Abstract
Sex bias in lupus incidence is thought to be due, in part, to the ability of estrogens to promote loss of tolerance. Previously, we showed that estrogens promote lupus via estrogen receptor α (ERα). C57BL/6 (B6) mice carrying the Sle1 lupus susceptibility locus (B6.Sle1) display loss of tolerance and develop anti-nuclear antibodies and immune cell hyperactivation. The incidence of loss of tolerance in B6.Sle1 females is greater than in males. Here, we show that a deficiency of either estrogens or ERα attenuates loss of tolerance and autoantibody development in B6.Sle1 females. Furthermore, we demonstrate that immune cell activation in B6.Sle1 mice shows sex bias and that ERα deficiency diminishes this phenotype in B6.Sle1 females. Thus, estrogens, acting via ERα, control sex bias in the Sle1 phenotype. Furthermore, we show that ERα may impact the Sle1 phenotype by modulating the expression Pbx1, one of genes that underlies the Sle1 locus.
Keywords: Lupus, estrogen receptor alpha, sex bias, immunologic tolerance, immune cell activation
1. Introduction
Approximately 90% of lupus patients are women. This dramatic sex bias is thought to be due, in large part, to endogenous estrogens. Although abundant evidence links exposure to estrogens with enhanced lupus risk and increased lupus manifestations [1-5], relatively little is understood in regard to cellular and molecular mechanisms. Much of our understanding of how estrogens may promote lupus has come from studies in lupus prone mice, such as (NZB × NZW)F1 mice [6, 7]. Lupus development in these mice is influenced by sex; lupus incidence is higher and survival time is reduced in female (NZB X NZW)F1 mice relative to males [6, 7]. We demonstrated previously that disruption of estrogen receptor α (ERα) in (NZB X NZW)F1 mice eliminated the sex bias in the development of lupus in these mice [12]. The decreased incidence of lupus and increased survival in ERα deficient (NZB X NZW)F1 females was associated with a dramatic attenuation of loss of tolerance to nuclear antigens and development of pathogenic autoantibodies[8]. These data indicate that the sex bias in lupus pathogenesis in this model is due, at least in part, to estrogens, acting via ERα. These data also suggest that estrogens, acting through ERα, may augment the effects of at least some lupus susceptibility loci, leading to an enhanced lupus phenotype in females.
Loss of tolerance to chromatin is thought to represent an initial step in the development of lupus [9, 10]. In (NZB × NZW)F1 mice, the loss of tolerance to chromatin is controlled by NZB- and NZW- derived lupus susceptibility alleles [11-13]. Among these loci, the NZW-derived Sle1 locus is the best characterized [14-16]. B6.Sle1 congenic mice, in which the NZW- derived Sle1 allele is carried on the non-autoimmune C57BL/6 (B6) genetic background, spontaneously lose tolerance to chromatin and develop anti-chromatin IgG [12]. The incidence of loss of tolerance in B6.Sle1 females is significantly greater than in males, suggesting that sex hormones may impact the actions of Sle1 [11, 17]. Sle1 also leads to increased activation in B and T cells [18, 19], although it is not known if these aspects of the Sle1 phenotype also display a sex bias. Virtually nothing is known about how hormones, including estrogens, influence Sle1 action.
The Sle1 interval represents at least three distinct subloci, Sle1a, Sle1b and Sle1c, each of which acts via distinct pathways to independently contribute to loss of tolerance and the development of anti-chromatin IgG [17]. Analysis of congenic strains carrying these individual subloci has shown that Sle1b-induced loss of tolerance to chromatin displays a robust and significant female sex bias [17]. Consistent with the idea that Sle1b enhances loss of tolerance preferentially in females, a recent study indicates that B cell activation and proliferation is more robust in B6.Sle1b congenic females than males [20]. Both Sle1a- and Sle1c- induced loss of tolerance to chromatin also appear to be more pronounced in female mice than in male mice, although these differences fall short of statistical significance [17].
We postulate that estrogens, acting via ERα-dependent pathways, synergize with the pathways controlled by certain lupus susceptibility loci to preferentially enhance loss of tolerance and the development of lupus in females. Given the fact that development of anti-chromatin IgG in B6.Sle1 congenic mice shows a clear sex bias, we hypothesize that the effects of Sle1 are likely to be influenced by estrogens via ERα signaling. To test this hypothesis, we examined the impact of a targeted mutation in ERα on the phenotype in B6.Sle1 congenic mice. We observed that ERα deficiency attenuated loss of tolerance and the development of anti-chromatin IgG in B6.Sle1 congenic females but not males. ERα deficiency significantly decreased Sle1-induced immune cell hyperactivation in females, and to a lesser extent, in males. These effects of ERα deficiency were associated with a decrease in the relative expression of Pbx1a, an isoform of one of the genes that underlies Sle1. The impact of removal of the ovaries, the primary source of estrogens, on B6.Sle1 congenic females was similar to that associated with ERα deficiency. Furthermore, the phenotype of ERα deficient B6.Sle1 congenic females was similar to that in ERα wildtype B6.Sle1 congenic males, suggesting that the sex bias in the phenotype of B6.Sle1 congenic mice is both estrogen- and ERα-dependent.
2. Materials and Methods
2.1. Care and Treatment of Mice
The Institutional Animal Care and Use Committee of the University of Nebraska Medical Center approved all procedures involving live animals. The ERα knockout strain (B6.129-Esr1tm1Ksk or B6.ERα) [21] was originally obtained from Dennis Lubahn. The B6.Sle1 congenic strain [12, 22] was provided by Laurence Morel. Animals were housed under controlled temperature, humidity, and 12h light/12h dark lighting conditions in a facility accredited by the American Association for Accreditation of Laboratory Animal Care and operated in accordance with the standards outlined in Guide for the Care and Use of Laboratory Animals (The National Academies Press, 1996). Mice were provided Harlan irradiated rodent diet 7904 (Harlan Teklad, Madison, WI), which contains soy, milk, and meat-based protein sources, and allowed to feed ad libitum.
B6 females heterozygous for targeted disruption of the ERα gene (ERα+/−) were crossed to B6.Sle1 congenic males. The resulting ERα+/− males were backcrossed to B6.Sle1 females. Resulting ERα+/− offspring were genotyped at markers (D1Mit47, D1Mit159, D1Mit111, D1Mit206, D1Mit426, and D1Mit17) that are polymorphic between the NZW and B6 strains and span the Sle1 congenic interval to identify mice that were homozygous for NZW alleles throughout the interval [23]. These mice were interbred to generate the experimental mice. PCR-based genotyping was performed as described previously [8]. For the ovariectomy studies, mice were randomized to the sham and ovariectomy groups at 5-6 weeks of age and subjected to either a sham procedure in which the ovaries are externalized and then returned to the abdominal cavity or removed, respectively. Success of these surgical procedures was confirmed by measuring serum estradiol levels using a quantitative ELISA assay (Alpha Diagnostics International, San Antonio, TX) from serum collected prior to sacrifice. Testosterone levels were also measured by using a quantitative ELISA assay (Alpha Diagnostics International, San Antonio, TX).
2.2. Serological Analysis
Autoantibody levels were assessed by ELISA using serum isolated from blood collected monthly via the saphenous vein and stored at −80C. Samples were assayed in duplicate for each ELISA. The anti-chromatin IgG and anti-dsDNA IgG concentrations were determined using plates prepared as described previously [8, 24]. Autoantibody levels in these samples were quantitated in arbitrary ELISA units (U/μl) based upon a standard curve generated by serial dilution of a positive control sample that was made by pooling serum from a group of (NZB × NZW)F1 females with heavy albuminuria. The threshold for a positive autoantibody titer in the experimental mice was set at 2 standard deviations above the mean of a group of age-matched control B6 mice [17]. Total serum concentrations of antibodies of each isotype were determined using the clonotyping kit (Southern Biotech, Birmingham, AL) according to the manufacturer’s instructions. All optimal density measurements were made using a BioRad 680 Microplate reader and Microplate Manager software, version 5.2.1 (Hercules, CA). Sera were diluted serially from 1:100 to 1:2000 for measurement of autoantibody concentrations and to 1:50,000 for measurement of total serum immunoglobulins.
2.3. Flow cytometry
The antibodies (BD Biosciences, San Jose, CA) used for flow cytometry were: CD4-PE (RM4-5), CD4-v450 (RM4-5), CD8-APC (53-6.7), CD69-FITC (H1.2F3), CD134-Biotin (OX-86), CD62L-APC (MEL-14), B220-APC (RA3-6B2), CD86-PE (GL1), CD22-PE (Cy34.1), CD25-APC.Cy7 (PC61), and FoxP3-PE (150D). Biotinylated antibodies were detected using FITC-conjugated streptavidin (BD Biosciences). Flow cytometric analysis was performed using various combinations of these antibodies on single cell suspensions of splenocytes. For intracellular FoxP3 staining, surface-stained cells were treated with fixation/permeabilization buffer and stained with FoxP3-PE (150D) using the BioLegend FoxP3 flow kit, following the manufacturer’s protocol (BioLegend). Stained cells were analyzed in the UNMC Flow Cytometry Research Facility using the BD LSR II flow cytometer. Data were analyzed using FACSDiva software, version 6.1.2 (BD Biosciences). For analysis of T regulatory cells, splenocytes were isolated from mice that were 3–4 months of age. For all other flow cytometry analyses, splenocytes were collected from mice that were 6–12 months of age.
