Abstract
Sarcoidosis is a granulomatous inflammatory disease, diagnosed through tissue biopsy of involved organs in the absence of other causes such as tuberculosis (TB). No specific serologic test is available to diagnose and differentiate sarcoidosis from TB. Using a high throughput method, we developed a T7 phage display cDNA library derived from mRNA isolated from bronchoalveolar lavage (BAL) cells and leukocytes of sarcoidosis patients. This complex cDNA library was biopanned to obtain 1152 potential sarcoidosis antigens and a microarray was constructed to immunoscreen two different sets of sera from healthy controls and sarcoidosis. Meta-analysis identified 259 discriminating sarcoidosis antigens, and multivariate analysis identified 32 antigens with a sensitivity of 89% and a specificity of 83% to classify sarcoidosis from healthy controls. Additionally, interrogating the same microarray platform with sera from subjects with TB, we identified 50 clones that distinguish between TB, sarcoidosis and healthy controls. The top 10 sarcoidosis and TB specific clones were sequenced and homologies were searched in the public database revealing unique epitopes and mimotopes in each group. Here, we show for the first time that immunoscreenings of a library derived from sarcoidosis tissue differentiates between sarcoidosis and tuberculosis antigens. These novel biomarkers can improve diagnosis of sarcoidosis and TB, and may aid to develop or evaluate a TB vaccine.
Keywords: T7 Phage library, Sarcoidosis, Tuberculosis, IgG, Microarray, Immunoscreening
Highlights
-
•
Immunity plays a major role in a vast array of human diseases.
-
•
Sarcoidosis shares similarities with non-infectious and infectious granulomatous diseases, including tuberculosis.
-
•
A highly sensitive and specific T7 phage library discriminates the immune signature between sarcoidosis patients and TB.
1. Introduction
Sarcoidosis is an inflammatory granulomatous disease of unknown etiology affecting multiple organs, such as the lungs, skin, CNS, and eyes (Costabel, 2001, Costabel and Hunninghake, 1999, Iannuzzi et al., 2007, Hunninghake et al., 1999). Common features shared by patients with sarcoidosis are the presence of non-caseating granuloma, a lack of cutaneous reaction to tuberculin skin testing (PPD) and increased local and circulating inflammatory cytokines (Costabel, 2001, Costabel and Hunninghake, 1999, Iannuzzi et al., 2007). In addition, there is evidence of abnormal immune function accompanied by hypergammaglobulinemia (Iannuzzi et al., 2007). Sarcoidosis shares striking clinical and pathological similarities with infectious granulomatous diseases, especially Mycobacteria tuberculosis (MTB) (Iannuzzi et al., 2007, Prince et al., 2003). Although there is mounting evidence of the presence of nonviable bacterial components (including MTB and Propionibacterium acnes) in sarcoidosis tissue (Gupta et al., 2007, Chen et al., 2010, Negi et al., 2012), all attempts to isolate viable MTB or other microbial pathogens from sarcoidosis tissue have failed (Hunninghake et al., 1999, Chen et al., 2008).
Intradermal injection of the Kveim–Siltzbach suspension (a granulomatous splenic tissue suspension) induces granuloma formation weeks later in sarcoidosis patients suggesting presence of antigen(s) in granuloma tissue and host immunoreactivity to these antigens (Dubaniewicz, 2010, Hajizadeh et al., 2007, Hiramatsu et al., 2003, Fox et al., 1983, Munro and Mitchell, 1987, Siltzbach and Ehrlich, 1954). Several studies using state-of-the-art technologies have attempted to identify sarcoidosis antigens or to identify the underlying genetic and environmental factors (Hajizadeh et al., 2007, Chen and Moller, 2007, Zhang et al., 2013), yet unifying environmental or genetic factors as initiators of this disease have not been found (Hunninghake et al., 1999, Dubaniewicz, 2010, Eishi et al., 2002, Oswald-Richter and Drake, 2010). These studies reported a number of markers or variations in gene expression signatures, however failed to discriminate between sarcoidosis and other inflammatory or granulomatous diseases (Koth et al., 2011, Maertzdorf et al., 2012). This is partly due to the fact that several inflammatory diseases may respond to various antigens with activation of a similar transcriptome and/or inflammatory gene expression profiles (Koth et al., 2011, Maertzdorf et al., 2012).
