SUMMARY
The CREB-regulated transcription coactivator CRTC2 stimulates CREB target gene expression and has a well-established role in modulating glucose and lipid metabolism. Here we find, unexpectedly, that loss of CRTC2, as well as CREB1 and its coactivator CREB-binding protein (CBP), results in a deficiency in DNA mismatch repair (MMR) and a resultant increased mutation frequency. We show that CRTC2, CREB1 and CBP are transcriptional activators of well-established MMR genes, including EXO1, MSH6, PMS1 and POLD2. Mining of expression profiling databases and analysis of patient samples reveal that CRTC2 and its target MMR genes are down-regulated in specific T-cell lymphoma subtypes, which are microsatellite unstable. The levels of acetylated histone H3 on the CRTC2 promoter are significantly reduced in lymphoma compared to normal tissue, explaining the decreased CRTC2 expression. Our results establish a role for CRTC2 as a lymphoma tumor suppressor gene that preserves genome integrity by stimulating transcription of MMR genes.
INTRODUCTION
In response to a variety of extracellular signals, the transcription factor CREB regulates diverse cellular responses by modulating the expression of genes containing a cAMP-response element (CRE) (Shaywitz and Greenberg, 1999). CREB works in concert with its coactivator, CREB-binding protein (CBP; also called CREBBP), and, depending on cellular context, with a family of coactivators called CREB-regulated transcription coactivators (CRTCs) (Altarejos and Montminy, 2011). Of the CRTC family members, CRTC2 is thought to be the major mediator of CREB target gene expression (Conkright et al., 2003).
DNA mismatch repair (MMR) is an evolutionarily conserved process that is responsible for recognizing and repairing single-base mismatches and small insertion/deletion loops that are formed during DNA replication (Jiricny, 2006). Cells deficient for MMR have higher mutation rates relative to normal cells. Consequently, genetic and epigenetic modifications that impair the expression of genes required for MMR, especially MSH2, MSH6, and MLH1, cause susceptibility to certain types of cancer, and have a so-called mutator phenotype that can be detected in eukaryotic cells as microsatellite instability (MSI) (Kolodner, 1996; Modrich and Lahue, 1996). Mutations in MMR genes are frequently found in MSI-positive hereditary nonpolyposis colorectal cancer (Peltomaki, 2003). A significant fraction of non-hereditary colorectal cancers are also MSI-positive but do not carry a detectable mutation or epigenetic modification of known MMR genes (Peltomaki, 2003), suggesting that in these cases other molecular aberrations are the basis of MSI. Here we report that CRTC2 plays an important role in maintaining genome integrity, and functions by promoting transcription of MMR genes.
RESULTS AND DISCUSSION
Loss of CRTC2 Results in Defective MMR
Several previous findings prompted us to analyze the possible role of CRTC2 as a tumor suppressor that functions in MMR. First, mining of the COSMIC (Catalogue of Somatic Mutations in Cancer) database (Forbes et al., 2011) revealed that CRTC2 is recurrently mutated in several cancer types (Table S1). Second, a search of the Oncomine cancer profiling database (Rhodes et al., 2007) revealed that CRTC2 is down-regulated in multiple cancers (Figure S1). Third, analysis of a previous genome-wide occupancy study (Zhang et al., 2005) revealed that CREB1 was bound to several genes with well-established roles in MMR (see below), raising the possibility that CRTC2 may also bind to and regulate MMR gene expression.
To investigate the possible role of CRTC2 in MMR, we constructed CRTC2 knockout (KO) cell lines using a CRISPR/Cas9 genome editing strategy. The experiments were performed in HeLa cells, a commonly used MMR proficient, microsatellite stable cell line. Immunoblot analysis shows that CRTC2 was undetectable in two independently derived CRTC2 KO HeLa clones (Figure 1A), and sequencing confirmed that both alleles in each cell line were disrupted (Figure S2A).
