomyelitis Disease Susceptibility.
(A) Disease severity was scored on a daily basis starting at day 7 after immunization. Mean score ± SEM is plotted (WT = 9 mice; Bmal1T-KO = 7 mice). Mice were analyzed on d22 post immunization.
(B) Immune infiltration of the CNS (pooled brain and spinal chord) was assessed by counting CD45+ lymphocyte infiltrates at d22 post immunization.
(C) Frequencies of CNS-infiltrating CD4+, CD8+ and Treg cells are comparable between WT and Bmal1T-KO mice.
(D) Representative FACS plot of cytokine production of CD4+Foxp3− T cells infiltrating the CNS (αCD3/αCD28 stimulation).
(E, F) IFNγ- and IL-17A- production by CNS-derived T cells upon αCD3/αCD28 (E) or MOG peptide stimulation (F). Frequencies are represented in the upper panels, total numbers in the lower panels.
(B, C and E, F) Error bars represent mean ± SD. All data are representative of two individual experiments.
T Cells Show Only Minor Oscillation of the Molecular Circadian Clock
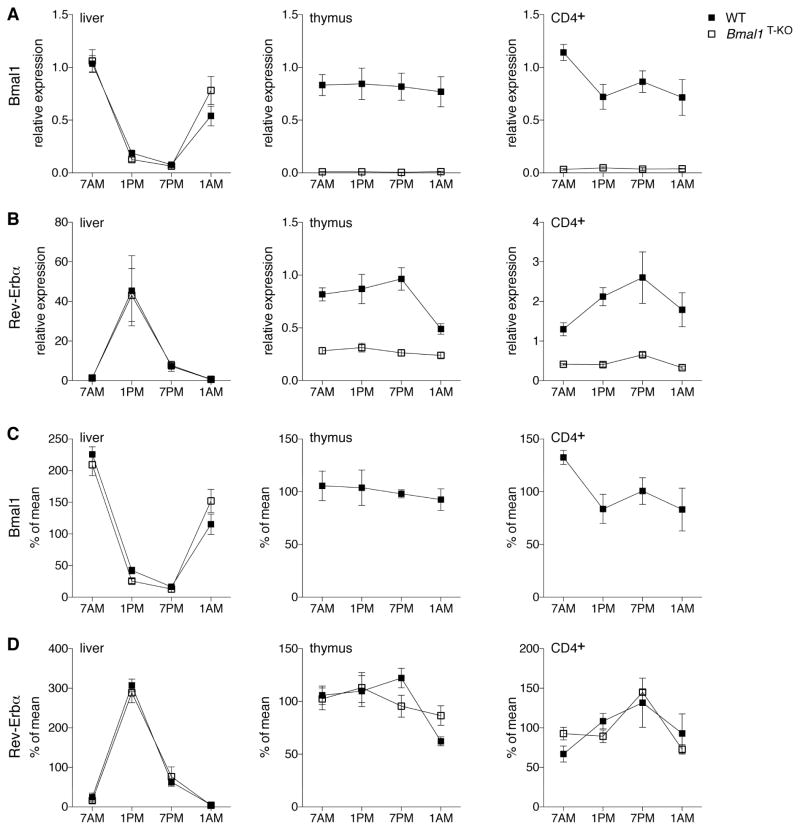
Since BMAL1 deficiency had no discernable impact on lymphocyte differentiation, we took a closer look at the molecular components of the circadian machinery in T lymphocytes and how they are impacted by the loss of BMAL1. We analyzed expression of the core components of the circadian clock in thymus and splenic CD4+ T cells over a 24h period in control and Bmal1T-KO animals. As a positive control, we analyzed time dependent modulation of clock regulated transcripts in the liver. As expected, we detected robust cycling of all clock-related transcripts in the liver (Figure 3A–D left panel, Figure S4). In contrast, analysis of the expression of the clock machinery in the thymus did not reveal robust oscillation of any of the transcripts. Nevertheless, BMAL1 deficiency significantly reduced expression of Rev-Erbα and Rev-Erbβ (Figure 3B and S4B). Splenic CD4+ T cells displayed minor oscillations of the Bmal1 and Rev-Erbα transcripts (Figure 3A, C and 3B, D). Similar to the thymus, the expression of Rev-Erbα and Rev-Erbβ was severely reduced in CD4+ T cells purified from Bmal1T-KO mice; however, the remaining Rev-Erbα transcript maintained its oscillation in a BMAL1-independent manner (Figure 3D). This minor oscillation of the Rev-Erbα transcript was maintained in the absence of BMAL1, likely due to extrinsic circadian factors.
