Abstract
Celiac disease is a common, lifelong autoimmune disorder for which dietary control is the only accepted form of therapy. A strict gluten-free diet is burdensome to patients and can be limited in efficacy, indicating there is an unmet need for novel therapeutic approaches to supplement or supplant dietary therapy. Many molecular events required for disease pathogenesis have been recently characterized and inspire most current and emerging drug-discovery efforts. Genome-wide association studies (GWAS) confirm the importance of human leukocyte antigen genes in our pathogenic model and identify a number of new risk loci in this complex disease. Here, we review the status of both emerging and potential therapeutic strategies in the context of disease pathophysiology. We conclude with a discussion of how genes identified during GWAS and follow-up studies that enhance susceptibility may offer insight into developing novel therapies.
Keywords: Celiac disease, Genome-wide association study, Gluten-free diet, Investigational therapies, Pathogenesis
Introduction
Celiac disease is a lifelong autoimmune disease characterized by an aberrant inflammatory response to dietary gluten in genetically susceptible individuals. Currently affecting 0.5-1% in most parts of the world, it is one of the most common chronic digestive disorders, with studies showing the prevalence of the disease is increasing [1,2]. The genetic predisposition to celiac disease is strong but complex. Ninety-five percent of patients are HLA-DQ2 or -DQ8 positive, but the presence of these alleles has a low positive predictive value [3]. The majority of the genetic component of celiac disease, as much as 65%, may be caused by over 50 non-HLA genes, with each gene slightly contributing to the risk of celiac disease development [4].
The celiac lesion is characterized by villous atrophy, crypt hyperplasia, and infiltration of inflammatory cells, both in the small intestinal epithelium and in the lamina propria. The only current treatment for the disease is strict, lifelong adherence to a gluten-free diet, but many celiac patients experience persistent symptoms and enteropathy despite their best efforts to avoid dietary gluten. Additionally, patients with chronic undetected and untreated celiac disease are at an increased risk for developing enteropathy-associated T-cell lymphoma, small bowel adenocarcinoma, and other gastrointestinal cancers [5-7]. Thus, there is an unmet need for novel, non-dietary therapies that improve both health and quality of life for celiac patients [8-10].
Celiac Disease Pathogenesis
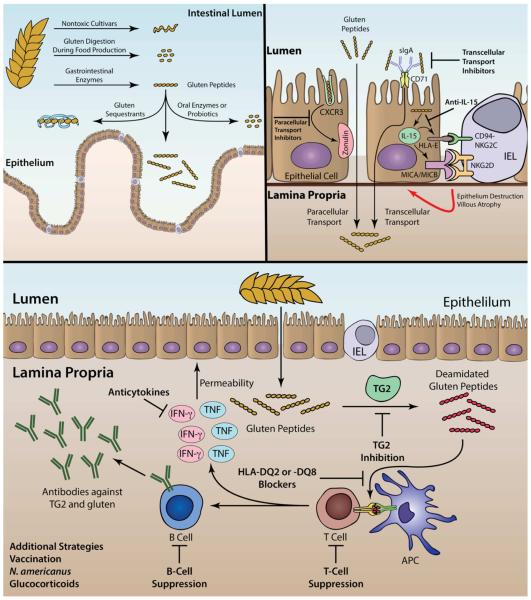
The design of novel non-dietary therapies to treat celiac disease requires a mechanistic understanding of disease pathogenesis (Fig. 1). At the broadest level, intestinal enteropathy in celiac disease is caused by genetic, immunological, and environmental factors. Gluten, a proline and glutamine rich glycoprotein, is the most critical environmental driver of the disease, while both human leukocyte antigen (HLA) and non-HLA genes are predisposing hereditary factors. MHC locus is the single most important genetic factor of the disease, with the majority of patients carrying a particular variant of HLA-DQ2 (DQA1*05:01, DQB1*02:01; also known as DQ2.5) [4]. Those who are not DQ2.5+ almost all carry HLA-DQ8 (DQA1*03, DQB1*03:02) or another variant of HLA-DQ2 (DQA1*02:01, DQB1*02:02; also known as DQ2.2) [11]. HLA-DQ2 and HLA-DQ8 predispose patients to celiac disease by preferential presentation of gluten peptides to CD4+ helper T cells in the lamina propria. Activation of these T cells with gluten-derived peptides induces the secretion of various inflammatory cytokines dominated by interferon (IFN)-γ [12]. This, in turn, triggers a cascade of inflammatory reactions that leads to the hallmark intestinal enteropathy of celiac disease.
Fig. 1.
Investigational approaches targeting the factors contributing to celiac disease pathogenesis. Long, Pro- and Gln-rich fragments of gluten survive gastrointestinal breakdown by luminal and brush border enzymes and consequently enter the lamina propria. Production of non-toxic cereals lacking antigenic peptides or sourdough fermentation during baking could avoid the ensuing immune response. Alternatively, gluten-sequestering polymers, oral proteases, and probiotics may reduce the exposure to immunogenic gluten peptides. At the intestinal epithelium, compromised epithelial barrier function enables paracellular and transcellular transport of gluten peptides. Antagonists of zonulin and inhibitors of the peptide-sIgA-CD71 transport pathway could reduce this paracellular and transcellular permeability, respectively. Gluten peptides induce epithelial and other cells to secrete IL-15, resulting in an increase in the number of IELs. These IELs are subsequently activated by epithelial MICA/B-NKG2D and CD94/NKG2C-HLA-E interactions that stimulating cytotoxic effects on epithelial cells. Neutralizing IL-15-mediated effects could provide a therapeutic benefit for celiac patients. Most gluten peptides survive gastric digestion and are excellent substrates of TG2 in the lamina propria. The resulting deamidated gluten peptides are then recognized by CD4+ T cells in the context of HLA-DQ2 or –DQ8 on antigen presenting cells. Thus, either TG2 inhibitors or HLA blockers are potential drug candidates. The T-cell response in the mucosa could be suppressed by anti-CD3-antibodies or peptide vaccination, or by blocking T-cell homing with anti-integrin α4β7 or CCR9 antagonists. Upon activation, gluten reactive T cells secrete inflammatory cytokines including IFN-γ and TNF that contribute to enteropathy. Anti-IFN-γ– and –TNF-antibodies could be considered as therapeutic targets. Finally, through interactions with T cells, B cells differentiate into plasma cells that produce autoantibodies against TG2. B-cell depletion with anti-CD20 antibodies is yet another possible therapeutic strategy. Further means to prevent or treat celiac disease in the future is modifying the inflammatory immune response by hookworm infection or with steroid therapy. Abbreviations: sIgA, secretory IgA. TG2, transglutaminase 2; HLA, human leukocyte antigen; IFN-γ, interferon gamma; TNF, tumor necrosis factor.
Given that virtually all patients with celiac disease carry particular HLA variants, HLA can be considered a necessary, but not a sufficient, factor for disease development. This claim is substantiated by the fact that while 40% of Caucasians possess one of the two predisposing haplotypes, only 3% of them develop celiac disease [13]. In addition to HLA genes, genome-wide association studies (GWAS) have identified 57 associated non-HLA variants located in 26 regions, with each locus contributing modestly to the overall genetic risk [14]. The majority of these non-HLA loci identified in GWAS harbor genes involved in the biology of T cells and antigen presenting cells. The story that emerges from the genetics of celiac disease bodes well for ongoing drug discovery efforts, given that most are based on the assumption that gluten-reactive T cells play a central role in controlling disease onset and severity. The other genes identified in GWAS reveal additional potential targets for future celiac disease drug discovery efforts.
The principal environmental driver, dietary gluten, contains a number of distinct disease-specific T-cell epitopes. A common feature of these epitopes is the presence of multiple Pro and Gln residues, with the high Pro content rendering these peptides resistant to proteolytic breakdown by gastric, pancreatic, and intestinal digestive proteases [15]. The result is an elevated intestinal concentration of potentially immunoreactive peptides following gluten ingestion. Some of the Gln residues of these immunoreactive peptides can be deamidated by the enzyme transglutaminase 2 (TG2), which is also the dominant autoantigen of celiac disease [16]. Deamidation enhances gluten peptide immunogenicity by increasing the affinity of the interactions between the immunoreactive peptides and specific pockets in the ligand-binding sites of HLA-DQ2 or HLA-DQ8 [17,18].
While the HLA-mediated response to gluten-derived antigens in patients with celiac disease is well understood, several features of gluten enteropathy in celiac disease remain unclear at present. First, dietary gluten reversibly increases small intestinal permeability in many patients with celiac disease. It has been proposed that enhanced paracellular intestinal permeability is the consequence of increased expression of zonulin, a protein released by the small intestinal mucosa after gliadin challenge [19]. Additionally, this phenomenon may be caused by IFN-γ and other cytokines produced by gluten-activated CD4+ T cells [12]. Genomic studies of patients with celiac disease also report involvement of genes that control intestinal permeability, including MAGI2, MYO9B, and PARD3 [20,21]. Secondly, dietary gluten also appears to activate the innate immune system in patients with celiac disease, leading to production of interleukin-15 (IL-15) both in the lamina propria and in the epithelium. Elevated IL-15 drives two primary effects—expansion of intra-epithelial lymphocytes (IELs) and increased NKG2D expression on IELs, which interacts with MICA and MICB displayed on epithelial cells [22,22,23]. Additionally, unlike IELs in the normal intestine, IELs in patients with celiac disease express another NK receptor called CD94/NKG2C [24]. CD94/NKG2C recognizes HLA-E, a protein that is upregulated in epithelial cells in response to IFN-γ. The interaction of NKG2D with MICA/B and CD94/NKG2C with HLA-E activates the IELs and triggers them to destroy the epithelial cells. Finally, gluten consumption also induces anti-TG2 autoantibody production in individuals with celiac disease. While these antibodies are used to diagnose celiac disease, the cause and pathogenic consequences of autoantibody production remain unclear. Further pursuit of these lines of investigation may reveal important new targets for celiac disease therapy.
Insight into celiac disease pathogenesis has inspired the evaluation of a range of therapeutic strategies (Fig. 1). This chapter will first discuss the approaches undergoing clinical evaluation and then focus on therapeutic modes in the development stage (Table 1), concluding with discussion of potential targets identified during genome-wide association studies.
Table 1.