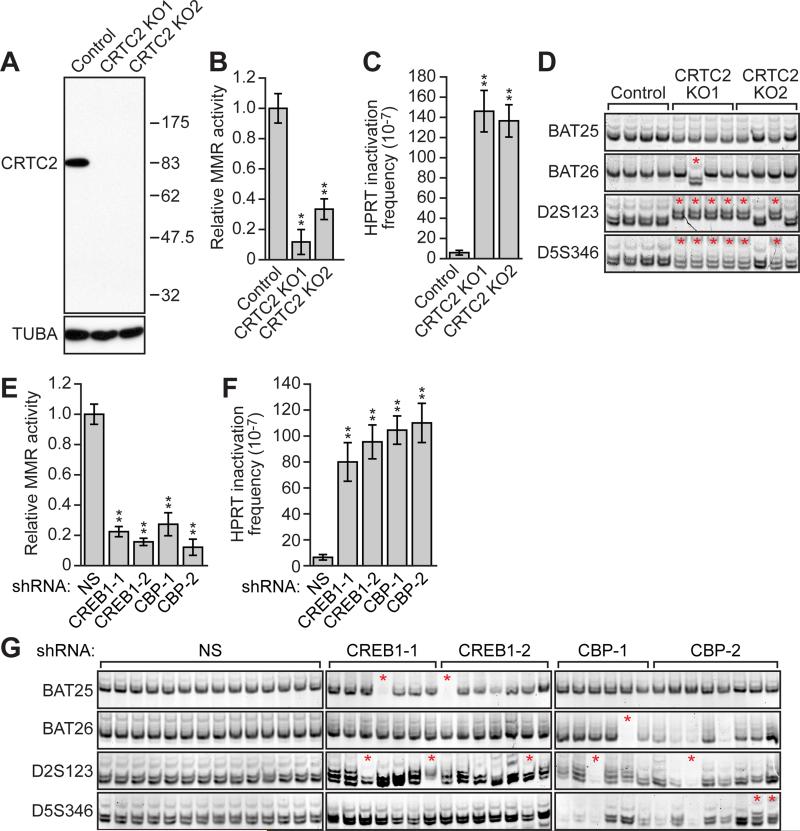
Figure 1. Loss of CRTC2, CREB1 or CBP Results in Defective MMR and a Mutator Phenotype.
(A) Immunoblot analysis monitoring CRTC2 levels in control or CRTC2 KO HeLa cells. α-tubulin (TUBA) was monitored as a loading control.
(B,C) MMR activity assay (B) and spontaneous mutation frequency (C) in control or CRTC2 KO HeLa cells.
(D) MSI analysis. Control HeLa cells and CRTC2 KO HeLa subclones were analyzed for MSI at BAT25, BAT26, D2A123 and D5S346. Red asterisks indicate new microsatellite species or microsatellite deletions.
(E,F) MMR activity assay (E) and spontaneous mutation frequency (F) in HeLa cells expressing a non-silencing (NS), CREB1 or CBP shRNA. For all graphs, error bars indicate SD. **P<0.01.
(G) MSI analysis in HeLa cells expressing a NS, CREB1 or CBP shRNA.
See also Figures S1 and S2.
To determine whether CRTC2 KO HeLa cells had reduced MMR activity, we performed an in vivo MMR activity assay that monitored repair of a single-base mismatch in an enhanced green fluorescent protein (EGFP) reporter gene (Zhou et al., 2009). Cells that are proficient for MMR correctly repair the mismatch and therefore express EGFP. The results show that CRTC2 KO HeLa cells had greatly reduced MMR activity compared to control HeLa cells (Figures 1B and S2B). Ectopic expression of CRTC2 in CRTC2 KO HeLa cells largely restored MMR activity (Figures S2C and S2D), ruling out the possibility that the reduced MMR activity we observed was due to an off-target effect.
To determine whether MMR-defective CRTC2 KO HeLa cells had a mutator phenotype, we measured the spontaneous mutation frequency of the hypoxanthine guanine phosphoribosyltransferase (HPRT) gene. The results show that the spontaneous mutation frequency in CRTC2 KO HeLa cells was ~25-fold higher than in control HeLa cells (Figure 1C). We also tested whether CRTC2 KO HeLa cells had MSI by analyzing four loci, BAT25, BAT26, D2S123 and D5S346, which are commonly used to diagnose MSI (Boland et al., 1998; Parsons et al., 1993). As expected, we observed no evidence of MSI in control HeLa cells, whereas all four CRTC2 KO1 HeLa subclones and two of the four CRTC2 KO2 HeLa subclones examined showed new microsatellite species, indicative of MSI (Figure 1D).
Loss of CREB1 or CBP Leads to Decreased MMR and a Mutator Phenotype
To determine whether loss of CREB1 or CBP produced a phenotype similar to that of CRTC2 KO, we performed an in vivo MMR activity assay in HeLa cells expressing a control non-silencing (NS) short hairpin RNA (shRNA) or one of two unrelated CREB1 or CBP shRNAs. MMR activity was significantly reduced in CREB1 knockdown (KD) and CBP KD cells compared to control NS HeLa cells (Figures 1E and S2E-S2G). Likewise, CREB1 or CBP KD cells had a higher spontaneous mutation frequency than control NS cells (Figure 1F). Finally, five out of 14 CREB1 or CBP KD cell lines produced new microsatellite species or microsatellite deletions, indicative of MSI (Figure 1G).