2.4. Cell isolation and Quantitative real-time PCR
Splenic B cells were isolated from mice at 5-9 months of age using the B Cell Isolation Kit (Miltenyi Biotec, Auburn, CA) and collected using MACS columns and the VarioMACS separator (Miltenyi Biotec). Splenic CD4+ T cells were isolated from mice at 5-9 months of age using the CD4+ T Cell Isolation Kit II (Miltenyi Biotec) and collected using MACS columns and either the VarioMACS or AutoMACS separator (Miltenyi Biotec). Flow cytometry was used to confirm the purity of the isolated cell population. RNA was prepared from isolated cells was extracted using the Absolutely RNA Miniprep Kit (Stratagene Corporation, La Jolla, CA). cDNA was generated using either the High Capacity RNA-to-cDNA Master Mix (Applied Biosystems, Carlsbad, CA) or SuperScript VILO Master Mix (Invitrogen, Carlsbad, CA ) according the manufacturer’s instructions. Quantitative RT-PCR (qRT-PCR) was performed using Power SYBR Green PCR Master Mix, the ABI 7500 Real-Time PCR System, and Sequence Detection Software, version 1.4 (Applied Biosystems). Quantitative data regarding gene expression was extracted from the PCR data and the abundance of each gene transcript was normalized to that of Gapdh for each sample.
2.5. Immunoprecipitation and SDS-PAGE Analysis
CR1/CR2 expression and glycosylation were quantified using a modification of the method described previously by Boackle and colleagues [25]. Briefly, single cell suspensions of splenocytes from mice at 5-9 months of age were depleted of RBC with ACK Lysing Buffer (Life Technologies, Carlsbad, CA) and surface biotinylated with the EZ-LINK Sulfo-NHS-LC-Biotin (Thermo Scientific, Waltham, MA). Cells were lysis in RIPA Buffer (1XPBS, 1% Nonidet P-40 or Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS.) supplemented with a protease inhibitor cocktail (Sigma, St. Louis, MO) diluted to final concentrations of 1 mM AEBSF, 0.8 μM Aprotinin, 40 μM Bestatin, 14 μM E-64, 20 μM Leupeptin, and 15 μM Pepstatin A. Lysates were incubated with anti-mouse CR1/CR2 antibody (7E9) for 30 min on ice. After the addition of Protein G Plus agarose (Santa Cruz Biotechnology, Santa Cruz, CA), samples were incubated on a rotating mixer at 4° C. Samples were electrophoresed through an 8% non-reducing gel and transferred to a nitrocellulose membrane. Membranes were incubated with streptavidin-Alexa Fluor® 700 conjugate (Life Technologies) and imaged and quantified using the Li-Cor Odyssey Imaging system (Li-Cor, Lincoln, NE)
2.6. Statistical methods
Comparisons were performed using Fishers exact test, independent or paired samples t-test, or one-way ANOVA with Tukey’s post hoc test where appropriate. Statistical analyses were performed using SPSS software (version 19.0). A two-sided P ≤ 0.05 was considered significant. Two-sided p-values are provided. Mean ± standard error of the mean is presented.
3. Results
3.1. Estrogen receptor α deficiency attenuates Sle1-induced loss of tolerance and epitope spreading in females
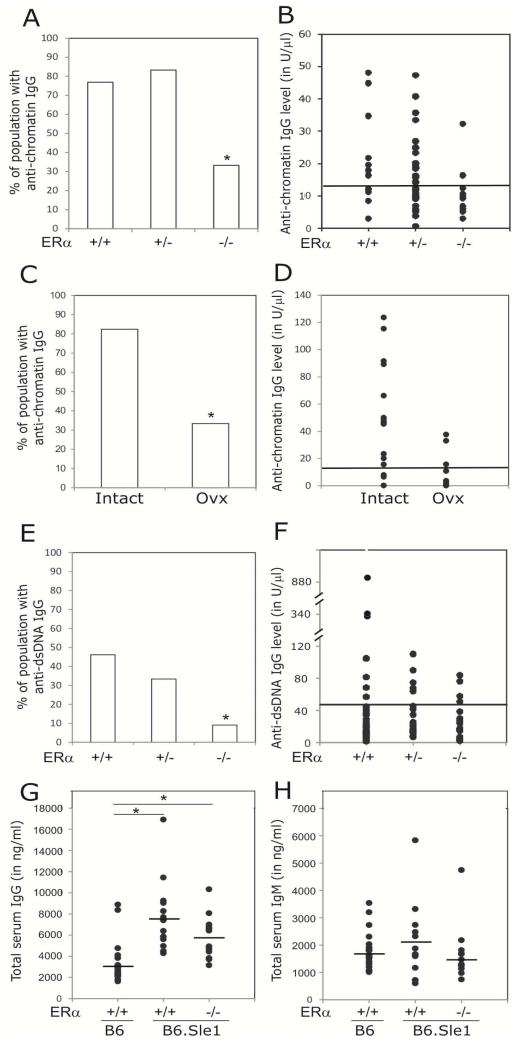
To test the hypothesis that estrogens, acting via ERα, promote loss of tolerance in B6.Sle1 female mice, we intercrossed mice from the B6.ERα knockout strain, which are heterozygous for a targeted disruption of ERα, with B6.Sle1 congenic mice to produce B6.Sle1 female mice that were wildtype (ERα+/+), heterozygous (ERα+/−) or homozygous null (ERα−/−) at ERα. As expected, a significant proportion (77%) of B6.Sle1;ERα+/+ females developed anti-chromatin antibodies (Figure 1A-B). Heterozygosity for ERα did not impact the development of anti-chromatin antibodies (Figure 1A-B). However, the proportion of B6.Sle1;ERα−/− female mice that developed anti-chromatin antibodies (33%) was significantly lower than that observed in B6.Sle1;ERα+/+ females (Figure 1A-B; P ≤ 0.005). These results suggest that ERα signaling promotes loss of tolerance in B6.Sle1 females.
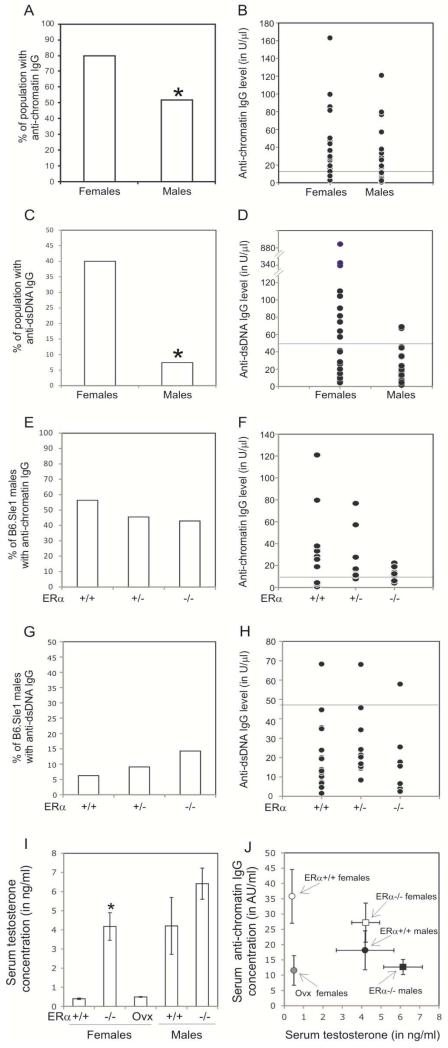
Figure 1. Attenuation of Sle1-induced loss of tolerance by ERα deficiency and ovariectomy.
ELISAs were used to assess the abundance of anti-chromatin IgG (A–D), anti-dsDNA IgG (E-F),total serum IgG (G) and IgM (H) in 5-9 month old female mice. The proportion of B6.Sle1;ERα+/+ (N=13), B6.Sle1;ERα+/− (N=12), and B6.Sle1;ERα−/− (N=12) females that developed anti-chromatin IgG (A) and anti-dsDNA IgG (E) is shown. The proportion of sham-treated, ovary-intact B6.Sle1 (N=10) and ovariectomized B6.Sle1 (N=16) females that develop anti-chromatin IgG (C) is shown. The absolute levels of anti-chromatin IgG (B,D) and anti-dsDNA IgG (F) are also shown. For panels G,H, the horizontal bar denotes the mean. In panels B, D and F, the horizontal line represents the threshold for a positive autoantibody titer in the experimental mice. This threshold was set at 2 standard deviations above the mean of a group of age-matched control B6 mice as has been described previously [17]. The mean (± standard error of the mean) abundance of total serum IgG (G) and IgM (H) in B6 (N=20), B6.Sle1;ERα+/+ (N=13), and B6.Sle1;ERα−/− (N=12) females is shown. The * indicates a P<0.05 compared to B6.Sle1;ERα+/+ females (A and E), intact females (C), and B6 females (G).
In a parallel study, we examined the impact of removing the ovaries, the primary source of estrogens, on loss of tolerance in B6.Sle1 females. In sham-operated B6.Sle1 females, the proportion of mice developing anti-chromatin IgG was 82% (Figure 1C-D). By contrast, just 33% of ovariectomized B6.Sle1 females developed anti-chromatin IgG (Figure 1C-D; P ≤ 0.05). Interestingly, the proportion of ovarectomized B6.Sle1 females that developed anti-chromatin IgG was identical to that observed in B6.Sle1;ERα−/− females. These data suggest that in terms of impact on Sle1-induced loss of tolerance, removal of endogenous estrogens was equivalent to loss of ERα signaling. The fact that genetic disruption of ERα signaling had essentially the same impact on Sle1-induced loss of tolerance as ovariectomy, which removes the primary source of endogenous estrogens, suggests that ERα-dependent estrogen action promotes Sle1-induced loss of tolerance in females.
