Abstract
The mechanisms by which Xist RNA associates with the X chromosome to mediate alterations in chromatin structure remain mysterious. Recent genome-wide Xist RNA distribution studies suggest that this long noncoding RNA uses 3-dimensional chromosome contacts to move to its sites of action.
In organisms with XY sex chromosomes, chromatin modifications are directed to the X chromosomes (X) to equalize X-linked gene dosage between males and females. In flies and worms, the dosage compensation complexes are localized to the X by sequence specific binding to high affinity sites and subsequent spread to nearby lower affinity sites [1]. Despite over five decades of X chromosome inactivation (XCI) research, little is understood of the mechanisms controlling the localization of the mammalian dosage compensation machinery to the X. In XCI, a long noncoding RNA (lcnRNA), Xist RNA, recruits chromatin modifying complexes to the X. The Xist gene is encoded in the X-inactivation center (Xic), an X-linked cis-element that is essential for XCI. Xist RNA spreads from the Xic to coat the X and contributes to the initial establishment of silencing and subsequent maintenance of XCI [2]. In two recent studies, Engreitz et al. and Simon et al. used genome-wide approaches to map the DNA associated with Xist RNA to provide insight into how this lncRNA spreads [3,4].
Both groups utilized pools of antisense oligonucleotides complementary to the 17 kb Xist RNA to enrich for Xist RNA-associated genomic sequences in crosslinked cells. Comparison of Xist RNA distribution with data sets for genomic features provided clues about the mechanisms of Xist RNA localization and spread. During the maintenance stage of XCI the same pattern of Xist RNA distribution emerged in both studies. Xist RNA enrichment was observed across the entire X relative to autosomes. There was variability across the X, with gene-dense regions exhibiting the highest representation in the pools of DNA pulled down with Xist RNA. A small number of genes escape XCI, and these escapees had the lowest representation on average. Previous cytological analyses suggest that escapees are more likely to make forays outside the Xist RNA domain, raising the possibility that this lower representation reflects unique dynamics of the escapees [5,6].
Comparison of Xist RNA distribution with genome feature data sets revealed a significant correlation between Xist RNA enrichment and histone H3 lysine 27 trimethylation and EZH2, the methyltransferase that mediates this modification. This correlation is consistent with Xist RNA-dependent recruitment of EZH2 to the X [7,8]. Both studies also report an anti-correlation with Long Interspersed Elements (LINEs), which may be a consequence of gene density as LINEs are preferentially located in gene poor regions that show low Xist RNA accumulation. These analyses provide the highest resolution of Xist RNA distribution to date, complementing cytological approaches.
A powerful way to gain further insight into Xist RNA dynamics would be to apply these high-resolution approaches to establishment of XCI, when Xist RNA spreads in cis from the Xic to coat the X for the first time. Establishment of XCI can be recapitulated ex vivo by differentiation of female mouse embryonic stem cells (ESCs). However, XCI is highly asynchronous in this system, complicating temporal analysis. To obtain a population of cells synchronously establishing silencing, Engreitz et al. utilized a male ESC line containing inducible Xist expressed from its endogenous location in the Xic. Analysis was carried out at four time points: before Xist induction; one hour after induction, when there is a strong focal point of Xist RNA accumulation around the Xic; three hours after induction, when there is a small cloud of Xist RNA; and six hours after induction, when Xist RNA fully coats the X. At six hours the Xist RNA distribution closely resembled that of cells in the maintenance stage of XCI. At the one-hour time point there was moderate enrichment of Xist RNA at gene rich regions. Superimposed on this broad Xist RNA distribution was a very strong peak of Xist RNA enrichment over the Xist locus and significant enrichment at 28 distal sites. Comparison to over 250 genomic annotations revealed that these Xist RNA-enriched regions showed the strongest correlation with the domains topologically associated with the Xic in ESCs. This correlation suggests that Xist RNA first contacts regions closest to the Xic in 3-dimensional (3D) space, rather than tracking linearly along the X.
To test whether this artificial system accurately reflects Xist RNA dynamics during bona fide XCI, Engreitz et al. also interrogated Xist RNA distribution in differentiating female ESCs. Patterns at early differentiation time points in female ESCs were highly similar to those seen in inducible Xist male ESCs: general enrichment at gene-rich regions with higher enrichment at the Xic and 3D contact sites. Simon et al. also saw a modest correlation between Xist RNA enrichment sites and regions spatially associated with the Xic at later time points in female ESC differentiation. These results reinforce the hypothesis that Xist RNA’s initial mode of localization is predominantly through proximity-mediated interactions. Engreitz et al. tested this hypothesis using a male ESC line expressing inducible Xist from the Hprt locus, a site ~ 50 Mb distal to the Xic [9]. Shortly after Xist induction, an enrichment pattern appeared that almost perfectly mirrored the 3D contacts of the Hprt locus in ESCs. These analyses of Xist RNA distribution during establishment of XCI indicate that the first step of Xist RNA coating exploits the 3D organization of the X.
