Abstract
Iron-sulfur clusters act as important cofactors for a number of transcriptional regulators in bacteria, including many mammalian pathogens. The sensitivity of iron-sulfur clusters to iron availability, oxygen tension, and reactive oxygen and nitrogen species enables bacteria to use such regulators to adapt their gene expression profiles rapidly in response to changing environmental conditions. In this review, we discuss how the [4Fe-4S] or [2Fe-2S] cluster-containing regulators FNR, Wbl, aconitase, IscR, NsrR, SoxR, and AirSR contribute to bacterial pathogenesis through control of both metabolism and classical virulence factors. In addition, we briefly review mammalian iron homeostasis as well as oxidative/nitrosative stress to provide context for understanding the function of bacterial iron-sulfur cluster sensors in different niches within the host.
Iron and Iron-Sulfur Clusters
Iron
Iron is an essential nutrient for almost all organisms examined, with some unique exceptions including Lactobacillus plantarum and Borrelia burgdorferi1, 2. The importance of this element is underscored by the requirement of iron as a cofactor for a variety of processes including energy generation, DNA replication, and oxygen transport. The most common forms of iron under physiological conditions are the reduced ferrous form (Fe2+) and the oxidized ferric form (Fe3+). While iron is critical for almost all forms of life, its unregulated accumulation in the presence of oxygen or reactive oxygen species (ROS) is extremely toxic. Iron amplifies ROS production through the Fenton and Haber-Weiss reactions, leading to the production of hydroxyl radicals that damage biological macromolecules including DNA3. Thus, organisms must balance sufficient iron utilization for maintaining optimal growth rates while preventing excess oxidative stress. Bacteria achieve this through use of oxidative stress response pathways as well as through mechanisms such as coordinated iron uptake, storage, and detoxification to ensure proper iron homeostasis4. Many bacterial pathogens encounter environments with varying iron availability (see Section on Iron, Oxygen, and Nitric Oxide in the Mammalian Host Environment below), creating a need to sense the amount of intrabacterial iron and couple this information to control of gene expression.
Iron-sulfur (Fe-S) clusters
Iron-sulfur (Fe-S) clusters were first identified over 50 years ago, when their role in electron transfer was discovered5. There are various Fe-S clusters that act as prosthetic groups; however, those most commonly found in nature are [2Fe-2S] and [4Fe-4S]. Biologically, Fe-S clusters are not randomly generated from free Fe2+/Fe3+ and S2− components, as these would be toxic. As such, there are dedicated Fe-S cluster biosynthesis pathways (Isc, Suf, and Nif) that generate Fe-S clusters from Fe2+ and L-cysteine substrates. These Fe-S clusters are typically coordinated to proteins through conserved cysteine residues, although aspartate, histidine, serine, or backbone amides have also been shown to play a role6.
Fe-S clusters have a unique ability to delocalize electron density to both the Fe and S atoms, which explains their prominent role in respiratory and photosynthetic electron transport7, 8. A variety of regulatory proteins utilize Fe-S clusters in order to sense iron, environmental oxidants, or nitric oxide9. Fe-S clusters can be reversibly oxidized in the presence of oxygen or ROS, leading to conversion of the cluster to a distinct oxidation state or to complete loss of the cluster from the Fe-S cluster-coordinating protein. In addition, nitric oxide can also damage Fe-S clusters10, 11. These alterations to the Fe-S cluster are thought to cause a conformational change in the regulatory protein, leading to altered activity.
In this review, we will focus on bacterial Fe-S cluster-coordinating regulatory proteins important for the ability of mammalian pathogens to cause disease. These regulators use their Fe-S cluster to sense environmental cues such as oxygen and iron availability to control expression of bacterial genes important for virulence. In this review, we summarize key findings from five decades of literature on bacterial Fe-S cluster regulation as it pertains to pathogenesis of the mammalian host.
Iron, Oxygen, and Nitric Oxide in the Mammalian Host Environment
Mammalian pathogens occupy a diverse set of niches within the host organism. These niches vary in iron and oxygen availability as well as ROS and reactive nitrogen species (RNS) concentration, and are therefore predicted to impact Fe-S cluster sensors differently. In this section, we briefly discuss mammalian iron homeostasis and oxidative/nitrosative stress, reviewed in more detail elsewhere12-15, in order to gain perspective on host factors that influence the activity of bacterial Fe-S cluster sensors during infection13, 16-19.
Mammals regulate iron on both a systemic and cellular level20. These distinct but overlapping regulatory circuits impact iron availability for extracellular pathogens, vacuolar pathogens, and cytosolic pathogens in different ways. In addition, inflammation limits iron availability through a number of mechanisms. Lastly, the composition of the microbiota may alter the availability of iron in the gut.
Up to 95% of the human daily iron need is obtained from recycling of senescent red blood cells by macrophages,13 with the remainder absorbed from the diet in the duodenum and colon21. Nramp2/DMT1 transports iron from the intestinal lumen across the duodenum brush border, while ferroportin transports iron across the basolateral membrane of enterocytes and into the bloodstream20, 22. Similarly, in macrophages, Nramp2/DMT1 transports iron recycled from red blood cells across the endosomal membrane and ferroportin transports the iron across the plasma membrane and into the bloodstream. However, a vanishingly small amount of free iron is present in mammalian tissues (10-24 M) due to the concerted action of a number of iron-binding proteins21. Transferrin is the main iron carrier in the bloodstream and transferrin-bound iron is taken up into cells through transferrin receptor 1 (TfR1). Levels of ferroportin are controlled by hepcidin, a peptide hormone produced by the liver23, controlling iron absorption and recycling. Hepcidin levels are regulated in response to iron levels through a mechanism that involves TfR1 and the MHC class I protein HFE. In addition, production of cytokines such as IL-6 in response to innate immune stimuli induces hepcidin expression, thereby decreasing ferroportin levels and promoting hypoferremia13, 14. During chronic and/or severe inflammation associated with infection or cancer, this response can result in prolonged hypoferremia referred to as anemia of inflammation24, 25.
Mutations in the HFE gene are associated with hereditary hemochromatosis (HH), a genetic iron overload disorder common among people of Northern European descent26. In a subset of HH individuals, transferrin saturation is increased and iron overload occurs in the liver and other tissues, leading to damage most likely as a result of oxidative stress26, 27. HH-associated HFE mutations lead to decreased hepcidin levels and increased ferroportin. In turn, increased ferroportin leads to elevated absorption of iron from the gut as well as elevated transport of iron recycled from red blood cells out of macrophages. Paradoxically, while HH is characterized by overall iron overload, macrophages from HH individuals are iron poor28, 29. This may at least partially explain why hepcidin plays a protective role against the extracellular pathogens Vibrio vulnificus and Yersinia spp., by reducing extracellular bioavailable iron, but promotes growth of pathogens such as Salmonella and Mycobacteria whose primary growth niche is intracellular30. It will be important to examine how disorders in host iron metabolism might impact intracellular and extracellular pathogens that use Fe-S cluster sensors to control virulence gene expression. Iron that is transported into the cell cytoplasm either enters the labile iron pool (LIP), and is used to metallate cytoplasmic or mitochondrial components, or is stored in ferritin31. Approximately 80-90% of the LIP is in the Fe(II) reduced state and is bound to molecules like glutathione and poly C binding proteins (PCBPs), iron chaperones that interact with ferritin31. Some cytosolic pathogens utilize the LIP while some utilize ferritin-iron32. In phagosomes and neutrophils, an Nramp2 paralog called Nramp1 is induced by pattern recognition receptors (PRRs) or proinflammatory cytokines. Nramp1 pumps iron and other metals out of the phagosome, limiting phagosomal iron concentration and influencing survival of vacuolar pathogens such as Salmonella14.
It is tempting to assume that the intestinal lumen has a high iron availability for bacterial pathogens, and indeed excess iron can be measured in feces21. However, while total luminal iron content can be high, the majority of the iron does not appear to be readily available to microbes, as much of it is bound to food, certain members of the microbiota, or host iron-binding proteins21. Indeed, production of siderophores by the microbiota serves as evidence that conditions in the intestinal lumen are iron limiting, because bacteria only produce siderophore-based iron acquisition systems when they are iron starved21. This is true particularly in the inflamed gut, as a result of production of host defense molecules such as lipocalin-2, which sequesters certain siderophores14, 33. However, Salmonella enterica serovar Typhimurium (S. Typhimurium) produces lipocalin-2-resistant siderophores, and so can access iron more effectively during inflammation than gut microbes producing only lipocalin-2-sensitive siderophores34.
Production of ROS and RNS by macrophages, neutrophils, as well as other cell types such as intestinal epithelial cells is of great importance to innate immune defense, as exemplified by chronic granulomatous disease patients with defects in NADPH oxidase activity15, 32, 35. NADPH oxidase is the main source for the antimicrobial oxidative burst of macrophages and neutrophils and is induced by innate immune recognition of pathogen-associated molecular patterns, such as lipopolysaccharide, through pattern recognition receptors such as Toll-like receptor 436, 37. In addition to host-derived sources of ROS such as NADPH oxidase, aerobic respiration can generate ROS and iron overload can amplify ROS production. Hence, bacteria encounter a number of environments within the mammalian host where ROS and RNS could impact the function of Fe-S cluster regulators. Likewise, different niches with the mammalian host differ in oxygen tension38. For example, the lumen of the large intestine is devoid of oxygen as a result of the collective action of facultative anaerobes within the microbiota. Yet near the apical surface of colonic epithelial cells, the oxygen concentration increases as a result of diffusion from the intestinal barrier capillary network (see below). Additionally, influx of neutrophils during intestinal inflammation leads to localized depletion of oxygen creating a hypoxic microenvironment for invading pathogens39. This response has been suggested to play a role in modulating oxygen tensions in other environments that accumulate large volumes of neutrophils such as during uropathogenic Escherichia coli urinary tract infections40. Thus, in order to understand how Fe-S cluster regulators impact bacterial virulence, it is important to consider the specific conditions a given pathogen will encounter during the course of infection.
Iron-Sulfur Cluster Regulators and their Role in Bacterial Pathogenesis
As described above, the mammalian host environment contains a diverse array of niches with variable amounts of iron and oxygen as well as oxidative and nitrosative stresses. As these conditions can have profound effects on iron-sulfur cluster homeostasis, bacterial pathogens typically encode one or more Fe-S sensing regulators that act to modulate gene transcription in response to the changing host environment. This section details those Fe-S sensors with characterized roles in the virulence of bacterial pathogens of mammals (Table 1).
Table 1.