CRTC2, CREB1 and CBP Bind to the Promoters and Stimulate Transcription of Select MMR Genes
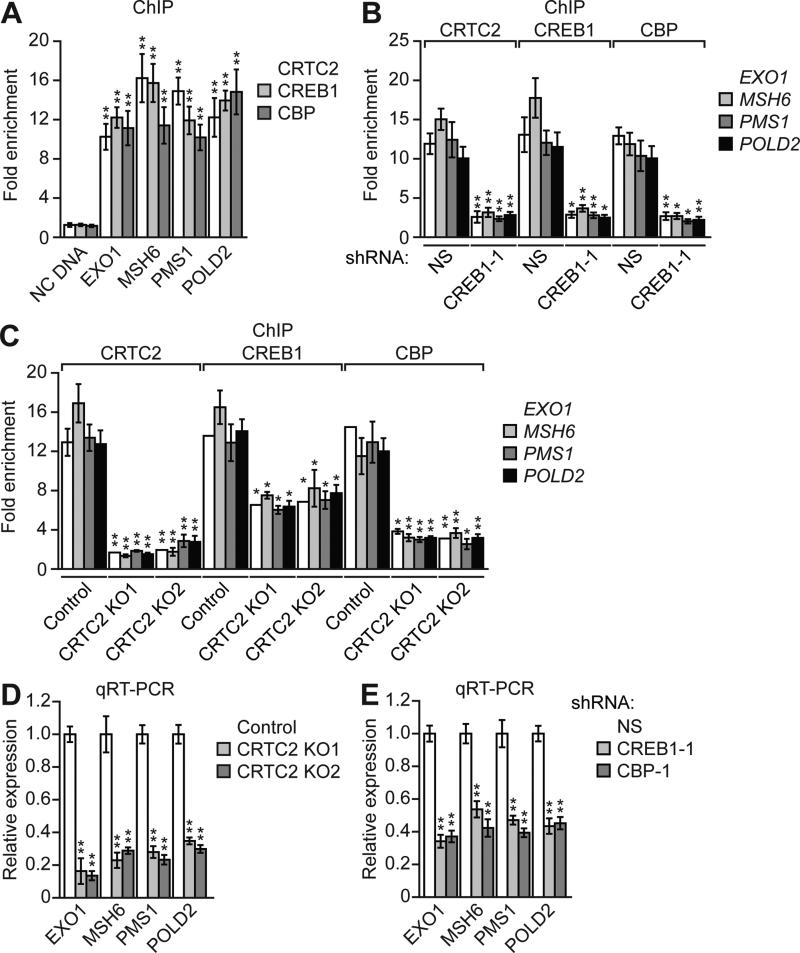
We next sought to investigate the mechanism by which CRTC2, CREB1 and CBP promote MMR. We mined a published CREB1 genome-wide occupancy study (Zhang et al., 2005) and identified four MMR genes that are bound by CREB1: EXO1, MSH6, PMS1, and POLD2 (Jiricny, 2006). Motif search analysis confirmed the presence of a CRE in the promoters of these MMR genes (Figure S3A). To confirm and extend the results of the genome-wide occupancy study, we performed chromatin immunoprecipitation (ChIP) assays. In HeLa cells, CRTC2, CREB1 and CBP were directly bound to the promoters of EXO1, MSH6, PMS1, and POLD2 at the predicted CRE sites (Figure 2A), indicating that these genes are direct targets of CRTC2, CREB1 and CBP. Therefore, we refer to these four MMR genes as “target MMR genes” below. As expected, binding of CRTC2 and CBP was substantially reduced in CREB1 KD cells (Figures 2B and S3B). Notably, in CRTC2 KO cells, CBP recruitment was substantially decreased and CREB1 binding was also diminished (Figure 2C).
Figure 2. CRTC2, CREB1 and CBP Bind to the Promoters and Stimulate Transcription of MMR Genes.
(A) ChIP analysis monitoring occupancy of CRTC2, CREB1 and CBP on the promoters of EXO1, MSH6, PMS1 and POLD2 or an irrelevant negative control (NC) DNA region.
(B) ChIP analysis monitoring occupancy of CRTC2, CREB1 and CBP on the promoters of EXO1, MSH6, PMS1 and POLD2 in HeLa cells expressing a NS or CREB1 shRNA.
(C) ChIP analysis monitoring occupancy of CRTC2, CREB1 and CBP on the promoters of EXO1, MSH6, PMS1 and POLD2 in control or CRTC2 KO HeLa cells.
(D,E) qRT-PCR monitoring expression of EXO1, MSH6, PMS1 and POLD2 in control or CRTC2 KO HeLa cells (D) or in HeLa cells expressing a NS, CREB1 or CBP shRNA (E). For all graphs, error bars indicate SD. *P<0.05, **P<0.01.
See also Figure S3.
We next investigated whether CRTC2, CREB1 and CBP are transcriptional activators of the target MMR genes. The quantitative real-time RT-PCR (qRT-PCR) analysis shows that CRISPR/Cas9-mediated knockout of CRTC2 substantially decreased target MMR gene expression (Figure 2D), which was largely restored by ectopic expression of CRTC2 (Figure S3C). Immunoblot analysis confirmed reduced expression of EXO1. MSH6, PMS1 and POLD2 at the protein level in CRTC2 KO cells (Figure S3D). Finally, shRNA-mediated knockdown of CRTC2 (Figures S3E and S3F) or CREB1 or CBP (Figures 2E and S3G) also substantially decreased target MMR gene expression.