Figure 3. Circadian Oscillation of the Molecular Clock in T Cells is Minor Compared to Liver.
Liver (left panel), thymus (middle panel) and CD4+ splenocytes (right panel) were harvested from control and Bmal1T-KO mice at the indicated time-points. RNA was processed, reverse transcribed and analyzed by quantitative PCR. Expression (2−ddCt) of Bmal1 (A) and Rev-Erbα (B) is shown relative to the WT 7AM time-point. Expression of Rev-Erbα is significantly lower in thymus and CD4+ splenocytes at all time-points analyzed. To visualize oscillation of the transcripts independent of gene expression, the data from (A) and (B) was calculated as % of mean for each individual mouse and time-point for Bmal1 (C) and Rev-Erbα (D). Data are comprised from 3–6 mice per group. Error bars represent mean ± SEM. See also Figure S4.
These data indicate that lymphocytes do not have a synchronized circadian clock. However, it remained possible that the clock could still be functional on the single cell level and become synchronized during an inflammatory challenge when a coordinated response might be beneficial.
Time of Infection Has a Minor Impact on the Acute Antiviral Response of BMAL1-sufficient and -deficient T Cells
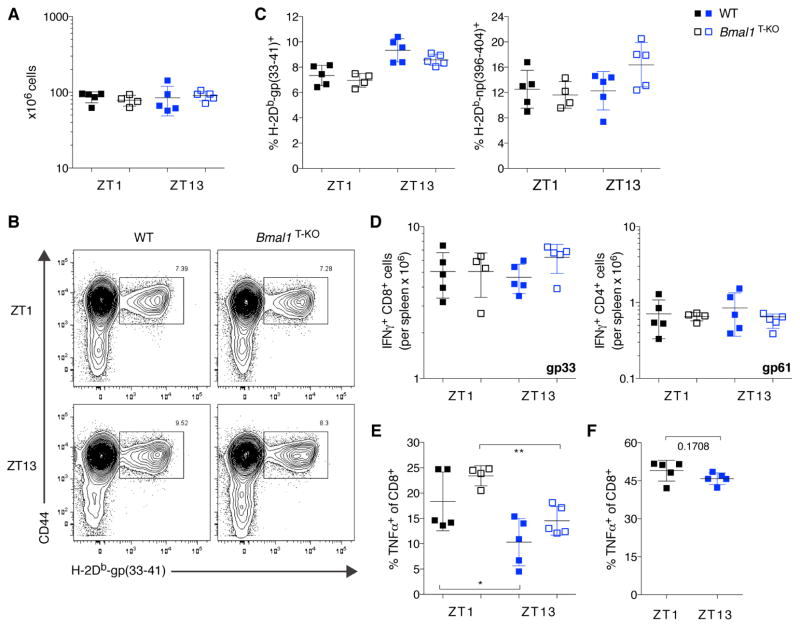
Therefore, we sought to compare T cell responses to acute viral infection in these mice. We infected WT and Bmal1T-KO mice with lymphocytic choriomeningitis virus (LCMV) either at ZT1 or at ZT13 and analyzed the animals at the peak of infection. Splenic cellularity was comparable between the experimental groups and was independent of time of infection (Figure 4A). We assessed the virus-specific CD8+ T cell responses by tetramer staining and flow cytometric analysis (Figure 4B). The magnitude of the antiviral response was comparable for all experimental groups for both the gp33 and the np396 epitopes (Figure 4C). Cytokine production by virus-specific T cells was characterized by ex vivo re-stimulation with virus-specific peptides. No differences were observed in IFNγ, TNFα or IL-2 production between the experimental groups (Figure 4D and data not shown). Polyclonal stimulation with PMA and ionomycin revealed a slight, but statistically significant decrease in TNFα-producing CD8+ T cells when mice had been infected at ZT13 as compared to ZT1 (Figure 4E). This decrease was dependent on the time of infection, but independent of Bmal1 expression in T cells. This circadian time-dependent reduction in TNFα production was not observed in uninfected animals (Figure 4F). No such effects were observed for IFNγ or IL-2 production by T cells (Figure S5A–C).
Figure 4. Circadian Time of Infection has a Minor Effect on Acute Antiviral T cell Response.
(A) Total splenocyte counts on d7 post LCMV-Armstrong infection at ZT1 and ZT13 in WT or Bmal1T-KO mice.