Bacterial Fe-S sensors important for mammalian pathogenesis.
| CLUSTER | REGULAT OR | ORGANISM(S) | PROPOSED ROLE IN PATHOGENESIS |
|---|---|---|---|
| [4FE-4S] | FNR | Shigella flexneri | Regulates the Mxi-Spa T3SS in response to changes in colonic O2 |
| Neisseria meningitidis | Regulates genes involved in denitrification and sugar metabolism | ||
| Salmonella enterica serovar Typhimurium | Regulates the SPI-1 T3SS and other virulence-associated genes | ||
| Uropathogenic Escherichia coli (UPEC) | Regulates type I and P fimbrae and other virulence-associated genes | ||
| (ANR) | Pseudomonas aeruginosa | Regulates plcH and is active in lung surfactant | |
| Bacillus cereus | Regulates toxin production in response to O2 concentrations | ||
| Wbl | Mycobacterium tuberculosis | Important for virulence lipid production in response to reducing equivalents | |
| Aconitase | Staphylococcus aureus | Important for production of both secreted and cell-associated virulence factors | |
| Pseudomonas aeruginosa | Inversely correlated with exotoxin A synthesis | ||
| [2FE-2S] | IscR | Yersinia pseudotuberculosis | Regulates the virulence-associated Ysc T3SS |
| Vibrio vulnificus | Regulates virulence determinants in response to host ROS production | ||
| Pseudomonas aeruginosa | Protects against ROS through regulation of katA | ||
| Burkholderia mallei | Defends against reactive nitrogen species209 | ||
| Shigella flexneri | Essential for invasion of host epithelial cells210 | ||
| ([4FE-4S])a | NsrR | Salmonella enterica serovar Typhimurium | Regulates NO detoxifying flavohaemoglobin, hmp |
| Enterohemorrhagic Escherichia coli (EHEC) | Regulates T3SS and other LEE-encoded genes in response to NO | ||
| SoxR | Pseudomonas aeruginosa | Activates quorum-sensing, efflux pumps and a monooxygenase | |
| Vibrio vulnificus | Defends against host ROS production through activation of sodA | ||
| AirSR | Staphylococcus aureus | Regulates agr expression in response to oxygen and oxidative/NO stresses |
Homologs of NsrR have been shown to contain either a [2Fe-2S] cluster or [4Fe-4S] cluster depending on the organism.
[4Fe-4S] Cluster Containing Regulators
FNR
The regulatory protein, FNR (fumarate and nitrate reduction) is one example of a global regulator whose function is modulated by the coordination of an [4Fe-4S] cluster41. FNR has been well characterized in E. coli where it has been shown to control gene expression in response to oxygen42, 43. Interestingly, FNR is produced, but is not an active transcription factor, under aerobic conditions44. FNR is constitutively expressed, leading to continued generation of apo-FNR, which is either degraded via the ATP-dependent protease ClpXP or converted to [4Fe-4S]-FNR45, 46. Activity of FNR is modulated in response to oxygen levels through oxidation of the [4Fe-4S] cluster to [2Fe-2S]47. With extended exposure to oxygen, the [2Fe-2S] cluster is lost and apo-FNR begins to accumulate. Under these conditions, FNR is not an active transcription factor; however, as the oxygen levels decrease, FNR is loaded with a [4Fe-4S] cluster mainly by the Isc biosynthesis pathway48. In this holo form, FNR dimerizes leading to increased DNA-binding at a consensus motif consisting of a symmetrical dyad (TTGAT X4 ATCAA). This increased transcriptional activity leads, in E. coli, to upregulation of approximately 125 genes involved in anaerobiosis42, 43. Thus, FNR acts as a molecular switch to regulate energy metabolism in response to fluctuating oxygen levels.
FNR is expressed by many facultative anaerobes that must survive the transition between aerobic and anaerobic lifestyles or by aerobes that can supplement growth by using alternative metabolic pathways under oxygen limiting conditions49, 50. This includes a number of pathogens that encounter changes in oxygen tension during the course of infection of a host organism. For example, FNR is important for Neisseria meningitidis virulence in rodent models of infection, regulating a number of genes involved in denitrification as well as sugar metabolism and fermentation51. In addition to controlling metabolic pathways important for pathogens to adapt to changes in oxygen availability, FNR has also been co-opted to regulate expression of virulence genes (see below).
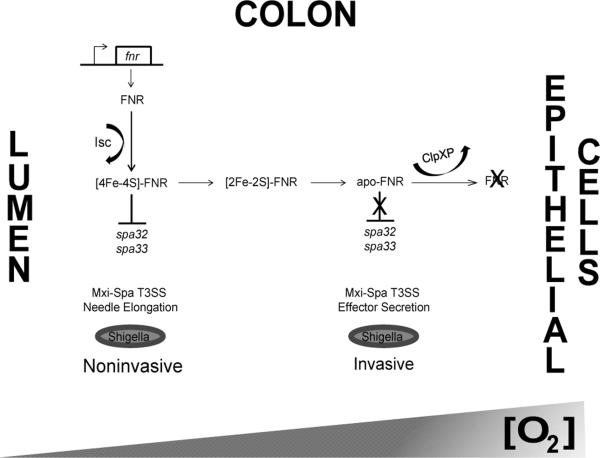
Shigella flexneri infects the human intestine and causes dysentery. A major Shigella virulence factor is the plasmid-encoded Mxi-Spa type III secretion system (T3SS), which enables bacterial internalization into colonic epithelial cells52, 53. Expression of the Shigella T3SS is regulated by pH, osmolarity, temperature, and oxygen availability, enabling optimal timing of T3SS deployment. Shigella uses FNR not only to adapt metabolically to anaerobic conditions, but to control expression of the T3SS in response to changing oxygen availability, as depicted in Figure 1. Importantly, in the absence of FNR, S. flexneri is unable to colonize the intestine54. Under anaerobic conditions such as those found in the lumen of the colon, FNR is bound to a [4Fe-4S] cluster and acts as a repressor of two genes essential for proper T3SS function, spa32 and spa3354. Spa32 mediates the switch between secretion of needle components and effector proteins55-57, while Spa33 is an essential component of the T3SS C-ring where it plays a role in recruiting and exporting T3SS-associated proteins58. As a result of holo-FNR repression of spa32 and spa33 in the low oxygen environment of the colonic lumen, T3SS needles are elongated and ‘primed’, yet effector secretion is suppressed54. The oxygen concentration at the surface of intestinal epithelial cells is thought to be elevated compared to the lumen as a result of diffusion out of the capillary network at the tips of villi54. This increased oxygen concentration is likely to oxidize the FNR [4Fe-4S] cluster upon interaction of S. flexneri with intestinal epithelial cells. This cluster loss leads to alleviation of repression and reversal of the anaerobic block of effector secretion, allowing appropriately timed cell invasion through activation of the T3SS.
Figure 1. The mechanism of oxygen sensing by FNR during Shigella flexneri colonic infection.
A model of Mxi-Spa T3SS regulation by FNR in response to the oxygen concentration gradient in the human colon. It has been demonstrated in E. coli that transcription of fnr is constitutive both in the presence and absence of oxygen and the resulting FNR protein is loaded with a [4Fe-4S] cluster via the Isc Fe-S cluster biosynthesis pathway44. In S. flexneri, under anaerobic conditions such as the lumen of the colon, the [4Fe-4S] cluster is stable and [4Fe-4S]-FNR represses transcription of spa32 and spa3354. Spa32 is essential for proper T3SS function as it mediates the switch between secretion of needle components and effector proteins55-57, while Spa33 is an essential component of the T3SS C-ring where it plays a role in recruiting and exporting T3SS-associated proteins58. Due to these repressive effects of FNR, the Mxi-Spa T3SS needles are elongated and primed, but S. flexneri is unable to secrete T3SS effector proteins and invade host cells. However, in areas surrounding host colonic epithelial cells, oxygen levels are increased due to diffusion from the capillary network of cell villi54. Under these conditions, the [4Fe-4S]-FNR cluster is oxidized to [2Fe-2S] and eventually lost. This leads to accumulation of the inactive form, apo-FNR, which in E. coli is degraded by ClpXP, allowing for derepression of spa32 and spa3345, 54. Induction of Spa32 and Spa33 leads to a switch from secretion of needle components to effector proteins allowing S. flexneri to invade host colonic epithelial cells54.
The Gram-positive organism Bacillus cereus also encodes an FNR homolog. B. cereus is a facultative anaerobic organism that causes food-borne diarrheal syndrome in humans59. During infection, B. cereus colonizes the small intestine, where it secretes a number of virulence factors including hemolysin BL (Hbl), nonhemolytic enterotoxin (Nhe), and cytotoxin (CytK)60. Nhe and Hbl are pore-forming toxins each comprised of three protein components NheA, NheB, NheC and Hbl-B, Hbl-L1 and Hbl-L2 respectively61. FNR positively regulates expression of the nheABC and hbl operons and forms a ternary complex with ResD, the response regulator of the redox sensing two-component system ResDE and the virulence regulator PlcR62, 63. Under environments rich in oxygen, the oxygen-labile [4Fe-4S] cluster is lost resulting in accumulation of apo-FNR. Unlike E. coli and many other FNR containing organisms, apo-FNR of B. cereus is an active transcription factor with a binding affinity similar to [4Fe-4S]-FNR for certain promoters including the nheABC and hbl operons62, 63. However, under conditions of anaerobic growth, such as in the mammalian small intestine, the [4Fe-4S] cluster is stable and [4Fe-4S]-FNR binds with higher affinity to the fnr promoter region62. As such, there is an increase in FNR production, which subsequently leads to increased expression of the nheABC and hbl operons for maximal toxin production during B. cereus infection of the small intestine62. The ternary complex formed between FNR, ResD, and PlcR is believed to play a role in modulating toxin expression; however, the exact mechanism is not fully understood.
FNR is also important for the virulence of S. Typhimurium64, 65. In this pathogen, FNR regulates a similar cohort of genes as in E. coli, including several metabolic pathways and flagellar motility. Interestingly, Salmonella FNR also regulates ethanolamine utilization as well as the ttr operon encoding tetrathionate reductase65-67, both of which were recently shown to provide a growth advantage to S. Typhimurium in the inflamed intestinal lumen68, 69. In addition, Salmonella FNR is involved in regulation of the SPI-1 T3SS essential for bacterial invasion into intestinal epithelial cells, as well as several virulence-associated genes that may promote intracellular growth65. Thus, it appears that FNR aids Salmonella in reprogramming metabolic gene expression under the anaerobic conditions of the intestinal lumen to compete with the microbiota and grow within that niche, while inducing expression of virulence factors that promote entry inside intestinal epithelial cells and intracellular survival should the bacteria encounter host cells.
In uropathogenic Escherichia coli (UPEC), the causative agent of the majority of urinary tract infections, FNR is essential for virulence in a mouse urinary tract infection model and for adherence to and invasion of bladder and kidney epithelial cells70, 71. UPEC FNR is a global regulator controlling gene expression of type I and P fimbriae (important for adherence to bladder and kidney cells), motility (which plays a role in UPEC ascension to the upper urinary tract), as well as other virulence-associated genes such as a hemolysin and a novel pathogenicity island not found in other commensal or intestinal E. coli (which enables utilization of α-ketoglutarate)70. Alpha-ketoglutarate is an intermediate in the TCA cycle and is an abundant metabolite in renal proximal tubule cells, an infection site of UPEC72, 73. The ability of UPEC to utilize host-derived α-ketoglutarate under anaerobic conditions has been demonstrated to be essential for colonization of the bladder and kidneys74. Collectively, FNR facilitates UPEC host cell contact and metabolic adaptation to promote growth in the urinary tract.