The CREB-CRTC2 Pathway is Stimulated by DNA Damage, Resulting in Up-Regulation of Target MMR Genes
We next sought to investigate whether CRTC2, CREB1 and CBP could promote expression of target MMR genes under conditions of DNA damage. In most cell types, CRTC2 and its family members are phosphorylated by LKB1-AMP-activated protein kinase (AMPK) signaling (Shackelford and Shaw, 2009) and are sequestered in the cytoplasm until cAMP triggers dephosphorylation and nuclear entry (Bittinger et al., 2004; Screaton et al., 2004). However, HeLa cells lack functional LKB1 (Katoh et al., 2006) and, as a result, have a constitutively active CREB-CRTC2 pathway that makes them unsuitable for studying possible regulation of the pathway by DNA damage. Therefore, we instead used as a model system the LKB1-positive human embryonic kidney cell line, HEK293T. DNA damage by UV irradiation results in phosphorylation of CREB1 at S133 and increased CREB1 transcriptional activity (Iordanov et al., 1997). We therefore reasoned that induction of DNA damage in HEK293T cells would activate the CREB-CRTC2-mediated pathway of target MMR gene activation.
As a positive control in these experiments, we used forskolin, an adenylyl cyclase agonist that increases intracellular levels of cAMP, resulting in both phosphorylation of CREB1 at S133 and dephosphorylation of CRTC2 (Screaton et al., 2004). Like forskolin, UV irradiation of HEK293T cells promoted CRTC2 dephosphorylation (Figure S3H), and resulted in increased binding of CREB1, CRTC2 and CBP to target MMR gene promoters (Figure S3I) as well as increased target MMR gene expression (Figure S3J). Consistent with these results, ectopic expression of a constitutively active CRTC2 phosphorylation defective mutant (CRTC2-S171A) in HEK293T cells also resulted in increased target MMR gene expression (Figure S3K). By contrast, in UV-treated HEK293T cells, ectopic expression of a CREB1 phosphorylation defective mutant (CREB1-S133A) (Figure S3L) did not stimulate target MMR gene expression (Figure S3M). Collectively, these results show that following DNA damage, CREB-CRTC2 pathway activity is increased through regulation of both CREB1 and CRTC2, resulting in increased expression of MMR target genes.
Decreased Expression of CRTC2 in Specific Lymphoma Subtypes
Several MMR genes, such as MSH2 and MLH1, are known tumor suppressor genes (Li, 2008). Our finding that CRTC2 is required for expression of MMR genes raised the possibility that CRTC2 may be a tumor suppressor gene. We therefore sought to determine whether loss of CRTC2 would transform cells. For these experiments we used LKB1-positive mouse NIH 3T3 fibroblasts, which are immortalized but not transformed and can be rendered tumorigenic by a wide range of oncogenic events (see, for examples, Di Fiore et al., 1987; Smith et al., 1990; Stacey and Kung, 1984; Velu et al., 1987). As expected, in NIH 3T3 cells expression of the four CRTC2 target MMR genes was greatly reduced upon shRNA-mediated knockdown of Crtc2 (Figures S4A and S4B). Most importantly, knockdown of Crtc2, or a target MMR gene (Exo1 or Msh6; Figures S4C and S4D), promoted anchorage-independent growth of NIH 3T3 cells, as evidenced by increased colony formation in a soft agar assay (Figures 3A and S4E).
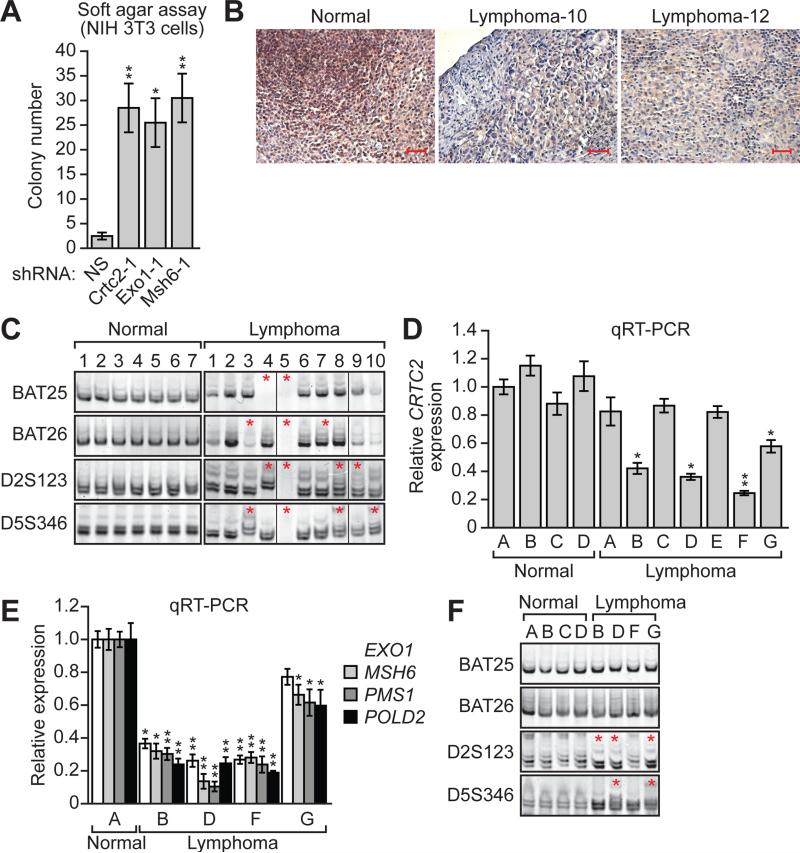
Figure 3. Decreased Expression of CRTC2 in Specific Lymphoma Subtypes.