Between 30% and 80% of B6.Sle1 congenic mice develop anti-dsDNA IgG as a result of epitope spreading [12, 22]. However, the level of anti-dsDNA IgG in B6.Sle1congenic mice is low, and just 10% of these mice show evidence of mild nephritis [22]. To examine the impact of ERa deficiency on epitope spreading, we evaluated the impact of ERα genotype on the development of anti-dsDNA IgG autoantibodies (Figure 1E-F). In our study, the proportion of B6.Sle1;ERα+/+ females with measurable levels of anti-dsDNA IgG was 46%, and this proportion was not significantly altered by heterozygosity for ERα (Figure 1E). By contrast, the proportion of B6.Sle1;ERα−/− female mice that developed anti-dsDNA IgG autoantibodies was 9%, which is significantly lower than that observed in B6.Sle1;ERα+/+ females (Figure 1E-F; P ≤ 0.05).
In addition to developing autoantibodies of the IgG isotype, B6.Sle1 congenic mice also exhibit significantly greater total serum IgG levels compared to B6 controls [11]. Consistent with these previously published studies, we observed that total serum IgG levels were significantly elevated in B6.Sle1;ERα+/+ females compared to age- and sex-matched B6.ERα+/+ controls (Figure 1G; P < 0.05). Total serum IgG levels in B6.Sle1;ERα+/+ females was significantly different than that in B6.ERα+/+ controls (Figure 1G; P < 0.05) but not different from that in B6.Sle1;ERα+/+ females (Figure 1G; P = 0.09). This observation is consistent with the fact that the increase in total serum IgG in B6.Sle1 mice shows no sex bias [11]. Furthermore, this result suggests that the attenuated development of anti-chromatin IgG in B6.Sle1;ERα−/− females is not due a generalized defect in IgG production or maturation of the immune response. No significant differences were seen among the groups with respect to total serum IgM levels (Figure 1H), which are reported to be increased in B6.Sle1 congenic mice in some, but not all, studies [11, 17].
3.2. ERα deficiency attenuates Sle1-induced B and T cell activation in females
Sle1 is associated with the hyperactivation of both B cells and T cells [17]. To determine the impact of ERα deficiency on immune cell activation in B6.Sle1 female mice, flow cytometry was used to assess the expression of B and T cell activation markers (Table 1). As predicted, the proportion of B220+ CD86+ B cells was significantly greater in B6.Sle1;ERα+/+ congenic females compared to B6.ERα+/+ females (P < 0.05). Strikingly, in B6.Sle1;ERα−/− females, the percentage of B220+CD86+ B cells was significantly less than that in B6.Sle1;ERα+/+ females (P < 0.01), but not different than that in either B6.ERα+/+ or B6.ERα−/− controls (P > 0.05). These results indicate that ERα is required for Sle1-dependent B cell hyperactivation in females.
Table 1.
Impact of ERα genotype on lymphocyte activation in female B6.Sle1 micea
| B6 |
B6.Sle1 |
|||
|---|---|---|---|---|
| Cell type | ERα +/+ | −/− | +/+ | −/− |
| B220+b | 54.7±2.0 | 55.2±3.1 | 49.1±2.4e | 49.3±2.7 |
| B220+CD86+c | 8.6±1.3 | 9.3±2.5 | 12.9±2.5e | 6.2±1.4g |
|
| ||||
| CD4+b | 17.2±1.0 | 18.2 ±1.3 | 17.8±0.8 | 17.8±1.2 |
| CD4+CD69+d | 13.3±2.3 | 12.8±4.3 | 22.6±3.2f | 8.8±1.4g |
| CD4+CD134+d | 13.5±3.6 | 9.4±3.8 | 24.8±4.1e | 11.9±3.2f |
| CD4+CD62L+d | 40.6±4.7 | 59.2±4.7g | 15.0±1.4f | 25.2±5.2f,g |
| CD4+CD25+d | 2.4±0.2 | 2.7±0.2 | 1.9±0.2e | 1.9±0.3e |
Splenocytes from each mouse (6-12 months of age) were analyzed individually by flow cytometry (N=12-24 per group). The values presented represent the mean ± SEM for the samples in each group.
Values represent the mean percentage of total splenocytes.
Values represent the mean percentage of B220+ splenocyte
Values represent the mean percentage of CD4+ splenocytes.
Significant difference compared to B6 mice of the same ERα genotype are denoted by (P≤0.05)
Significant difference compared to B6 mice of the same ERα genotype are denoted by (P≤0.01)
Significant difference compared to ERα+/+ mice of the same Sle1 genotype are denoted by (P≤0.01)
B6.Sle1 mice also display a prominent T cell hyperactivation phenotype. Compared to B6.ERα+/+ females, a significantly smaller fraction of CD4+ T cells in B6.Sle1;ERα+/+ females expressed the naïve T cell marker CD62L whereas a significantly greater percentage of CD4+ T cells expressed the activation markers CD69 and CD134 (Table 1; P < 0.05). Sle1-induced T cell activation was largely abolished by ERa deficiency in females (Table 1); The proportion of activated CD4+ CD69+ and CD4+ CD134+ T cells in B6.Sle1; ERα−/− females was significantly less than that in B6.Sle1; ERα+/+ females (P < 0.05). Consistent with this observation, the proportion of naïve CD4+CD62L+ T cells in B6.Sle1; ERα−/− females was significantly greater than that in B6.Sle1; ERα+/+ females (P < 0.01). However, we noted that the fraction of CD4+ CD62L+ T cells in B6.Sle1; ERα−/− females remained significantly less than that in B6.ER+/+ or B6.ERα−/− control females (P < 0.01), indicating that ERα deficiency did not completely impede Sle1-induced T cell activation in females. Interestingly, the proportion of CD4+CD62L+ naïve T cells in B6.ER−/− control females was significantly greater than that in B6.ERα+/+ control females, suggesting that ERα may promote T cell activation in females independently of Sle1. However, there was no significant difference between B6.ERα−/− and B6.ERα+/+ control females with respect to the proportion of CD4+ T cells expressing CD69 or CD134 (P > 0.05). Altogether, these results suggest that ERα deficiency attenuates Sle1-induced T cell hyperactivation
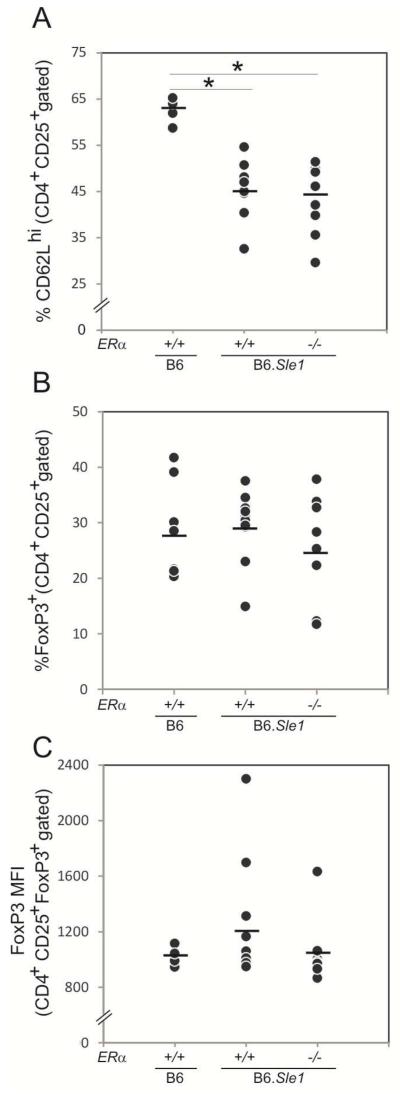
We also noted that the relative abundance of CD4+CD25+ T cells in B6.Sle1;ERα+/+ congenic females was significantly less than that in B6.ERα+/+ (P > 0.05). The proportion of CD4+CD25+ T cells in B6.Sle1; ERα−/− females was significantly less than that in B6.ERα−/− females (P < 0.05), but not different than that in B6.Sle1; ERα+/+ females. Because Sle1 has been reported to decrease the relative abundance of CD4+CD25+ Treg cells [26, 27], we examined the impact of ERα deficiency on the relative abundance of this particular subset of CD4+CD25+ T cells. Consistent with the previous studies, the proportion of CD4+CD25+ T cells that were CD62Lhi (CD4+CD25+ CD62Lhi Tregs) in B6.Sle;ERα+/+ females was significantly less than that in B6. ERα+/+ controls (Figure 2A;P < 0.01). However, the relative abundance of CD4+CD25+ CD62Lhi Tregs in B6.Sle1.ERα−/− females did not differ from that in B6.Sle1.ERα+/+ females, indicating that ERα deficiency does not impact the Sle1-associated decrease in the size of the Treg population (Figure 2A). Consistent with this observation, ERα deficiency also did not impact the relative size of the CD4+CD25+FoxP3+ Treg population or FoxP3 expression level (Figure 2B-C). These observations suggest that ERα deficiency attenuates Sle1-induced T cell hyperactivation but does not remediate the Sle1-associated decrease in the Treg population.
Figure 2. Impact of ERα genotype on the T regulatory population in B6.Sle1 congenic mice.