Pseudomonas aeruginosa is an important opportunistic pathogen that can cause life-threatening infections in immunocompromised hosts. The FNR homolog ANR (anaerobic regulator of arginine deiminase and nitrate reductase) of P. aeruginosa is important for colonization of airway epithelial cells and is essential for virulence in a murine model of acute-phase pneumonia75-77. Similar to FNR, ANR is an active transcription factor under low oxygen conditions when coordinated with a [4Fe-4S] cluster and recognizes a 5′-TTGATNNNNATCAA-3 consensus motif77, 78. Interestingly, ANR is active in lung surfactant-containing medium despite elevated oxygen levels, which occurs in a hemolytic phospholipase C (PlcH)-dependent manner76. PlcH, is a secreted virulence factor of P. aeruginosa that cleaves host-associated phosphotidylcholine (PC) and sphingomyelin located in eukaryotic membranes and host lung surfactant79, 80. The release of choline by PlcH is believed to stimulate ANR activity leading to enhanced biofilm production and host airway colonization in the presence of oxygen, by an as-yet unidentified mechanism76. Under anaerobic conditions, ANR represses transcription of plcH in a negative feedback loop81. This example highlights the interplay between metabolism and virulence driven by Fe-S cluster sensing regulators.
Wbl
WhiB-like (Wbl) proteins in Actinobacteria have been demonstrated to play diverse roles in morphogenesis, cell division, virulence, and metabolism as well as in antibiotic resistance82, 83. The well characterized WhiB3 of Mycobacterium tuberculosis has been shown to coordinate a [4Fe-4S] cluster and its DNA-binding activity is altered in response to fluctuations in the concentration of NO and O2 as well as redox stress84, 85. When WhiB3 is exposed to high levels of O2 or NO, the [4Fe-4S]1+ is oxidized to [4Fe-4S]2+ and further converted to a [3Fe-4S]1+ cluster. After prolonged exposure, the Fe-S cluster is eventually lost, enabling apo-WhiB3 to bind DNA with high affinity. NifS is able to restore the [4Fe-4S] cluster to apo-WhiB3, leading to generation of holo-WhiB3 and a reduction in DNA-binding affinity85. Uniquely, the exposed cysteine residues of apo-WhiB3 are susceptible to redox stress and as such serve as an additional level of regulation85. During exposure to thiol-specific oxidants, the four conserved cysteines undergo formation of two intramolecular disulphide bonds, resulting in enhanced DNA-binding activity. This activity can be abolished by exposure to thiol-specific reductants and loss of the disulphide bonds. Thus, sensing by WhiB3 is bi-phasic such that the [4Fe-4S] cluster senses NO and O2 stresses to modulate DNA-binding activity, while in the absence of cluster ligation the exposed cysteines further influence DNA-binding activity in response to reductive stress84, 85.
M. tuberculosis encodes seven Wbl proteins, WhiB1-WhiB7. These regulatory proteins respond to a number of environmental stimuli including exposure to detergents, acid, heat, and variable concentrations of nutrients, ethanol, oxygen, NO, and iron86-89. WhiB3 is essential for full virulence in mammalian tuberculosis models by maintaining redox homeostasis and promoting lipid biosynthesis during macrophage infection, the favored niche of M. tuberculosis90. Approximately 60% of the M. tuberculosis cell wall is comprised of lipids and its lipid profile is altered during infection in order to defend against the host immune system91. A number of M. tuberculosis lipids act as virulence factors. For example, sulfolipid-1 (SL-1) is a tetraacylated glycolipid that modulates host immune responses through inhibition of phagasome-lysosome fusion and modulation of cytokine and host ROS production92-100. Other important glycolipids include the di-, tri- and polyacyltrehaloses (DAT, TAT and PAT, respectively), which hinder host cell phagocytosis101. Another prominent cell wall lipid, trehalose dimycolate (TDM), also known as cord factor, is toxic to mammalian cells and functions by inhibiting phospholipid vesicle fusion and neutrophil migration91. Removal of M. tuberculosis lipids leads to decreased persistence within macrophages and a reduction in the host immune response, demonstrating the importance of these lipids in M. tuberculosis pathogenesis102.
During infection, M. tuberculosis is exposed to oxygen and NO stress, which influence stability of the WhiB3 [4Fe-4S] cluster84, 103. In the presence of these stressors, the [4Fe-4S] cluster is degraded leading to an accumulation of apo-WhiB3. Interestingly, this form of WhiB3 is able to undergo further post-translational modifications in response to redox stress, modulating DNA-binding activity85, 103. M. tuberculosis experiences redox stress during infection as a result of NADPH production following fatty acid β-oxidation103, 104. Reducing equivalents such as NADPH can undergo autoxidation, leading to increased ROS production105. The exact mechanism by which apo-WhiB3 senses redox stress is unknown. However, it is hypothesized that accumulation of NADPH during infection increases oxidative stress, leading to accumulation of oxidized apo-WhiB3 containing intraprotein disulphide bonds between the four cluster-coordinating cysteines85. The oxidized form of apo-WhiB3 exhibits strong DNA-binding activity leading to upregulation of SL-1, PAT/DAT and TDM lipid synthesis85, 103. Additionally, excess NADPH generated by M. tuberculosis via fatty acid β-oxidation is consumed in the production of these cell wall lipids, serving as a feedback loop for maintaining redox homeostasis84, 85, which is illustrated in Saini et al.103. Oxygen and NO sensing by the [4Fe-4S] cluster serve as an important first step in WhiB3 regulation, as exposure of [4Fe-4S]-WhiB3 to redox stresses does not influence DNA-binding activity. Collectively, WhiB3 functions through a unique mechanism in order to regulate redox homeostasis during macrophage infection and induce the proper cell wall lipid composition required for host immune evasion.
Aconitase
Aconitases are highly conserved enzymes in both eukaryotes and prokaryotes that convert citrate to isocitrate in the TCA cycle and contain a labile [4Fe-4S] cluster that is essential for enzymatic activity106. In addition to their catalytic function, the eukaryotic aconitase, IRP-1 (iron regulatory protein 1), is located in the cytosol and is a bifunctional protein that acts as an RNA-binding protein in an iron-dependent manner107-109. IRP-1 recognizes specific sequences on the mRNA transcript termed iron-responsive elements (IREs), which are stem-loop structures located in either the 5’ or 3’ untranslated regions (UTR) of mRNAs encoding iron metabolism proteins106, 110. The location of the IRE dictates the effect that binding of IRP-1 will have on the mRNA transcript. Specifically, binding of IRP-1 to IREs located in the 5’ UTR will decrease protein production through inhibition of translation. Conversely, binding in the 3’ UTR will increase protein levels through stabilization of the mRNA transcript. In the presence of iron, the [4Fe-4S] cluster is bound to IRP-1 resulting in inhibition of RNA-binding activity. Under iron depleted conditions, the [4Fe-4S] cluster is lost and IRP-1 is able to bind to IREs and coordinately regulate protein production111-114. Thus the IRP-1 aconitase has a dual role in modulating iron metabolism and contributing to energy generation. Most pathogenic bacteria have IRP-1 homologs that are believed to play a role similar to that of their eukaryotic counterparts. For example, aconitase of B. subtilis is a bifunctional protein, demonstrated to possess both enzymatic activity through its role in converting citrate to isocitrate in the TCA cycle as well as mRNA-binding activity through recognition of IRE-like sequences115-117.
Aconitase has been linked to synthesis of an important exotoxin that is central to P. aeruginosa disease causation. This organism encodes a plethora of secreted and cell-associated virulence determinates including proteases, toxins, phospholipases, pili, rhamnolipids and the exopolysaccharide alginate118. The most toxic of these virulence factors to mammalian cells is the secreted enzyme, exotoxin A (ETA). ETA is able to inhibit protein synthesis through its ADP-ribosylating activity leading to host cell death, as it catalyzes the transfer of ADP-ribose from NAD to eukaryotic elongation factor 2119, 120. Interestingly, Somerville et al., demonstrated an inverse correlation between aconitase activity and synthesis of this important exotoxin121. In the absence of iron, aconitase activity was decreased and the gene encoding ETA, toxA, transcribed. Furthermore, fluorocitrate, an aconitase-specific inhibitor, reduced toxA transcription. The exact mechanism by which aconitase influences ETA synthesis is not fully understood; however, the study by Somerville et al., suggests a role for aconitase in contributing to virulence factor synthesis in P. aeruginosa.
Aconitase of another important opportunistic pathogen, Staphylococcus aureus, has also been implicated in the production of secreted virulence factors as well as cell-associated adhesion factors122. S. aureus utilizes a vast arsenal of virulence determinants to successfully colonize an abundance of niches within the host. In the absence of aconitase, there is a decrease in production of glycerol ester hydrolase, a lipase that hinders phagocytic killing by granulocytes123, a type C enterotoxin that may play a role in food poisoning124, as well as α- and β-toxins, two cytolytic toxins that target host cells122, 125. Production of cytolytic toxins by S. aureus has been shown to play a role in modulating immune responses as well as scavenging nutrients such as iron from erythrocytes125, 126. Therefore, S. aureus likely utilizes virulence factor production in order to combat iron deprivation in the host environment. Indeed the iron regulator, Fur coordinates production of S. aureus hemolysins and cytotoxins, including α-toxin, in response to iron availability127. As such, it is tempting to speculate that S. aureus utilizes aconitase as an additional level of regulation to sense and respond to iron limiting conditions within the host. Interestingly, mutation of aconitase does not drastically alter the severity of S. aureus infection after intraperitoneal infection. However, in a murine wound formation model, mice infected with aconitase mutants lost significantly more weight and displayed delayed onset of ulceration as well as delayed recovery at the infection site122. Whether aconitase is important for colonization of one or more of the other numerous host niches that S. aureus is capable of infecting remains to be determined. Although the mechanism by which aconitase coordinates energy generation with virulence factor expression during infection has yet to be elucidated, it is clear that aconitases play an important role in host-pathogen interactions.