(B) Representative FACS plots of the antiviral CD8+ T cell response assessed by gp33-specific tetramer staining at ZT1 and ZT13.
(C) The magnitude of the virus-specific CD8+ T cell response is comparable between WT and Bmal1T-KO cells and not affected by time of infection.
(D) Virus-specific IFNγ-production by CD8+ (left panel) and CD4+ T cells (right panel) is comparable in all groups analyzed.
(E) The time of infection impacts the CD8+ T cell specific TNFα response independent of BMAL1-expression in T cells. *p<0.05, **p<0.01; significantly different from ZT1 as per unpaired t-test.
(F) The CD8+ T cell specific TNFα response in uninfected mice is not affected by circadian time of analysis.
(A and C–F) Error bars represent mean ± SD. All data are representative of at least two individual experiments. See also Figure S5.
Circadian Effects on IL-2 Production during Listeria Infection are independent of Bmal1 Expression in T cells
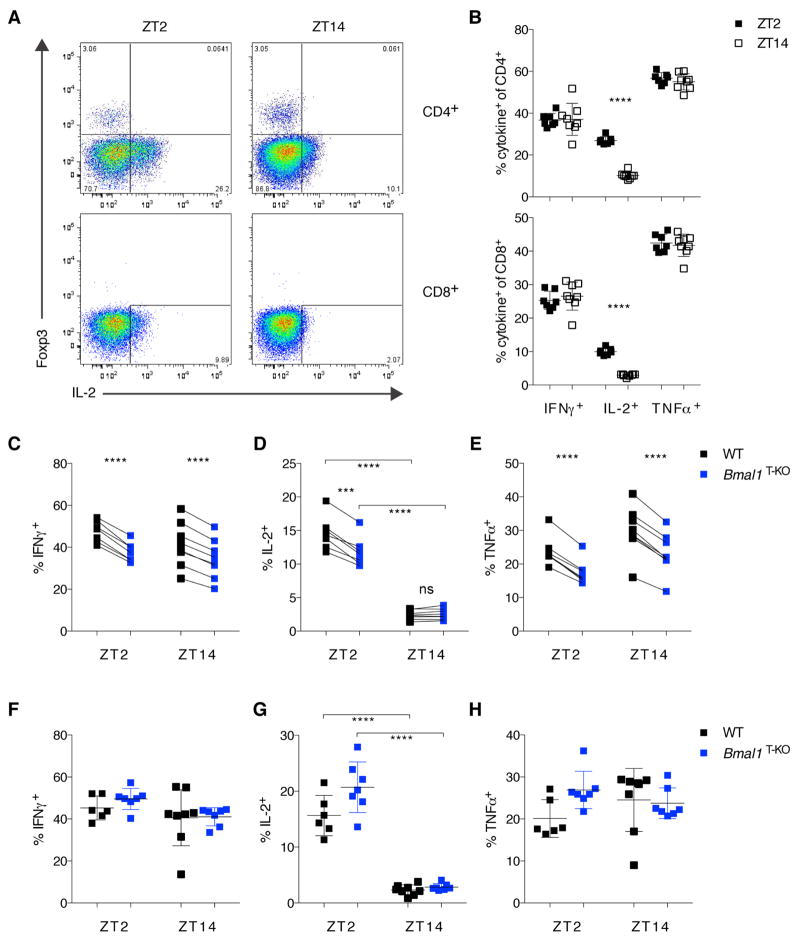
To further explore a role for cell-intrinsic clock deficiency in clonal expansion and effector function of antigen-specific CD8+ T cells, we generated OTI TCR transgenic Bmal1T-KO mice. We confirmed efficient Bmal1-deletion by quantitative PCR. BMAL1-deficient OTI cells displayed deregulated expression of the circadian clock genes Per2, Rev-Erbα and Rev-Erbβ (Figure S6A). To assess function of these cells in response to a bacterial pathogen, we co-transferred highly purified naïve BMAL1-sufficient and -deficient OTI CD8+ T cells into congenically marked recipients and infected them with a L. monocytogenes strain expressing OVA (Lm-OVA) at ZT2 and ZT14. All animals were analyzed seven days later (Figure S6B). This experimental setup allowed a pairwise comparison of OTI cells with and without a cell-intrinsic circadian clock in the same environment. At the same time, we were also able to study the effect of time of infection on the host response.
