Abstract
Copper (Cu) is a trace element essential for the growth and development of almost all organisms, including bacteria. However, Cu overload in most systems is toxic. Studies show Cu accumulates in macrophage phagosomes infected with bacteria, suggesting Cu provides an innate immune mechanism to combat invading pathogens. To counteract the host-supplied Cu, increasing evidence suggests that bacteria have evolved Cu resistance mechanisms to facilitate their pathogenesis. In particular, Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, has evolved multiple pathways to respond to Cu. Here, we summarize what is currently known about Cu homeostasis in Mtb and discuss potential sources of Cu encountered by this and other pathogens in a mammalian host.
Keywords: Mycobacterium tuberculosis, copper resistance, virulence
Mtb: a successful intracellular human pathogen
Mtb infects nearly one-third of the world’s population and kills 1.3 million people annually, making it one of the most devastating infectious agents on earth (http://www.who.int/tb/publications/global_report/en/). Tuberculosis is transmitted by the inhalation of aerosolized droplets containing Mtb bacilli coughed or sneezed by an infected person. Once bacteria are inhaled, they localize to the alveoli, where macrophages and dendritic cells phagocytose them. Mtb has evolved to survive within macrophages that have multiple antimicrobial activities, including the production of reactive nitrogen and oxygen intermediates, phagosomal acidification and iron limitation.1–3 Numerous labs around the world are working to understand how Mtb is able to respond to this hostile environment, the knowledge of which may help in the development of improved tuberculosis treatments.
Evidence for Cu in the host response to mycobacterial infections
Cu is essential for the development of almost all aspects of mammalian physiology, thus defects in Cu homeostasis almost certainly impact immune responses to microbial infections. Dietary Cu-deficiency in farm animals is linked to a higher incidence of bacterial infections,4 perhaps because Cu-deficient diets reduce the number of antibody-producing cells in mice.5 In a study that was key to the realization that Cu and other metals may be important for impacting the outcome of tuberculosis infections, Bermudez and coworkers found that the concentration of Cu markedly increases in phagolysosomes of peritoneal-derived mouse macrophages after infection with several Mycobacterium species.6 In a later study, it was found that dietary supplementation with Cu results in the accumulation of Cu in lung granulomas of Mtb-infected guinea pigs, and coincides with a reduction in bacterial burden.7 Collectively, these data suggest that Cu is mobilized in mammals to control bacterial growth.
Further supporting these observations, several studies show a link between Cu transporting machinery and antimicrobial activity. Petris and co-workers showed Cu enhances the bactericidal activity of RAW264.7 murine macrophage cells toward E. coli. Treatment of RAW264.7 macrophages with proinflammatory factors, such as IFN-γ or lipopolysaccharide, is associated with increased levels of the high affinity Cu importer CTR1 at the plasma membrane8 and the P1B-type ATPase ATP7A.9 P-type ATPases are integral membrane proteins that use ATP to either transport molecules in or out of a cell and are found in both eukaryotes and prokaryotes.10 Significantly, ATP7A is trafficked from the trans-Golgi network into the phagosomal compartment of IFN-γ activated macrophages, providing a possible mechanism by which bactericidal levels of Cu may be delivered into phagolysosomes.9 Moreover, IFN-γ stimulated RAW264.7 macrophages kill E. coli lacking the Cu efflux pump CopA more effectively than wild type bacteria, and this effect is reduced by silencing the ATP7A gene.9 These data show that mammalian Cu transporters can mobilize Cu into phagosomal compartments to control the growth of at least one bacterial species. It remains to be determined if macrophage-associated ATP7A is also required for controlling mycobacterial or other infections.
Copper is both essential and toxic for Mtb
Although Cu is antimicrobial, it is also essential. Cu can undergo reversible oxidation states between reduced Cu+ and oxidized Cu2+ and has a high redox potential, making it a critical cofactor of enzymes used for electron transfer reactions in the presence of oxygen. In Mtb, the most prominent Cu binding enzymes include cytochrome c oxidase and the Cu/Cu superoxide dismutase11, which contributes to resistance to oxidative stress.12 Thus, like for most life forms, Cu is essential for Mtb viability.