[2Fe-2S] Cluster Containing Regulators
IscR
In a separate group are regulators that coordinate a [2Fe-2S] cluster to modulate gene transcription. This group includes the iron-sulfur cluster regulator, IscR, belonging to the Rrf2 family of winged helix-turn-helix transcription factors128, 129. IscR has been extensively characterized in E. coli where its DNA-binding activity is modulated based on the coordination of a [2Fe-2S] cluster through three conserved cysteines and a histidine129-133. In a mechanism distinct from that of FNR, IscR is an active transcription factor both in the apo-IscR and holo-IscR forms. Fe-S cluster loading of IscR occurs through the activity of the cotranscribed Isc Fe-S biosynthesis pathway. Holo-IscR directly represses transcription of the iscRSUA operon in order to maintain proper Fe-S cluster homeostasis; however, IscR also regulates gene expression beyond the isc operon. This occurs through the ability of IscR to recognize two distinct binding motifs: type 1 motifs (ATASYYGACTRwwwYAGTCRRSTAT), which are recognized solely by holo-IscR, and type 2 motifs (AxxxCCxxAxxxXxxxTAxGGxxxT), which are bound by both holo- and apo-IscR133, 134. The holo-IscR/apo-IscR ratio is affected by iron availability, oxidative stress, and oxygen limitation; thus, these environmental stimuli are believed to effect gene expression through IscR131-133.
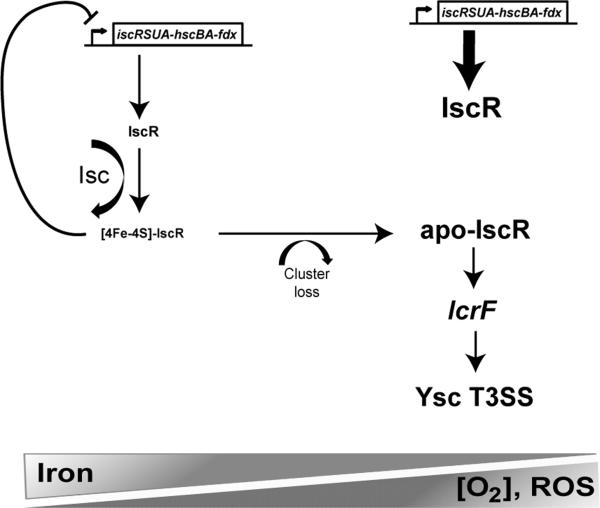
Enteropathogenic Yersinia, Y. enterocolitica and Y. pseudotuberculosis, cause gastrointestinal disease in healthy individuals and more serious disseminated infection in immunocompromised hosts or those with iron overload disorders such as hereditary hemochromatosis135-138. Interestingly, IscR was recently shown to be critical for Y. pseudotuberculosis pathogenesis139. Y. pseudotuberculosis can cross the small intestinal barrier to enter the bloodstream and deeper tissues140-142, where its Ysc T3SS is required for full virulence143. Yersinia utilize their Ysc T3SS to inject a series of effector proteins into host cells that collectively inhibit bacterial uptake into phagocytic cells and dampen other host defense responses such as ROS production. Expression and function of the T3SS is tightly regulated in Yersinia and there are several environmental cues that are known to mediate T3SS control including temperature, calcium concentration, and host cell contact144. Y. pseudotuberculosis IscR was recently shown to directly regulate the Ysc T3SS139. In tissues with low iron availability, sufficient oxygen tension, and/or oxidative stress, loss of the [2Fe-2S] cluster on IscR may lead to derepression of the iscRSUA operon and subsequent increase in IscR levels (Figure 2). Elevated apo-IscR induces transcription from type II motif-containing promoters, which includes the gene encoding the Ysc T3SS master regulator, LcrF139. As such, it is hypothesized that Y. pseudotuberculosis uses IscR to sense iron, O2, and/or ROS concentration, in addition to temperature and host cell contact, in order to optimize T3SS expression during infection.
Figure 2. The mechanism of IscR-dependent Ysc T3SS regulation during enteropathogenic Yersinia infection.
A model of IscR control of the Yersinia Ysc T3SS under differing iron availability, oxygen tension, and ROS concentration. Under anaerobic, low ROS conditions where Yersinia is able to obtain iron, such as the gut lumen, transcription of the isc operon should be limited due to sufficient [2Fe-2S] cluster loading onto IscR (holo-IscR)208, which recognizes a type 1 DNA-binding motif in the isc promoter to repress transcription in a negative feedback loop139. Ysc T3SS expression is predicted to be low under such conditions. However, in tissues that are iron-poor (such as the blood), rich in ROS (such as inflamed tissue), or high in oxygen tension, apo-IscR is predicted to accumulate, leading to stimulation of type II motif-containing promoters including the promoter upstream of the gene encoding LcrF, the Ysc T3SS master regulator139. This upregulation may allow increased T3SS expression in niches where Y. pseudotuberculosis requires its T3SS to inhibit uptake and killing by phagocytic cells.
Vibrio vulnificus is capable of causing food poisoning as well as wound infections in mammalian hosts, typically through contamination of a preexisting laceration during swimming in warm coastal waters145, 146. IscR of V. vulnificus is induced in the presence of host epithelial cells as a result of reactive oxygen species production147. Based on the documented biochemistry of E. coli [2Fe-2S]-IscR, this increase in iscR expression is likely a result of [2Fe-2S] cluster loss following its oxidation, leading to apo-IscR derepression of the isc operon. In V. vulnificus, this increased apo-IscR also leads to induction of several virulence-associated pathways. Specifically, Vibrio IscR is required for appropriate expression of two genes encoding proteins with putative antioxidant properties, peroxiredoxin (Prx) and glutaredoxin 2 (Grx2). Both of these proteins in other organisms have been shown to be important for detoxifying the host environment following antibacterial defenses elicited by the immune system148, 149. Furthermore, IscR of V. vulnificus is essential for proper regulation of the vvhBA operon, encoding a putative cytolysin secretory protein VvhB and the cytolysin VvhA, a potent toxin that targets erythrocytes150, 151. Interestingly, iron has been shown to influence both the expression and secretion of this hemolysin151. This suggests that low iron and exposure to host ROS may act as signals for IscR-mediated gene regulation in order to acquire iron and detoxify oxidative stresses during V. vulnificus infection.
IscR also plays an important role in the virulence of P. aeruginosa, which can be exposed to high levels of ROS resulting from the macrophage oxidative burst152, 153. In order to circumvent this host defense strategy, P. aeruginosa employs the highly stable catalase, KatA, to detoxify ROS, as demonstrated by the inability of P. aeruginosa katA mutants to cause disease in a mouse peritonitis model154. Interestingly, expression of katA is not effected by IscR; however, decreased activity of KatA is observed in an iscR mutant. Furthermore, mutation of iscR also leads to decreased pathogenesis in a peritonitis model152. Kim et al., hypothesized that the regulatory effect of IscR on KatA activity may occur through disruptions in the intracellular pool of iron available to generate heme, an essential cofactor for KatA152, 155.
NsrR
The nitric oxide sensing Rrf2-type transcriptional repressor NsrR belongs to the winged helix superfamily and is structurally similar to IscR with three conserved cysteines in the C-terminal region that serve to coordinate the Fe-S cluster. In E. coli, NsrR binds DNA at a 23 base pair (bp) palindrome that is arranged as two 11 bp inverted sequences (AANATGCATTT) separated by a single nucleotide156. The regulatory activity of NsrR is dependent on the reversible coordination of an oxygen-insensitive Fe-S cluster through the three conserved cysteines157-159. Interestingly, while NsrR of Streptomyces coelicolor, S. Typhimurium, Escherichia coli, and Neisseria gonorrhoeae were found to contain [2Fe-2S] clusters, studies in Bacillus subtilis demonstrated it to harbor a [4Fe-4S] containing NsrR157, 160, 161. In Neisseria gonorrhoeae and other organisms, the regulatory activity of NsrR is modulated by NO stress. Specifically, nitrosylation of [2Fe-2S]-NsrR leads to cluster destabilization, thereby abolishing DNA-binding activity157-160, 162, 163.
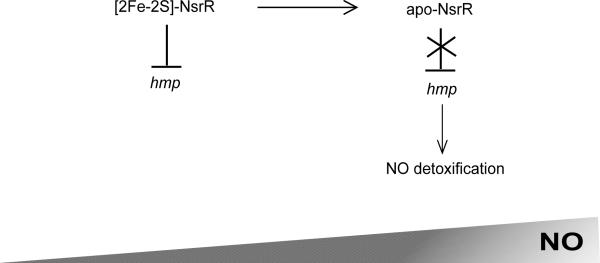
The enteric pathogen S. Typhimurium is a leading cause of human gastroenteritis. In order for S. Typhimurium to cause invasive disease, it must be able to persist within macrophages164. Macrophages serve as an important niche for S. Typhimurium, yet they utilize a number of mechanisms to inhibit invading organisms including the production of RNS165, 166. As such, it is essential that S. Typhimurium be able to sense host NO production and respond in order to coordinate the virulence factors necessary to subvert the host immune response. NsrR of S. Typhimurium contains a NO-sensitive [2Fe-2S] cluster, which in the absence of nitrosative stress represses transcription of hmp encoding a NO detoxifying flavohaemoglobin (Figure 3)161, 167. When S. Typhimurium is exposed to NO stress, such as in the intracellular environment of the macrophage, the [2Fe-2S] cluster is destabilized leading to derepression of hmp161, 167. Flavohaemoglobin subsequently converts NO to N2O or to nitrate (NO −3), enabling S. Typhimurium to resist nitric oxide killing by host macrophages161, 168.
Figure 3. The mechanism of S. Typhimurium NsrR-dependent hmp regulation.
A model of NsrR control of hmp encoding a NO detoxifying flavohaemoglobin under increasing nitric oxide stress. During growth of S. Typhimurium when concentrations of nitric oxide are minimal, such as in the environment outside of the mammalian host, the NsrR [2Fe-2S] cluster is stable. Under these conditions, [2Fe-2S]-NsrR is a functional DNA-binding protein and represses transcription of hmp encoding a nitric oxide detoxifying flavohaemoglobin. However, when S. Typhimurium is exposed to NO stress, such as that generated by host macrophages where S. Typhimurium survives and proliferates during mammalian infection, the NsrR [2Fe-2S] cluster is nitrosylated. These conditions lead to cluster destabilization and abolish NsrR DNA-binding activity. As such, hmp expression is no longer repressed and the resulting NO detoxification by flavohaemoglobin allows for S. Typhimurium persistence within host macrophages.