Potential therapies for celiac disease.
| Mode of action | Compound | Compound Class | Company/University | Status |
|---|---|---|---|---|
| Topical steroid | Budesonide | Small molecule | Generic drug | Approved |
| Topical steroid | Prednisolone | Small molecule | Generic drug | Phase II |
| Glutenase | ALV003 | Enzyme | Alvine, USA | Phase IIb |
| Glutenase | AN-PEP | Enzyme | DSM, Netherlands | Phase I + II |
| Glutenase | STAN1 (enzyme supplements) |
Enzyme | Heim Pal Childrens Hospital, Hungary |
Phase I + II |
| Zonulin antagonist | AT-1001 | Peptide | Alba, USA | Phase IIb |
| CCR9 antagonist | CCX282-B | Small molecule | ChemoCentryx, USA | Phase II |
| Immune modulation | Necator americanus | Parasite | Princess Alexandra Hospital, Australia |
Phase II |
| Peptide vaccination | Nexvax2 | Peptide | Nexpep, Australia | Phase I |
| Anti-IL-15 | AMG 714 | Monoclonal antibody | Amgen, USA | Phase II in RA, psoriasis, (discont.) |
| Anti-IFN-γ | Fontolizumab | Monoclonal antibody | PDL and Biogen Idec, USA | Phase II in IBD, (discont.) |
| Anti-CD3 | Visilizumab | Monoclonal antibody | Facet, USA | Phase II in UC, GvHD (discont.) |
| Anti-CD3 | Teplizumab | Monoclonal antibody | MacroGenics, USA | Phase II in T1D |
| Anti-CD3 | Otelixizumab | Monoclonal antibody | Tolerx, USA | Phase III in T1D |
| Anti-CD20 | Rituximab | Monoclonal antibody | Biogen Idec, USA | Approved |
| Anti-CD20 | Tositumab | Monoclonal antibody | GlaxoSmithKline, USA | Approved |
| Anti-CD20 | Ibritumomab | Monoclonal antibody | Spectrum, USA | Approved |
| Anti-TNF-α | Infliximab | Monoclonal antibody | Janssen Biotech, USA | Approved |
| Anti-TNF-α | Certolizumab | Monoclonal antibody | UCB, USA | Approved |
| Anti-TNF-α | Adalimumab | Monoclonal antibody | AbbVie, USA | Approved |
| Anti-integrin α4β7 | Vedolizumab | Monoclonal antibody | Millennium Pharmaceuticals, USA |
Approved |
| IL-2/IL-15R Beta | Hu-Mik- Beta-1 | Monoclonal antibody | National Cancer Institute, USA | Phase I |
| Anti-gluten | AGY | Polyclonal antibody | Igy, Canada | Phase I |
| Pancrelipase | Creon | Enzyme cocktail | Sheffield Teaching Hospitals, UK | Phase 4 |
| TG2 inhibitor | Dihydroisoxazoles | Small molecule | Sitari Pharmaceuticals, USA | Discovery |
| TG2 inhibitor | ZED-101 | Small molecule | Zedira, Germany | Discovery |
| TG2 inhibitor | Cinnamoyl triazoles | Small molecule | University of Montreal, Canada | Discovery |
| HLA-DQ2 blocker | Dimeric analogue of gluten peptide |
Peptide | Stanford University, USA & University of Oslo, Norway |
Discovery |
| HLA-DQ2 blocker | Azidoproline analogue of gluten peptide |
Peptide | Leiden University, Netherlands | Discovery |
| Gluten tolerization | Bifidobacterium Infantis | Probiotic | Universidad de Buenos Aires, Argentina |
Exploratory |
| Gluten tolerization | Genetically modified Lactococcus lactis |
Probiotic | ActoGeniX, Belgium | Discovery |
|
Gluten-sequestering
polymers |
P(HEMA-co-SS) (BL-7010) | Polymer resin | University of Montreal, Canada | Phase I + II |
RA, rheumatoid arthritis; HLA, human leucocyte antigen; IBD, inflammatory bowel disease; UC, ulcerative colitis; GvHD, graft versus host disease; TG2, transglutaminase 2; T1D, type 1 diabetes; P(HEMA-co-SS), poly(hydroxyethyl methacrylate-co-styrene sulphonate); Approved, Approved in other diseases.
Therapies in Clinical Trials
Glucocorticoids with Low Systemic Bioavailability
Glucocorticoids are frequently used to induce a remission in or reduce the morbidity of immune-mediated diseases including asthma and Crohn’s disease. They elicit their therapeutic effects by induction of transient lymphopenia and immunosuppression. While significant side effects of systemic glucocorticoids limit their utility in the treatment of lifelong disorders such as celiac disease, it may be possible to utilize topically active glucocorticoids with pharmacological effects that are localized to the gut mucosa. One candidate is budesonide, a glucocorticoid with high-first pass metabolism and poor oral bioavailability that is currently used to treat Crohn’s disease. Three pilot studies in celiac disease demonstrated that budesonide may provide clinical benefit to those patients with both refractory and non-refractory celiac disease [25-27]. In a separate Phase II pilot study using prednisolone, a glucocorticoid with higher oral bioavailability, celiac patients receiving a 4-week course of the drug experienced a rapid reduction in epithelial apoptosis but a simultaneous suppression of villous regeneration, suggesting short courses of oral prednisolone could benefit specific patient groups [28]. While oral prednisolone may not have acceptable safety characteristics for use in patients with active celiac disease, oral budesonide may; in patients with primary biliary cirrhosis, 6 mg budesonide has been administered daily for up to 3 years with no change in budesonide pharmacokinetics and only minor changes in bone mineral density [29]. One drawback of current formulations of oral budesonide, however, is that they are used to treat illnesses of the lower intestine thereby making them unsuitable for celiac disease. Thus, for oral budesonide to have the greatest therapeutic benefit, a novel formulation is required.
Oral Proteases for Gluten Detoxification
The gluten degrading ability of various bacteria, fungi, and plants has been exploited to develop oral protease therapies for celiac disease. The stability and immunogenicity of gliadin peptides is largely attributable to their high Gln and Pro content—a characteristic that confers resistance to breakdown by pepsin, pancreatic proteases, and intestinal brush border membrane peptidases [15]. Both in people with and without celiac disease, the stability of these epitopes derives primarily from the inability of gastric and pancreatic endoproteases to cleave after Pro or Gln residues coupled with the inability of dipeptidyl peptidase IV and dipeptidyl carboxypeptidase I to cleave long peptides in the brush border membrane. The accumulation of long gliadin peptides in the small intestinal lumen is in turn responsible for eliciting an HLA-DQ2- or -DQ8-restricted T-cell response in patients with celiac disease.
Given that the high Pro content of gluten partially confers resistance to proteases, administration of prolyl endopeptidases (PEPs) has been considered as a strategy for detoxifying gluten peptides given their ability to cleave peptides at proline residues [30]. While PEPs are widely expressed in both mammals and microbes, the levels expressed in humans are insufficient for detoxifying gluten peptides. In contrast, recombinant PEPs from a variety of bacteria and fungi including Aspergillus niger, Flavobacterium meningosepticum, Myxococcus xanthus, and Sphingomonas capsulata are able to proteolyze gliadin peptides both in vitro and in vivo [31,32]. Importantly, these enzymes maintain both structure and function in the pH ranges of the human gastrointestinal tract [33]. In a study with celiac patient-derived gluten-specific T-cell clones, digestion of gluten peptides with a PEP from A. niger was able to abrogate T-cell expansion [34]. More significantly, in a randomized, double-blind, cross-over study, pretreatment with PEP prevented gluten-induced fat or carbohydrate malabsorption in over half of patients with celiac disease [35].
The high Gln content of gluten peptides provides an alternative target for oral protease therapy. A Gln-specific cysteine endoprotease B, isoform 2 (EP-B2) from germinating barley seeds is able to rapidly proteolyze gliadin peptides into short polypeptides [36]. Like the fungal and bacterial PEPs, EP-B2 maintains its structure and activity under gastrointestinal conditions [37]. Early proof-of-concept studies demonstrated EP-B2 effectively digested gluten both in the rat stomach and in gluten-sensitive rhesus macaques in a dose- and time-dependent manner [38].
Given that PEP and EP-B2 have complementary roles in proteolyzing gliadin peptides, one strategy that has been pursued for accelerating gluten detoxification is the administration of both enzymes as a combination therapy. To this end, one potential therapeutic in clinical trials is ALV003, an orally administered mixture of barley EP-B2 and Sphingomonas capsulata PEP [39]. The results of two Phase I single, escalating-dose clinical trials demonstrated that all doses of ALV003 were well tolerated, with no observed serious adverse events or allergic reactions [40]. The results of an initial Phase II trial indicated that ALV003 attenuated gluten-induced small intestinal mucosal injury in patients with celiac disease in the context of an everyday gluten-free diet, although symptoms did not significantly differ from the control group [41]. To further investigate efficacy and safety, a Phase IIb, randomized, double-blind, placebo-controlled dose ranging study is being conducted in celiac disease patients with persisting signs and symptoms despite a gluten-free diet.
Clinical trials have been pursued for other proteases as well. A PEP from Aspergillus niger (AN-PEP) was shown to degrade gluten in an artificial gastrointestinal model [32]. While a Phase I/II pilot clinical trial in celiac patients showed AN-PEP was both safe and well tolerated, additional trials have not been pursued [42]. Another enzyme cocktail has been shown to have a modest capacity to detoxify gluten [43]. A clinical trial is also underway for the use of Creon, an enzyme cocktail of pancrelipase, for the treatment of celiac patients with low faecal pancreatic elastase.
Gluten-Sequestering Polymers
A distinct strategy for attenuating the immunotoxicity of gliadin peptides utilizes orally administered polymeric resins, which sequester gliadin peptides in the small intestinal lumen before they can elicit their immunotoxic effects in the lamina propria. One such polymer is poly(hydroxyethyl methacrylate-co-styrene sulfonate) (P[HEMA-co-SS], BL-7010), which has been shown to bind to gluten proteins under simulated gastric and intestinal conditions in vitro [44]. P[HEMA-co-SS] is also able to reverse gliadin-induced alterations to intestinal epithelial cells and to reduce the secretion of the inflammatory cytokine tumor necrosis factor-α (TNF-α) ex vivo in mucosal biopsy specimens from patients with celiac disease [45]. The same studies also concluded that P[HEMA-co-SS] could reduce paracellular intestinal permeability and attenuate the systemic immune response to gluten in a variety of mouse models. These data support the clinical evaluation of a luminal polymeric binder as an effective adjunctive therapy to a gluten-free diet. A randomized, double bind Phase I/II clinical trial is currently recruiting celiac patients to evaluate the safety and systemic exposure of single escalating administrations and repeated administration of BL-7010. Much like the oral protease strategy, an important question that remains to be answered is the gluten dose that can be effectively detoxified in vivo by a given dose of P[HEMA-co-SS].
Zonulin Antagonists
Following partial gluten digestion in the intestinal lumen, gliadin peptides encounter the epithelial layer. The intestinal epithelial tight junctions function as a barrier that is critical for controlling foreign particle entry into the lamina propria. While the epithelial layer is typically impermeable to macromolecules, both clinical and experimental studies have demonstrated enhanced intestinal permeability upon gluten exposure in untreated celiac patients resulting from abnormal epithelial tight junction protein expression. Compromised barrier function enables elevated transport of the gliadin peptides into the lamina propria. One epithelial tight junction protein that has been implicated in enhancing barrier permeability is zonulin (prehaptoglobin-2) [19,46]. Through the interaction of gliadin peptides with the CXCR3 receptor, gluten is thought to induce overexpression of zonulin in intestinal tissue of patients with celiac disease and simultaneously activate zonulin signaling in tight junctions between epithelial cells, thereby leading to increased intestinal permeability [47]. When exposed to luminal gliadin peptides, intestinal biopsies from celiac patients in remission exhibited sustained luminal zonulin release and increased intestinal permeability, while a far less prominent effect was observed in control biopsies [48]. As a result, antagonists of zonulin have been proposed as a possible therapeutic option for the treatment of celiac disease.