(A) Soft agar assay. NIH 3T3 cells stably expressing a NS, Crtc2, Exo1 or Msh6 shRNA were analyzed for colony formation, which was normalized to that obtained in cells expressing NS shRNA, which was set to 1.
(B) Representative examples of immunohistochemistry for CRTC2 in normal (tonsil) and two independent lymphoma tissue samples. Scale bars, 100 μM.
(C) MSI analysis at BAT25, BAT26, D2A123 and D5S346 in normal and in lymphoma samples. Non-adjacent lanes from the same gel were spliced together as indicated by the dividing lines.
(D) qRT-PCR monitoring CRTC2 expression in normal and lymphoma samples.
(E) qRT-PCR analysis monitoring expression of EXO1, MSH6, PMS1 and POLD2 in normal and lymphoma samples with loss of CRTC2 expression. For all graphs, error bars indicate SD. *P<0.05, **P<0.01.
(F) MSI analysis at BAT25, BAT26, D2A123 and D5S346 in normal and lymphoma samples with loss of CRTC2 expression.
See also Figure S4.
Mining of the Oncomine database revealed that CRTC2 is down-regulated in multiple cancer types, with the most significant down-regulation occurring in several subtypes of lymphoma such as anaplastic large cell lymphoma (ALCL), angioimmunoblastic T-cell lymphoma (ATCL) and peripheral T-cell lymphoma unspecified (PTCL/U) (Piccaluga et al., 2007) (see Figure S1). We therefore elected to focus on the role of CRTC2 as a tumor suppressor in lymphoma.
To confirm that CRTC2 is down-regulated in lymphoma, we analyzed CRTC2 expression in formalin-fixed, paraffin-embedded ALCL, ATCL and PTCL/U lymphoma patient samples by immunohistochemistry. CRTC2 staining was strong in lymphocytes from normal tonsils but was greatly reduced in 23 of 24 lymphomas analyzed (Figure 3B and Table S2). Notably, seven of 10 samples analyzed displayed new species of microsatellite or microsatellite deletions (Figure 3C), confirming MSI and verifying that lymphomas with decreased CRTC2 are deficient in MMR.
To extend our findings, we analyzed CRTC2 expression in seven snap-frozen ALCL, ATCL and PTCL/U lymphoma patient samples, from which we could isolate mRNA. The qRT-PCR results show that the levels of CRTC2 mRNA in normal tonsil tissues were roughly equivalent, however, they were significantly reduced in four of seven human lymphoma samples analyzed (Figure 3D; see also Table S2). qRT-PCR analysis in the four lymphoma samples with reduced CRCT2 expression confirmed that expression of EXO1, MSH6, PMS1 and POLD2 was down-regulated (Figure 3E). Finally, three of four lymphomas with reduced CRTC2 expression produced new microsatellite species, indicative of MSI (Figure 3F).
Reduced CRTC2 Expression in Lymphoma Cell Lines and Patient Samples Due to Decreased Acetylation of Histone H3
Tumor suppressor genes are frequently inactivated at the transcriptional level due to epigenetic silencing resulting from DNA hypermethylation or alterations in histone modifications such as loss of histone H3 acetylation (Baylin and Jones, 2011). To investigate whether CRTC2 was epigenetically silenced in lymphoma we analyzed two previously described patient-derived ALCL cell lines, Karpas 299 and SUDHL-1 (Quintanilla-Martinez et al., 2006). We first confirmed that CRTC2 expression, as well as that of EXO1, MSH6, PMS1 and POLD2, was reduced in the two ALCL cell lines relative to Jurkat cells, a cell line derived from a patient with T-cell leukemia (Schneider et al., 1977) (Figure 4A). As expected, we observed no evidence of MSI in Jurkat cells, whereas all four Karpas 299 subclones and SUDHL-1 subclones examined showed new microsatellite species or microsatellite deletions, indicative of MSI (Figure 4B).