Of course, too much Cu is toxic to Mtb.7, 13, 14 Several mechanisms have been ascribed to the toxicity of Cu. Under aerobic conditions, Cu can react with hydrogen peroxide (H2O2) to create hydroxyl radical (•OH) and hydroxyl anion (OH−) via a Fenton-like reaction:15
These molecules can react with and irreversibly damage many macromolecules including proteins, lipids and nucleic acids, potentially leading to cell death.16 However, it has not yet been definitively proven that the production of these reactive products is the mechanism of Cu-induced cytotoxicity in any bacterial system. In fact, Imlay and colleagues showed that a mutant E. coli strain that hyper-accumulates Cu is more resistant to H2O2 stress than bacteria without accumulated Cu.17 Furthermore, Cu treatment is surprisingly associated with less, not more, oxidative DNA damage, even though the production of reactive oxygen species (ROS) is apparent in these bacteria.17 Although Imlay and colleagues tested several hypotheses to explain the protective effects of Cu, they could not identify a mechanism to explain their observations. The authors of this study speculated that an alteration in Cu accumulation results in an adaptation that either sequesters or otherwise prevents Cu from interacting with hydrogen peroxide near DNA, a hypothesis that remains to be tested.17
Because the creation of ROS does not explain why Cu is toxic to E. coli, Macomber et al tested the hypothesis that Cu displaces iron-sulfur (Fe-S) clusters from important metabolic enzymes to inactivate bacterial growth.18 Indeed, a study found Cu targets isopropylmalate dehydratase, which is needed for the synthesis of branched chain amino acids. An E. coli strain lacking several Cu homeostasis proteins is very Cu sensitive, but the addition of several branched-chain amino acids restores some growth during Cu treatment.18 Initially it was thought that Cu increases the amount of intracellular H2O2, which might directly inactivate several amino acid biosynthetic pathways; however, the presence of oxygen (and thus H2O2) is not needed to see this effect. Furthermore, growth inhibition occurs at the same time as the displacement of Fe atoms from the solvent-exposed cluster of dehydratases, suggesting that Cu inactivates these enzymes by liganding to coordinating S atoms.18 Unlike what was observed in E. coli, the addition of branched-chain amino acids to a Cu susceptible Salmonella strain culture does not rescue Cu toxicity, suggesting the mechanisms of Cu toxicity may be multifactorial and vary from organism to organism.19
Another target of Cu toxicity has been discovered in the obligate human pathogen Neisseria gonorrheae. Cu is predicted to disrupt Fe-S clusters of the enzyme HemN, which is required for the heme biosynthesis. Failure to equip hemoproteins such as catalases, peroxideses and nitric oxide (NO) reductases with their cognate prosthetic group may lead to the increased toxicity of reactive species and thus decreased bacterial survival.20
Interestingly, Nathan and colleagues showed that NO can reduce Cu2+ to Cu+.21 Macrophages infected with Mtb and other pathogens stimulate the production NO, which can kill invading microbes by different mechanisms, including the production of reactive nitrogen and oxygen species, the displacement of metal co-factors from enzymes, and in ways that have yet to be identified.22 Perhaps another mechanism of NO mediated toxicity is via the reduction of Cu to make it more toxic to invading microbes.
Copper homeostasis in Mtb: a tale of three pathways
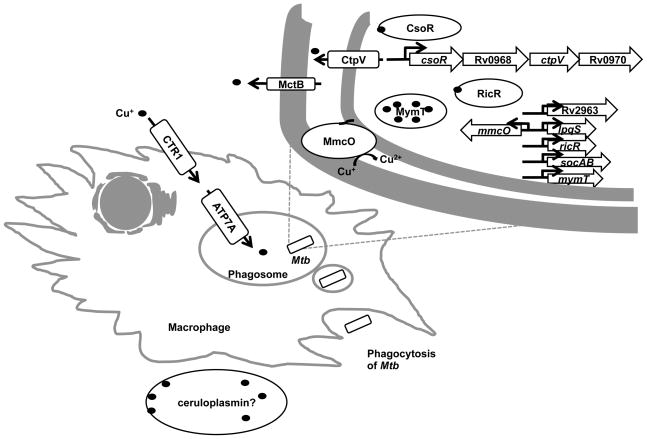
Several independent lines of study converged upon the conclusion that Mtb expresses Cu resistance pathways in order to successfully persist in a mammalian host. Talaat and colleagues found the first Cu responsive operon in Mtb after following up on the identification of a unique genomic island called the in vivo expressed genomic island or iVEGI that is highly induced in bacteria isolated from mouse lungs but not in bacteria that are grown in broth culture.23 Analysis of this island for potential transcriptional units uncovered the presence of an operon (Rv0967-Rv0970) including ctpV, a cation P-type ATPase that is predicted to transport Cu out of Mtb (Fig. 1).24 Moreover, the operon is highly induced upon Cu treatment, therefore, it was named the copper sensitive operon (cso).25 The organization of the genes in the cso operon is similar to that of the cmt (cadmium/lead metal transporter) operon, in which the first gene encodes a cadmium and lead-sensing transcriptional regulator, CmtR, controlling the whole operon.26 Liu et al hypothesized the first gene in the operon, Rv0967, was a Cu responsive transcriptional regulator. Indeed, Rv0967, later named CsoR (copper sensitive operon repressor), binds to DNA as a dimer of dimers in the absence of Cu.25 At elevated Cu+ concentrations the binding affinity of CsoR to the promoter of the cso operon is reduced due to a conformational change in the CsoR-DNA complex caused by Cu+. Giedroc and colleagues concluded that this was the mechanism of derepression of the cso operon observed in Cu-treated Mtb cultures.24
Fig. 1. Model of Cu-mediated control of Mtb in activated macrophages.