Larazotide acetate (AT-1001) is an octapeptide derived from the zonula occludens toxin from Vibrio cholerae that locally inhibits the opening of tight junctions in epithelial cells in the small intestine [49]. Sharing a motif with zonulin, this compound presumably antagonizes zonulin action and is thus believed to prevent gliadin-induced permeability both ex vivo and in vivo [48,50]. A randomized, placebo-controlled Phase I clinical trial demonstrated AT-1001 was both safe and well tolerated in celiac patients, and suggested AT-1001 was capable of reducing gluten-induced intestinal permeability [51]. Despite this promising initial result, two subsequent clinical trials failed to demonstrate AT-1001 could reduce intestinal permeability. Specifically, in a 2-week and a 6-week study on celiac patients on a gluten-free diet, AT-1001 did not affect gluten-induced intestinal permeability, whereas it did alleviate the severity of gastrointestinal symptoms [52,53]. Recently, Alba Pharmaceuticals completed a 12-week Phase IIb clinical study with 342 patients evaluating the efficacy and safety of larazotide acetate in the treatment of celiac patients with persistent symptoms despite being on a gluten-free diet [54]. This study met its primary endpoint, with a 0.5 mg dose of larazotide acetate improving signs and symptoms of celiac disease compared with placebo by Modified Intention to Treat. The 0.5 mg dose also showed effect on exploratory endpoints, reducing patient reported outcome symptomatic days, average abdominal pain, and non-GI symptoms of headache and tiredness as well as increasing improved symptom days. These results warrant further clinical evaluation of larazotide acetate.
Gluten Tolerization
In contrast to therapies targeting a specific event in celiac disease pathogenesis, an alternative strategy is aimed at inducing gluten tolerance through vaccination. A variety of peptide-based desensitization therapies are undergoing clinical evaluation for the treatment of allergic diseases, including Cat-PAD for cat allergy and AllerT for birch pollen allergy. In the case of Cat-PAD, peptide vaccination induces CD4+ T cells with regulatory activity, suggesting induction of regulatory T cells is likely the mode of action of this treatment [55,56]. In the case of celiac disease, knowledge of the major HLA-DQ2-restricted T-cell epitopes of gluten has enabled the development of Nexvax2, a vaccine based on immunodominant gluten peptides that is currently undergoing clinical evaluation. A Phase I clinical trial demonstrated Nexvax2 was well tolerated in healthy, HLA-DQ2 positive celiac patients [57]. ImmunsanT, the company developing Nexvax2, is reportedly planning a Phase II clinical study to evaluate efficacy. Key questions that could be addressed by such studies include: (1) does the peptide vaccination tolerize gluten-reactive T cells in the intestinal mucosa, and (2) to what extent does this acquired immunity extend to less immunodominant epitopes? Notably, a distinct vaccine would be required for HLA-DQ8-positive celiac patients, given that immunodominant HLA-DQ2- and HLA-DQ8-restricted T-cell epitopes are non-overlapping.
Probiotics for Gluten Proteolysis
Enteric bacteria serve important immunological roles including the development of gut-associated lymphoid tissues, the polarization of gut-specific immune responses, and the prevention of colonization by pathogens [58]. Dysbiosis has been associated with a variety of autoimmune inflammatory disorders of the intestine, including Crohn’s disease, irritable bowel syndrome, and celiac disease [59-61]. Abnormalities in the gut microbiome of celiac patients, the association of specific microorganisms in the regulation of intestinal barrier function and immunity, and the ability of a variety of microbes to enzymatically degrade gluten are factors that have motivated investigation of intestinal bacteria as probiotics for the treatment of celiac disease. Species of Bifidobacterium are of interest, given the reduced prevalence of this genus in duodenal biopsies and feces of active as well as well-controlled celiac patients compared to individuals without the disease [61]. The results of in vitro studies that demonstrate various Bifidobacteria are able to hydrolyze gliadin peptides into less immunogenic peptides [62]. An exploratory clinical study concluded that while the Bifidobacterium infantis natren strain had no effect on elevated intestinal permeability or inflammatory protein abnormalities in patients with untreated celiac disease, it may alleviate symptoms in these patients [63].
Hookworm Infection
An unusual approach that has been pursued for the treatment of a variety of inflammatory conditions is infection with parasitic helminthes—with efforts primarily focused on the use of either the pig whipworm (Trichuris suis) or the human hookworm (Necator americanus)—given their ability to modulate host immune responses [64,65]. For autoimmune diseases, parasitic helminthes have also been pursued because of the correlation between the disappearance of intestinal parasites and the increasing prevalence of autoimmune diseases in developed countries. In the context of celiac disease, clinical studies have demonstrated that infection with N. americanus alone does not suppress disease symptoms sufficiently for it to be a viable therapy, with most subjects developing symptoms and mucosal inflammation immediately following reintroduction of gluten, irrespective of treatment group [66]. Given the ability of N. americanus to suppress gluten-induced IFN-γ, IL-17, and IL-23 expression and upregulate IL-10, TGF-β, and IL-22 expression in celiac patients, a 52-week pilot clinical Phase I/II study was undertaken with twelve patients to evaluate whether N. americanus infection could tolerize subjects to trace amounts of gluten [67]. The results indicated that celiac patients inoculated with N. americanus did not experience a decline in median villous height-to-crypt depth ratios upon a 1-gram gluten challenge, although they experienced a decrease in mean TG2 IgA titers upon exposure to 3-grams of gluten. It was argued that the approach merited further investigation in celiac patients experiencing occasional gluten exposure.
Preclinical targets
Blocking Transcellular Gliadin Transport
While paracellular intestinal permeability is enhanced in celiac patients, gliadin peptides also undergo transcellular transport across the epithelium. This process is hypothesized to be mediated by secretory IgA (sIgA) via binding to the transferrin receptor CD71, a protein upregulated in the intestinal epithelium of celiac patients [68]. Significantly, the ability of IgA from celiac patients to enhance transcellular passage of gliadin peptides was abolished by inhibition of TG2 enzymatic activity [69]. Thus, inhibition of the gliadin-sIgA-CD71 transport pathway, either through TG2 inhibition or by another mechanism, could be a therapeutic strategy for celiac disease.
Blocking IL-15
The cytokine IL-15 plays a critical role in celiac disease pathogenesis through dysregulation of IELs that leads to villous atrophy. In a transgenic mouse that overexpresses IL-15 in the lamina propria, antibody targeting of IL-15 reversed intestinal damage, suggesting this form of therapy may warrant evaluation in celiac patients [70]. In experiments using small intestinal mucosal biopsies from celiac patients, anti-IL-15 antibodies abrogated gluten-induced overexpression of MICA by epithelial cells and neutralized enterocyte apoptosis [22,71]. Additionally, anti-IL-15 antibodies down-regulated the adaptive immune response in the lamina propria [72]. All of these observations suggest anti-IL-15 therapy could benefit patients with celiac disease, particularly those with a refractory condition. A Phase I clinical study is currently recruiting patients to evaluate the efficacy of the humanized Mik-Beta-1 monoclonal antibody that blocks IL-15 action by binding to the β-subunit of the IL-2/IL-15 receptor in patients with refractory celiac disease. In the past, an anti-IL-15 monoclonal antibody, AMG 714, had undergone Phase II clinical studies in rheumatoid arthritis and psoriasis, however, its development was discontinued given limited efficacy [73].
Inhibition of TG2
Catalytic activity of the primary celiac disease autoantigen transglutaminase 2 (TG2) is an essential step in generating gliadin peptides with high HLA affinity. Inhibition of TG2 thus has potential as a therapy for attenuating the inflammatory response in celiac patients. A number of classes of TG2 inhibitors have been developed, including both active site-directed irreversible inhibitors such as thiadiazoles, epoxides, α,β-unsaturated amides, and dihydroisoxazoles, as well as reversible inhibitors such as thienopyrimidines, cinnamoyl compounds, β-aminoethyl ketones, and acylidine oxindoles. Perhaps the most extensively evaluated are the weakly electrophilic 3-bromo-4,5-dihydroisoxazole class of inhibitors [74-81]. In vivo experiments using a prototypical inhibitor ERW1041E blocked elevated TG2 activity in pulmonary tissue in a mouse model of pulmonary hypertension [82]. In the same study, twice-daily intraperitoneal dosing of this inhibitor at 50 mg/kg was well tolerated over the course of several weeks. Given that extracellular TG2 is predominantly inactive under normal physiologic conditions, and that TG2 knockout mice are developmentally and reproductively normal, TG2 inhibition warrants clinical evaluation as an oral therapy for celiac disease [83,84]. Notably, TG2 is one of nine transglutaminase homologs that catalyze posttranslational modifications of selected Gln residues on target peptides or proteins; therefore, the design of an isozyme-selective TG2 inhibitor is a challenging undertaking. Minimizing cross-reactivity with other transglutaminases could be important, given that (1) TG1 mutations give rise to skin barrier dysfunctions, (2) Factor XIIIa is essential in the blood clotting cascade and is responsible for cross-linking fibrin to form stable clots, and (3) TG3 mutations have been associated with an increased risk of basal cell carcinoma [85-87]. While ERW1041E does not possess sufficient selectivity, other analogs show improved potency and isoform selectivity [88].
Blocking HLA-DQ2 or HLA-DQ8
The presence of either HLA-DQ2 or -DQ8 on antigen presenting cells is the most significant genetic factor in predisposing an individual to celiac disease, although neither haplotype is sufficient for disease development. Since homozygosity for HLA alleles does not enhance susceptibility to infection, blocking gliadin peptide presentation by either HLA-DQ2 or -DQ8 represents a potential strategy for reducing the severity of the toxic effects of gluten seen in celiac patients. Efforts to block HLA in other immune-mediated conditions—including rheumatoid arthritis, multiple sclerosis, and type I diabetes—have met with limited success, presumably due to ineffective drug delivery to the disease-relevant antigen presenting cells. If so, then topical delivery to the small intestine in celiac patients should be a considerably easier task. A number of peptides have been synthesized to target HLA-DQ2 [89,90]. While these initial efforts utilized a gliadin peptide scaffold, the ligands showed limited efficacy in reducing T-cell activation. More recent strategies have sought to optimize each amino acid’s interaction with the HLA binding cleft [91,92]. Although some of these ligands bind to HLA-DQ2 with greater than 50-fold affinity compared to the immunodominant gluten epitope of DQ2-α-I-gliadin, this affinity may be insufficient for attenuating T-cell activation. Moreover, applicability of this approach to all celiac patients would require the development of analogous HLA-DQ8 blockers.