Flow cytometry was used to assess the T regulatory cell pool in female mice at 3-4 months of age. Splenocytes from each mouse were analyzed individually by flow cytometry (N=4-12 per group). The percentage of the CD4+CD25+ cells that were CD62Lhi T regulatory cells (A) and FoxP3+ T regulatory cells (B) is shown. The level of expression (mean fluorescence intensity or MFI) of FoxP3 in CD4+CD25+FoxP3+ T regulatory cells was also assessed (C). The horizontal bar in each panel denotes the mean. The * indicates a P<0.05 compared to B6 females.
3.3. Impact of ERα deficiency on expression of Sle1-associated genes in female mice
The Sle1 subloci are associated with polymorphisms that impact the expression and/or splicing of one or more genes within the Sle1 interval. Altered expression of these genes is responsible for the immune dysfunction and loss of tolerance in B6.Sle1 congenic mice. Because ERα functions as a ligand-activated transcription factor, we postulated that ERα might promote Sle1-associated phenotypes in B6.Sle1 congenic females by regulating the expression of the genes that underlie this lupus susceptibility locus. Therefore, we examined the effect of ERα deficiency on the expression of the genes that underlie Sle1.
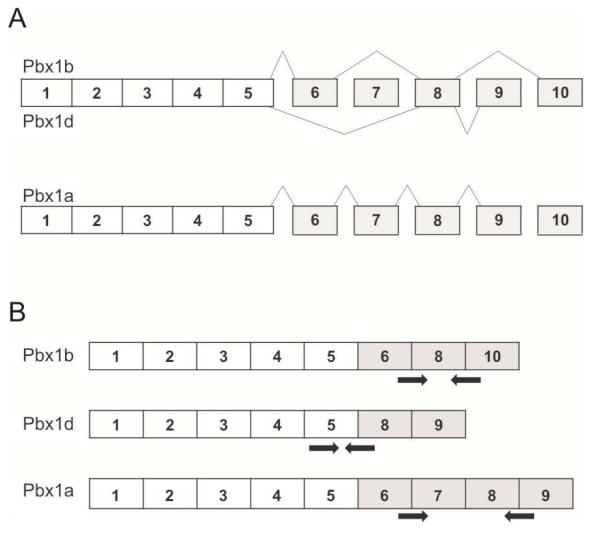
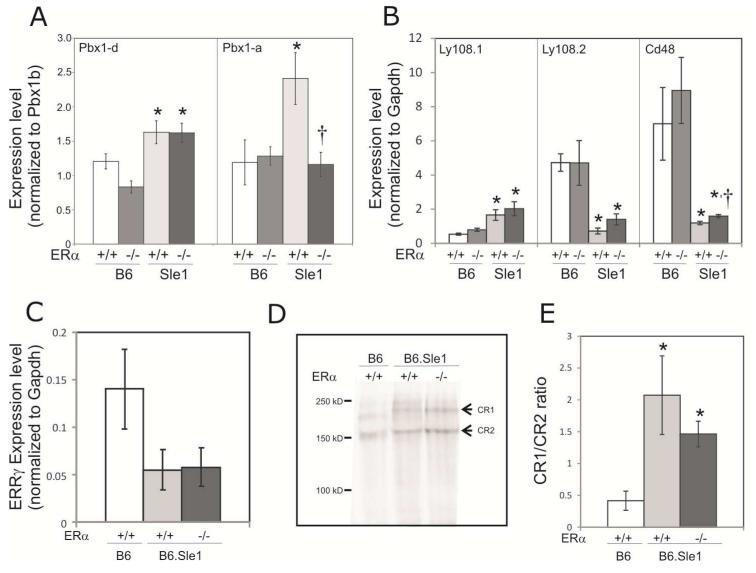
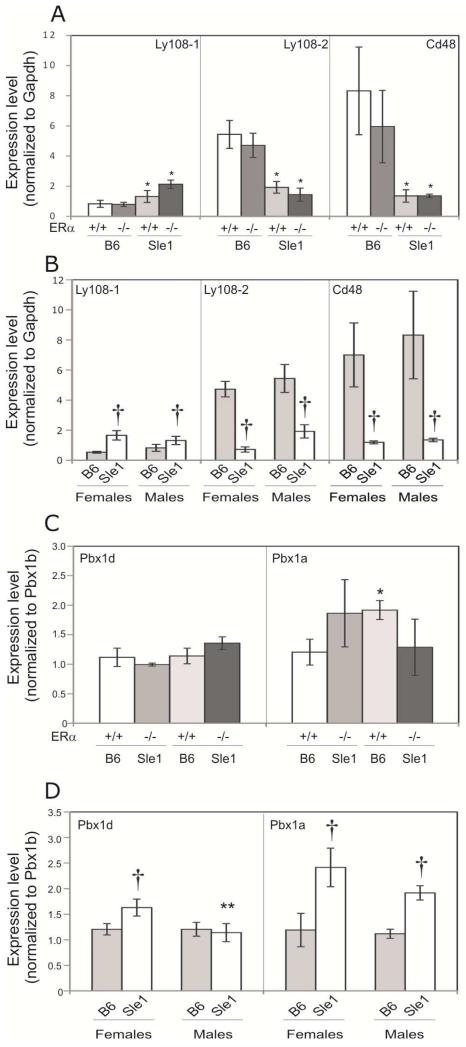
The Sle1a sublocus is comprised of multiple genes, including the pre-B-cell leukemia homoebox 1 (Pbx1) gene [28, 29]. Sle1a is associated with differential splicing of the Pbx1 transcript (Figure 3A), leading to an increase in the relative abundance of the lupus-associated Pbx1d mRNA isoform compared to the Pbx1b mRNA isoform [29]. To evaluate the impact of ERα genotype on the relative abundance of Pbx1d and Pbx1b isoforms, we developed isoform-specific PCR primers (Figure 3B) and performed q-RT-PCR. Consistent with previous reports, the Pbx1d/Pbx1b ratio in CD4+ T cells from B6.Sle1;ERα+/+ female mice was significantly greater than that in B6.ERα+/+ females (Figure 4A; P < 0.05). The Pbx1d/Pbx1b ratio in CD4+ T cells derived from B6.Sle1;ERα−/− female mice did not differ from that in B6.Sle1;ERα+/+ female mice (Figure 4A), indicating that ERα genotype did not impact the relative abundance of these transcripts. A third Pbx1 mRNA isoform, Pbx1a (Figure 3A), has been reported to be expressed in human T cells but not T cells in the B6.Sle1a.1 subcongenic strain [29]. However, using q-RT-PCR and isoform-specific PCR primers (Figure 3B), we were able to detect the Pbx1a isoform in splenic CD4+T cells from B6.Sle1 congenic mice, and thus we evaluated the impact of Sle1 and ERα on the relative expression of Pbx1a in female mice. The Pbx1a/Pbx1b ratio in CD4+ T cells from B6.Sle1;ERα+/+ female mice was significantly greater than that in B6.ERα+/+ females, indicating that expression of this Pbx1 isoform is also modulated by Sle1 genotype (Figure 4A; P < 0.05). Interestingly, the Pbx1a/Pbx1b ratio in CD4+ T cells form B6.Sle1;ERα−/− female mice was significantly less than that in B6.Sle1;ERα+/+ females, suggesting that the relative expression of this isoform is modulated by ERα signaling (Figure 4A; P < 0.05).
Figure 3. Alternative splicing of Pbx1 yields multiple isoforms at the mRNA and protein level.
Alternative splicing of exons 6-10 of the Pbx1 gene yields 3 unique isoforms, Pbx1b, Pbx1d and Pbx1a (A). PCR primers (indicated by arrows) were designed to independently quantify each transcript (B). These primers, with the exception of the Pbx1d forward primer, span exon junctions that are unique to each transcript.
Figure 4. Impact of ERα genotype on the expression of Sle1-associated genes in female mice.
The relative abundance of transcripts produced by genes that underlie Sle1 was assessed using quantitative RT-PCR. Data for panels A, B, C and E presented as mean ± standard error of the mean. The expression of Pbx1d and Pbx1a relative to Pbx1b in T cells (A) and Ly108-1, Ly108-2 and CD48 relative to Gapdh in B cells (B) isolated from 5-9 month old B6.ERα+/+, B6.ERα−/−, B6.Sle1;ERα+/+ and B6.Sle1;ERα−/− females (N=5-9 per genotype) is shown. Expression of Esrrg in T cells isolated from B6.ERα+/+, B6.Sle1;ERα+/+ and B6.Sle1;ERα−/− females (N=5-7 per genotype) is shown (C). The relative molecular weight of CR1 and CR2 (indicated by arrows) expressed in splenocytes from B6.ERα+/+, B6.Sle1;ERα+/+ and B6.Sle1;ERα−/− females (N=5-7 per genotype) was assessed by western blotting following immunoprecipitation and native gel electrophoresis. A representative image of each genotype is shown (D). CR1 and CR2 bands were quantified and the mean ratio (± standard error of the mean) for each genotype (N=5-7) is shown (E). The * indicates a P<0.05 compared to B6 females of the same ERα genotype (A and B) or B6.ERα+/+ females (E). The † indicates a P<0.05 compared to B6.Sle1;ERα+/+ females (A and B).