Another food-borne pathogen, Enterohemorrhagic Escherichia coli (EHEC) O157:H7, which causes diarrhea, hemorrhagic colitis, and even renal failure, encodes a NO-sensing NsrR. In order for EHEC to colonize the host, it must adhere to intestinal epithelial cells169. During this attachment, EHEC subverts host cytoskeletal processes in order to form attaching and effacing (A/E) lesions170. The ability of EHEC to form these A/E lesions and associate with the plasma membrane of host intestinal epithelial cells is a direct result of a chromosomally-encoded pathogenicity island termed the locus of enterocyte effacement (LEE)171. This chromosomal pathogenicity island is largely organized in 5 major operons (LEE1-LEE5). The LEE1, LEE2, and LEE3 operons encode T3SS secreted proteins, chaperones, and regulators including the main activator, Ler, encoded on LEE1172. LEE4 encodes genes that comprise the T3SS translocon and a syringe, while LEE5 encodes the adhesin intimin as well as the intimin receptor Tir173. NO production is an innate immune response of intestinal mucosa, as such EHEC are exposed to nitrosative stress during attachment and invasion of epithelial cells. Interestingly, NsrR directly activates transcription of the LEE1, LEE4, and LEE5 operons in the absence of NO171. Counterintuitively, T3SS-dependent EHEC adhesion to host cells is inhibited in the presence of NO in an NsrR-dependent manner171. It has been hypothesized by Branchu et al., that this NO directed regulation of LEE may serve to limit EHEC colonization of the stomach lining in order to promote colonic infection171. Furthermore, EHEC has been shown to inhibit RNS production by human enterocytes, which may ultimately serve to promote host cell attachment and invasion171, 174.
SoxR
The E. coli superoxide response regulator SoxR utilizes a [2Fe-2S] cluster to sense superoxide stress in order to coordinately regulate gene transcription. Like IscR and NsrR, SoxR belongs to the Rrf2 family of winged helix-turn-helix regulators, whose function is modulated based on the coordination of an Fe-S cluster. Coordination of the [2Fe-2S] cluster occurs through a conserved sequence, CysX2CysXCysX5Cys, in the carboxy-terminus of SoxR homologs in E. coli, P. aeruginosa, and Streptomyces coelicolor175-177. SoxR activity is distinct from that of IscR and NsrR in that it functions as a regulator solely when the [2Fe-2S] cluster is in the oxidized form as a result of exposure to the superoxide anion O2•-178. Additionally, E. coli SoxR has been shown to be activated in the presence of NO stress through nitrosylation of the Fe-S cluster179. As demonstrated in Figure 4, upon sensing redox stress, SoxR activates expression of soxS encoding an AraC-type regulator180, 181. SoxS subsequently upregulates genes involved in redox homeostasis and repair182. Interestingly, SoxRS regulatory activity appears to be confined to members of the Enterobacteriaceae, as SoxS homologs are only present in enteric bacteria. In other bacteria, SoxR alone has been shown to directly regulate a small set of genes183. While SoxRS of Enterobacteriaceae seem to play a role in protection against exogenous redox-cycling compounds, SoxR of P. putida, P. aeruginosa, and S. coelicolor are believed to protect against endogenously generated antibiotics175, 177, 184-188.
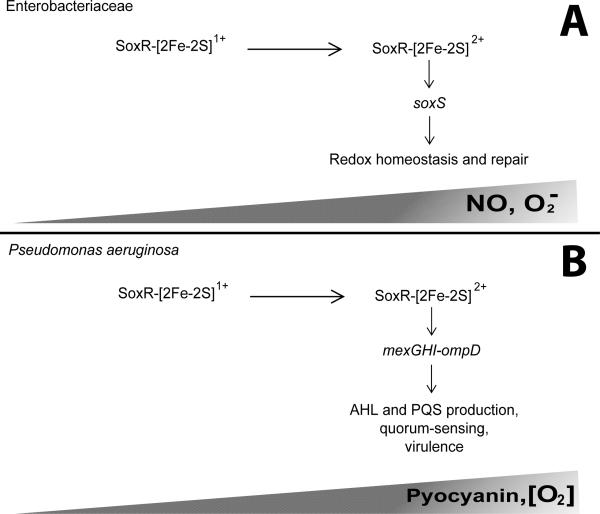
Figure 4. The mechanism of SoxR regulation in the family Enterobacteriaceae and P aeruginosa.
A model of SoxR regulatory activity in members of the Enterobacteriaceae and P. aeruginosa. (A) SoxR of Enterobacteria coordinates a NO- and superoxide anion-sensitive [2Fe-2S] cluster. During growth under minimal NO and oxidative stress conditions, the SoxR-[2Fe-2S] cluster is in the reduced form (1+) resulting in abolished DNA-binding activity. However, as the concentration of NO and/or superoxide anion increases, such as within the mammalian host, the [2Fe-2S] cluster is oxidized (2+). Under these conditions, SoxR is an active transcription factor and functions solely to upregulate soxS encoding an AraC-type regulator, which then functions to coordinate gene expression to maintain redox homeostasis. (B) The activity of SoxR of P. aeruginosa is slightly different from that described for Enterobacteria. Specifically, the [2Fe-2S] cluster is oxidized is the presence of pyocyanin, which accumulates during stationary phase, and high oxygen concentrations. Additionally, SoxS is absent from P. aeruginosa and other non-Enterobacteria. As such, the oxidized form, SoxR-[2Fe-2S]2+ directly regulates a small subset of genes, mexGHI-ompD. Among other proteins, this operon encodes a multidrug efflux pump required for secretion of the quorum sensing quoromones, N-acylhomoserine lactone (AHL) and 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS). As quorum sensing is required for both acute and chronic infections, SoxR plays an important role in P. aeruginosa pathogenesis.
SoxR of P. aeruginosa is essential for full virulence in mammalian hosts, as demonstrated by a notable decrease in the ability of soxR mutants to avoid killing by macrophages, increased survival of mice following pulmonary challenge with a soxR mutant, as well as diminished systemic dissemination in mice infected with a soxR mutant185, 189. Accumulation of the endogenous redox-active small molecule pyocyanin during stationary phase growth leads to activation of SoxR in a superoxide-independent manner187. Additionally, SoxR has been demonstrated to respond to oxygen-induced stress185. As P. aeruginosa is not a member of the family Enterobacteriaceae, it does not encode a soxS gene. Instead, SoxR of P. aeruginosa directly regulates expression of mexGHI-ompD, an operon consisting of a multidrug efflux pump involved in quorum-sensing, as well as genes encoding a putative efflux pump and a monooxygenase185, 187, 190. Quorum-sensing is a mechanism utilized by many bacteria in order to sense the surrounding population density and coordinately regulate gene expression as a type of cell-to-cell communication. Typically, a quoromone signal is produced, and in the case of P. aeruginosa there are two, the N-acylhomoserine lactone (AHL) and 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS). A quoromone threshold is commonly sensed by a two component signal transduction system leading to global regulatory changes, often in virulence gene expression. In P. aeruginosa, quorum sensing is active in vivo and is essential for both acute and chronic infections, demonstrated through numerous infection models. Mutation of the mexGHI-ompD encoded pump leads to decreased AHL and PQS production and subsequently reduced virulence in a rat lung infection model191. During the course of infection, P. aeruginosa is exposed to oxidative stresses generated by the innate immune response; therefore, it can be hypothesized that this exposure leads to oxidation of [2Fe-2S]-SoxR and subsequent upregulation of the mexGHI-ompD encoded pump. As this pump is necessary for appropriate AHL and PQS quorum-sensing, this mechanism of SoxR regulation likely contributes to the pathogenesis of P. aeruginosa through properly timed virulence determinant expression (Figure 4).
SoxR has also been implicated in the virulence of V. vulnificus192. Integral to the capacity of V. vulnificus to cause food-borne disease is an ability to tolerate the acidic pH of the stomach. Survival occurs through the use of an acid-neutralizing system cadBA, which encodes a lysine-cadaverine antiporter (CadB) and a lysine decarboxylase (CadA)193. Furthermore, the gene encoding the manganese superoxide dismutase sodA is also an important component of the acid response and has been shown to promote survival under low pH194, 195. SoxR is essential for transcription of both sodA and cadBA and thus may sense ROS in the stomach, enabling protection from acid stress and facilitating disease causation194, 196. Interestingly, mice that have been infected via intraperitoneal injection with either a soxR or sodA mutant display reduced virulence and complementation of the soxR mutant with sodA restores this defect192. This suggests a requirement for SoxR-dependent regulation of sodA in the disease causation of V. vulnificus beyond survival within acidic gastric fluids.
AirSR
The anaerobic iron-sulfur cluster-containing redox sensor regulator AirSR, also known as YhcSR, is an [2Fe-2S]-containing two-component signal transduction system that regulates S. aureus virulence gene expression in response to oxidative and redox stresses197. As illustrated by Sun et al., the membrane bound histidine kinase AirS coordinates an [2Fe-2S] cluster, which influences its kinase activity197. Specifically, during growth in the absence of ROS or under low oxygen concentrations, the [2Fe-2S] cluster is reduced ([2Fe-2S]1+) and phosphorylation of the response regulator AirR is limited197. Oxidation of the cluster to [2Fe-2S]2+ leads to fully active AirS; however, over exposure to oxidative stress leads to cluster loss and inhibition of AirS kinase activity197. Moreover, exposure to NO stress results in dinitrosyl-iron-dithiol complex formation with the AirS [2Fe-2S] cluster, leading to protein inactivation197. Under anaerobic conditions, AirR represses transcription of a number of genes encoding S. aureus virulence factors including the two-component system SaeRS, which regulates many extracellular proteins in response to environmental stimuli198-200, an immunoglobulin binding surface protein Spa, which aids in immune evasion201, and Agr197. Agr is a quorum sensing, two-component system central to the pathogenesis of S. aureus202-205. The agr locus is expressed as growth progresses from exponential to stationary phase, where there is a shift in gene expression profiles from surface proteins, to secreted proteases and toxins203-205. These secreted and surface-associated virulence determinants play an important role in the success of S. aureus as a pathogen, as these factors allow for adhesion, immune evasion, and dissemination206, 207. Collectively, these data suggest that AirRS plays an important role in coordinating appropriately timed virulence determinant production in response to the host environment. Specifically, host generated ROS/RNS likely leads to AirS [2Fe-2S] cluster loss, inhibiting kinase activity. Inactivation of AirS then leads to accumulation of inactive, unphosphorylated AirR, thereby alleviating repression of agr. This increased agr activity would subsequently lead to increased production of secreted proteases and toxins allowing for evasion of the host immune response as well as dissemination.
Concluding remarks
The examples listed above as well as other reports not covered in this review have established the importance of Fe-S cluster coordinating regulators in bacterial pathogenesis, making them potential therapeutic targets for development of novel antimicrobials. However, targeting Fe-S cluster coordination in general would not be a viable option given the importance of these prosthetic groups in eukaryotes. The unique ability of Fe-S cluster coordinating regulators to respond to changes in iron availability, oxygen tension, and ROS/RNS levels places them in an ideal position to enable bacteria to adapt their gene expression profiles to optimize survival within the often hostile host environment. It will be important to more concretely link our knowledge of how Fe-S cluster coordinating regulators control gene expression and virulence with the nature of the environmental conditions encountered by bacterial pathogens inside and outside the host.
Supplementary Material
Acknowledgements
The authors acknowledge the National Institutes of Health (R21AI099747 to V.A.) for support.