Suppression of T Cells
Suppression of gluten-specific T-cell mediated immune responses through antibody-based therapies could prove useful in certain circumstances for the treatment of celiac disease. Antibodies against TNF including infliximab, certolizumab and adalimumab have been used in the treatment of Crohn’s disease, rheumatoid arthritis, and IBD. Two case studies have evaluated the efficacy of the anti-TNF monoclonal antibody infliximab in treating refractory celiac disease, with both patients experiencing histological improvements [93,94]. Larger scale clinical trials are necessary to demonstrate clinical utility of anti-TNF antibodies for the treatment of refractory celiac disease.
Antibodies against interferon-γ, the dominant inflammatory cytokine produced by gluten reactive T cells, may also prove useful for the treatment of celiac disease. While doses of anti-IFN-γ antibody fontolizumab were well tolerated by patients with Crohn’s disease, development was discontinued given limited efficacy [95]. Nevertheless, evaluation of anti-IFN-γ antibodies may still be justifiable for certain patient groups with celiac disease.
A third antibody-based approach to modulate the T-cell response targets the CD3 protein complex, a co-receptor for the T-cell receptor. Given that anti-CD3 therapy is thought to function by both eliminating effector T cells and inducing regulatory T cells and that it is primarily effective in the context of a primed and ongoing immune response, anti-CD3 antibodies are currently being evaluated for the treatment of Crohn's disease, ulcerative colitis, and type 1 diabetes [96-98]. For these same reasons, it may prove to be an effective treatment of celiac disease. One cause for reservation about the potential use of this approach is the fact that effector T cells with active celiac patients become resistant to suppression by regulatory T cells.
Targeting B Cells
In addition to suppressing disease-specific T cells, therapies targeting disease-specific B cells have potential for the treatment of celiac disease. Earlier studies have demonstrated a clinical benefit of depleting B cells with anti-CD20 antibodies in the context of other HLA-associated disorders including rheumatoid arthritis, multiple sclerosis, and type I diabetes [99-101]. B cells may play a role in the pathogenesis of celiac disease, since gluten-specific and TG2-specific B cells should be able to present gliadin peptides to T cells, making CD20 an appealing target [102]. Approaches targeting CD20 in celiac disease have been largely unsuccessful, however, demonstrating that anti-CD20 antibodies are unable to abrogate IgA plasma cells generation in the gut mucosa despite peripheral B-cell depletion [103]. While a number of anti-CD20 antibodies are clinically used, if antibodies produced in the mucosa are pathogenically relevant to celiac disease, they are unlikely to provide a therapeutic benefit to celiac patients.
Blocking Intestinal Homing
Lymphocyte homing to the gastrointestinal mucosa is aided by the chemokine receptor CCR9 and integrin α4β7 in a process mediated by their chemokine ligands CCL25 and MADCAM1, respectively [104]. Celiac patients have elevated CCR9-expressing peripheral blood T cells and MADCAM1 augmentation in the duodenum, suggesting targeting either of these homing mechanisms could be used to treat celiac patients [105,106]. CCX282-B (GSK1605786A) is a selective, orally bioavailable antagonist of human CCR9 that is undergoing clinical trials in Crohn’s disease and celiac disease [107]. Numerous clinical trials evaluating CCX282-B for the treatment of Crohn’s disease have been withdrawn or terminated early, and a Phase II trial of celiac disease patients initiated in 2007 has been completed but the results have not yet been disclosed. A monoclonal antibody against integrin α4β7, vedolizumab (Entyvio), was recently approved for the treatment of moderately to severely active ulcerative colitis and Crohn’s disease, and could be useful for the treatment of subsets of celiac disease patients [108,109]. If any of these drugs are effective therapeutics for celiac disease, increased monitoring for gastrointestinal infections will be necessary given that this mode of action is not antigen specific.
Future Targets
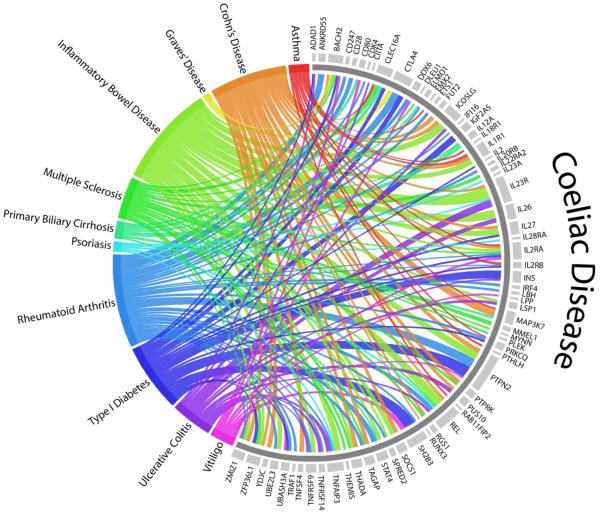
As mentioned previously, celiac disease is a polygenic disorder having the most important genetic susceptibility determinant located in the class II MHC locus, with HLA-DQ2 and/or HLA-DQ8 being necessary but not sufficient for disease development. These susceptibility alleles only explain 35% of genetic risk, however, and several other genes contribute to celiac disease pathogenesis [110]. Two GWAS and Immunochip analyses performed on celiac disease patients have revealed 39 regions of genetic susceptibility to the disease [4,110]. Many of the genes identified in these studies have been implicated in numerous other immune-modulated diseases (Fig. 2). These studies suggest several possible etiological candidates, with the majority of them related either to T-cell maturation or immune response pathways. Given these genes may modulate gluten sensitivity, future therapies may stem from investigation into the role of these genes in celiac disease pathogenesis. The coding alleles THEMIS, PTPRK, FUT2, BACH2, and RGS1 are reviewed here.
Fig. 2.
Non-HLA susceptibility regions shared between celiac disease and select immune-modulated diseases [144,145]. A genome-wide significance threshold was set at P = 5 × 10−8. FUT2 was included despite not being identified in a celiac disease-specific GWAS.
THEMIS
Healthy individuals mount immune responses to exogenous pathogens while avoiding autoimmune attacks on normal tissue. In this regard, the ability to recognize self from non-self is essential. A diverse pool of functional T cells with immunological tolerance is generated through positive selection while overly self-reactive T cells are removed by negative selection during ontogeny. Thymocytes arrive at these cell fate decisions through interactions with ligands, where a narrow range of affinities distinguishes positive from negative selection. At this selection threshold, a small increase in ligand affinity for the T-cell receptor leads to the induction of negative selection. In celiac disease-specific GWAS, the peak on 6q22.33 includes the gene thymocyte-expressed molecule involved in selection (THEMIS). An association study in a north Indian population showed a SNP on intron 2 of THEMIS was associated with celiac disease, although the functional significance remains unknown [111]. THEMIS encodes a 73-kDa protein expressed in the thymus, spleen, and lymph nodes that is involved in T-cell maturation and in regulating lineage commitment of thymocytes into CD4+ or CD8+ cells [112]. More specifically, THEMIS enables thymocyte positive selection through its ability to attenuate TCR signals via SHP1 recruitment and activation in response to low- but not high-affinity TCR engagement [113]. Functionally it converts TCR affinity into a selection outcome and thus determines the affinity threshold for activation. Recent work in a THEMIS knockout rat has linked the function of this gene to the suppressive function of CD4+ regulatory T cells and suggested that the defect is involved in intestinal inflammation [114]. Previous studies have demonstrated that impaired thymic negative selection can result in autoimmune disease. THEMIS has also been implicated in Crohn’s disease and multiple sclerosis [115,116]. In celiac patients, work evaluating the expression levels of THEMIS in duodenal mucosa of active and treated celiac disease patients and in controls found higher expression in active patients compared with treated patients and controls [117]. It is thus possible that higher expression of THEMIS in celiac patients could lead to dysregulation of thymocyte selection, thereby contributing to the pathogenesis of the disease.
PTPRK
In addition to THEMIS, the celiac GWAS peak on 6q22.33 also included the gene protein tyrosine phosphatase, receptor type, kappa (PTPRK) encoding RPTPκ. RPTPκ is a ubiquitous transmembrane protein tyrosine phosphatase whose expression is induced by TGF-β [118]. One hypothesis is that RPTPκ is involved in the pathogenesis of celiac disease through the modulation of T-cell development. An important role for RPTPκ in T-cell development was hypothesized following the discovery that PTPRK is deleted in the LEC rat, a model characterized by hypoplasia of the thymus and spleen, reduced levels of IgG, selective deficiency in CD4+ T cells, and strongly reduced T helper function [119]. Of note, however, was the discovery that the LEC rat also lacks the THEMIS gene calling into question the role of PTPRK in T-cell development [120]. A more plausible role for RPTPκ in celiac disease is through the dysregulation of intestinal cell junctions. Numerous studies have validated the role of the extracellular domain of PTPRK in promoting cell-cell adhesion through homophilic binding, a process that is critical for intestinal barrier function [121]. The GWAS showed that two independently associated sets of SNPs were finely localized around the 3’ untranslated region of PTPRK, with one located in a predicted binding site for the microRNA hsa-miR-1910 [14]. Consistent with this observation was the finding that PTPRK showed lower expression in active celiac disease compared with treated patients and controls [117]. In addition to celiac disease, GWAS have also implicated PTPRK in other autoimmune diseases characterized by intestinal permeability including Crohn’s disease and multiple sclerosis [115,122]. Future studies are warranted to elucidate the role of PTPRK in celiac disease and to determine if this gene could represent a novel therapeutic target.
FUT2
The FUT2 gene present on 19q13.33 encodes α-1,2-fucosyltransferase, an enzyme that controls the expression of A-, B- and H- blood group antigens in mucus and other body secretions [123]. Specifically, it catalyzes the conversion of type 1 precursor to H antigen type 1 by adding a terminal α-1,2-linked fucose to galactose on type 1 precursor, where the H antigen type 1 is the precursor for the further synthesis of A and B blood group antigens. These antigens are also expressed in the gastrointestinal mucosa, serving as anchors for various microbes. Previous studies have demonstrated that FUT2 can influence the composition of gut microbiota, in turn protecting the host from pathogenic microbes and affect the development of the mucosal immune system [124]. In 20% of Caucasians, FUT2 is rendered nonfunctional by a nonsense mutation (rs601338-AA) that introduces a premature stop codon, where individuals homozygous for this mutation are called non-secretors. While a number of studies have linked secretor status to either risk or protection from a variety of disease, important here is the evidence that the FUT2 non-secretor status has been associated with increased risk for celiac disease as well as Crohn’s disease and type I diabetes [125-127]. In the context of celiac disease, past studies have demonstrated that secretor status is strongly associated with the diversity and composition of intestinal microbiota while others have shown microbiota can induce marked changes in monocyte derived dendritic cell morphology and simultaneously increase the production of inflammatory cytokines including IFN-γ, TNF-α, and IL-12 [124,128-130]. Taken together, these results suggest FUT2 may indirectly, by means of microbiota composition, be involved in the manifestation of celiac disease.