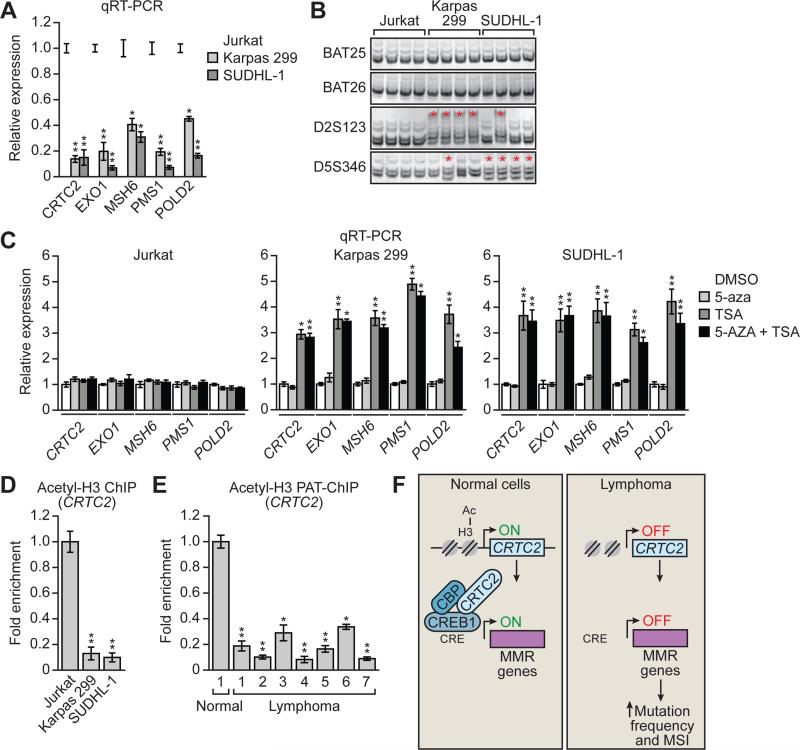
Figure 4. Reduced CRTC2 Expression in Lymphoma Cell Lines and Patient Samples Due to Decreased Acetylation of Histone H3.
(A) qRT-PCR analysis monitoring expression of CRTC2, EXO1, MSH6, PMS1 and POLD2 in Jurkat, Karpas 299 and SUDHL-1 cells.
(B) MSI analysis at BAT25, BAT26, D2A123 and D5S346 in Jurkat, Karpas 299 and SUDHL-1 cells.
(C) qRT-PCR analysis monitoring expression of CRTC2, EXO1, MSH6, PMS1 and POLD2 in Jurkat, Karpas 299 and SUDHL-1 cells treated with 5-aza, TSA or both.
(D) ChIP analysis monitoring histone H3 acetylation (acetyl-H3) levels on the CRTC2 promoter in Jurkat, Karpas 299 and SUDHL-1 cells.
(E) PAT-ChIP assay monitoring acetyl-H3 levels of the CRTC2 promoter in normal and lymphoma samples. For all graphs, error bars indicate SD. *P<0.05, **P<0.01.
(F) Schematic model.
To determine the mechanism by which CRTC2 was silenced, we treated Karpas 299 and SUDHL-1 cells with 5-azacytidine (5-aza), a DNA methyltransferase inhibitor, trichostatin A (TSA), a histone deactylase inhibitor, or both drugs, and monitored CRTC2 expression by qRT-PCR. Figure 4C shows that treatment of Karpas 299 or SUDHL-1 cells with 5-aza had no effect on CRTC2 expression, whereas treatment with TSA significantly increased CRTC2 expression. By contrast, treatment of Jurkat cells with either 5-aza or TSA had no effect on CRTC2 expression. Similar results were obtained for EXO1, MSH6, PMS1 and POLD2.
To confirm these pharmacological results, we performed ChIP analysis for acetylated histone H3. Figure 4D shows that the level of H3 acetylation on the CRTC2 promoter was much higher in Jurkat cells than in Karpas 299 or SUDHL-1 cells. Finally, to determine the clinical relevance of these cell culture results, we performed pathology-tissue (PAT)-ChIP (Fanelli et al., 2011) for acetylated H3 in formalin-fixed, paraffin-embedded lymphoma patient samples that showed reduced CRTC2 expression. Figure 4E shows that levels of acetylated H3 on the CRTC2 promoter were significantly lower in lymphomas than in the control normal tonsil tissue.
In this report, we describe an unanticipated role for the CREB transcriptional activator CRTC2, as well as CREB1 and CBP, in MMR, which is summarized in the model of Figure 4F. We find that in certain subtypes of lymphoma, CRTC2 is epigenetically silenced, resulting in down-regulation of several well-known MMR genes and MS1. We further show that knockdown of CREB1 or CBP also leads to decreased MMR and MSI. Consistent with our findings, previous studies have shown CBP is mutated in a high percentage of MSI-positive colon cancer cell lines, and loss of CBP correlates with inactivation of MMR (Ionov et al., 2004). These previous findings, in conjunction with the results of our study, suggest that in certain cancers alterations of transcription coactivators CRTC2 and CBP lead to the decreased expression of MMR genes and MSI. Our results help to explain why genetic or epigenetic abnormities in MMR genes have not been observed in some MSI-positive cancers (Peltomaki, 2003).