Sle1b consists of several linked polymorphisms in genes within the SLAM/CD2 cluster [30]. As a result of these polymorphisms, the expression of the CD48 gene is significantly altered. Furthermore, there is differential splicing of the Ly108 gene, leading to increased expression of the autoimmune Ly108-1 isoform and decreased expression of the “normal” Ly108-2 isoform. Consistent with previous reports, B cells derived from female B6.Sle1;ERα+/+ mice expressed significantly greater levels of Ly108-1 and significantly lower levels of both Ly108-2 and CD48 compared to female B6.ERα+/+ mice (Figure 4B; P < 0.05). ERα deficiency did not significantly impact the expression of Ly108-1, Ly108-2, or CD48 in female B6 mice (Figure 4B). Likewise, in female B6.Sle1;ERα−/− mice, the level of expression of Ly108-1 and Ly108-2 did not differ significantly from that in B6.Sle1;ERα+/+ mice (Figure 4B). Although B cells from female B6.Sle1;ERα−/− mice did express significantly more CD48 than that in female B6.Sle1;ERα+/+ mice, the magnitude of this difference was quite small and ERα deficiency in B6.Sle1 mice was insufficient to restore CD48 levels to that seen in B6.ERα+/+ or B6.ERα−/− controls (Figure 4B; P ≤ 0.01). Expression of CD48 and Ly108-2 is also downregulated in CD4+ T cells from B6.Sle1 mice [30]. The expression of Ly108-2 and CD48 in CD4+ T cells derived from female B6.Sle1;ERα−/− mice did not differ from that in B6.Sle1;ERα+/+ females (data not shown). Although some previous studies indicate that CD84 is also differentially expressed in B6.Sle1 mice compared to B6 controls, we did not find that Sle1or ERα genotype had any impact on CD84 expression (data not shown).
Sle1c is associated with multiple genes, including Estrogen Related Receptor γ (Esrrg) and Complement Receptor 1/2 (Cr2) [25, 31-33]. T cells from B6.Sle1c subcongenic mice have been reported to express lower levels of Esrrg compared to B6 CD4+ T cells [33]. Consistent with these reports, we found that B6.Sle1 CD4+ T cells expressed lower levels of Esrrg compared to B6 in female mice, although this difference fell short of statistical significance (P = 0.1). ERα genotype had no impact on the abundance of the Esrrg transcript (Figure 4C). B6.Sle1c splenocytes also exhibit differential glycosylation of Complement receptor 2 (CR2) and Complement receptor 1 (CR1), which is produced by alternative splicing from the Cr2 gene in mice. Similar to previous reports, the CR1 and CR2 expressed on splenocytes from B6.Sle1 mice migrated at a higher molecular weight than CR1 and CR2 expressed on splenocytes from B6 mice (Figure 3D). CR1 and CR2 from B6.Sle1;ERα+/+ and B6.Sle1;ERα−/− appear to have equivalent molecular weights, suggesting that ER genotype does not have a major impact on the glycosylation of CR1 or CR2 (Figure 4C). It has also been reported that the Sle1c allele of Cr2 undergoes differential splicing, leading to changes in the relative abundance of CR1 and CR2 [25]. Therefore, we examined the impact of ERα deficiency on the relative abundance of the CR1 and CR2 proteins. Consistent with what has been reported previously, we found that female B6.Sle1;ERα+/+ had a significantly higher CR1/CR2 ratio compared to B6.ERα+/+ mice (Figure 4E; P< 0.05). B6.Sle1;ERα+/+ and B6.Sle1;ERα−/− females had similar ratios of CR1/CR2 expression, indicating that ERα genotype does not affect the splicing of Cr2 (Figure 4E).
3.4. ERα deficiency does not impact loss of tolerance to chromatin in B6.Sle1 congenic male mice
Our previous studies in lupus-prone (NZB × NZW)F1 mice demonstrate that the low levels of endogenous estrogens, acting through ERα, promote loss of tolerance to chromatin and development of autoantibodies in male mice [8]. Although this effect in males was measurable, it was significantly less dramatic than that observed in female mice [8]. Based upon these results, we postulated that ERα deficiency might also attenuate to some degree the development of autoantibodies in B6.Sle1 congenic males. As has been reported previously, we observed that the proportion of B6.Sle1 congenic males that develop autoantibodies is significantly less than that in B6.Sle1 congenic females (Figure 5A-D; P <0.05). However, among those mice that had a positive anti-chromatin IgG titer, sex did not appear to impact the absolute levels of anti-chromating IgG (Figure 5B); this observation is also consistent with previous studies [12]. Interestingly, ERα deficiency did not have a significant impact on the proportion of B6.Sle1 males that developed anti-chromatin IgG (55% versus 43%; Figure 5E-F; P >0.05) or anti-dsDNA IgG (6% versus 14%; Figure 5G-H; P >0.05). Overall, these results, which indicated that ERα deficiency did not have a major impact on autoantibody development in B6.Sle1 males, were in sharp contrast to what was observed in B6.Sle1 females.
Figure 5. ERα deficiency does not impact loss tolerance in B6.Sle1 males.
ELISAs were used to assess the abundance of anti-chromatin IgG (A,B,E,F), anti-dsDNA IgG (C, D,G, H) in 5-9 month old mice. The proportion of B6.Sle1;ERα+/+ females (N=25) and B6.Sle1;ERα+/+ males (N=27) that develop anti-chromatin IgG (A,B) and anti-dsDNA IgG (C,D) is shown. The proportion of B6.Sle1;ERα+/+ (N=16), B6.Sle1;ERα+/− (N=11) and B6.Sle1;ERα−/− (N=10) males that develop anti-chromatin IgG (E,F) and anti-dsDNA IgG (G,H) is shown. For panels B,D,F and H, the horizontal bar denotes the mean Mean serum testosterone levels (I) in mice of the indicated genotypes is shown (N=9-11 of each sex per genotype). For each group shown in panel I, the mean concentration of anti-chromatin IgG (in arbitrary units/ml) was plotted in relation to the mean serum testosterone concentration (J) to determine is these two parameters were correlated. For panels I and J, the error bars represent the standard error of the mean. The * indicates a P<0.05 compared to B6.Sle1;ERα+/+ females (A, C, I).
3.5. Estrogens and ERα control sex bias in Sle1-induced loss of tolerance
We noted that the proportion of B6.Sle1 males, irrespective of ERα genotype, that developed anti-chromatin IgG, was not different than that observed in either B6.Sle1;ERα−/− females or ovariectomized B6.Sle1 females (P > 0.05). These observations indicate that removing either estrogens or ERα eliminates the sex bias in Sle1-induced loss of tolerance and suggest that estrogens are the major drivers of sex bias in the Sle1 phenotype. Nevertheless, because ERα deficiency leads to elevated serum testosterone levels [34, 35], we considered the possibility that androgens, which can also attenuate the development of autoantibodies in (NZB × NZW)F1 mice[36, 37] might also contribute to the sex bias in B6.Sle1 congenic mice. Similar to what has been reported previously in B6.ERα−/− female mice, we found that B6.Sle1;ERα−/− females have serum testosterone levels that are significantly higher than that B6.Sle1;ERα+/+ female mice but similar to that seen in intact B6.Sle1 male mice (Figure 5I). Thus, it seemed plausible that the attenuated loss of tolerance in B6.Sle1;ERα−/− females could be attributable, at least in part, to increased serum testosterone levels. However, the fact that the incidence of loss of tolerance in B6.Sle1;ERα−/− females is identical to that in ovariectomized B6.Sle1 females, which have very low levels of serum androgen levels (Figure 5I), does not support this possibility. We also observed no evidence of an inverse correlation between serum anti-chromatin levels and serum testosterone levels in these mice (Figure 5J). Altogether, these data suggest that estrogens, acting via ERα are responsible for the sex bias in Sle1-induced loss of tolerance.
3.6. ERα deficiency has a modest impact on Sle1-induced T cell activation in males
Although ERα deficiency did not impact Sle1-induced loss of tolerance and development of autoantibodies in males, we considered the possibility that ERα might nevertheless impact Sle1-associated immune cell hyperactivation in males [17]. Therefore, we wished to examine the impact of ERα deficiency on this aspect of the Sle1 phenotype in B6.Sle1 congenic males. Although no sex bias in Sle1-induced immune cell hyperactivation phenotype has been reported previously, we first performed some preliminary studies to explicitly examine this issue. We were surprised to find that in contrast to what was seen in B6.Sle1 females, B6.Sle1 males showed no evidence of B cell hyperactivation; there was no significant difference in the proportion of B220+CD86+ B cells in B6.Sle1;ERα+/+ males compared to B6.ERα+/+ males (Table 2). This observation was confirmed by analysis with CD25, a second marker of B cell activation (~12.9% B220+CD25+ in B6.ERα+/+ versus ~13.5% ±1.0 B220+CD25+ in B6.ERα+/+; P >0.05). Compared to B6.ERα+/+ males, B6.Sle1 males possessed a higher proportion of CD4+CD69+ and CD4+CD134+ T cells and a lower proportion of CD4+CD62L+ T cells, indicating that B6.Sle1 males do exhibit a T cell activation phenotype. Although B6.Sle1 congenic males exhibited T cell hyperactivation, the relative increase in activation compared to sex-matched B6 mice was somewhat less than that observed in female mice. Furthermore, the absolute level of activation, as indicated by CD62L expression, in B6.Sle1 males was somewhat less than that in B6.Sle1 females (Table 2). Altogether, these data indicate that Sle1-induced immune cell hyperactivation is a sex-biased phenotype.
Table 2.