BACH2
Polymorphisms within the gene encoding BTB and CNC homolog 2 (BACH2) on 6q15 have been associated with numerous allergic and autoimmune diseases including including asthma, Crohn’s disease, vitiligo, multiple sclerosis, type I diabetes, autoimmune thyroid disease, and celiac disease [110,122,131-134]. BACH2 is a BTB-leucine zipper family transcription factor that has been identified as a B cell-specific transcriptional repressor responsible for regulating plasma cell differentiation, immunoglobulin class switching, and somatic hypermutation [135]. More recent work has demonstrated that BACH2 is expressed in T cells and is critical for the formation of regulatory T cells and maintenance of naive T-cell state [136]. Importantly, BACH2 represses genes with effector-lineage-specific functions in a manner that constrains full effector differentiation within TH1, TH2, and TH17 cell lineages. Consistent with this observation is the fact that absence of BACH2 during regulatory T-cell polarization results in inappropriate diversion to effector lineages. BACH2 knockout mice have limited viability resulting from severe inflammation in the gut, lungs, and other tissues [136]. Taken together, these results identify BACH2 as an essential regulator of T-cell differentiation that can prevent inflammatory disease by maintaining immune homeostasis. Concordant with this conclusion, the SNP identified in the celiac disease GWAS is associated with BACH2 down-regulation [137]. While specific targeting of BACH2 represents a significant challenge, further investigation into the role of this gene in celiac disease pathogenesis may identify novel therapeutic targets.
RGS1
SNPs in RGS1 have been associated with a variety of autoimmune diseases including type I diabetes, multiple sclerosis, and celiac disease [138-140]. In celiac disease, the SNPs for RGS1 show association in the 5’-UTR region—specifically in the first exon and 10 kb upstream of it—suggesting these SNPs affect its transcriptional regulation. RGS1 encodes regulator of G-protein signaling 1, a protein that modulates chemokine-induced GPCR activity. It has long been recognized that RGS1 can regulate B cell chemotaxis, although splenic and lymph node T cells from RGS1−/− mice show seemingly normal T-cell chemotaxis [141,142]. Recent work has demonstrated that RGS1 mRNA is highly enriched in murine gut versus lymphoid T cells, and that RGS1 expression is substantially higher in T cells from the human gut compared to peripheral blood in a manner that is exacerbated during intestinal inflammation [143]. A significant observation has been that elevated RGS1 levels markedly reduce T-cell migration in response to lymphoid-homing cytokines, while RGS1 depletion enhances gut T-cell egress in a manner that impairs colitogenic potential [143]. While it cannot be predicted whether RGS1 is necessarily involved in either celiac disease initiation and/or progression, given the critical role the IELs play in the villous atrophy characteristic of celiac disease, further investigation into the therapeutic potential of modulating RGS1 is warranted.
Conclusions
The importance of developing alternative, non-dietary therapies for the treatment of celiac disease is underscored by the increasing prevalence of the disease and the growing awareness that a substantial fraction of treated celiac patients show lingering evidence of disease activity. Despite the pursuit of numerous strategies for combating the disease, development of successful therapies has proved to be challenging given the complexity of this autoimmune disorder. Recent GWAS and follow-up studies have increased our understanding of the genetic factors that enhance susceptibility to celiac disease that offer insight into the underlying molecular pathways and mechanisms. Evidence linking polymorphisms in these genes to this and other autoimmune and inflammatory conditions warrants investigation into their target for therapeutic relief.
Practice Points.
Celiac disease is a lifelong autoimmune disease, where intestinal enteropathy is caused by genetic, immunological, and environmental factors
Dietary control through the avoidance of gluten is the only accepted form of therapy
Previous and ongoing clinical trials for celiac disease include treatment with glucocorticoids, oral proteases, gluten-sequestering polymers, zonulin antagonists, gluten vaccines, probiotics, and hookworms
Genome-wide association studies have identified a number of new risk loci
Research Agenda.
The lack of a non-dietary therapy demands pursuit of therapies currently undergoing clinical evaluation
Strategies including blocking transcellular gliadin transport, IL-15, HLA–DQ2 or –DQ8, and intestinal homing in addition to inhibiting TG2, suppressing T cells, and targeting B cells are promising preclinical targets
GWAS and Immunochip analyses suggest several possible etiological candidates that warrant investigation into their value as future targets for therapeutic relief
Acknowledgment
Research on celiac disease in the authors’ laboratories has been supported by a grant from the NIH (R01 DK063158). C.K. is a stockholder in Alvine Pharmaceuticals and Sitari Pharmaceuticals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rubio-Tapia A, Kyle RA, Kaplan EL, Johnson DR, Page W, Erdtmann F, et al. Increased Prevalence and Mortality in Undiagnosed Celiac Disease. Gastroenterology. 2009;137(1):88–93. doi: 10.1053/j.gastro.2009.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang JY, Kang AHY, Green A, Gwee KA, Ho KY. Systematic review: worldwide variation in the frequency of coeliac disease and changes over time. Aliment Pharmacol Ther. 2013;38(3):226–245. doi: 10.1111/apt.12373. [DOI] [PubMed] [Google Scholar]
- 3.van der Windt DAWM, Jellema P, Mulder CJ, Kneepkens CMF, van der Horst HE. Diagnostic testing for celiac disease among patients with abdominal symptoms: a systematic review. JAMA. 2010;303(17):1738–1746. doi: 10.1001/jama.2010.549. [DOI] [PubMed] [Google Scholar]
- 4.van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39(7):827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viljamaa M, Kaukinen K, Pukkala E, Hervonen K, Reunala T, Collin P. Malignancies and mortality in patients with coeliac disease and dermatitis herpetiformis: 30-year population-based study. Dig Liver Dis. 2006;38(6):374–380. doi: 10.1016/j.dld.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Smedby KE, Akerman M, Hildebrand H, Glimelius B, Ekbom A, Askling J. Malignant lymphomas in coeliac disease: evidence of increased risks for lymphoma types other than enteropathy-type T cell lymphoma. Gut. 2005;54(1):54–59. doi: 10.1136/gut.2003.032094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Y, Kristinsson SY, Goldin LR, Björkholm M, Caporaso NE, Landgren O. Increased Risk for Non-Hodgkin Lymphoma in Individuals With Celiac Disease and a Potential Familial Association. Gastroenterology. 2009;136(1):91–98. doi: 10.1053/j.gastro.2008.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sollid LM, Khosla C. Novel therapies for coeliac disease. J Intern Med. 2011;269:604–613. doi: 10.1111/j.1365-2796.2011.02376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaukinen K, Lindfors K, Mäki M. Advances in the treatment of coeliac disease: an immunopathogenic perspective. Nature Publishing Group. 2013;11(1):36–44. doi: 10.1038/nrgastro.2013.141. [DOI] [PubMed] [Google Scholar]
- 10.Castillo NE, Theethira TG, Leffler DA. The present and the future in the diagnosis and management of celiac disease. Gastroenterology Report. 2014:1–9. doi: 10.1093/gastro/gou065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sollid LM, Lie BA. Celiac disease genetics: current concepts and practical applications. Clin Gastroenterol Hepatol. 2005;3(9):843–851. doi: 10.1016/s1542-3565(05)00532-x. [DOI] [PubMed] [Google Scholar]
- 12.Nilsen EM, Lundin KE, Krajci P, Scott H, Sollid LM, Brandtzaeg P. Gluten specific, HLA-DQ restricted T cells from coeliac mucosa produce cytokines with Th1 or Th0 profile dominated by interferon gamma. Gut. 1995;37(6):766–776. doi: 10.1136/gut.37.6.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green PHR, Jabri B. Celiac disease. Annu Rev Med. 2006;57:207–221. doi: 10.1146/annurev.med.57.051804.122404. [DOI] [PubMed] [Google Scholar]
- 14.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43(12):1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shan L, Molberg Ø , Parrot I, Hausch F, Filiz F, Gray GM, et al. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297(5590):2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 16.Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken EO, et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med. 1997;3(7):797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- 17.Kim C-Y, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci U.S.A. 2004;101(12):4175–4179. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson KN, Tye-Din JA, Reid HH, Chen Z, Borg NA, Beissbarth T, et al. A Structural and Immunological Basis for the Role of Human Leukocyte Antigen DQ8 in Celiac Disease. Immunity. 2007;27(1):23–34. doi: 10.1016/j.immuni.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Fasano A, Not T, Wang W, Uzzau S, Berti I, Tommasini A, et al. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet. 2000;355(9214):1518–1519. doi: 10.1016/S0140-6736(00)02169-3. [DOI] [PubMed] [Google Scholar]
- 20.Monsuur AJ, de Bakker PIW, Alizadeh BZ, Zhernakova A, Bevova MR, Strengman E, et al. Myosin IXB variant increases the risk of celiac disease and points toward a primary intestinal barrier defect. Nat Genet. 2005;37(12):1341–1344. doi: 10.1038/ng1680. [DOI] [PubMed] [Google Scholar]
- 21.Wapenaar MC, Monsuur AJ, van Bodegraven AA, Weersma RK, Bevova MR, Linskens RK, et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. 2008;57(4):463–467. doi: 10.1136/gut.2007.133132. [DOI] [PubMed] [Google Scholar]
- 22.Hüe S, Mention J-J, Monteiro RC, Zhang S, Cellier C, Schmitz J, et al. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity. 2004;21(3):367–377. doi: 10.1016/j.immuni.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 23.Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, Krausz TN, et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21(3):357–366. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Meresse B, Curran SA, Ciszewski C, Orbelyan G, Setty M, Bhagat G, et al. Reprogramming of CTLs into natural killer-like cells in celiac disease. J Exp Med. 2006;203(5):1343–1355. doi: 10.1084/jem.20060028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daum S, Ipczynski R, Heine B, Schulzke J-D, Zeitz M, Ullrich R. Therapy with budesonide in patients with refractory sprue. Digestion. 2006;73(1):60–68. doi: 10.1159/000092639. [DOI] [PubMed] [Google Scholar]