When we analyzed the effect of time of infection on the host CD4+ and CD8+ T cell responses, we noticed a striking difference in IL-2 secretion by both cell types upon in vitro re-stimulation. The frequency of IL-2 producers was more than two-fold higher at ZT2 when compared to ZT14 (Figure 5A and B). Re-stimulation with PMA and ionomycin or with OVA and LLO peptides yielded similar results (Figure S7A–D). In contrast to IL-2, IFNγ and TNFα production by T cells was not affected by time of infection (Figure 5B). Analysis of cytokine production by the transferred OTI cells in response to re-stimulation with OVA peptide, revealed a slight competitive disadvantage for BMAL1-deficient OTI cells when compared to BMAL1-sufficient cells in the same host in two out of three experiments (Figure 5C–E). In a third experiment, there was no competitive disadvantage between Bmal1-deficient OTI and control cells, suggesting that the effect of Bmal1 deficiency is at best very minor and, if observable, is limited to an experimental competitive setting. Importantly, the circadian gating of the IL-2 response was independent of BMAL1 in these experiments in agreement with the analysis of polyclonal T cell responses to LCMV infection (Figure 5D).
Figure 5. L. monocytogenes-Elicited IL-2 Production is Sensitive to Circadian Time of Infection but Independent of Bmal1 Expression in T Cells.
(A) Representative FACS plots of IL-2 production by CD4+ (upper panels) and CD8+ T cells (lower panels) at d7 post Lm-OVA infection at ZT2 versus ZT14.
(B) Summary of data from (A). Cytokine response was measured after αCD3/αCD28 stimulation ex vivo. (Upper panel: CD4+ T cells; lower panel: CD8+ T cells).
(C–D) Paired analysis of co-transferred WT (black squares) or Bmal1T-KO OTI cells (blue squares) at d7 post Lm-OVA infection after re-stimulation with OVA-peptide. (C, E) Paired comparison of co-transferred cells showed significant differences in IFNγ and TNFα production (paired t-test). (D) Pairwise comparison revealed significant differences in IL-2-production at ZT2 (paired t-test), as well as significant differences in IL-2 production at ZT2 versus ZT14 (unpaired t-test).
(F–H) Single-transfer of WT or Bmal1T-KO OTI cells were analyzed at d7 post Lm-OVA infection for IFNγ (F), IL-2 (G), and TNFα production (H). IL-2 production was significantly different at ZT2 versus ZT14 (unpaired t-test).
(B–H) Error bars represent mean ± SD. All data are representative of at least two individual experiments. ***p<0.001, ****p<0.0001. See also Figures S6–7.
We also analyzed single-transfer of WT and Bmal1T-KO OTI cells into congenically marked hosts followed by Lm-OVA infection at ZT2 and ZT14. In this experimental setting, we also observed the circadian gating of the IL-2 response (Figure 5G); no differences in IFNγ or TNFα production between WT and Bmal1-deficient OTI cells were detected (Figure 5F–H). These studies showed that the cell-intrinsic circadian clock does not affect generation of effector T cells and their responses to pathogens and that circadian gating of IL-2 production is likely a consequence of diurnal oscillations of clock activity in other immune or non-immune cell types.
DISCUSSION
The circadian oscillator emerged in unicellular organisms to enable anticipation of environmental changes within a 24-hour light-dark cycle and their coordination with metabolic processes. In mammals, daily rhythms are driven by signals emanating from a circadian pacemaker in the suprachiasmatic nucleus (SCN) of the hypothalamus. Synchronization of the SCN neurons, which is a prerequisite for efficient entrainment by 24h-light-dark cycles, is achieved through robust intercellular coupling partially through neurotransmitters (Aton and Herzog, 2005; Gonze et al., 2005; Yamaguchi et al., 2003). This central pacemaker entrains ubiquitously expressed peripheral clocks operating across different cell types (Dibner et al., 2010; Mohawk et al., 2012; Reppert and Weaver, 2002).
The pervasive circadian control of diverse biological processes in mammals includes immunity to infections and inflammation (Curtis et al., 2014; Scheiermann et al., 2013). This regulation can be executed by both the central clock and the peripheral clocks. The molecular components of the circadian oscillators are expressed in primary and secondary lymphoid tissues including thymus, spleen and lymph nodes indicative of the presence of functional peripheral clocks (Bollinger et al., 2011; Keller et al., 2009). Indeed, using a Per1Venus fluorescent clock reporter, we found that cells of the adaptive and innate immune system express functional circadian clock machinery in agreement with previous reports by others. Consistent with these observations, selective deletion of a Bmal1 conditional allele in myeloid cells revealed a cell intrinsic requirement for the circadian clock in inflammatory monocyte maintenance patterns. Disruption of the clock abolished diurnal oscillation of Ly6chi monocytes and led to exacerbated tissue damage in acute and chronic inflammation (Nguyen et al., 2013). These observations raised the possibility that circadian regulation of immunity involves the integration of signals from the central clock, peripheral clocks operating in innate and adaptive immune cell types as well as non-immune cells.