Impact gender on lymphocyte activation in B6.Sle1 micea
| Female |
Male |
|||
|---|---|---|---|---|
| Cell type | B6 | B6.Sle1 | B6 | B6.Sle1 |
| B220+b | 60.9±2.4 | 44.7±3.5e | 65.2±6.6 | 57.0±2.2h |
| B220+CD86+c | 4.7±1.3 | 13.8±2.5e | 3.4±0.3 | 4.0±0.5h |
|
| ||||
| CD4+b | 15.0±1.0 | 17.4±1.0 | 15.1±2.3 | 14.3±0.7 |
| CD4+CD69+d | 6.7±1.1 | 16.3±2.5f | 9.6±2.9 | 18.3±2.9e |
| CD4+CD134+d | 5.5±0.8 | 17.6±2.8e | 9.3±5.6 | 14.6±2.5e |
| CD4+CD62L+d | 40.5 ±7.1 | 13.9 ±1.7f | 62.3 ±9.8g | 35.6±3.4f,h |
| CD4+CD25+d | 2.4±0.2 | 1.9±0.1e | 2.7±0.1g | 2.0±0.1e |
Splenocytes from each mouse (6-12 months of age) were analyzed individually by flow cytometry (N=12-25 per group). The values presented represent the mean ± SEM for the samples in each group.
Values represent the mean percentage of total splenocytes.
Values represent the mean percentage of B220+ splenocytes
Values represent the mean percentage of CD4+ splenocytes.
Significant difference compared to B6 mice of same sex are denoted by (P≤0.05)
Significant difference compared to B6 mice of same sex are denoted by (P≤0.01)
Significant difference compared to female mice of the same Sle1 genotype are denoted by (P≤0.05)
Significant difference compared to female mice of the same Sle1 genotype are denoted by (P≤0.01)
ERα deficiency had no significant impact on the proportion of activated B cells in B6.Sle1 congenic males (Table 3). Sle1-induced T cell activation in males was somewhat attenuated by ERa deficiency. The proportion of CD4+CD69+ T cells in B6.Sle1;ERα−/− males was significantly less than that in the B6.Sle1;ERα+/+ males (Table 3; P < 0.05). The proportion of CD4+CD134+ T cells was reduced in B6.Sle1;ERα−/− males compared to B6.Sle1;ERα+/+ males, but this difference fell short of statistical significance (P = 0.1). Interestingly, ERα deficiency had no impact on the proportion of CD4+CD62L+ T cells (P > 0.05). These data indicate that Sle1-induced T cell hyperactivation in males is only modestly affected by ERα deficiency.
Table 3.
Impact of ERα genotype on lymphocyte activation male B6.Sle1 micea
| B6 |
B6.Sle1 |
|||
|---|---|---|---|---|
| Cell type | ERα +/+ | −/− | +/+ | −/− |
| B220+b | 57.3±2.2 | 52.9±3.7 | 58.6±2.0 | 59.4±2.2 |
| B220+CD86+c | 6.8±1.1 | 6.6±1.8 | 7.0±2.2 | 7.9±2.2 |
| CD4+b | 15.6±1.9 | 15.9±1.7 | 16.2±0.7 | 15.2±0.9 |
| CD4+CD69+d | 17.6±2.7 | 14.7±3.4 | 25.5±2.5 | 15.0±3.9f |
| CD4+CD134+d | 17.8±3.4 | 21.3±6.4 | 25.7±3.4 | 15.3±4.6 |
| CD4+CD62L+d | 52.9±6.9 | 55.9±6.3 | 31.4±3.0e | 33.7±4.6e |
| CD4+CD25+ | 2.3±0.2 | 2.5±0.3 | 2.2±0.1 | 2.3±0.2 |
Splenocytes from each mouse (6-12 months of age) were analyzed individually by flow cytometry (N=14-32 per group). The values presented represent the mean ± SEM for the samples in each group.
Values represent the mean percentage of total splenocytes.
Values represent the mean percentage of B220+ splenocytes
Values represent the mean percentage of CD4+ splenocytes.
Significant differences compared to Sle1B/B mice of same gender and ERα genotype are denoted by (P≤0.01)
Significant difference compared to ERα+/+mice of same gender and Sle1 genotype are denoted by (P≤0.05)
3.7. ERα deficiency does not alter the expression of Sle1-associated genes in male B6.Sle1 mice
As we observed in females, ERα genotype did not have a major impact on the expression of Ly108-1, Ly108-2, or CD48 in B6.Sle1 male mice (Figure 6A). Consistent with this observation, we also found that sex did not impact the expression of these genes (Figure 6B). Likewise, the magnitude of the effect of Sle1 on the expression of each of these genes was not different in females and males. ERα deficiency did not have a significant impact on the relative expression of either Pbx1d or Pbx1a in CD4+ T cells in B6.Sle1 male mice (Figure 6C; P>0.05). Interestingly, the relative expression of both Pbx1d and Pbx1a in CD4+ T cells was greater in B6.Sle1 congenic females compared to congenic males, but this difference achieved significance only for Pbx1d (Figure 6D). Consistent with our results in B6.Sle1 congenic female mice, ERα genotype had no impact on Esrrg transcript level in CD4+ T cells or CR1/CR2 ratios in B6.Sle1 congenic male mice (data not shown).
Figure 6. Impact of ERα genotype on the expression of Sle1-associated genes in male mice.
The relative abundance of transcripts produced by genes that underlie Sle1 was assessed using quantitative RT-PCR. Data is presented as mean ± standard error of the mean. The expression of Ly108-1, Ly108-2 and CD48 relative to Gapdh in B cells (A and B) and Pbx1d and Pbx1a relative to Pbx1b in T cells (C and D) is shown. The impact of Sle1 and ERα genotype on the expression of Ly108-1, Ly108-2 and CD48 is shown by comparing relative expression in B6.ERα+/+, B6.ERα−/−, B6.Sle1;ERα+/+ and B6.Sle1;ER−/− males (N=4-5 per genotype) (A). The impact of sex on the expression of Ly108-1, Ly108-2 and CD48 is shown by comparing expression in B6 and B6.Sle1 females and males (B). The impact of Sle1 and ERα genotype on the expression of Pbx1d and Pbx1a is shown by comparing relative expression in B6.ERα+/+, B6.ERα−/−, B6.Sle1;ERα+/+ and B6.Sle1;ERα−/− males (N=4-5 per genotype) (C). The impact of sex on the expression of Pbx1d and Pbx1a is shown by comparing expression in B6 and B6.Sle1 females and males (D). The * indicates a P<0.05 compared to B6 male mice of the same ERα genotype (A and C). The † indicates a P<0.05 compared to sex-matched B6 mice (B and D). The ** indicates a P<0.05 compared to female mice of the same Sle1 genotype (D).
4. Discussion
It has been shown previously that the effects of the lupus susceptibility locus Sle1 are more robust in female mice than male mice. However, the basis for this sex bias is not known. In the present study, we examined the impact of removing estrogens or ERα on the Sle1 phenotype, and our results indicate that sex bias in the Sle1 phenotype is due to estrogen-dependent, ERα-mediated processes. Our data do not provide any evidence to support the hypothesis that androgens, which are known to attenuate lupus in (NZB × NZW)F1 mice[36, 37], contribute to the sex bias associated with Sle1.
Based upon our results, we conclude that the Sle1 lupus susceptibility locus induces a basal level of loss of tolerance that is independent of both sex and ERα signaling. Sle1, via these basal mechanisms, results in loss of tolerance in ~50% of mice, the proportion of B6.Sle1 males the develop anti-chromatin IgG. Our observations also suggest that estrogen-dependent, ERα-mediated processes synergize with Sle1, increasing the proportion of B6.Sle1 females that develop anti-chromatin IgG to more than 80%. Thus ERα signaling results in a significant increase in the penetrance of Sle1 in females.
Sle1 also has been associated with hyperactivation of both B cells and T cells. We report here for the first time that Sle1-induced immune cell activation also shows a significant sex bias. Sle1-induced B cell hyperactivation was observed only in B6.Sle1 females and was completely abrogated by ERα deficiency, suggesting that this phenotype in is fully dependent upon ERα signaling. This observation is consistent with a previous report indicating that increased B cell activation is a feature of B6.Sle1b subcongenic female, but not male, mice [20].
By contrast, we found that Sle1-induced T cell activation was detectable in both B6.Sle1 females and males. However, we noted that this aspect of the Sle1 phenotype was more robust in B6.Sle1 females than males. We also observed that ERα deficiency attenuated but did not completely eliminate Sle1-induced T cell activation. Altogether, these results indicate that Sle1 induces some degree of T cell hyperactivation independent of sex and ERα. ERα-dependent signaling synergizes with Sle1 to induce a more profound T cell hyperactivation phenotype in females. Here again, our results regarding the impact of sex on Sle1-induced T cell activation are consistent with the findings of Wong et al [20], which indicate that Sle1b-induced T cell activation is more pronounced in female mice than male mice. We also noted that ERα deficiency alone slightly increased the proportion of naïve T cells, suggesting the ERα signaling can promote some degree of T cell activation. This effect was observed in females only and was independent of Sle1 genotype. Finally, we also observed that ERα deficiency had no impact on the decrease in the number of regulatory T cells that is observed in B6.Sle1 congenic mice.