- 26.Brar P, Lee S, Lewis S, Egbuna I, Bhagat G, Green PHR. Budesonide in the Treatment of Refractory Celiac Disease. Am J Gastroenterol. 2007;102(10):2265–2269. doi: 10.1111/j.1572-0241.2007.01380.x. [DOI] [PubMed] [Google Scholar]
- 27.Ciacci C, Maiuri L, Russo I, Tortora R, Bucci C, Cappello C, et al. Efficacy of budesonide therapy in the early phase of treatment of adult coeliac disease patients with malabsorption: An in vivo/ in vitropilot study. Clin Exp Pharmacol P. 2009;36(12):1170–1176. doi: 10.1111/j.1440-1681.2009.05211.x. [DOI] [PubMed] [Google Scholar]
- 28.Shalimar, Das P, Sreenivas V, Datta Gupta S, Panda SK, Makharia GK. Effect of addition of short course of prednisolone to gluten-free diet on mucosal epithelial cell regeneration and apoptosis in celiac disease: a pilot randomized controlled trial. Dig Dis Sci. 2012;57(12):3116–3125. doi: 10.1007/s10620-012-2294-1. [DOI] [PubMed] [Google Scholar]
- 29.Rautiainen H, Färkkilä M, Neuvonen M, Sane T, Karvonen A-L, Nurmi H, et al. Pharmacokinetics and bone effects of budesonide in primary biliary cirrhosis. Aliment Pharmacol Ther. 2006;24(11-12):1545–1552. doi: 10.1111/j.1365-2036.2006.03155.x. [DOI] [PubMed] [Google Scholar]
- 30.Hausch F, Shan L, Santiago NA, Gray GM, Khosla C. Intestinal digestive resistance of immunodominant gliadin peptides. Am J Physiol Gastrointest Liver Physiol. 2002;283(4):G996–G1003. doi: 10.1152/ajpgi.00136.2002. [DOI] [PubMed] [Google Scholar]
- 31.Shan L, Marti T, Sollid LM, Gray GM, Khosla C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for coeliac sprue. Biochem J. 2004;383:311–318. doi: 10.1042/BJ20040907. Pt 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitea C, Havenaar R, Drijfhout JW, Edens L, Dekking L, Koning F. Efficient degradation of gluten by a prolyl endoprotease in a gastrointestinal model: implications for coeliac disease. Gut. 2008;57(1):25–32. doi: 10.1136/gut.2006.111609. [DOI] [PubMed] [Google Scholar]
- 33.Gass J, Khosla C. Prolyl Endopeptidases. Cell Mol Life Sci. 2007;64:345–355. doi: 10.1007/s00018-006-6317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stepniak D, Spaenij-Dekking L, Mitea C, Moester M, de Ru A, Baak-Pablo R, et al. Highly efficient gluten degradation with a newly identified prolyl endoprotease: implications for celiac disease. Am J Physiol Gastrointest Liver Physiol. 2006;291(4):G621–629. doi: 10.1152/ajpgi.00034.2006. [DOI] [PubMed] [Google Scholar]
- 35.Pyle GG, Paaso B, Anderson BE, Allen DD, Marti T, Li Q, et al. Effect of pretreatment of food gluten with prolyl endopeptidase on gluten-induced malabsorption in celiac sprue. Clin. Gastroenterol. Hepatol. 2005;3(7):687–694. doi: 10.1016/s1542-3565(05)00366-6. [DOI] [PubMed] [Google Scholar]
- 36.Bethune MT, Strop P, Tang Y, Sollid LM, Khosla C. Heterologous Expression, Purification, Refolding, and Structural-Functional Characterization of EP-B2, a Self-Activating Barley Cysteine Endoprotease. Chem Biol. 2006;13(6):637–647. doi: 10.1016/j.chembiol.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Gass J. Effect of Barley Endoprotease EP-B2 on Gluten Digestion in the Intact Rat. J Pharmacol Exp Ther. 2006;318(3):1178–1186. doi: 10.1124/jpet.106.104315. [DOI] [PubMed] [Google Scholar]
- 38.Bethune MT, Borda JT, Ribka E, Liu M-X, Phillippi-Falkenstein K, Jandacek RJ, et al. A Non-Human Primate Model for Gluten Sensitivity. PLoS ONE. 2008;3(2):e1614. doi: 10.1371/journal.pone.0001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tye-Din JA, Anderson RP, Ffrench RA, Brown GJ, Hodsman P, Siegel M, et al. The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin Immunol. 2010;134(3):289–295. doi: 10.1016/j.clim.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Siegel M, Garber M, Spencer A, Botwick W. Safety, Tolerability, and Activity of ALV003: Results from Two Phase 1 Single, Escalating-Dose Clinical Trials. Dig Dis Sci. 2012;57:440–450. doi: 10.1007/s10620-011-1906-5. [DOI] [PubMed] [Google Scholar]
- 41.Lähdeaho M-L, Kaukinen K, Laurila K, Vuotikka P, Koivurova O-P, Kärjä-Lahdensuu T, et al. Glutenase ALV003 Attenuates Gluten-Induced Mucosal Injury in Patients With Celiac Disease. Gastroenterology. 2014;146(7):1649–1658. doi: 10.1053/j.gastro.2014.02.031. [DOI] [PubMed] [Google Scholar]
- 42.Tack GJ, van de Water JMW, Bruins MJ, Kooy-Winkelaar EMC, van Bergen J, Bonnet P, et al. Consumption of gluten with gluten-degrading enzyme by celiac patients: a pilot-study. World J Gastroenterol. 2013;19(35):5837–5847. doi: 10.3748/wjg.v19.i35.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ehren J, Morón B, Martin E, Bethune MT, Gray GM, Khosla C. A food-grade enzyme preparation with modest gluten detoxification properties. PLoS ONE. 2009;4(7):e6313. doi: 10.1371/journal.pone.0006313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinier M, Verdu EF, Eddine MN, David CS, Vézina A, Rivard N, et al. Polymeric Binders Suppress Gliadin-Induced Toxicity in the Intestinal Epithelium. YGAST. 2009;136(1):288–298. doi: 10.1053/j.gastro.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 45.Pinier M, Fuhrmann G, Galipeau HJ, Rivard N, Murray JA, David CS, et al. The Copolymer P(HEMA-co-SS) Binds Gluten and Reduces Immune Response in Gluten-Sensitized Mice and Human Tissues. Gastroenterology. 2012;142(2):316–325. doi: 10.1053/j.gastro.2011.10.038. e12. [DOI] [PubMed] [Google Scholar]
- 46.Tripathi A, Lammers KM, Goldblum S, Shea-Donohue T, Netzel-Arnett S, Buzza MS, et al. Identification of human zonulin, a physiological modulator of tight junctions, as prehaptoglobin-2. Proc Natl Acad Sci. 2009;106(39):16799–16804. doi: 10.1073/pnas.0906773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lammers KM, Lu R, Brownley J, Lu B, Gerard C, Thomas K, et al. Gliadin Induces an Increase in Intestinal Permeability and Zonulin Release by Binding to the Chemokine Receptor CXCR3. Gastroenterology. 2008;135(1):194–204. doi: 10.1053/j.gastro.2008.03.023. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drago S, Asmar El R, Di Pierro M, Grazia Clemente M, Sapone ATA, Thakar M, et al. Gliadin, zonulin and gut permeability: Effects on celiac and non-celiac intestinal mucosa and intestinal cell lines. Scand J Gastroenterol. 2006;41(4):408–419. doi: 10.1080/00365520500235334. [DOI] [PubMed] [Google Scholar]
- 49.Di Pierro M, Lu R, Uzzau S, Wang W, Margaretten K, Pazzani C, et al. Zonula occludens toxin structure-function analysis. Identification of the fragment biologically active on tight junctions and of the zonulin receptor binding domain. J Biol Chem. 2001;276(22):19160–19165. doi: 10.1074/jbc.M009674200. [DOI] [PubMed] [Google Scholar]
- 50.Clemente MG, De Virgiliis S, Kang JS, Macatagney R, Musu MP, Di Pierro MR, et al. Early effects of gliadin on enterocyte intracellular signalling involved in intestinal barrier function. Gut. 2003;52(2):218–223. doi: 10.1136/gut.52.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paterson B, Lammers K, Arrieta M. The safety, tolerance, pharmacokinetic and pharmacodynamic effects of single doses of AT-1001 in coeliac disease subjects: a proof of concept study. Aliment Pharmacol Ther. 2007;26:757–766. doi: 10.1111/j.1365-2036.2007.03413.x. [DOI] [PubMed] [Google Scholar]
- 52.Leffler DA, Kelly CP, Abdallah HZ, Colatrella AM, Harris LA, Leon F, et al. A Randomized, Double-Blind Study of Larazotide Acetate to Prevent the Activation of Celiac Disease During Gluten Challenge. Am J Gastroenterol. 2012;107(10):1554–1562. doi: 10.1038/ajg.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kelly C, Green P, Murray J. Larazotide acetate in patients with coeliac disease undergoing a gluten challenge: a randomised placebo-controlled study. Aliment Pharmacol Ther. 2013;37:252–262. doi: 10.1111/apt.12147. [DOI] [PubMed] [Google Scholar]
- 54.Leffler DA, Kelly CP, Green PHR, Fedorak RN, DiMarino A, Perrow W, et al. Larazotide Acetate for Persistent Symptoms of Celiac Disease Despite a Gluten-Free Diet: A Randomized Controlled Trial. Gastroenterology. 2015;1:58. doi: 10.1053/j.gastro.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oldfield WLG, Larché M, Kay AB. Effect of T-cell peptides derived from Fel d 1 on allergic reactions and cytokine production in patients sensitive to cats: a randomised controlled trial. Lancet. 2002;360(9326):47–53. doi: 10.1016/s0140-6736(02)09332-7. [DOI] [PubMed] [Google Scholar]
- 56.Verhoef A, Alexander C, Kay AB, Larché M. T cell epitope immunotherapy induces a CD4+ T cell population with regulatory activity. PLoS Med. 2005;2(3):e78. doi: 10.1371/journal.pmed.0020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown GJ, Daveson J, Marjason JK, Ffrench RA, Smith D, Sullivan M, et al. A Phase I Study to Determine Safety, Tolerability and Bioactivity of Nexvax2 in HLA DQ2+ Volunteers With Celiac Disease Following a Long-Term, Strict Gluten-Free Diet. YGAST. 2011;140(5):S–437–S–438. [Google Scholar]
- 58.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shanahan F. Crohn's disease. Lancet. 2002;359(9300):62–69. doi: 10.1016/S0140-6736(02)07284-7. [DOI] [PubMed] [Google Scholar]
- 60.Podolsky DK. The current future understanding of inflammatory bowel disease. Best Pract Res Cl Ha. 2002;16(6):933–943. doi: 10.1053/bega.2002.0354. [DOI] [PubMed] [Google Scholar]
- 61.Wacklin P, Laurikka P, Lindfors K, Collin P, Salmi T, hdeaho M-LLA, et al. Altered Duodenal Microbiota Composition in Celiac Disease Patients Suffering From Persistent Symptoms on a Long-Term Gluten-Free Diet. Am J Gastroenterol. 2014;109(12):1933–1941. doi: 10.1038/ajg.2014.355. [DOI] [PubMed] [Google Scholar]
- 62.Angelis MD, Rizzello CG, Fasano A, Clemente MG, Simone CD, Silano M, et al. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for Celiac Sprue probiotics and gluten intolerance. Biochim Biophys Acta. 2006;1762(1):80–93. doi: 10.1016/j.bbadis.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Smecuol E, Hwang HJ, Sugai E, Corso L, Cherñavsky AC, Bellavite FP, et al. Exploratory, randomized, double-blind, placebo-controlled study on the effects of Bifidobacterium infantis natren life start strain super strain in active celiac disease. J Clin Gastroenterol. 2013;47(2):139–147. doi: 10.1097/MCG.0b013e31827759ac. [DOI] [PubMed] [Google Scholar]