EXPERIMENTAL PROCEDURES
Cell Lines and Culture
HeLa cells (American Type Culture Collection; ATCC) were cultured in Dulbecco's Modified Eagle's Medium (DMEM). Karpas-299 and SUDHL-1 cells (provided by Dr. Youli Zu, Houston Methodist Hospital) and Jurkat cells (ATCC) were maintained in RPMI medium with 10% fetal bovine serum (FBS). Jurkat, Karpas-299 and SUDHL-1 cells were treated with 10 μM 5-aza-2’-deoxycytidine (5’-aza; Sigma) for 3 days, 500 nM trichostatin A (TSA; Sigma) for 12 hours, or a combination of both (where TSA treatment was administered for the last 12 hours of the 3-day 5-aza treatment).
CRISPR/Cas9-Mediated Knockout
Candidate target sequences for CRISPR were designed using CRISPR Design Tool (http://tools.genome-engineering.org). The guide RNA to target nucleotides 199-218 (ATGGGTATGGGGGTAACCGC) in the second exon of CRTC2 was cloned into pX459 (Addgene), and transfected into HeLa cells. Two days later, transfected cells were selected with 1.5 μg/ml puromycin for more than 3 weeks to generate stable knockout cell lines. Eight single colonies were randomly picked, expanded and confirmed for loss of CRTC2 expression by immunoblotting. Two of these clones were randomly selected to confirm the CRTC2 mutations by sequencing and used in all subsequent analyses.
shRNA-Mediated Knockdown
Cells were stably transduced with short hairpin RNA (shRNAs) viruses from Open Biosystems/Thermo Scientific and selected with puromycin.
Immunoblot Analysis
Protein extracts were prepared by lysis in a buffer containing 50 mM Tris-HCl (pH 7.4), 0.1% Triton X-100, 5 mM EDTA, 250 mM NaCl, 50 mM NaF, 0.1 mM Na3VO4, and protease inhibitor (Roche). Blots were probed with the following antibodies: anti-CRTC2 (Santa Cruz), anti-CREB1 (Cell Signaling), anti-α-tubulin (Sigma), and anti-EXO1, anti-MSH6, anti-PMS1 and anti-POLD2 (all from One World Lab).
Real-Time qRT-PCR
Total RNA was isolated, and reverse transcription was performed as described previously (Gazin et al., 2007), followed by real-time qPCR with Platinum SYBR green qPCR SuperMix-UDG with Rox (Invitrogen).
DNA Mismatch Repair Assay
The in vivo DNA mismatch repair assay was performed as previously described (Zhou et al., 2009). Briefly, an EGFP heteroduplexed reporter plasmid containing a mismatch loop was prepared using plasmids p111 and p189 (provided by LuZhe Sun, University of Texas Health Science Center at San Antonio) and co-transfected with pDsRed-N1 (Clontech) into cells. The percentage of EGFP+ and Ds-Red+ cells was analyzed by FACS using a Becton-Dickinson FACSCalibur flow cytometer. The normalized ratio of EGFP+ cells was calculated as the percentage of EGFP+, DsRed+ cells divided by the percentage of DsRed+ cells (i.e., the sum of EGFP+, DsRed+ cells and EGFP−, DsRed+ cells), which in control cells was set to 1.
Spontaneous Mutation Frequency
The HPRT mutation assay was conducted as described previously (Kat et al., 1993). The mutation frequency was determined by dividing the number of 6-TG-resistant colonies by the total number of cells plated after being corrected for colony-forming ability.
MSI Analysis
Genomic DNA was isolated from either single cell colonies picked or isolated in 96-well microtiter plates (for cell lines) or from patient samples. Microsatellite markers BAT25, BAT26, D2S123, and D5S346 were analyzed (Parsons et al., 1993) with improved primers as described (Umetani et al., 2000).
ChIP Assays
ChIP assays were performed as previously described (Raha et al., 2005) using an anti-CRTC2 (Bethyl Laboratories), anti-CREB1 (Cell Signaling Technology), anti-CBP (Bethyl Laboratories) or anti-histone H3 (acetyl K9+K14+K18+K23+K27) (Abcam, ab47915) antibody. ChIP products were analyzed by qPCR. Samples were quantified as percentage of input, and then normalized to an irrelevant region in the genome (~3.2 kb upstream from the transcription start site of GCLC). Fold enrichment was calculated by setting the IgG control IP sample to 1.
Lymphoma Sample Analysis
Formalin-fixed paraffin-embedded tissue sections of normal tonsil (n=7) and lymphoma (n=24), and snap-frozen samples of normal tonsil (n=4) and lymphoma (n=7) were obtained from the archived pathology files of the Pathology Department at the University of Massachusetts Medical School (UMMS), and the lymphoma diagnosis was made by a UMMS pathologist (see Table S2). This study was approved by the institutional review board at UMMS.Immunohistochemical analysis was carried out as previously described (Wajapeyee et al., 2008), using a CRTC2 antibody (1:150 dilution; Santa Cruz). For PAT-ChIP, sections were deparaffinized, rehydrated, and processed as previously described (Fang et al., 2014) using an anti-histone H3 (acetyl K9+K14+K18+K23+K27) (Abcam, ab47915) antibody.