Circadian control of antigen-specific immune responses has been suggested by several studies. Time of inoculation with a TLR9 ligand-based vaccine had an impact on the magnitude of the adaptive response (Silver et al., 2012b). Similarly, the timing of immunization with peptide-loaded dendritic cells had a significant impact on the magnitude of the T cell response (Fortier et al., 2011). Although these studies have not directly explored the contribution of extrinsic vs. intrinsic clocks in immune cells to the diurnal modulation of T cell responses, they implied that the observed effects are dependent on the T cell-intrinsic clock in agreement with the aforementioned notion.
However, it is also possible that the functional coupling of the cell intrinsic clock in cells of the adaptive immune system, i.e. T and B lymphocytes, to the central clock or to peripheral clocks operating elsewhere is dispensable. For example, it was long considered that clocks operating in different plant tissues are uncoupled or only weakly coupled (Thain et al., 2000; Wenden et al., 2012) although a very recent study revealed asymmetric regulation of clocks in the vasculature and mesophyll tissue in Arabidopsis (Endo et al., 2014).
To address these questions and to explore potential roles of cell intrinsic circadian clocks in cells of the adaptive immune system, we generated mice with B- and T-cell specific deletions of a conditional Bmal1 allele, respectively. Even though we observed pronounced regulation of the molecular clock activity during B cell and T cell differentiation, cell-type specific deletion of BMAL1, the core transcription factor of the clock, did not perturb B lymphocyte maturation in the bone marrow and had no effect on T cell differentiation in the thymus. Consistent with these observations, peripheral subsets of T and B cells were unaffected by BMAL1 deficiency. Furthermore, T cells exhibited only minor circadian oscillation of the clock-dependent transcripts despite expression of all components of the molecular clock in a BMAL1-dependent fashion. We also observed that immune responses to both viral and bacterial infections were indistinguishable in mice with a T cell-specific deletion of Bmal1. During infection we found comparable expansion and IFNγ production by BMAL1-sufficient and -deficient CD4 and CD8 T cells specific for LCMV and Listeria monocytogenes antigens. In addition, differentiation of virus-specific memory T cells was also unaltered (data not shown). Furthermore, we found comparable antigen-specific antibody responses in mice with a B cell-specific deletion of Bmal1. These findings suggest that the cell intrinsic circadian clock is dispensable for normal T and B lymphocyte function.
Recently, it has been proposed that TH17 cell differentiation in the gut is subject to circadian regulation (Yu et al., 2013). This study suggested that transcription factor NFIL3 directly represses transcription of Rorc1, whose product - nuclear receptor RORγt - serves as a lineage specification factor of TH17 cells. NFIL3 itself is a part of the circadian clock and NFIL3 diurnal oscillation is regulated through its direct repression by the BMAL1:CLOCK target REV-ERBα. NFIL3-deficient mice were shown to exhibit elevated splenic and small intestine lamina propria TH17 cell numbers and NFIL3-deficient T cells transfer of naïve T cells into Rag1-deficient mice revealed enhanced differentiation of NFIL3-deficient T cells into TH17 cells (Yu et al., 2013). However, we found that T cell-specific BMAL1 deficiency had no effect on TH17 differentiation in the small intestine, the main site of TH17 generation in unchallenged mice, and in a setting of TH17-mediated autoimmune inflammation in the CNS, i.e. EAE induced upon MOG peptide immunization. These results suggest that TH17 differentiation is independent of the cell-intrinsic clock activity in T cells and that the reported repression of TH17 differentiation by NFIL3 is most likely independent of its diurnal regulation.