- 64.Bager P, Arnved J, Ronborg S, Wohlfahrt J, Poulsen LK, Westergaard T, et al. Trichuris suis ova therapy for allergic rhinitis: A randomized, double-blind, placebo-controlled clinical trial. J Allergy Clin Immunol. 2010;125(1):123–123. doi: 10.1016/j.jaci.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 65.Croese J, O'neil J, Masson J, Cooke S, Melrose W, Pritchard D, et al. A proof of concept study establishing Necator americanus in Crohn's patients and reservoir donors. Gut. 2006;55(1):136–137. doi: 10.1136/gut.2005.079129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daveson A, Jones D, Gaze S, McSorley H. Effect of Hookworm Infection on Wheat Challenge in Celiac Disease – A Randomised Double-Blinded Placebo Controlled Trial. PLoS ONE. 2011;6(3):e17366. doi: 10.1371/journal.pone.0017366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Croese J, Giacomin P, Navarro S, Clouston A, McCann L, Dougall A, et al. Experimental hookworm infection and gluten microchallenge promote tolerance in celiac disease. J Allergy Clin Immunol. 2015;135(2):508–516. doi: 10.1016/j.jaci.2014.07.022. e5. [DOI] [PubMed] [Google Scholar]
- 68.Matysiak-Budnik T, Moura IC, Arcos-Fajardo M, Lebreton C, Ménard S, Candalh C, et al. Secretory IgA mediates retrotranscytosis of intact gliadin peptides via the transferrin receptor in celiac disease. J Exp Med. 2008;205(1):143–154. doi: 10.1084/jem.20071204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rauhavirta T, Qiao SW, Jiang Z, Myrsky E, Loponen J, Korponay-Szabó IR, et al. Epithelial transport and deamidation of gliadin peptides: a role for coeliac disease patient immunoglobulin A. Clin Exp Immunol. 2011;164(1):127–136. doi: 10.1111/j.1365-2249.2010.04317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yokoyama S, Watanabe N, Sato N, Perera P-Y, Filkoski L, Tanaka T, et al. Antibody-mediated blockade of IL-15 reverses the autoimmune intestinal damage in transgenic mice that overexpress IL-15 in enterocytes. Proc Natl Acad Sci. 2009;106(37):15849–15854. doi: 10.1073/pnas.0908834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malamut G, Machhour RE, Montcuquet N, Martin-Lanneree S, Dusanter-Fourt I, Verkarre V, et al. IL-15 triggers an antiapoptotic pathway in human intraepithelial lymphocytes that is a potential new target in celiac disease–associated inflammation and lymphomagenesis. J Clin Invest. 2010;120(6):2131. doi: 10.1172/JCI41344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sarra M, Cupi ML, Monteleone I, Franzé E, Ronchetti G, Di Sabatino A, et al. IL-15 positively regulates IL-21 production in celiac disease mucosa. Mucosal Immunol. 2012;6(2):244–255. doi: 10.1038/mi.2012.65. [DOI] [PubMed] [Google Scholar]
- 73.Lebrec H, Horner MJ, Gorski KS, Tsuji W, Xia D, Pan W-J, et al. Homeostasis of Human NK Cells Is Not IL-15 Dependent. J Immunol. 2013;191:5551–5558. doi: 10.4049/jimmunol.1301000. [DOI] [PubMed] [Google Scholar]
- 74.Marrano C, de Macédo P, Gagnon P, Lapierre D, Gravel C, Keillor JW. Synthesis and evaluation of novel dipeptide-bound 1, 2, 4-thiadiazoles as irreversible inhibitors of guinea pig liver transglutaminase. Bioorg Med Chem. 2001;9(12):3231–3241. doi: 10.1016/s0968-0896(01)00228-0. [DOI] [PubMed] [Google Scholar]
- 75.de Macédo P, Marrano C, Keillor JW. Synthesis of dipeptide-bound epoxides and alpha,beta-unsaturated amides as potential irreversible transglutaminase inhibitors. Bioorg Med Chem. 2002;10(2):355–360. doi: 10.1016/s0968-0896(01)00292-9. [DOI] [PubMed] [Google Scholar]
- 76.Choi K, Siegel M, Piper J, Yuan L, Cho E, Strnad P, et al. Chemistry and Biology of Dihydroisoxazole Derivatives: Selective Inhibitors of Human Transglutaminase 2. Chem Biol. 2005;12(4):469–475. doi: 10.1016/j.chembiol.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 77.Watts RE, Siegel M, Khosla C. Structure-activity relationship analysis of the selective inhibition of transglutaminase 2 by dihydroisoxazoles. J Med Chem. 2006;49(25):7493–7501. doi: 10.1021/jm060839a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duval E, Case A, Stein RL, Cuny GD. Structure–activity relationship study of novel tissue transglutaminase inhibitors. Bioorg Med Chem Lett. 2005;15(7):1885–1889. doi: 10.1016/j.bmcl.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 79.Pardin C, Pelletier JN, Lubell WD, Keillor JW. Cinnamoyl inhibitors of tissue transglutaminase. J Org Chem. 2008;73(15):5766–5775. doi: 10.1021/jo8004843. [DOI] [PubMed] [Google Scholar]
- 80.Ozaki S, Ebisui E, Hamada K, Goto J-I, Suzuki AZ, Terauchi A, et al. Potent transglutaminase inhibitors, aryl β-aminoethyl ketones. Bioorg Med Chem Lett. 2010;20(3):1141–1144. doi: 10.1016/j.bmcl.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 81.Klöck C, Jin X, Choi K, Khosla C, Madrid PB, Spencer A, et al. Acylideneoxoindoles: A new class of reversible inhibitors of human transglutaminase 2. Bioorg Med Chem Lett. 2011;21(9):2692–2696. doi: 10.1016/j.bmcl.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Diraimondo TR, Klöck C, Warburton R, Herrera Z, Penumatsa K, Toksoz D, et al. Elevated transglutaminase 2 activity is associated with hypoxia-induced experimental pulmonary hypertension in mice. ACS Chem Biol. 2014;9(1):266–275. doi: 10.1021/cb4006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nanda N. Targeted Inactivation of Gh/Tissue Transglutaminase II. J Biol Chem. 2001;276(23):20673–20678. doi: 10.1074/jbc.M010846200. [DOI] [PubMed] [Google Scholar]
- 84.De Laurenzi V, Melino G. Gene Disruption of Tissue Transglutaminase. Mol Cell Biol. 2001;21(1):148–155. doi: 10.1128/MCB.21.1.148-155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Herman ML, Farasat S, Steinbach PJ, Wei M-H, Toure O, Fleckman P, et al. Transglutaminase-1 gene mutations in autosomal recessive congenital ichthyosis: summary of mutations (including 23 novel) and modeling of TGase-1. Hum Mutat. 2009;30(4):537–547. doi: 10.1002/humu.20952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muszbek L, Bereczky Z, Bagoly Z, Komáromi I, Katona É . Factor XIII: a coagulation factor with multiple plasmatic and cellular functions. Physiol Rev. 2011;91(3):931–972. doi: 10.1152/physrev.00016.2010. [DOI] [PubMed] [Google Scholar]
- 87.Stacey SN, Sulem P, Gudbjartsson DF, Jonasdottir A, Thorleifsson G, Gudjonsson SA, et al. Germline sequence variants in TGM3 and RGS22 confer risk of basal cell carcinoma. Hum Mol Genet. 2014;23(11):3045–3053. doi: 10.1093/hmg/ddt671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klöck C, Herrera Z, Albertelli M, Khosla C. Discovery of potent and specific dihydroisoxazole inhibitors of human transglutaminase 2. J Med Chem. 2014;57(21):9042–9064. doi: 10.1021/jm501145a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xia J, Bergseng E, Fleckenstein B, Siegel M, Kim C-Y, Khosla C, et al. Cyclic and dimeric gluten peptide analogues inhibiting DQ2-mediated antigen presentation in celiac disease. Bioorg Med Chem. 2007;15(20):6565–6573. doi: 10.1016/j.bmc.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kapoerchan VV, Wiesner M, Overhand M, van der Marel GA, Koning F, Overkleeft HS. Design of azidoproline containing gluten peptides to suppress CD4+ T-cell responses associated with Celiac disease. Bioorg Med Chem. 2008;16(4):2053–2062. doi: 10.1016/j.bmc.2007.10.091. [DOI] [PubMed] [Google Scholar]
- 91.Jüse U, van de Wal Y, Koning F, Sollid LM, Fleckenstein B. Design of new high-affinity peptide ligands for human leukocyte antigen-DQ2 using a positional scanning peptide library. HIM. 2010;71(5):475–481. doi: 10.1016/j.humimm.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 92.Huan J, Meza-Romero R, Mooney JL, Vandenbark AA, Offner H, Burrows GG. Single-chain recombinant HLA-DQ2.5/peptide molecules block α2-gliadin-specific pathogenic CD4+ T-cell proliferation and attenuate production of inflammatory cytokines: a potential therapy for celiac disease. Mucosal Immunol. 2010;4(1):112–120. doi: 10.1038/mi.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gillett HR, Arnott IDR, McIntyre M, Campbell S, Dahele A, Priest M, et al. Successful infliximab treatment for steroid-refractory celiac disease: A case report. Gastroenterology. 2002;122(3):800–805. doi: 10.1053/gast.2002.31874. [DOI] [PubMed] [Google Scholar]
- 94.Costantino G, Torre della A, Presti Lo MA, Caruso R, Mazzon E, Fries W. Treatment of life-threatening type I refractory coeliac disease with long-term infliximab. Digest Liver Dis. 2008;40(1):74–77. doi: 10.1016/j.dld.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 95.Reinisch W, de Villiers W, Bene L, Simon L, Rácz I, Katz S, et al. Fontolizumab in moderate to severe Crohn’s disease: A phase 2, randomized, double-blind, placebo-controlled, multiple-dose study. Inflamm Bowel Dis. 2010;16(2):233–242. doi: 10.1002/ibd.21038. [DOI] [PubMed] [Google Scholar]
- 96.van der Woude CJ, Stokkers P, van Bodegraven AA, Van Assche G, Hebzda Z, Paradowski L, et al. Phase I, double-blind, randomized, placebo-controlled, dose-escalation study of NI-0401 (a fully human anti-CD3 monoclonal antibody) in patients with moderate to severe active Crohn's disease. Inflamm Bowel Dis. 2010;16(10):1708–1716. doi: 10.1002/ibd.21252. [DOI] [PubMed] [Google Scholar]
- 97.Herold KC, Gitelman S, Greenbaum C, Puck J, Hagopian W, Gottlieb P, et al. Treatment of patients with new onset Type 1 diabetes with a single course of anti-CD3 mAb teplizumab preserves insulin production for up to 5 years. Clin Immunol. 2009;132(2):166–173. doi: 10.1016/j.clim.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wiczling P, Rosenzweig M, Vaickus L, Jusko WJ. Pharmacokinetics and pharmacodynamics of a chimeric/humanized anti-CD3 monoclonal antibody, otelixizumab (TRX4), in subjects with psoriasis and with type 1 diabetes mellitus. J Clin Pharmacol. 2010;50(5):494–506. doi: 10.1177/0091270009349376. [DOI] [PubMed] [Google Scholar]
- 99.Edwards JCW, Szczepanski L, Szechinski J, Filipowicz-Sosnowska A, Emery P, Close DR, et al. Efficacy of B-cell-targeted therapy with rituximab in patients with rheumatoid arthritis. N Engl J Med. 2004;350(25):2572–2581. doi: 10.1056/NEJMoa032534. [DOI] [PubMed] [Google Scholar]