Upon activation, Cu uptake in macrophages is increased due to the elevated levels of the Cu importer CTR1 and ATP7A. ATP7A traffics to the phagosome, potentially leading to the increased concentration of Cu in that compartment. To combat the toxicity of excess Cu, Mtb has at least three independent Cu resistance pathways as described in the text. The Cu-binding ferroxidase ceruloplasmin49, 50 may contribute a source of Cu to control Mtb and other infections.
In order to determine the contribution of the cso operon on Cu resistance, Talaat and colleagues characterized CtpV, a predicted P-type ATPase.24 Overexpression of the Mtb cso operon in Mycobacterium smegmatis (M. smegmatis), a non-pathogenic relative of Mtb that does not have a cso operon, shows lower levels of intracellular Cu accumulation suggesting CtpV functions as an efflux pump.13 This study also showed that ctpV is the most highly Cu-induced ctp gene of 11 predicted ctp genes in the Mtb genome.13 In vitro, an Mtb ctpV mutant is hyper-sensitive to Cu, and this phenotype can be partially complemented in trans. Mice infected with a ctpV mutant survive significantly longer than mice infected with wild type Mtb, however this effect cannot be rescued when ctpV is added back at another site on the chromosome. Furthermore, in guinea pig infections, the numbers of ctpV mutant bacteria is significantly lower than those of wild type bacteria at 21 days after infection, however, no colonization defect is observed at a later time point.13 Taken together, CtpV appears to be required for resistance of Cu toxicity, but its contribution to Mtb virulence is unclear.
In another study, Nathan and colleagues discovered the first metallothionein in Mtb named MymT (mycobacterial metallothionein).21 Metallothioneins are small, cysteine-rich proteins with the ability to bind metal ions. Their biological roles include metal detoxification, intracellular distribution and defense against oxidative stress.27 MymT harbors several cysteines (Cys) arranged in Cys-X-Cys or Cys-X-His motifs that allow it to coordinate metal ions.21 Expression of mymT is highly induced by several metals including Cu, cadmium, cobalt, nickel and zinc, with the strongest induction by Cu.21 Mass spectrometry analysis shows that MymT can bind with up to six Cu+ ions in a solvent-shielded thiolate core.21 Although deletion of mymT from Mtb leads to increased Cu sensitivity in vitro, MymT does not appear to be essential for virulence in a murine infection model.21
Several years after the discovery of CsoR, a second locus required for Cu resistance was characterized. Mycobacterial copper transport protein B (MctB/Rv1698) (Fig. 1) was initially identified as a putative outer membrane protein by a genome-wide secondary structure prediction study of Mtb.28 Mycobacteria do not have canonical outer membranes as observed in Gram-negative bacteria but instead have what is termed a “mycomembrane”, which is composed of mycolic acids and other components unique to mycobacteria.29 Like Gram-negative bacteria, mycobacteria have what appear to be porins,30–33 thus it was predicted that MctB might have this function.34 Deletion of mctB or its homologue in M. smegmatis, ms3747, leads to Cu accumulation in the bacterial cytoplasm and increased Cu sensitivity, demonstrating an essential role of MctB in maintaining low Cu concentrations in mycobacteria.7
Niederweis and colleagues used two models to test the role of MctB during infections. In mice, an Mtb mctB mutant is modestly attenuated compared to wild type Mtb.7 It was presumed that Cu would be more effective if it encountered Mtb in a hypoxic environment, where Cu+ would remain reduced. The authors of this study noted that mice do not form hypoxic granulomas like humans, and that guinea pigs represent a better model for human infections.35 Wolschendorf et al tested the fitness of an mctB deletion mutant in both animal models. In BALBc mice, wild type Mtb grows to about 10 times higher numbers than an mctB mutant. This difference is further exacerbated by the addition of Cu sulfate (CuSO4) to the animals’ drinking water, suggesting the attenuation is due to a Cu specific effect.7 In guinea pigs, the difference growth defect between the wild type and mctB strains is more pronounced than what is observed in mice.7 This may be consistent with the notion that a more hypoxic environment may more effectively use Cu to control bacterial growth.