References
- 1.Archibald F. FEMS microbiology letters. 1983;19:29–32. [Google Scholar]
- 2.Posey JE, Gherardini FC. Science. 2000;288:1651–1653. doi: 10.1126/science.288.5471.1651. [DOI] [PubMed] [Google Scholar]
- 3.Touati D. Archives of biochemistry and biophysics. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 4.Andrews SC, Robinson AK, Rodriguez-Quinones F. FEMS microbiology reviews. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 5.Mortenson LE, Valentine RC, Carnahan JE. Biochemical and biophysical research communications. 1962;7:448–452. doi: 10.1016/0006-291x(62)90333-9. [DOI] [PubMed] [Google Scholar]
- 6.Moulis JM, Davasse V, Golinelli MP, Meyer J, Quinkal I. Journal of Biological Inorganic Chemistry. 1996;1:2–14. [Google Scholar]
- 7.Glaser T, Hedman B, Hodgson KO, Solomon EI. Accounts of chemical research. 2000;33:859–868. doi: 10.1021/ar990125c. [DOI] [PubMed] [Google Scholar]
- 8.Noodleman L, Case DA. Adv Inorg Chem. 1992;38:423–+. [Google Scholar]
- 9.Mettert EL, Kiley PJ. Biochimica et biophysica acta. 2014 doi: 10.1016/j.bbamcr.2014.11.018. DOI: 10.1016/j.bbamcr.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beinert H, Kiley PJ. Current opinion in chemical biology. 1999;3:152–157. doi: 10.1016/S1367-5931(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 11.Johnson DC, Dean DR, Smith AD, Johnson MK. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 12.Loreal O, Cavey T, Bardou-Jacquet E, Guggenbuhl P, Ropert M, Brissot P. Frontiers in pharmacology. 2014;5:128. doi: 10.3389/fphar.2014.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nairz M, Haschka D, Demetz E, Weiss G. Frontiers in pharmacology. 2014;5:152. doi: 10.3389/fphar.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cassat JE, Skaar EP. Cell Host Microbe. 2013;13:510–520. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang FC. Nature reviews. Microbiology. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 16.Cassat JE, Skaar EP. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nairz M, Schroll A, Demetz E, Tancevski I, Theurl I, Weiss G. Immunobiology. 2015;220:280–294. doi: 10.1016/j.imbio.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Mladenka P, Simunek T, Hubl M, Hrdina R. Free radical research. 2006;40:263–272. doi: 10.1080/10715760500511484. [DOI] [PubMed] [Google Scholar]
- 19.Circu ML, Aw TY. Free radical biology & medicine. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherayil BJ, Ellenbogen S, Shanmugam NN. Current opinion in gastroenterology. 2011;27:523–528. doi: 10.1097/MOG.0b013e32834a4cd1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kortman GA, Raffatellu M, Swinkels DW, Tjalsma H. FEMS microbiology reviews. 2014;38:1202–1234. doi: 10.1111/1574-6976.12086. [DOI] [PubMed] [Google Scholar]
- 22.Forbes JR, Gros P. Blood. 2003;102:1884–1892. doi: 10.1182/blood-2003-02-0425. [DOI] [PubMed] [Google Scholar]
- 23.Ganz T, Nemeth E. Biochimica et biophysica acta. 2012;1823:1434–1443. doi: 10.1016/j.bbamcr.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivera S, Ganz T. Semin Hematol. 2009;46:351–357. doi: 10.1053/j.seminhematol.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal N, Prchal JT. Acta Haematol. 2009;122:103–108. doi: 10.1159/000243794. [DOI] [PubMed] [Google Scholar]
- 26.Vujic M. Frontiers in pharmacology. 2014;5:42. doi: 10.3389/fphar.2014.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barton JC. The American journal of the medical sciences. 2013;346:403–412. doi: 10.1097/MAJ.0000000000000192. [DOI] [PubMed] [Google Scholar]
- 28.Cairo G, Recalcati S, Montosi G, Castrusini E, Conte D, Pietrangelo A. Blood. 1997;89:2546–2553. [PubMed] [Google Scholar]
- 29.Moura E, Noordermeer MA, Verhoeven N, Verheul AF, Marx JJ. Blood. 1998;92:2511–2519. [PubMed] [Google Scholar]
- 30.Fang FC, Weiss G. Cell Host Microbe. 2014;15:515–516. doi: 10.1016/j.chom.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philpott CC, Ryu MS. Frontiers in pharmacology. 2014;5:173. doi: 10.3389/fphar.2014.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paiva CN, Bozza MT. Antioxidants & redox signaling. 2014;20:1000–1037. doi: 10.1089/ars.2013.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherayil BJ. Immunol Res. 2011;50:1–9. doi: 10.1007/s12026-010-8199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Baumler AJ. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nathan C, Shiloh MU. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal AW. Annual review of immunology. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeya R, Sumimoto H. Molecules and cells. 2003;16:271–277. [PubMed] [Google Scholar]
- 38.Siegemund M, van Bommel J, Ince C. Intensive care medicine. 1999;25:1044–1060. doi: 10.1007/s001340051011. [DOI] [PubMed] [Google Scholar]
- 39.Campbell EL, Bruyninckx WJ, Kelly CJ, Glover LE, McNamee EN, Bowers BE, Bayless AJ, Scully M, Saeedi BJ, Golden-Mason L, Ehrentraut SF, Curtis VF, Burgess A, Garvey JF, Sorensen A, Nemenoff R, Jedlicka P, Taylor CT, Kominsky DJ, Colgan SP. Immunity. 2014;40:66–77. doi: 10.1016/j.immuni.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subashchandrabose S, Hazen TH, Brumbaugh AR, Himpsl SD, Smith SN, Ernst RD, Rasko DA, Mobley HL. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18327–18332. doi: 10.1073/pnas.1415959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lambden PR, Guest JR. Journal of general microbiology. 1976;97:145–160. doi: 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- 42.Kiley PJ, Beinert H. FEMS microbiology reviews. 1998;22:341–352. doi: 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 43.Sawers RG, Zehelein E, Bock A. Arch Microbiol. 1988;149:240–244. doi: 10.1007/BF00422011. [DOI] [PubMed] [Google Scholar]
- 44.Unden G, Duchene A. Arch Microbiol. 1987;147:195–200. doi: 10.1007/BF00415284. [DOI] [PubMed] [Google Scholar]
- 45.Mettert EL, Kiley PJ. Journal of molecular biology. 2005;354:220–232. doi: 10.1016/j.jmb.2005.09.066. [DOI] [PubMed] [Google Scholar]
- 46.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Molecular cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 47.Green J, Bennett B, Jordan P, Ralph ET, Thomson AJ, Guest JR. Biochemical Journal. 1996;316:887–892. doi: 10.1042/bj3160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz CJ, Djaman O, Imlay JA, Kiley PJ. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9009–9014. doi: 10.1073/pnas.160261497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rock JD, Mahnane MR, Anjum MF, Shaw JG, Read RC, Moir JW. Molecular microbiology. 2005;58:800–809. doi: 10.1111/j.1365-2958.2005.04866.x. [DOI] [PubMed] [Google Scholar]
- 50.Unden G, Becker S, Bongaerts J, Schirawski J, Six S. Antonie van Leeuwenhoek. 1994;66:3–22. doi: 10.1007/BF00871629. [DOI] [PubMed] [Google Scholar]
- 51.Bartolini E, Frigimelica E, Giovinazzi S, Galli G, Shaik Y, Genco C, Welsch JA, Granoff DM, Grandi G, Grifantini R. Molecular microbiology. 2006;60:963–972. doi: 10.1111/j.1365-2958.2006.05163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marteyn B, Gazi A, Sansonetti P. Gut microbes. 2012;3:104–120. doi: 10.4161/gmic.19325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schroeder GN, Hilbi H. Clinical microbiology reviews. 2008;21:134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prevost MC, Sansonetti P, Tang CM. Nature. 2010;465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magdalena J, Hachani A, Chamekh M, Jouihri N, Gounon P, Blocker A, Allaoui A. Journal of bacteriology. 2002;184:3433–3441. doi: 10.1128/JB.184.13.3433-3441.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamano K, Katayama E, Toyotome T, Sasakawa C. Journal of bacteriology. 2002;184:1244–1252. doi: 10.1128/JB.184.5.1244-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watarai M, Tobe T, Yoshikawa M, Sasakawa C. The EMBO journal. 1995;14:2461–2470. doi: 10.1002/j.1460-2075.1995.tb07243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morita-Ishihara T, Ogawa M, Sagara H, Yoshida M, Katayama E, Sasakawa C. The Journal of biological chemistry. 2006;281:599–607. doi: 10.1074/jbc.M509644200. [DOI] [PubMed] [Google Scholar]
- 59.Stenfors Arnesen LP, Fagerlund A, Granum PE. FEMS microbiology reviews. 2008;32:579–606. doi: 10.1111/j.1574-6976.2008.00112.x. [DOI] [PubMed] [Google Scholar]
- 60.Kotiranta A, Lounatmaa K, Haapasalo M. Microbes and infection / Institut Pasteur. 2000;2:189–198. doi: 10.1016/s1286-4579(00)00269-0. [DOI] [PubMed] [Google Scholar]
- 61.Schoeni JL, Wong AC. Journal of food protection. 2005;68:636–648. doi: 10.4315/0362-028x-68.3.636. [DOI] [PubMed] [Google Scholar]
- 62.Esbelin J, Jouanneau Y, Duport C. BMC microbiology. 2012;12:125. doi: 10.1186/1471-2180-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zigha A, Rosenfeld E, Schmitt P, Duport C. Journal of bacteriology. 2007;189:2813–2824. doi: 10.1128/JB.01701-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rollenhagen C, Bumann D. Infection and immunity. 2006;74:1649–1660. doi: 10.1128/IAI.74.3.1649-1660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fink RC, Evans MR, Porwollik S, Vazquez-Torres A, Jones-Carson J, Troxell B, Libby SJ, McClelland M, Hassan HM. Journal of bacteriology. 2007;189:2262–2273. doi: 10.1128/JB.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price-Carter M, Tingey J, Bobik TA, Roth JR. Journal of bacteriology. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. Molecular microbiology. 1999;32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- 68.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbieri NL, Nicholson B, Hussein A, Cai W, Wannemuehler YM, Dell'Anna G, Logue CM, Horn F, Nolan LK, Li G. Infection and immunity. 2014;82:5086–5098. doi: 10.1128/IAI.02315-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ronald A. Disease-a-month : DM. 2003;49:71–82. doi: 10.1067/mda.2003.8. [DOI] [PubMed] [Google Scholar]
- 72.Pritchard JB. The Journal of pharmacology and experimental therapeutics. 1995;274:1278–1284. [PubMed] [Google Scholar]
- 73.Uhlen P, Laestadius A, Jahnukainen T, Soderblom T, Backhed F, Celsi G, Brismar H, Normark S, Aperia A, Richter-Dahlfors A. Nature. 2000;405:694–697. doi: 10.1038/35015091. [DOI] [PubMed] [Google Scholar]