The NZW-derived allelic variants that underlie the Sle1 lupus susceptibility locus lead to differential expression and/or splicing of genes within the Sle1 interval, and the altered expression of these genes is responsible for the immune dysfunction and loss of tolerance in B6.Sle1 congenic mice. The impact of sex on expression and splicing of these genes has not been described. We postulated that ERα may impact the Sle1 phenotype by directly regulating the expression of these same genes. Using qRT-PCR, we found that the relative expression of the Pbx1d isoform of Pbx1 was increased in B6.Sle1 congenic females but not males. However, Pbx1d expression was not attenuated in B6.Sle1;ERα−/− females, indicating that the expression of this isoform is not ERα dependent. We note that the present study, the relative proportion of Pbx1d to Pbx1b in non-autoimmune B6 control mice was somewhat higher than that reported previously [29]. However, the Pbx1d:Pbx1b ratio in our other set of non-autoimmune control mice, the B6.ERα−/− mice, was not significantly different from that in our B6.ERα+/+ controls and was also very similar to that reported by Cuda et al [29]. It is likely that any differences in the relative expression of Pbx1 isoforms among control mice in these two studies reflects the differences in the methods used to evaluate and quantify gene expression. Nevertheless, despite differences in methods and relative expression in control samples, we observe that Sle1 is indeed associated with an increase in the relative abundance of Pbx1d; this result is fully consistent with the findings of Cuda et al [29].
We also detected Pbx1a, another isoform of Pbx1, in T cells from both B6 and B6.Sle1 congenic mice. Although Pbx1a was reported to the undetectable by conventional RT-PCR in both the B6 strain and the B6.Sle1a.1 subcongenic strain [29], we were able to detect this isoform using an isoform specific Q-RT-PCR assay. The relative expression of the Pbx1a isoform was also increased in B6.Sle1 congenic mice compared to B6 mice, but sex did not appear to impact Pbx1a expression. Surprisingly, Pbx1a expression was significantly reduced in B6.Sle1;ERα−/− congenic females. These observations suggest that ERα signaling may impact the Sle1 phenotype, in part, by modulating the expression the Pbx1a isoform. It remains to be determined if the effects of ERα on Pbx1 isoform expression are due to direct effects on Pbx1 transcription and/or splicing or due to indirect effects. Neither sex nor ERα deficiency had an impact on the expression of the other genes that underlie Sle1. In this context, it is particularly noteworthy that neither sex nor ERα deficiency had an impact on the expression of Ly108-1, Ly108-2 or Cd48, which are associated with Sle1b, the sublocus that displays the most dramatic sex bias [11, 12].
Pbx1 is a member of the three amino acid loop extension family of transcription factors ([38, 39]). Pbx1 proteins form homodimers and heterodimers with other homeodomain containing proteins. These Pbx1-containing dimers form ternary complexes with other transcription factors, such as Hox proteins and nuclear steroid hormone receptors, and modulate the ability of these other transcription factors to bind to specific binding sites in the genome [40-43]. Evidence suggests that Pbx1-containing complexes impact transcription factor binding by recruiting co-repressors and co-activators and thus modulating the accessibility of specific binding sites [43-45]. In this context, it is of particular interest that Pbx1 has been shown to modulate the binding of ERα to specific estrogen response elements in ER+ breast cancers [46]. However, the impact of Pbx1 on either the accessibility of estrogen response elements or ERα function in other cell types, including those relevant to lupus susceptibility, has not been examined.
Our results suggest that sex and ERα impact the relative expression of Pbx1 isoforms. Differential expression of these isoforms has been shown previously to correlate with lupus susceptibility [29], but the basis for this effect is not fully clear. The Pbx1a isoform encodes a protein that contains the central domains required for DNA and homeodomain-containing protein binding as well as the c-terminal activation domain and co-repressor binding domain [45, 47-49]. The Pbx1b protein product has a c-terminal truncation that eliminates the c-terminal activation domain and co-repressor binding domains [45, 48]. The Pbx1d peptide contains an internal deletion that eliminates the amino acids required for optimal Hox protein and DNA binding and is thus postulated to function as a dominant negative [29]. Pbx1d also lacks a portion of the c-terminal activation domain and co-repressor binding domain [29]. It is likely that the Pbx1a, Pbx1b and Pbx1d isoforms differ dramatically in their ability to interact with co-repressors, co-activator and transcription factors, and thus, in their ability to modulate transcription in different cellular contexts. Given the fact that Pbx1 is a critical regulator of self-renewal and lineage commitment in the hematopoietic and lymphoid lineage [50-52], changes in the relative expression of the Pbx1 isoforms could have a dramatic impact on autoimmunity. Further studies are required to determine the mechanism through which different Pbx1 isoforms modulate lupus susceptibility and to determine the impact of ERα signaling of these processes.
Acknowledgments
This work was supported by the National Institutes of Health R01 AI075167 (KAG). We gratefully acknowledge the assistance of the staff of The UNMC Flow Cytometry Research Facility. This facility is administrated through the Office of the Vice Chancellor for Research and is supported by state funds from the Nebraska Research Initiative (NRI) and The Fred and Pamela Buffet Cancer Center’s National Cancer Institute Cancer Support Grant. Major instrumentation has been provided by the Office of the Vice Chancellor for Research, The University of Nebraska Foundation, the Nebraska Banker’s Fund, and by the NIH-NCRR Shared Instrument Program.
Footnotes
Abbreviations used: Estrogen receptorα (ERα)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lahita RG, Bradlow HL. Klinefelter’s syndrome: hormone metabolism in hypogonadal males with systemic lupus erythematosus. J. Rheumatol. 1987;14(Suppl 13):154–157. [PubMed] [Google Scholar]
- [2].Lahita RG, Bradlow HL, Kunkel HG, Fishman J. Alterations of estrogen metabolism in systemic lupus erythematosus. Arthritis Rheum. 1979;22:1195–1198. doi: 10.1002/art.1780221106. [DOI] [PubMed] [Google Scholar]
- [3].Lahita RG, Bradlow HL, Kunkel HG, Fishman J. Increased 16 alpha-hydroxylation of estradiol in systemic lupus erythematosus. J. Clin. Endocrinol. Metab. 1981;53:174–178. doi: 10.1210/jcem-53-1-174. [DOI] [PubMed] [Google Scholar]
- [4].Liu ZH, Cheng ZH, Gong RJ, Liu H, Liu D, Li LS. Sex differences in estrogen receptor gene polymorphism and its association with lupus nephritis in Chinese. Nephron. 2002;90:174–180. doi: 10.1159/000049039. [DOI] [PubMed] [Google Scholar]
- [5].Lee YJ, Shin KS, Kang SW, Lee CK, Yoo B, Cha HS, Koh EM, Yoon SJ, Lee J. Association of the oestrogen receptor alpha gene polymorphisms with disease onset in systemic lupus erythematosus. Ann. Rheum. Dis. 2004;63:1244–1249. doi: 10.1136/ard.2003.012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Howie JB, Helyer BJ. The immunology and pathology of NZB mice. Adv. Immunol. 1968;9:215–266. doi: 10.1016/s0065-2776(08)60444-7. [DOI] [PubMed] [Google Scholar]
- [7].Andrews BS, Eisenberg RA, Theofilopoulos AN, Izui S, Wilson CB, McConahey PJ, Murphy ED, Roths JB, Dixon FJ. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J. Exp. Med. 1978;148:1198–1215. doi: 10.1084/jem.148.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bynote KK, Hackenberg JM, Korach KS, Lubahn DB, Lane PH, Gould KA. Estrogen receptor-alpha deficiency attenuates autoimmune disease in (NZB × NZW)F1 mice. Genes Immun. 2008;9:137–152. doi: 10.1038/sj.gene.6364458. [DOI] [PubMed] [Google Scholar]
- [9].Burlingame RW, Boey ML, Starkebaum G, Rubin RL. The central role of chromatin in autoimmune responses to histones and DNA in systemic lupus erythematosus. J. Clin. Invest. 1994;94:184–192. doi: 10.1172/JCI117305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Burlingame RW, Rubin RL, Balderas RS, Theofilopoulos AN. Genesis and evolution of antichromatin autoantibodies in murine lupus implicates T-dependent immunization with self antigen. J. Clin. Invest. 1993;91:1687–1696. doi: 10.1172/JCI116378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mohan C, Morel L, Yang P, Watanabe H, Croker B, Gilkeson G, Wakeland EK. Genetic dissection of lupus pathogenesis: a recipe for nephrophilic autoantibodies. J. Clin. Invest. 1999;103:1685–1695. doi: 10.1172/JCI5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mohan C, Alas E, Morel L, Yang P, Wakeland EK. Genetic dissection of SLE pathogenesis. Sle1 on murine chromosome 1 leads to a selective loss of tolerance to H2A/H2B/DNA subnucleosomes. J. Clin. Invest. 1998;101:1362–1372. doi: 10.1172/JCI728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gubbels MR, Jorgensen TN, Metzger TE, Menze K, Steele H, Flannery SA, Rozzo SJ, Kotzin BL. Effects of MHC and gender on lupus-like autoimmunity in Nba2 congenic mice. J. Immunol. 2005;175:6190–6196. doi: 10.4049/jimmunol.175.9.6190. [DOI] [PubMed] [Google Scholar]