Although MctB was initially described as a putative mycomembrane protein, recent studies suggest MctB may be anchored to the inner membrane.36 Thus it is less clear that MctB forms a channel to export Cu and may have a different activity to maintain Cu homeostasis in mycobacteria. Furthermore, because mctB expression is not Cu inducible,24 it may have functions that are important for reasons beyond Cu resistance.
The final pathway to be involved in Cu homeostasis was identified in an attempt to understand the link between proteasome function and Mtb pathogenesis. The Mtb proteasome, and ATP-dependent chambered protease, is required for causing lethal infections in animals for reasons that have largely been mysterious.37–40 Using microarray analysis, Festa et al tested the hypothesis that the proteasome impacts transcriptional regulation in Mtb.14 The analysis identified a new regulon unique to Mtb that includes lpqS (encoding a putative lipoprotein), mmcO (encoding a mycobacterial multicopper oxidase), Rv2963 (encoding a possible permease), mymT, socAB (small ORF induced by copper A and B), and ricR, which encodes a homologue of CsoR.14 All genes are Cu-inducible suggesting RicR represses gene expression under Cu-deplete concentrations (Fig. 1).14, 21, 24
A ricR null mutant, which overexpresses all genes in the RicR regulon, is hyper-resistant to Cu in vitro, suggesting that one or more RicR-regulated genes are important in combating Cu toxicity.14 The only RicR-regulon gene products that have known functions are MmcO, MymT and RicR itself. MmcO can oxidize Fe2+ to Fe3+ and perhaps also convert toxic Cu+ into Cu2+.41 Although MmcO is required for Cu resistance in vitro, it alone does not significantly contribute to Mtb virulence in a mouse model of infection.41, 42 Similarly, deletion and/or disruption of any single RicR-regulated gene or even simultaneous deletion of the two major Cu-protective genes (mymT and mmcO) has a minimal impact on the growth of Mtb in mice at time points of up to eight weeks after infection.42 However, an Mtb strain with a mutant allele of RicR that cannot respond to Cu and constitutively represses all of the genes of the RicR regulon is highly sensitive to Cu and attenuated for growth in mice.42 Taken together, it appears that deletion of two or more RicR-regulated genes will be necessary to observe a robust in vivo phenotype. Alternatively, the contribution of individual RicR-regulated genes may need to be ascertained in an infection model more closely resembling that of humans.
Conclusions and Prospects
It is notable that deletion of any single gene associated with Cu resistance, with the exception of mctB, does not significantly affect Mtb virulence, perhaps suggesting that the loss of one Cu-responsive gene may be compensated by the induction of expression of another. It is also worth mentioning that all of the above systems lack a key component: some Cu chaperone. Cu is assumed to always bind to proteins or otherwise be liganded to small molecules, thus it remains to be determined how Cu is transferred to the various Cu binding proteins and efflux pumps described here. It is tempting to speculate that there may be one or more Cu chaperones shared by all three systems that are critical for their function. It is also possible that some thiol-containing small molecule, such as mycothiol,43 the major thiol found in mycobacteria, may perform a Cu exchange function given its high affinity for Cu+.44 However, it is hard to envision how this could be regulated in a controlled manner. In addition to the search for a Cu chaperone, numerous questions still need to be addressed. For example, how does Cu kill Mtb? What are the sources of Cu during a tuberculosis infection? Ceruloplasmin is a serum ferroxidase that contains more than 95% of the Cu found in plasma.45 It is tempting to speculate that ceruploplasmin might play a critical role in Cu mobilization and antimicrobial activity. Can Cu resistance mechanisms in bacteria be targeted for drug development? A recent high-throughput drug screen identified compounds that possess Cu-dependent anti-Mtb activities, the mechanisms of which have not been elucidated.46 Intriguingly, another screen for compounds that inhibit the ability of Mtb to kill cultured cells found a small molecule, a benzyloxybenzylidene-hydrazine compound called BBH7, which strongly induces the RicR regulon.47 BBH7 was selected for analysis because it inhibits the secretion of a major virulence protein called EsxA.47 Based on these studies, it is tempting to speculate that there is a link between Cu homeostasis and virulence protein secretion in Mtb.