- 100.Hauser SL, Waubant E, Arnold DL, Vollmer T, Antel J, Fox RJ, et al. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N Engl J Med. 2008;358(7):676–688. doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- 101.Pescovitz MD, Bloom R, Pirsch J, Johnson J, Gelone S, Villano SA. A randomized, double-blind, pharmacokinetic study of oral maribavir with tacrolimus in stable renal transplant recipients. Am J Transplant. 2009;9(10):2324–2330. doi: 10.1111/j.1600-6143.2009.02768.x. [DOI] [PubMed] [Google Scholar]
- 102.Sollid LM, Molberg O, McAdam S, Lundin KE. Autoantibodies in coeliac disease: tissue transglutaminase--guilt by association? Gut. 1997;41(6):851–852. doi: 10.1136/gut.41.6.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mei H, Frölich D, Giesecke C, Loddenkemper C. Steady-state generation of mucosal IgA+ plasmablasts is not abrogated by B-cell depletion therapy with rituximab. Blood. 2010;116(24):5181–5190. doi: 10.1182/blood-2010-01-266536. [DOI] [PubMed] [Google Scholar]
- 104.Mora JR, Andrian von UH. Differentiation and homing of IgA-secreting cells. Mucosal Immunol. 2008;1(2):96–109. doi: 10.1038/mi.2007.14. [DOI] [PubMed] [Google Scholar]
- 105.Olaussen RW, Karlsson MR, Lundin KEA, Jahnsen J, Brandtzaeg P, Farstad IN. Reduced Chemokine Receptor 9 on Intraepithelial Lymphocytes in Celiac Disease Suggests Persistent Epithelial Activation. Gastroenterology. 2007;132(7):2371–2382. doi: 10.1053/j.gastro.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 106.Di Sabatino AD, Rovedatti L, Rosado MM, Carsetti R, Corazza GR, MacDonald TT, et al. Increased expression of mucosal addressin cell adhesion molecule 1 in the duodenum of patients with active celiac disease is associated with depletion of integrin α4β7-positive T cells in blood. Human Pathology. 2009;40(5):699–704. doi: 10.1016/j.humpath.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 107.Walters MJ, Wang Y, Lai N, Baumgart T, Zhao BN, Dairaghi DJ, et al. Characterization of CCX282-B, an orally bioavailable antagonist of the CCR9 chemokine receptor, for treatment of inflammatory bowel disease. J Pharmacol Exp Ther. 2010;335(1):61–69. doi: 10.1124/jpet.110.169714. [DOI] [PubMed] [Google Scholar]
- 108.Sandborn WJ, Feagan BG, Rutgeerts P, Hanauer S, Colombel JF, Sands BE, et al. Vedolizumab as Induction and Maintenance Therapy for Crohn's Disease. N Engl J Med. 2013;369(8):711–721. doi: 10.1056/NEJMoa1215739. [DOI] [PubMed] [Google Scholar]
- 109.Feagan BG, Rutgeerts P, Sands BE, Hanauer S, Colombel JF, Sandborn WJ, et al. Vedolizumab as Induction and Maintenance Therapy for Ulcerative Colitis. N Engl J Med. 2013;369(8):699–710. doi: 10.1056/NEJMoa1215734. [DOI] [PubMed] [Google Scholar]
- 110.Dubois PCA, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42(4):295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Senapati S, Gutierrez-Achury J, Sood A, Midha V, Szperl A, Romanos J, et al. Evaluation of European coeliac disease risk variants in a north Indian population. Eur J Hum Genet. 2014:1–6. doi: 10.1038/ejhg.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lesourne R, Uehara S, Lee J, Song K-D, Li L, Pinkhasov J, et al. Themis, a T cell–specific protein important for late thymocyte development. Nat Immunol. 2009;10(8):840–847. doi: 10.1038/ni.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fu G, Casas J, Rigaud S, Rybakin V, Lambolez F, Brzostek J, et al. Themis sets the signal threshold for positive and negative selection in T-cell development. Nature. 2013;504(7480):441–445. doi: 10.1038/nature12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chabod M, Pedros C, Lamouroux L, Colacios C, Bernard I, Lagrange D, et al. A Spontaneous Mutation of the Rat Themis Gene Leads to Impaired Function of Regulatory T Cells Linked to Inflammatory Bowel Disease. PLoS Genet. 2012;8(1):e1002461. doi: 10.1371/journal.pgen.1002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sawcer S, Hellenthal G, Pirinen M, Spencer CCA, Patsopoulos NA, Moutsianas L, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bondar C, Plaza-Izurieta L, Fernandez-Jimenez N, Irastorza INA, Withoff S, et al. THEMIS and PTPRK in celiac intestinal mucosa: coexpression in disease and after in vitro gliadin challenge. Eur J Hum Genet. 2013;22(3):358–362. doi: 10.1038/ejhg.2013.136. CEGEC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang SE, Wu FY, Shin I, Qu S, Arteaga CL. Transforming growth factor {beta} (TGF-{beta})-Smad target gene protein tyrosine phosphatase receptor type kappa is required for TGF-{beta} function. Mol Cell Biol. 2005;25(11):4703–4715. doi: 10.1128/MCB.25.11.4703-4715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Asano A, Tsubomatsu K, Jung C-G, Sasaki N, Agui T. A deletion mutation of the protein tyrosine phosphatase kappa (Ptprk) gene is responsible for T-helper immunodeficiency (thid) in the LEC rat. Mamm Genome. 2007;18(11):779–786. doi: 10.1007/s00335-007-9062-0. [DOI] [PubMed] [Google Scholar]
- 120.Iwata R, Sasaki N, Agui T. Contiguous gene deletion of Ptprk and Themis causes T-helper immunodeficiency (thid) in the LEC rat. Biomedical Research. 2010;31(1):83–87. doi: 10.2220/biomedres.31.83. [DOI] [PubMed] [Google Scholar]
- 121.Sap J, Jiang YP, Friedlander D, Grumet M, Schlessinger J. Receptor tyrosine phosphatase R-PTP-kappa mediates homophilic binding. Mol Cell Biol. 1994;14(1):1–9. doi: 10.1128/mcb.14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.International Multiple Sclerosis Genetics Consortium. Wellcome Trust Case Control Consortium 2. Sawcer S, Hellenthal G, Pirinen M, Spencer CCA, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem. 1995;270(9):4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- 124.Wacklin P, Mäkivuokko H, Alakulppi N, Nikkilä J, Tenkanen H, Räbinä J, et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE. 2011;6(5):e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Parmar AS, Alakulppi N, Paavola-Sakki P, Kurppa K, Halme L, Färkkilä M, et al. Association study of FUT2(rs601338) with celiac disease and inflammatory bowel disease in the Finnish population. Tissue Antigens. 2012;80(6):488–493. doi: 10.1111/tan.12016. [DOI] [PubMed] [Google Scholar]
- 126.McGovern DPB, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet. 2010;19(17):3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smyth DJ, Cooper JD, Howson JMM, Clarke P, Downes K, Mistry T, et al. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes. 2011;60(11):3081–3084. doi: 10.2337/db11-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schippa S, Iebba V, Barbato M, Di Nardo G, Totino V, Checchi MP, et al. A distinctive “microbial signature” in celiac pediatric patients. BMC Microbiol. 2010;10:175. doi: 10.1186/1471-2180-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wacklin P, Kaukinen K, Tuovinen E, Collin P, Lindfors K, Partanen J, et al. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm Bowel Dis. 2013;19(5):934–941. doi: 10.1097/MIB.0b013e31828029a9. [DOI] [PubMed] [Google Scholar]
- 130.De Palma G, Kamanova J, Cinova J, Olivares M, Drasarova H, Tuckova L, et al. Modulation of phenotypic and functional maturation of dendritic cells by intestinal bacteria and gliadin: relevance for celiac disease. J Leukoc Biol. 2012;92(5):1043–1054. doi: 10.1189/jlb.1111581. [DOI] [PubMed] [Google Scholar]
- 131.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souëf P, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378(9795):1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Franke A, McGovern DPB, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL, et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44(6):676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40(12):1399–12401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Igarashi K, Ochiai K, Itoh-Nakadai A, Muto A. Orchestration of plasma cell differentiation by Bach2 and its gene regulatory network. Immunol Rev. 2014;261(1):116–125. doi: 10.1111/imr.12201. [DOI] [PubMed] [Google Scholar]
- 136.Roychoudhuri R, Hirahara K, Mousavi K, Clever D, Klebanoff CA, Bonelli M, et al. BACH2 represses effector programs to stabilize Treg-mediated immune homeostasis. Nature. 2013;498(7455):506–510. doi: 10.1038/nature12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kumar V, Gutierrez-Achury J, Kanduri K, Almeida R, Hrdlickova B, Zhernakova DV, et al. Systematic annotation of celiac disease loci refines pathological pathways and suggests a genetic explanation for increased interferon-gamma levels. Hum Mol Genet. 2014;24(2):397–409. doi: 10.1093/hmg/ddu453. [DOI] [PubMed] [Google Scholar]
- 138.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JHM, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359(26):2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.International Multiple Sclerosis Genetics Consortium (IMSGC) Bush WS, Sawcer SJ, De Jager PL, Oksenberg JR, McCauley JL, et al. Evidence for polygenic susceptibility to multiple sclerosis--the shape of things to come. Am J Hum Genet. 2010;86(4):621–625. doi: 10.1016/j.ajhg.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hunt KA, Zhernakova A, Turner G, Heap GAR, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40(4):395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Moratz C, Kang VH, Druey KM, Shi CS, Scheschonka A, Murphy PM, et al. Regulator of G protein signaling 1 (RGS1) markedly impairs Gi alpha signaling responses of B lymphocytes. J Immunol. 2000;164(4):1829–1838. doi: 10.4049/jimmunol.164.4.1829. [DOI] [PubMed] [Google Scholar]
- 142.Moratz C, Hayman JR, Gu H, Kehrl JH. Abnormal B-cell responses to chemokines, disturbed plasma cell localization, and distorted immune tissue architecture in Rgs1−/− mice. Mol Cell Biol. 2004;24(13):5767–5775. doi: 10.1128/MCB.24.13.5767-5775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gibbons DL, Abeler-Dörner L, Raine T, Hwang I-Y, Jandke A, Wencker M, et al. Cutting Edge: Regulator of G protein signaling-1 selectively regulates gut T cell trafficking and colitic potential. J Immunol. 2011;187(5):2067–2071. doi: 10.4049/jimmunol.1100833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hindorff L, MacArthur J, Morales, J, Junkins H, Hall P, Klemm A, et al. A Catalog of Published Genome-Wide Association Studies. (European Bioinformatics Institute) (European Bioinformatics Institute) Available at: www.genome.gov/gwastudies; Accessed 2/14/2015.
- 145.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]