- [14].Morel L, Rudofsky UH, Longmate JA, Schiffenbauer J, Wakeland EK. Polygenic control of susceptibility to murine systemic lupus erythematosus. Immunity. 1994;1:219–229. doi: 10.1016/1074-7613(94)90100-7. [DOI] [PubMed] [Google Scholar]
- [15].Morel L, Yu Y, Blenman KR, Caldwell RA, Wakeland EK. Production of congenic mouse strains carrying genomic intervals containing SLE-susceptibility genes derived from the SLE-prone NZM2410 strain. Mamm. Genome. 1996;7:335–339. doi: 10.1007/s003359900098. [DOI] [PubMed] [Google Scholar]
- [16].Morel L, Mohan C, Yu Y, Schiffenbauer J, Rudofsky UH, Tian N, Longmate JA, Wakeland EK. Multiplex inheritance of component phenotypes in a murine model of lupus. Mamm. Genome. 1999;10:176–181. doi: 10.1007/s003359900964. [DOI] [PubMed] [Google Scholar]
- [17].Morel L, Blenman KR, Croker BP, Wakeland EK. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc. Natl. Acad. Sc.i U. S. A. 2001;98:1787–1792. doi: 10.1073/pnas.031336098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sobel ES, Satoh M, Chen Y, Wakeland EK, Morel L. The major murine systemic lupus erythematosus susceptibility locus Sle1 results in abnormal functions of both B and T cells. J. Immunol. 2002;169:2694–2700. doi: 10.4049/jimmunol.169.5.2694. [DOI] [PubMed] [Google Scholar]
- [19].Sobel ES, Mohan C, Morel L, Schiffenbauer J, Wakeland EK. Genetic dissection of SLE pathogenesis: adoptive transfer of Sle1 mediates the loss of tolerance by bone marrow-derived B cells. J. Immunol. 1999;162:2415–2421. [PubMed] [Google Scholar]
- [20].Wong EB, Khan TN, Mohan C, Rahman ZS. The Lupus-Prone NZM2410/NZW Strain-Derived Sle1b Sublocus Alters the Germinal Center Checkpoint in Female Mice in a B Cell-Intrinsic Manner. J. Immunol. 2012;189:5667–5681. doi: 10.4049/jimmunol.1201661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lubahn DB, Moyer JS, Golding TS, Couse JF, Korach KS, Smithies O. Alteration of reproductive function but not prenatal sexual development after insertional disruption of the mouse estrogen receptor gene. Proc. Natl. Acad. Sci. U. S A. 1993;90:11162–11166. doi: 10.1073/pnas.90.23.11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Morel L, Mohan C, Yu Y, Croker BP, Tian N, Deng A, Wakeland EK. Functional dissection of systemic lupus erythematosus using congenic mouse strains. J. Immunol. 1997;158:6019–6028. [PubMed] [Google Scholar]
- [23].Dietrich W, Katz H, Lincoln SE, Shin HS, Friedman J, Dracopoli NC, Lander ES. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yuan F, Tabor DE, Nelson RK, Yuan H, Zhang Y, Nuxoll J, Bynote KK, Lele SM, Wang D, Gould KA. A dexamethasone prodrug reduces the renal macrophage response and provides enhanced resolution of established murine lupus nephritis. PLoS One. 2013;8:e81483. doi: 10.1371/journal.pone.0081483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Boackle SA, Holers VM, Chen X, Szakonyi G, Karp DR, Wakeland EK, Morel L. Cr2, a candidate gene in the murine Sle1c lupus susceptibility locus, encodes a dysfunctional protein. Immunity. 2001;15:775–785. doi: 10.1016/s1074-7613(01)00228-x. [DOI] [PubMed] [Google Scholar]
- [26].Cuda CM, Wan S, Sobel ES, Croker BP, Morel L. Murine lupus susceptibility locus Sle1a controls regulatory T cell number and function through multiple mechanisms. J. Immunol. 2007;179:7439–7447. doi: 10.4049/jimmunol.179.11.7439. [DOI] [PubMed] [Google Scholar]
- [27].Chen Y, Cuda C, Morel L. Genetic determination of T cell help in loss of tolerance to nuclear antigens. J. Immunol. 2005;174:7692–7702. doi: 10.4049/jimmunol.174.12.7692. [DOI] [PubMed] [Google Scholar]
- [28].Cuda CM, Zeumer L, Sobel ES, Croker BP, Morel L. Murine lupus susceptibility locus Sle1a requires the expression of two sub-loci to induce inflammatory T cells. Genes Immun. 2010;11:542–553. doi: 10.1038/gene.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cuda CM, Li S, Liang S, Yin Y, Potula HH, Xu Z, Sengupta M, Chen Y, Butfiloski E, Baker H, Chang LJ, Dozmorov I, Sobel ES, Morel L. Pre-B cell leukemia homeobox 1 is associated with lupus susceptibility in mice and humans. J. Immunol. 2012;188:604–614. doi: 10.4049/jimmunol.1002362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR, Jr., Morel L, Wakeland EK. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21:769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- [31].Chen Y, Perry D, Boackle SA, Sobel ES, Molina H, Croker BP, Morel L. Several genes contribute to the production of autoreactive B and T cells in the murine lupus susceptibility locus Sle1c. J. Immunol. 2005;175:1080–1089. doi: 10.4049/jimmunol.175.2.1080. [DOI] [PubMed] [Google Scholar]
- [32].Xu Z, Potula HH, Vallurupalli A, Perry D, Baker H, Croker BP, Dozmorov I, Morel L. Cyclin-dependent kinase inhibitor Cdkn2c regulates B cell homeostasis and function in the NZM2410-derived murine lupus susceptibility locus Sle2c1. J. Immunol. 2011;186:6673–6682. doi: 10.4049/jimmunol.1002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Perry DJ, Yin Y, Telarico T, Baker HV, Dozmorov I, Perl A, Morel L. Murine lupus susceptibility locus Sle1c2 mediates CD4+ T cell activation and maps to estrogen-related receptor gamma. J. Immunol. 2012;189:793–803. doi: 10.4049/jimmunol.1200411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Couse JF, Yates MM, Walker VR, Korach KS. Characterization of the hypothalamic-pituitary-gonadal axis in estrogen receptor (ER) Null mice reveals hypergonadism and endocrine sex reversal in females lacking ERalpha but not ERbeta. Mol. Endocrinol. 2003;17:1039–1053. doi: 10.1210/me.2002-0398. [DOI] [PubMed] [Google Scholar]
- [35].Eddy EM, Washburn TF, Bunch DO, Goulding EH, Gladen BC, Lubahn DB, Korach KS. Targeted disruption of the estrogen receptor gene in male mice causes alteration of spermatogenesis and infertility. Endocrinology. 1996;137:4796–4805. doi: 10.1210/endo.137.11.8895349. [DOI] [PubMed] [Google Scholar]
- [36].Roubinian JR, Talal N, Greenspan JS, Goodman JR, Siiteri PK. Effect of castration and sex hormone treatment on survival, anti-nucleic acid antibodies, and glomerulonephritis in NZB/NZW F1 mice. J. Exp. Med. 1978;147:1568–1583. doi: 10.1084/jem.147.6.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Roubinian JR, Papoian R, Talal N. Androgenic hormones modulate autoantibody responses and improve survival in murine lupus. J. Clin. Invest. 1977;59:1066–1070. doi: 10.1172/JCI108729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kamps MP, Murre C, Sun XH, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- [39].Nourse J, Mellentin JD, Galili N, Wilkinson J, Stanbridge E, Smith SD, Cleary ML. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- [40].Chang CP, Brocchieri L, Shen WF, Largman C, Cleary ML. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol. Cell. Biol. 1996;16:1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- [42].Subramaniam N, Campion J, Rafter I, Okret S. Cross-talk between glucocorticoid and retinoic acid signals involving glucocorticoid receptor interaction with the homoeodomain protein Pbx1. Biochem J. 2003;370:1087–1095. doi: 10.1042/BJ20020471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Laurent A, Bihan R, Omilli F, Deschamps S, Pellerin I. PBX proteins: much more than Hox cofactors. In.t J. Dev. Biol. 2008;52:9–20. doi: 10.1387/ijdb.072304al. [DOI] [PubMed] [Google Scholar]
- [44].Saleh M, Rambaldi I, Yang XJ, Featherstone MS. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 2000;20:8623–8633. doi: 10.1128/mcb.20.22.8623-8633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Asahara H, Dutta S, Kao HY, Evans RM, Montminy M. Pbx-Hox heterodimers recruit coactivator-corepressor complexes in an isoform-specific manner. Mol. Cell. Biol. 1999;19:8219–8225. doi: 10.1128/mcb.19.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERalpha signaling underlying progression in breast cancer. PLoS Genet. 2011;7:e1002368. doi: 10.1371/journal.pgen.1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Peltenburg LT, Murre C. Specific residues in the Pbx homeodomain differentially modulate the DNA-binding activity of Hox and Engrailed proteins. Development. 1997;124:1089–1098. doi: 10.1242/dev.124.5.1089. [DOI] [PubMed] [Google Scholar]
- [48].Di Rocco G, Mavilio F, Zappavigna V. Functional dissection of a transcriptionally active, target-specific Hox-Pbx complex. EMBO J. 1997;16:3644–3654. doi: 10.1093/emboj/16.12.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chang CP, de Vivo I, Cleary ML. The Hox cooperativity motif of the chimeric oncoprotein E2a-Pbx1 is necessary and sufficient for oncogenesis. Mol. Cell. Biol. 1997;17:81–88. doi: 10.1128/mcb.17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Riddell J, Gazit R, Garrison BS, Guo G, Saadatpour A, Mandal PK, Ebina W, Volchkov P, Yuan GC, Orkin SH, Rossi DJ. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ficara F, Crisafulli L, Lin C, Iwasaki M, Smith KS, Zammataro L, Cleary ML. Pbx1 restrains myeloid maturation while preserving lymphoid potential in hematopoietic progenitors. J. Cell Sci. 2013;126:3181–3191. doi: 10.1242/jcs.125435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2:484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]