- 74.Cai W, Wannemuehler Y, Dell'anna G, Nicholson B, Barbieri NL, Kariyawasam S, Feng Y, Logue CM, Nolan LK, Li G. PLoS pathogens. 2013;9:e1003428. doi: 10.1371/journal.ppat.1003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye RW, Haas D, Ka JO, Krishnapillai V, Zimmermann A, Baird C, Tiedje JM. Journal of bacteriology. 1995;177:3606–3609. doi: 10.1128/jb.177.12.3606-3609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jackson AA, Gross MJ, Daniels EF, Hampton TH, Hammond JH, Vallet-Gely I, Dove SL, Stanton BA, Hogan DA. Journal of bacteriology. 2013;195:3093–3104. doi: 10.1128/JB.02169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Galimand M, Gamper M, Zimmermann A, Haas D. Journal of bacteriology. 1991;173:1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoon SS, Karabulut AC, Lipscomb JD, Hennigan RF, Lymar SV, Groce SL, Herr AB, Howell ML, Kiley PJ, Schurr MJ, Gaston B, Choi KH, Schweizer HP, Hassett DJ. The EMBO journal. 2007;26:3662–3672. doi: 10.1038/sj.emboj.7601787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA. PLoS pathogens. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berka RM, Vasil ML. Journal of bacteriology. 1982;152:239–245. doi: 10.1128/jb.152.1.239-245.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jackson AA, Daniels EF, Hammond JH, Willger SD, Hogan DA. Microbiology. 2014;160:2215–2225. doi: 10.1099/mic.0.081158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Microbiology and molecular biology reviews : MMBR. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chater KF, Chandra G. FEMS microbiology reviews. 2006;30:651–672. doi: 10.1111/j.1574-6976.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- 84.Singh A, Guidry L, Narasimhulu KV, Mai D, Trombley J, Redding KE, Giles GI, Lancaster JR, Jr., Steyn AJ. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11562–11567. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. PLoS pathogens. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Larsson C, Luna B, Ammerman NC, Maiga M, Agarwal N, Bishai WR. PloS one. 2012;7:e37516. doi: 10.1371/journal.pone.0037516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Geiman DE, Raghunand TR, Agarwal N, Bishai WR. Antimicrobial agents and chemotherapy. 2006;50:2836–2841. doi: 10.1128/AAC.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morris RP, Nguyen L, Gatfield J, Visconti K, Nguyen K, Schnappinger D, Ehrt S, Liu Y, Heifets L, Pieters J, Schoolnik G, Thompson CJ. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12200–12205. doi: 10.1073/pnas.0505446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burian J, Ramon-Garcia S, Sweet G, Gomez-Velasco A, Av-Gay Y, Thompson CJ. The Journal of biological chemistry. 2012;287:299–310. doi: 10.1074/jbc.M111.302588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Steyn AJ, Collins DM, Hondalus MK, Jacobs WR, Jr., Kawakami RP, Bloom BR. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rajni, Rao N, Meena LS. Biotechnology research international. 2011;2011:274693. doi: 10.4061/2011/274693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goren MB, D'Arcy Hart P, Young MR, Armstrong JA. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:2510–2514. doi: 10.1073/pnas.73.7.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kato M, Goren MB. Infection and immunity. 1974;10:733–741. doi: 10.1128/iai.10.4.733-741.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goren MB, Vatter AE, Fiscus J. Journal of leukocyte biology. 1987;41:111–121. doi: 10.1002/jlb.41.2.111. [DOI] [PubMed] [Google Scholar]
- 95.Guerardel Y, Maes E, Briken V, Chirat F, Leroy Y, Locht C, Strecker G, Kremer L. The Journal of biological chemistry. 2003;278:36637–36651. doi: 10.1074/jbc.M305427200. [DOI] [PubMed] [Google Scholar]
- 96.Vergne I, Chua J, Deretic V. The Journal of experimental medicine. 2003;198:653–659. doi: 10.1084/jem.20030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pabst MJ, Gross JM, Brozna JP, Goren MB. J Immunol. 1988;140:634–640. [PubMed] [Google Scholar]
- 98.Zhang L, English D, Andersen BR. J Immunol. 1991;146:2730–2736. [PubMed] [Google Scholar]
- 99.Zhang L, Goren MB, Holzer TJ, Andersen BR. Infection and immunity. 1988;56:2876–2883. doi: 10.1128/iai.56.11.2876-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brozna JP, Horan M, Rademacher JM, Pabst KM, Pabst MJ. Infection and immunity. 1991;59:2542–2548. doi: 10.1128/iai.59.8.2542-2548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rousseau C, Neyrolles O, Bordat Y, Giroux S, Sirakova TD, Prevost MC, Kolattukudy PE, Gicquel B, Jackson M. Cellular microbiology. 2003;5:405–415. doi: 10.1046/j.1462-5822.2003.00289.x. [DOI] [PubMed] [Google Scholar]
- 102.Indrigo J, Hunter RL, Jr., Actor JK. Microbiology. 2002;148:1991–1998. doi: 10.1099/00221287-148-7-1991. [DOI] [PubMed] [Google Scholar]
- 103.Saini V, Farhana A, Steyn AJ. Antioxidants & redox signaling. 2012;16:687–697. doi: 10.1089/ars.2011.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Farhana A, Guidry L, Srivastava A, Singh A, Hondalus MK, Steyn AJ. Advances in microbial physiology. 2010;57:43–117. doi: 10.1016/B978-0-12-381045-8.00002-3. [DOI] [PubMed] [Google Scholar]
- 105.Yan LJ, Levine RL, Sohal RS. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hentze MW, Kuhn LC. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tang Y, Guest JR. Microbiology. 1999;145(Pt 11):3069–3079. doi: 10.1099/00221287-145-11-3069. [DOI] [PubMed] [Google Scholar]
- 108.Viollier PH, Nguyen KT, Minas W, Folcher M, Dale GE, Thompson CJ. Journal of bacteriology. 2001;183:3193–3203. doi: 10.1128/JB.183.10.3193-3203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pantopoulos K. Annals of the New York Academy of Sciences. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- 110.Leibold EA, Guo B. Annual review of nutrition. 1992;12:345–368. doi: 10.1146/annurev.nu.12.070192.002021. [DOI] [PubMed] [Google Scholar]
- 111.Hirling H, Henderson BR, Kuhn LC. The EMBO journal. 1994;13:453–461. doi: 10.1002/j.1460-2075.1994.tb06280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kennedy MC, Mende-Mueller L, Blondin GA, Beinert H. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:11730–11734. doi: 10.1073/pnas.89.24.11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haile DJ, Rouault TA, Harford JB, Kennedy MC, Blondin GA, Beinert H, Klausner RD. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:11735–11739. doi: 10.1073/pnas.89.24.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Philpott CC, Klausner RD, Rouault TA. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7321–7325. doi: 10.1073/pnas.91.15.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alen C, Sonenshein AL. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10412–10417. doi: 10.1073/pnas.96.18.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Craig JE, Ford MJ, Blaydon DC, Sonenshein AL. Journal of bacteriology. 1997;179:7351–7359. doi: 10.1128/jb.179.23.7351-7359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dingman DW, Sonenshein AL. Journal of bacteriology. 1987;169:3062–3067. doi: 10.1128/jb.169.7.3062-3067.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gellatly SL, Hancock RE. Pathogens and disease. 2013;67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 119.Iglewski BH, Kabat D. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:2284–2288. doi: 10.1073/pnas.72.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Iglewski BH, Liu PV, Kabat D. Infection and immunity. 1977;15:138–144. doi: 10.1128/iai.15.1.138-144.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Somerville G, Mikoryak CA, Reitzer L. Journal of bacteriology. 1999;181:1072–1078. doi: 10.1128/jb.181.4.1072-1078.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, Musser JM. Infection and immunity. 2002;70:6373–6382. doi: 10.1128/IAI.70.11.6373-6382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rollof J, Braconier JH, Soderstrom C, Nilsson-Ehle P. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 1988;7:505–510. doi: 10.1007/BF01962601. [DOI] [PubMed] [Google Scholar]
- 124.Marr JC, Lyon JD, Roberson JR, Lupher M, Davis WC, Bohach GA. Infection and immunity. 1993;61:4254–4262. doi: 10.1128/iai.61.10.4254-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dinges MM, Orwin PM, Schlievert PM. Clinical microbiology reviews. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hammer ND, Skaar EP. Annual review of microbiology. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Torres VJ, Attia AS, Mason WJ, Hood MI, Corbin BD, Beasley FC, Anderson KL, Stauff DL, McDonald WH, Zimmerman LJ, Friedman DB, Heinrichs DE, Dunman PM, Skaar EP. Infection and immunity. 2010;78:1618–1628. doi: 10.1128/IAI.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. PLoS computational biology. 2006;2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shepard W, Soutourina O, Courtois E, England P, Haouz A, Martin-Verstraete I. The FEBS journal. 2011;278:2689–2701. doi: 10.1111/j.1742-4658.2011.08195.x. [DOI] [PubMed] [Google Scholar]
- 130.Fleischhacker AS, Stubna A, Hsueh KL, Guo Y, Teter SJ, Rose JC, Brunold TC, Markley JL, Munck E, Kiley PJ. Biochemistry. 2012;51:4453–4462. doi: 10.1021/bi3003204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu Y, Outten FW. Journal of bacteriology. 2009;191:1248–1257. doi: 10.1128/JB.01086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yeo WS, Lee JH, Lee KC, Roe JH. Molecular microbiology. 2006;61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 133.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. Molecular microbiology. 2006;60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 134.Nesbit AD, Giel JL, Rose JC, Kiley PJ. Journal of molecular biology. 2009;387:28–41. doi: 10.1016/j.jmb.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Abbott M, Galloway A, Cunningham JL. J Infection. 1986;13:143–145. doi: 10.1016/s0163-4453(86)92869-0. [DOI] [PubMed] [Google Scholar]
- 136.Abdelli N, Thiefin G, Chevalier P, Zeitoun P. Gastroenterologie clinique et biologique. 1996;20:212–213. [PubMed] [Google Scholar]
- 137.Galindo CL, Rosenzweig JA, Kirtley ML, Chopra AK. Journal of pathogens. 2011;2011:182051. doi: 10.4061/2011/182051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Harris S, Paraskevakis I. J Invest Med. 2012;60:364–364. [Google Scholar]
- 139.Miller HK, Kwuan L, Schwiesow L, Bernick DL, Mettert E, Ramirez HA, Ragle JM, Chan PP, Kiley PJ, Lowe TM, Auerbuch V. PLoS pathogens. 2014;10:e1004194. doi: 10.1371/journal.ppat.1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martin RB, Savory J, Brown S, Bertholf RL, Wills MR. Clinical chemistry. 1987;33:405–407. [PubMed] [Google Scholar]