Several studies have described a diurnal variation in inflammation-induced tissue pathology, with one aforementioned study showing a clear requirement for the cell intrinsic clock in myeloid cells (Bellet et al., 2013; Halberg et al., 1960; Nguyen et al., 2013). Thus, it was possible that although we did not detect a noticeable impact of the cell intrinsic clock in T and B lymphocytes, at least some lymphocyte responses to viral or bacterial pathogens might reveal circadian pattern due to clocks operating elsewhere. Indeed, our analysis of mice infected at different times of day revealed circadian gating of T cell mediated cytokine production. During LCMV infection, we observed a reproducible decrease in TNFα production by CD8+ T cells on d7 post infection when mice had been infected at ZT13 in comparison to ZT1 infection. Upon infection with Listeria monocytogenes, we also observed circadian gating of the IL-2 response in CD4 and in CD8 T cells. However, in both instances, the observed diurnal variation of cytokine production was independent of the T cell intrinsic expression of Bmal1. These results imply that an apparent diurnal pattern of some T cell responses is due to their integration into circadian oscillation by signals emanating from the central clock, like glucocorticoids, or from peripheral clocks operating in innate immune cells or non-immune cell types.
Thus, our findings support a model where the intrinsic clock in myeloid cells plays a prominent role in innate immunity likely because these responses take effect within minutes to hours after pathogen encounter, a time scale comparable to diurnal clock oscillations. These innate responses afford essential protection of the host upon pathogen encounter, which most likely occur during the active phase regulated by the central clock. In contrast, cells of the adaptive immune system operate on a much longer time scale. In this regard, thymic T cell maturation takes days to weeks (Yates, 2014), whereas differentiation of naïve T cells into pathogen-specific effector cells occurs on a scale of days (Iezzi et al., 1998). Therefore, it seems fitting that the adaptive responses of T and B cells are unaffected by their cell intrinsic clocks, but rather can be impacted by cell extrinsic diurnal signals affecting the numbers or activation status of antigen-presenting cells or soluble factors like cytokines and hormones.
EXPERIMENTAL PROCEDURES
Animals
Mice were housed under SPF conditions at Memorial Sloan Kettering Cancer Center in accordance with institutional guidelines. Unless otherwise noted, 6–10 week-old sex-matched mice were used for all experiments. Bmal1LoxP/LoxP, Cd4CRE, OTI TCR transgenic, and Rag2KO mice have been described and were purchased from Jackson Laboratories (Hao and Rajewsky, 2001; Hogquist et al., 1994; Lee et al., 2001; Storch et al., 2007). A. Tarakhovsky provided mb1CRE mice and K. Obrietan provided Per1VENUS mice (Cheng et al., 2009; Hobeika et al., 2006).
Lymphocyte Preparation
Single cell suspensions of spleen, lymph nodes, thymus, and Peyer’s patches were prepared by disruption of the tissue between glass slides and filtration through a 100μm membrane. Lymphocyte isolation from tissues was performed as previously described (Feng et al., 2014).
Flow Cytometry
The following fluorophore-conjugated antibodies were used: CD4 (RM4-5), CD8 (5H10; 53-6.7), CD45.1 (A20), CD45.2 (104), TCRVα2 (B20.1), TCRVβ5 (MR9-4), CD62L (MEL-14), CD44 (IM7), TCRβ (H57-597), B220 (RA3-6B2), CD19 (MB19-1; 1D3), IgM (11/41), IgD (11-26c), CD43 (1B11), Klrg1 (2F1), CD127 (A7R34), Foxp3 (FJK-16s), IL-17A (eBio17B7), IFNγ (XMG1.2), TNFα (MP6-XT22), IL-2 (JES6-5H4), and RORγt (Q31-378) (eBioscience, Life Technologies, BioLegend, BD Biosciences, Tonbo). MHC class I monomers of H-2Db complexed with NP396–404 and GP33–41 (NIH Tetramer Core) were tetramerized with PE-conjugated streptavidin (Molecular Probes). All intracellular staining was performed using eBioscience Fixation/Permeabilization buffers. All samples were stained with LIVE/DEAD Fixable Yellow Dead Cell Stain (Molecular Probes) to exclude dead cells. Samples were analyzed on a LSRII flow cytometer (BD Bioscience) and data was processed with FlowJo software (TreeStar). To analyze cytokine production, single cell suspensions were stimulated for 4–5h at 37C with soluble anti-CD3 (5μg/ml; 2C11; BioXCell) and anti-CD28 (5μg/ml; 37.51; BioXCell), or phorbol 12-myristate 13-acetate (PMA, 50ng/ml) and ionomycin (500ng/ml), or specific peptides (described later; 1μg/ml) in the presence of 1μg/ml brefeldin A (Sigma).