Statistics
All quantitative data were collected from experiments performed in at least triplicate, and expressed as mean ± standard deviation. Differences between groups were assayed using two-tailed Student’s t-test using Microsoft Excel. Significant differences were considered when P≤0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marc Montminy, LuZhe Sun and Youli Zu for reagents, the UMMS RNAi Core Facility for providing shRNAs, Xiaochun Zhu for assistance with the immunohistochemistry analysis, and Sara Deibler for editorial assistance. The results shown here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/). This work was supported by a grant from the NIH (R01GM033977) to M.R.G., who is also an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M.F. and M.R.G. designed the experiments; M.F. performed the experiments with the assistance of M.L.P. and L.C.; W.X. and H.Y. provided the lymphoma patient samples and interpreted the immunohistochemistry results; and M.F. and M.R.G. interpreted the data and wrote the manuscript.
REFERENCES
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell. Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr. Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol. Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Pierce JH, Kraus MH, Segatto O, King CR, Aaronson SA. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science. 1987;237:178–182. doi: 10.1126/science.2885917. [DOI] [PubMed] [Google Scholar]
- Fanelli M, Amatori S, Barozzi I, Minucci S. Chromatin immunoprecipitation and high-throughput sequencing from paraffin-embedded pathology tissue. Nat. Protoc. 2011;6:1905–1919. doi: 10.1038/nprot.2011.406. [DOI] [PubMed] [Google Scholar]
- Fang M, Ou J, Hutchinson L, Green MR. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG Island Methylator phenotype. Mol. Cell. 2014;55:904–915. doi: 10.1016/j.molcel.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionov Y, Matsui S, Cowell JK. A role for p300/CREB binding protein genes in promoting cancer progression in colon cancer cell lines with microsatellite instability. Proc. Natl. Acad. Sci. USA. 2004;101:1273–1278. doi: 10.1073/pnas.0307276101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanov M, Bender K, Ade T, Schmid W, Sachsenmaier C, Engel K, Gaestel M, Rahmsdorf HJ, Herrlich P. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 1997;16:1009–1022. doi: 10.1093/emboj/16.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Kat A, Thilly WG, Fang WH, Longley MJ, Li GM, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc. Natl. Acad. Sci. USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Takemori H, Lin XZ, Tamura M, Muraoka M, Satoh T, Tsuchiya Y, Min L, Doi J, Miyauchi A, et al. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. FEBS J. 2006;273:2730–2748. doi: 10.1111/j.1742-4658.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- Piccaluga PP, Agostinelli C, Califano A, Rossi M, Basso K, Zupo S, Went P, Klein U, Zinzani PL, Baccarani M, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J. Clin. Invest. 2007;117:823–834. doi: 10.1172/JCI26833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla-Martinez L, Pittaluga S, Miething C, Klier M, Rudelius M, Davies-Hill T, Anastasov N, Martinez A, Vivero A, Duyster J, et al. NPM-ALK-dependent expression of the transcription factor CCAAT/enhancer binding protein beta in ALK-positive anaplastic large cell lymphoma. Blood. 2006;108:2029–2036. doi: 10.1182/blood-2005-10-014258. [DOI] [PubMed] [Google Scholar]
- Raha T, Cheng SW, Green MR. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 2005;3:e44. doi: 10.1371/journal.pbio.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Kalyana-Sundaram S, Tomlins SA, Mahavisno V, Kasper N, Varambally R, Barrette TR, Ghosh D, Varambally S, Chinnaiyan AM. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9:443–454. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider U, Schwenk HU, Bornkamm G. Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int. J. Cancer. 1977;19:621–626. doi: 10.1002/ijc.2910190505. [DOI] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, 3rd, Takemori H, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Smith MR, Heidecker G, Rapp UR, Kung HF. Induction of transformation and DNA synthesis after microinjection of raf proteins. Mol. Cell Biol. 1990;10:3828–3833. doi: 10.1128/mcb.10.7.3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey DW, Kung HF. Transformation of NIH 3T3 cells by microinjection of Haras p21 protein. Nature. 1984;310:508–511. doi: 10.1038/310508a0. [DOI] [PubMed] [Google Scholar]
- Umetani N, Sasaki S, Watanabe T, Ishigami H, Ueda E, Nagawa H. Diagnostic primer sets for microsatellite instability optimized for a minimal amount of damaged DNA from colorectal tissue samples. Ann. Surg. Oncol. 2000;7:276–280. doi: 10.1007/s10434-000-0276-6. [DOI] [PubMed] [Google Scholar]
- Velu TJ, Beguinot L, Vass WC, Willingham MC, Merlino GT, Pastan I, Lowy DR. Epidermal-growth-factor-dependent transformation by a human EGF receptor proto-oncogene. Science. 1987;238:1408–1410. doi: 10.1126/science.3500513. [DOI] [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Huang C, Yang J, Lu J, Dong Q, Sun LZ. Preparation of heteroduplex enhanced green fluorescent protein plasmid for in vivo mismatch repair activity assay. Anal. Biochem. 2009;388:167–169. doi: 10.1016/j.ab.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.