- 141.Aisen P, Leibman A, Zweier J. The Journal of biological chemistry. 1978;253:1930–1937. [PubMed] [Google Scholar]
- 142.Kretchmar SA, Reyes ZE, Raymond KN. Biochimica et biophysica acta. 1988;956:85–94. doi: 10.1016/0167-4838(88)90301-9. [DOI] [PubMed] [Google Scholar]
- 143.Viboud GI, Bliska JB. Annual review of microbiology. 2005;59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- 144.Dewoody RS, Merritt PM, Marketon MM. Frontiers in cellular and infection microbiology. 2013;3:4. doi: 10.3389/fcimb.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Oliver JD. Epidemiology and infection. 2005;133:383–391. doi: 10.1017/s0950268805003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jones MK, Oliver JD. Infection and immunity. 2009;77:1723–1733. doi: 10.1128/IAI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lim JG, Choi SH. Infection and immunity. 2014;82:569–578. doi: 10.1128/IAI.01141-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kaihami GH, Almeida JR, Santos SS, Netto LE, Almeida SR, Baldini RL. PLoS pathogens. 2014;10:e1004442. doi: 10.1371/journal.ppat.1004442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chaves GM, Bates S, Maccallum DM, Odds FC. Genet Mol Res. 2007;6:1051–1063. [PubMed] [Google Scholar]
- 150.Choi HK, Park NY, Kim DI, Chung HJ, Ryu S, Choi SH. The Journal of biological chemistry. 2002;277:47292–47299. doi: 10.1074/jbc.M206893200. [DOI] [PubMed] [Google Scholar]
- 151.Kreger A, Lockwood D. Infection and immunity. 1981;33:583–590. doi: 10.1128/iai.33.2.583-590.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim SH, Lee BY, Lau GW, Cho YH. Journal of microbiology and biotechnology. 2009;19:1520–1526. doi: 10.4014/jmb.0906.06028. [DOI] [PubMed] [Google Scholar]
- 153.Iles KE, Forman HJ. Immunol Res. 2002;26:95–105. doi: 10.1385/IR:26:1-3:095. [DOI] [PubMed] [Google Scholar]
- 154.Lee JS, Heo YJ, Lee JK, Cho YH. Infection and immunity. 2005;73:4399–4403. doi: 10.1128/IAI.73.7.4399-4403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Deisseroth A, Dounce AL. Physiological reviews. 1970;50:319–375. doi: 10.1152/physrev.1970.50.3.319. [DOI] [PubMed] [Google Scholar]
- 156.Partridge JD, Bodenmiller DM, Humphrys MS, Spiro S. Molecular microbiology. 2009;73:680–694. doi: 10.1111/j.1365-2958.2009.06799.x. [DOI] [PubMed] [Google Scholar]
- 157.Tucker NP, Hicks MG, Clarke TA, Crack JC, Chandra G, Le Brun NE, Dixon R, Hutchings MI. PloS one. 2008;3:e3623. doi: 10.1371/journal.pone.0003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bodenmiller DM, Spiro S. Journal of bacteriology. 2006;188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tucker NP, Le Brun NE, Dixon R, Hutchings MI. Trends in microbiology. 2010;18:149–156. doi: 10.1016/j.tim.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 160.Isabella VM, Lapek JD, Jr., Kennedy EM, Clark VL. Molecular microbiology. 2009;71:227–239. doi: 10.1111/j.1365-2958.2008.06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gilberthorpe NJ, Lee ME, Stevanin TM, Read RC, Poole RK. Microbiology. 2007;153:1756–1771. doi: 10.1099/mic.0.2006/003731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Beaumont HJ, Lens SI, Reijnders WN, Westerhoff HV, van Spanning RJ. Molecular microbiology. 2004;54:148–158. doi: 10.1111/j.1365-2958.2004.04248.x. [DOI] [PubMed] [Google Scholar]
- 163.Rankin LD, Bodenmiller DM, Partridge JD, Nishino SF, Spain JC, Spiro S. Journal of bacteriology. 2008;190:6170–6177. doi: 10.1128/JB.00508-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nix RN, Altschuler SE, Henson PM, Detweiler CS. PLoS pathogens. 2007;3:e193. doi: 10.1371/journal.ppat.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fabrega A, Vila J. Clinical microbiology reviews. 2013;26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.MacMicking J, Xie QW, Nathan C. Annual review of immunology. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 167.Karlinsey JE, Bang IS, Becker LA, Frawley ER, Porwollik S, Robbins HF, Thomas VC, Urbano R, McClelland M, Fang FC. Molecular microbiology. 2012;85:1179–1193. doi: 10.1111/j.1365-2958.2012.08167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stevanin TM, Poole RK, Demoncheaux EA, Read RC. Infection and immunity. 2002;70:4399–4405. doi: 10.1128/IAI.70.8.4399-4405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bardiau M, Szalo M, Mainil JG. Veterinary research. 2010;41:57. doi: 10.1051/vetres/2010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kaper JB, Nataro JP, Mobley HL. Nature reviews. Microbiology. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 171.Branchu P, Matrat S, Vareille M, Garrivier A, Durand A, Crepin S, Harel J, Jubelin G, Gobert AP. PLoS pathogens. 2014;10:e1003874. doi: 10.1371/journal.ppat.1003874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Elliott SJ, Sperandio V, Giron JA, Shin S, Mellies JL, Wainwright L, Hutcheson SW, McDaniel TK, Kaper JB. Infection and immunity. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vareille M, Rannou F, Thelier N, Glasser AL, de Sablet T, Martin C, Gobert AP. J Immunol. 2008;180:5720–5726. doi: 10.4049/jimmunol.180.8.5720. [DOI] [PubMed] [Google Scholar]
- 175.Dela Cruz R, Gao Y, Penumetcha S, Sheplock R, Weng K, Chander M. Journal of bacteriology. 2010;192:6428–6438. doi: 10.1128/JB.00916-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bradley TM, Hidalgo E, Leautaud V, Ding H, Demple B. Nucleic acids research. 1997;25:1469–1475. doi: 10.1093/nar/25.8.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sheplock R, Recinos DA, Mackow N, Dietrich LE, Chander M. Molecular microbiology. 2013;87:368–381. doi: 10.1111/mmi.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gaudu P, Weiss B. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ding H, Demple B. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Amabile-Cuevas CF, Demple B. Nucleic acids research. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wu J, Weiss B. Journal of bacteriology. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pomposiello PJ, Bennik MH, Demple B. Journal of bacteriology. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dietrich LE, Teal TK, Price-Whelan A, Newman DK. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shin JH, Singh AK, Cheon DJ, Roe JH. Journal of bacteriology. 2011;193:75–81. doi: 10.1128/JB.00965-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Palma M, Zurita J, Ferreras JA, Worgall S, Larone DH, Shi L, Campagne F, Quadri LE. Infection and immunity. 2005;73:2958–2966. doi: 10.1128/IAI.73.5.2958-2966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Park W, Pena-Llopis S, Lee Y, Demple B. Biochemical and biophysical research communications. 2006;341:51–56. doi: 10.1016/j.bbrc.2005.12.142. [DOI] [PubMed] [Google Scholar]
- 187.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. Molecular microbiology. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 188.Singh AK, Shin JH, Lee KL, Imlay JA, Roe JH. Molecular microbiology. 2013;90:983–996. doi: 10.1111/mmi.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ha U, Jin S. Infection and immunity. 1999;67:5324–5331. doi: 10.1128/iai.67.10.5324-5331.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kobayashi K, Tagawa S. Journal of biochemistry. 2004;136:607–615. doi: 10.1093/jb/mvh168. [DOI] [PubMed] [Google Scholar]
- 191.Aendekerk S, Diggle SP, Song Z, Hoiby N, Cornelis P, Williams P, Camara M. Microbiology. 2005;151:1113–1125. doi: 10.1099/mic.0.27631-0. [DOI] [PubMed] [Google Scholar]
- 192.Kang IH, Kim JS, Lee JK. Journal of microbiology and biotechnology. 2007;17:1399–1402. [PubMed] [Google Scholar]
- 193.Rhee JE, Rhee JH, Ryu PY, Choi SH. FEMS microbiology letters. 2002;208:245–251. doi: 10.1111/j.1574-6968.2002.tb11089.x. [DOI] [PubMed] [Google Scholar]
- 194.Kim JS, Sung MH, Kho DH, Lee JK. Journal of bacteriology. 2005;187:5984–5995. doi: 10.1128/JB.187.17.5984-5995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kimoto R, Funahashi T, Yamamoto N, Miyoshi S, Narimatsu S, Yamamoto S. Microbiology and immunology. 2001;45:135–142. doi: 10.1111/j.1348-0421.2001.tb01281.x. [DOI] [PubMed] [Google Scholar]
- 196.Kim JS, Choi SH, Lee JK. Journal of bacteriology. 2006;188:8586–8592. doi: 10.1128/JB.01084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sun F, Ji Q, Jones MB, Deng X, Liang H, Frank B, Telser J, Peterson SN, Bae T, He C. Journal of the American Chemical Society. 2012;134:305–314. doi: 10.1021/ja2071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Giraudo AT, Mansilla C, Chan A, Raspanti C, Nagel R. Current microbiology. 2003;46:246–250. doi: 10.1007/s00284-002-3853-z. [DOI] [PubMed] [Google Scholar]
- 199.Goerke C, Fluckiger U, Steinhuber A, Bisanzio V, Ulrich M, Bischoff M, Patti JM, Wolz C. Infection and immunity. 2005;73:3415–3421. doi: 10.1128/IAI.73.6.3415-3421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Novick RP, Jiang D. Microbiology. 2003;149:2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- 201.Dossett JH, Kronvall G, Williams RC, Jr., Quie PG. J Immunol. 1969;103:1405–1410. [PubMed] [Google Scholar]
- 202.Novick RP. Molecular microbiology. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 203.Janzon L, Lofdahl S, Arvidson S. Molecular & general genetics : MGG. 1989;219:480–485. doi: 10.1007/BF00259623. [DOI] [PubMed] [Google Scholar]
- 204.Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. Molecular & general genetics : MGG. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 205.Janzon L, Arvidson S. The EMBO journal. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Powers ME, Bubeck Wardenburg J. PLoS pathogens. 2014;10:e1003871. doi: 10.1371/journal.ppat.1003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Foster TJ, Geoghegan JA, Ganesh VK, Hook M. Nature reviews. Microbiology. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Giel JL, Nesbit AD, Mettert EL, Fleischhacker AS, Wanta BT, Kiley PJ. Molecular microbiology. 2013;87:478–492. doi: 10.1111/mmi.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jones-Carson J, Laughlin J, Hamad MA, Stewart AL, Voskuil MI, Vazquez-Torres A. PloS one. 2008;3:e1976. doi: 10.1371/journal.pone.0001976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Runyen-Janecky L, Daugherty A, Lloyd B, Wellington C, Eskandarian H, Sagransky M. Infection and immunity. 2008;76:1083–1092. doi: 10.1128/IAI.01211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.