Quantitative PCR
To perform RT-qPCR, total RNA was prepared form FACS sorted thymocytes or naïve OTI cells using the TRIzol reagent (Life Technologies). The RNA was reverse transcribed into cDNA with qScript cDNA SuperMix (Quantas Biosciences, Inc.). Quantitative PCR was performed on an ABI PRISM7700 cycler (Applied Biosystems) using Power SYBR Green PCR master mix (Applied Biosystems) with the following primers (F, forward; R, reverse): Bmal1: F, GTGCTAAGGATGGCTGTTCA and R, CGGTCACATCCTACGACAAA; Clock: F, CCTGGTAACGCGAGAAAGAT and R, CGAATCTCACTAGCATCTGACTGT; RevErbβ : F, GGAGTTCATGCTTGTGAAGGCTGT and R, CAGACACTTCTTAAAGCGGCACTG; Hprt (Sandler et al., 2003); Per1, Per2, RevErbα, and Dbp (Hayashi et al., 2007).
LCMV Infection, Listeria monocytogenes Infection, and EAE Induction
Mice were infected intraperitoneally (i.p.) with 2×105 PFU LCMV Armstrong at ZT 1 or ZT13 and analyzed on day 7 after infection. Anti-viral T cell responses were monitored by measuring cytokine producing cells after re-stimulation with virus-derived peptides [GP33–41(KAVYNFATM), GP276–286(SGVENPGGYCL), GP61–80 (GLKGPDIYKGVYQFKSVEFD), NP396–404(FQPQNGQFI), NP309–328(SGEGWPYIACRTSVVGRAWE)].
Mice were infected intravenously (i.v.) with 5×104 to 1×105 CFU of Lm-OVA at ZT2 or ZT14 and analyzed on day 7 post infection. The pathogen-specific cytokine response was assessed by re-stimulation with OVA257–264 (SIINFEKL) or LLO190–201 (NEKYAQAYPNVS) peptides. 1×105 naïve OTI cells were transferred (i.v.) into congenically marked recipients at least seven hours prior to infection. OTI cells were enriched from pooled spleen and lymph nodes using the Dynabeads FlowComp mouse CD8 T cell kit (Life Technologies) and sort-purified on a FACS Aria II Cell Sorter (BD Bioscience).
EAE was induced by immunization with 0.1mg myelin oligodendrocyte protein-derived peptide 35–55 (MOG35–55, MEVGWYRSPFSRVVHLYRNGK, GenScript) emulsified in Complete Freund’s Adjuvant (Sigma) sub-cutaneously (s.c.), followed by two doses of 150ng pertussis toxin (List Biological Laboratories) i.p. on day 2 and day 4 post immunization. Mice were monitored for signs of disease as previously described. Animals were analyzed on day 21 post immunization (Stromnes and Goverman, 2006).
Highlights.
The cell-intrinsic circadian clock is dispensable for T and B cell development.
TH17 differentiation is normal in mice with a T cell-specific deletion of Bmal1.
Circadian gating of IL-2 secretion is independent of the cell-intrinsic clock.
Acknowledgments
We thank Karl Obrietan (Ohio State University) for provision of the Per1Venus mouse strain and Alexander Tarakhovsky for provision of the mb1CRE mice. We thank Yongqiang Feng, Takatoshi Chinen, Georg Gasteiger, Andrew Levine, and Bruno Moltedo for provision of reagents, technical assistance, and helpful discussion. This study was supported by NIH grant AI034206 and the Howard Hughes Medical Institute (A.Y.R.).