Analysis of early and late time points after Xist up-regulation suggests additional steps in Xist RNA spread beyond initial 3D contact sites. In the differentiating female ESC system Simon et al. report that Xist RNA accumulates first on gene-rich regions followed by gene-poor regions. When Xist RNA is depleted, the gene-poor regions lose Xist RNA association first, suggesting that these regions have lower affinity for Xist RNA. In the inducible system, Engreitz et al. provide additional resolution of gene-dense regions. Comparison of the one hour and three hour time points revealed that Xist RNA accumulates first at silent gene-dense regions and then progresses to active gene-dense regions. Together these studies suggest that Xist RNA coating is a multi-step process that depends on 3D chromosomal organization, gene density, and gene activity.
To determine whether any steps in Xist RNA spread are important for silencing, Engreitz et al. employed a male ESC line that inducibly expresses a mutant Xist defective in silencing due to deletion of a conserved element, the A-repeat (ΔAXist) [9]. ΔAXist RNA accumulated at 3D contacts and silent gene-dense regions but failed to efficiently accumulate over active gene-dense regions. Thus, Xist RNA interacts with active chromatin in a different manner than inactive chromatin. In earlier cytological studies ΔAXist RNA did not co-localize with active genes, though it still coated the X [10]. Together the high resolution and cytological studies highlight that silent and active regions of the X are sequestered in 3D space.
What is the relationship between nucleation at 3D contact points and the spread to encompass the entire X? The findings that Xist RNA preferentially associates with gene-dense regions, and that within these gene-dense regions it accumulates first on silent and then on active genes, indicate that underlying chromatin features associated with gene density and activity may affect Xist RNA affinity. These characteristics suggest two broad classes of models that may explain Xist RNA spread (Figure 1). One possibility is that while the Xic spends the most time at 3D contact sites, it samples the entire X over time, and that affinity of Xist RNA for chromatin features at each region determines how much Xist RNA accumulates at each region. Alternatively, it may be that once Xist RNA has moved from the Xic to a distal site it makes an alteration at the distal site to increase the affinity for and promote local cis-spread of Xist RNA.
Figure 1.
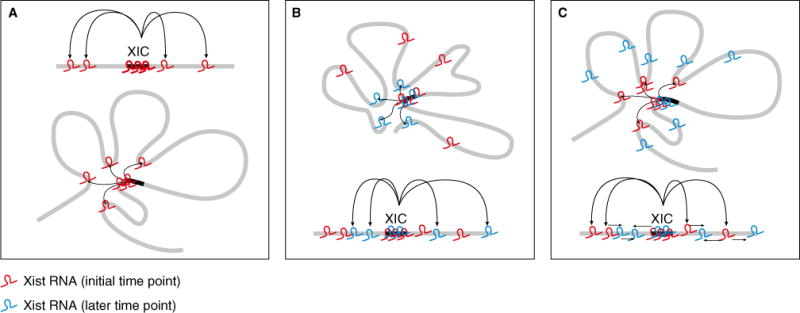
X chromosome topology directs Xist localization.
(A) Xist RNA (red) utilizes 3D contacts to localize to distal sites. (B) Model 1: the Xic samples the entire X over time and Xist RNA (red and blue at different time points) accumulation is determined by the affinity of Xist RNA for chromatin features at each region. (C) Model 2: Xist RNA (red) alters contact sites such that they have greater affinity for subsequently produced Xist RNA (blue), promoting its spread in cis along the X.
Together, Engreitz et al. and Simon et al. offer a new view of how Xist RNA coats the X. This lncRNA exploits the 3D contacts between the Xic and other regions of the X to access distal sites. This mode of spread contrasts with fly and worm dosage compensation complexes, in which defined sequence elements recruit the complexes to their sites of action. These new insights into Xist RNA spread will likely inform our understanding of how other lncRNAs can act at a distance.
References
- 1.Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2012;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gendrel AV, Heard E. Fifty years of X-inactivation research. Development. 2011;138:5049–5055. doi: 10.1242/dev.068320. [DOI] [PubMed] [Google Scholar]
- 3.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dietzel S, Schiebel K, Little G, Edelmann P, Rappold GA, Eils R, Cremer C, Cremer T. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity. Exp Cell Res. 1999;252:363–375. doi: 10.1006/excr.1999.4635. [DOI] [PubMed] [Google Scholar]
- 6.Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 7.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 8.Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 9.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 10.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]