Besides mycobacteria, can Cu also work as an antimicrobial weapon against other pathogens such as fungi, viruses, or eukaryotic parasites? Cu acquisition and detoxification pathways have been implicated in the virulence of the fungal pathogen Cryptococcus neoformans, showing Cu may also be used by the innate immune system to battle fungi.48 Further investigations into the biochemistry, genetics, and physiology of Cu homeostasis in both the host and pathogen will be essential to have a better understanding of the role of Cu in tuberculosis and other infectious diseases.
Supplementary Material
Acknowledgments
Copper research in the Darwin laboratory is supported by NIH grant HL92774.
Footnotes
Conflict of interest statement
The authors have no conflicts of interest to declare.
References
- 1.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pieters J. Cell Host Microbe. 2008;3:399–407. doi: 10.1016/j.chom.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Flynn JL, Chan J. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 4.Suttle N, Jones D. Proc Nutr Soc. 1986;45:317–325. doi: 10.1079/pns19860069. [DOI] [PubMed] [Google Scholar]
- 5.Lukasewycz O. Science. 1981;213:559–561. doi: 10.1126/science.7244654. [DOI] [PubMed] [Google Scholar]
- 6.Wagner D, Maser J, Lai B, Cai Z, Barry CE, Höner zu Bentrup K, Russell DG, Bermudez LE. J Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- 7.Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. Proc Natl Acad Sci U S A. 2011;108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puig S, Thiele DJ. Curr Opin Chem Biol. 2002;6:171–180. doi: 10.1016/s1367-5931(02)00298-3. [DOI] [PubMed] [Google Scholar]
- 9.White C, Lee J, Kambe T, Fritsche K, Petris MJ. J Biol Chem. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmgren MG, Axelsen KB. Biochim Biophys Acta. 1998;1365:37–45. doi: 10.1016/s0005-2728(98)00041-3. [DOI] [PubMed] [Google Scholar]
- 11.Spagnolo L, Törö I, D’orazio M, O’Neill P, Pedersen JZ, Carugo O, Rotilio G, Battistoni A, Djinović-Carugo K. J Biol Chem. 2004;279:33447–33455. doi: 10.1074/jbc.M404699200. [DOI] [PubMed] [Google Scholar]
- 12.Piddington DL, Fang FC, Laessig T, Cooper AM, Orme IM, Buchmeier NA. Infect Immun. 2001;69:4980–4987. doi: 10.1128/IAI.69.8.4980-4987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. Mol Microbiol. 2010;77:1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Festa RA, Jones MB, Butler-Wu S, Sinsimer D, Gerads R, Bishai WR, Peterson SN, Darwin KH. Mol Microbiol. 2011;79:133–148. doi: 10.1111/j.1365-2958.2010.07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liochev SI, Fridovich I. Redox Rep. 2002;7:55–57. doi: 10.1179/135100002125000190. [DOI] [PubMed] [Google Scholar]
- 16.Halliwell B, Gutteridge J. Mol Aspects Med. 1985;8:89–193. doi: 10.1016/0098-2997(85)90001-9. [DOI] [PubMed] [Google Scholar]
- 17.Macomber L, Rensing C, Imlay JA. J Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macomber L, Imlay JA. Proc Natl Acad Sci U S A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Achard ME, Tree JJ, Holden JA, Simpfendorfer KR, Wijburg OL, Strugnell RA, Schembri MA, Sweet MJ, Jennings MP, McEwan AG. Infect Immun. 2010;78:2312–2319. doi: 10.1128/IAI.01208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Djoko KY, McEwan AG. ACS Chem Biol. 2013;8:2217–2223. doi: 10.1021/cb4002443. [DOI] [PubMed] [Google Scholar]
- 21.Gold B, Deng H, Bryk R, Vargas D, Eliezer D, Roberts J, Jiang X, Nathan C. Nat Chem Biol. 2008;4:609–616. doi: 10.1038/nchembio.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacMicking J, Xie Q-w, Nathan C. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 23.Talaat AM, Lyons R, Howard ST, Johnston SA. Proc Natl Acad Sci U S A. 2004;101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward SK, Hoye EA, Talaat AM. J Bacteriol. 2008;190:2939–2946. doi: 10.1128/JB.01847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 26.Cavet JS, Graham AI, Meng W, Robinson NJ. J Biol Chem. 2003;278:44560–44566. doi: 10.1074/jbc.M307877200. [DOI] [PubMed] [Google Scholar]