Footnotes
AUTHOR CONTRIBUTIONS
S.H. and A.Y.R conceived of and designed the experiments, interpreted the results, and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aton SJ, Herzog ED. Come together, right…now: synchronization of rhythms in a mammalian circadian clock. Neuron. 2005;48:531–534. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet MM, Deriu E, Liu JZ, Grimaldi B, Blaschitz C, Zeller M, Edwards RA, Sahar S, Dandekar S, Baldi P, et al. Circadian clock regulates the host response to Salmonella. Proc Natl Acad Sci U S A. 2013;110:9897–9902. doi: 10.1073/pnas.1120636110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger T, Leutz A, Leliavski A, Skrum L, Kovac J, Bonacina L, Benedict C, Lange T, Westermann J, Oster H, Solbach W. Circadian clocks in mouse and human CD4+ T cells. PLoS One. 2011;6:e29801. doi: 10.1371/journal.pone.0029801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Alvarez-Saavedra M, Dziema H, Choi YS, Li A, Obrietan K. Segregation of expression of mPeriod gene homologs in neurons and glia: possible divergent roles of mPeriod1 and mPeriod2 in the brain. Hum Mol Genet. 2009;18:3110–3124. doi: 10.1093/hmg/ddp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Lee EJ, Yun S, Choe HK, Park SB, Son HJ, Kim KS, Dluzen DE, Lee I, Hwang O, et al. Impact of circadian nuclear receptor REV-ERBalpha on midbrain dopamine production and mood regulation. Cell. 2014;157:858–868. doi: 10.1016/j.cell.2014.03.039. [DOI] [PubMed] [Google Scholar]
- Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LA. Circadian clock proteins and immunity. Immunity. 2014;40:178–186. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dickmeis T. Glucocorticoids and the circadian clock. J Endocrinol. 2009;200:3–22. doi: 10.1677/JOE-08-0415. [DOI] [PubMed] [Google Scholar]
- Endo M, Shimizu H, Nohales MA, Araki T, Kay SA. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature. 2014;515:419–422. doi: 10.1038/nature13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, Rudensky AY. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 2014;158:749–763. doi: 10.1016/j.cell.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier EE, Rooney J, Dardente H, Hardy MP, Labrecque N, Cermakian N. Circadian variation of the response of T cells to antigen. J Immunol. 2011;187:6291–6300. doi: 10.4049/jimmunol.1004030. [DOI] [PubMed] [Google Scholar]
- Gibbs J, Ince L, Matthews L, Mei J, Bell T, Yang N, Saer B, Begley N, Poolman T, Pariollaud M, et al. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med. 2014;20:919–926. doi: 10.1038/nm.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonze D, Bernard S, Waltermann C, Kramer A, Herzel H. Spontaneous synchronization of coupled circadian oscillators. Biophys J. 2005;89:120–129. doi: 10.1529/biophysj.104.058388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halberg F, Johnson EA, Brown BW, Bittner JJ. Susceptibility rhythm to E. coli endotoxin and bioassay. Proc Soc Exp Biol Med. 1960;103:142–144. doi: 10.3181/00379727-103-25439. [DOI] [PubMed] [Google Scholar]
- Hao Z, Rajewsky K. Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J Exp Med. 2001;194:1151–1164. doi: 10.1084/jem.194.8.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haus E, Smolensky MH. Biologic rhythms in the immune system. Chronobiol Int. 1999;16:581–622. doi: 10.3109/07420529908998730. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Shimba S, Tezuka M. Characterization of the molecular clock in mouse peritoneal macrophages. Biol Pharm Bull. 2007;30:621–626. doi: 10.1248/bpb.30.621. [DOI] [PubMed] [Google Scholar]
- Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, Reth M. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Iezzi G, Karjalainen K, Lanzavecchia A. The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity. 1998;8:89–95. doi: 10.1016/s1074-7613(00)80461-6. [DOI] [PubMed] [Google Scholar]
- Janich P, Pascual G, Merlos-Suarez A, Batlle E, Ripperger J, Albrecht U, Cheng HY, Obrietan K, Di Croce L, Benitah SA. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, Kramer A, Maier B. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci U S A. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341:1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Sandler NG, Mentink-Kane MM, Cheever AW, Wynn TA. Global gene expression profiles during acute pathogen-induced pulmonary inflammation reveal divergent roles for Th1 and Th2 responses in tissue repair. J Immunol. 2003;171:3655–3667. doi: 10.4049/jimmunol.171.7.3655. [DOI] [PubMed] [Google Scholar]
- Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol. 2013;13:190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver AC, Arjona A, Hughes ME, Nitabach MN, Fikrig E. Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav Immun. 2012a;26:407–413. doi: 10.1016/j.bbi.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 2012b;36:251–261. doi: 10.1016/j.immuni.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, Weitz CJ. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromnes IM, Goverman JM. Active induction of experimental allergic encephalomyelitis. Nat Protoc. 2006;1:1810–1819. doi: 10.1038/nprot.2006.285. [DOI] [PubMed] [Google Scholar]
- Thain SC, Hall A, Millar AJ. Functional independence of circadian clocks that regulate plant gene expression. Curr Biol. 2000;10:951–956. doi: 10.1016/s0960-9822(00)00630-8. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenden B, Toner DL, Hodge SK, Grima R, Millar AJ. Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proc Natl Acad Sci U S A. 2012;109:6757–6762. doi: 10.1073/pnas.1118814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yates AJ. Theories and quantification of thymic selection. Front Immunol. 2014;5:13. doi: 10.3389/fimmu.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, Rothman PB, Takahashi JS, Hooper LV. TH17 cell differentiation is regulated by the circadian clock. Science. 2013;342:727–730. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]