- 27.Sato M, Bremner I. Free Radic Biol Med. 1993;14:325–337. doi: 10.1016/0891-5849(93)90029-t. [DOI] [PubMed] [Google Scholar]
- 28.Song H, Sandie R, Wang Y, Andrade-Navarro MA, Niederweis M. Tuberculosis. 2008;88:526–544. doi: 10.1016/j.tube.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan PJ, Nikaido H. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 30.Niederweis M, Ehrt S, Heinz C, Klöcker U, Karosi S, Swiderek KM, Riley LW, Benz R. Mol Microbiol. 1999;33:933–945. doi: 10.1046/j.1365-2958.1999.01472.x. [DOI] [PubMed] [Google Scholar]
- 31.Stahl C, Kubetzko S, Kaps I, Seeber S, Engelhardt H, Niederweis M. Mol Microbiol. 2001;40:451–464. doi: 10.1046/j.1365-2958.2001.02394.x. [DOI] [PubMed] [Google Scholar]
- 32.Faller M, Niederweis M, Schulz GE. Science. 2004;303:1189–1192. doi: 10.1126/science.1094114. [DOI] [PubMed] [Google Scholar]
- 33.Mahfoud M, Sukumaran S, Hülsmann P, Grieger K, Niederweis M. J Biol Chem. 2006;281:5908–5915. doi: 10.1074/jbc.M511642200. [DOI] [PubMed] [Google Scholar]
- 34.Siroy A, Mailaender C, Harder D, Koerber S, Wolschendorf F, Danilchanka O, Wang Y, Heinz C, Niederweis M. J Biol Chem. 2008;283:17827–17837. doi: 10.1074/jbc.M800866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowland JL, Niederweis M. Tuberculosis. 2012;92:202–210. doi: 10.1016/j.tube.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerda-Maira FA, Pearce MJ, Fuortes M, Bishai WR, Hubbard SR, Darwin KH. Mol Microbiol. 2010;77:1123–1135. doi: 10.1111/j.1365-2958.2010.07276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 39.Lamichhane G, Zignol M, Blades NJ, Geiman DE, Dougherty A, Grosset J, Broman KW, Bishai WR. Proc Natl Acad Sci U S A. 2003;100:7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darwin KH. Nat Rev Microbiol. 2009;7:485–491. doi: 10.1038/nrmicro2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowland JL, Niederweis M. J Bacteriol. 2013;195:3724–3733. doi: 10.1128/JB.00546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi X, Festa RA, Ioerger TR, Butler-Wu S, Sacchettini JC, Darwin KH, Samanovic MI. mBio. 2014;5:e00876–00813. doi: 10.1128/mBio.00876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newton GL, Buchmeier N, Fahey RC. Microbiol Mol Biol Rev. 2008;72:471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu Y, Chang F-MJ, Giedroc DP. Acc Chem Res. 2014 doi: 10.1021/ar500300n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellman NE, Gitlin JD. Annu Rev Nutr. 2002;22:439–458. doi: 10.1146/annurev.nutr.22.012502.114457. [DOI] [PubMed] [Google Scholar]
- 46.Speer A, Shrestha TB, Bossmann SH, Basaraba RJ, Harber GJ, Michalek SM, Niederweis M, Kutsch O, Wolschendorf F. Antimicrob Agents Chemother. 2013;57:1089–1091. doi: 10.1128/AAC.01781-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rybniker J, Chen JM, Sala C, Hartkoorn RC, Vocat A, Benjak A, Boy-Röttger S, Zhang M, Székely R, Greff Z. Cell Host Microbe. 2014;16:538–548. doi: 10.1016/j.chom.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Ding C, Festa RA, Chen YL, Espart A, Palacios Ò, Espín J, Capdevila M, Atrian S, Heitman J, Thiele DJ. Cell Host Microbe. 2013;13:265–276. doi: 10.1016/j.chom.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato M, Gitlin J. J Biol Chem. 1991;266:5128–5134. [PubMed] [Google Scholar]
- 50.Terada K, Kawarada Y, Miura N, Yasui O, Koyama K, Sugiyama T. Biochim Biophys Acta. 1995;1270:58–62. doi: 10.1016/0925-4439(94)00072-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.