Proteomics, genomics, transcriptomics, and high throughput technology clearly suggest that early immune reaction to diverse antigens is highly prevalent in a large number of rheumatic, neoplastic, and inflammatory diseases such as sarcoidosis (Koth et al., 2011, Maertzdorf et al., 2012, Bons et al., 2007, Stone et al., 2013). Since non-caseating granulomas, cutaneous anergy and hypergammaglobulinemia suggest an immune dysfunction in this disease, we hypothesize that sarcoidosis is triggered by a group of unknown antigens represented in the host immune cells. To identify the elusive antigen(s), we developed a heterologous cDNA library derived from bronchoalveolar cell (BAL) samples and total white blood cells (WBC) from sarcoidosis patients. We then combined both sarcoid-derived libraries with cultured human monocytes and embryonic lung fibroblast cDNA libraries to build a complex sarcoidosis library (CSL). Furthermore, we used antibody recognition and random plaque selection during biopanning of the cDNA libraries to minimize the confounding effects of autoantibodies unrelated to sarcoidosis. This approach has been successfully applied in biomarker discovery for the diagnosis of lung, head and neck and breast cancer (Fernandez-Madrid et al., 2004, Fernandez-Madrid et al., 1999, Lin et al., 2007). We tested whether this novel library represents relevant antigens and can specifically be recognized by high IgG titers present in sera of sarcoidosis subjects. The essential feature that distinguishes our method from previous studies is that we used the exquisite power of antibody recognition present in human sera to interrogate the potential antigens present in macrophages and monocytes.
The present study describes a novel approach to identify sarcoidosis antigens and to detect serum antibodies on high-throughput arrays. Sera from 3 cohorts (sarcoidosis, controls, and TB) were used for immunoscreening. Using bioinformatic tools, we have identified a large number of biomarkers with high sensitivity and specificity that can discriminate among the sera of patients with sarcoidosis, healthy controls and MTB. Using the integrative-analysis method that combines results from two independent trials, we have identified clones that significantly differentiate sarcoidosis from controls. Similarly, we identified clones that differentially react with TB sera and not with sarcoidosis or control sera. Furthermore, the top 10 discriminating antigens for TB and sarcoidosis were sequenced and homologies were identified in a public database. These data indicate that we have developed a unique library enabling the detection of highly significant antigens to discriminate between patients with sarcoidosis and tuberculosis.
2. Materials and Methods
2.1. Chemicals
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless specified otherwise. LeukoLOCK filters and RNAlater were purchased from Life Technologies (Grand Island, NY). The RNeasy Midi kit was obtained from Qiagen (Valencia, CA). The T7 mouse monoclonal antibody was purchased from Novagen (San Diego, CA). Alexa Fluor 647 goat anti-human IgG and Alex Fluor goat anti-mouse IgG antibodies were purchased from Life Technologies (Grand Island, NY).
2.2. Patient Selection
This study was approved by the institutional review board at Wayne State University and the Detroit Medical Center. Patients were recruited at the center for Sarcoidosis and Interstitial Lung Diseases (SILD), which is a referral center for patients with sarcoidosis and other ILDs. Three sources of patient derived materials have been used in this study: A) a BAL cDNA library was derived from BAL cells obtained during diagnostic bronchoscopy from newly diagnosed patients with sarcoidosis (n = 20); B) a leukocyte cDNA library were developed from a different cohort of sarcoidosis patients who were followed in an outpatient setting with various stages of sarcoidosis (n = 36); and C) sera collected from 3 groups: 1) healthy controls, who were volunteers recruited from the community; 2) subjects with biopsy confirmed sarcoidosis who were followed in an outpatient setting; and 3) sera from subjects with culture positive TB collected at the Detroit Department of Health and Wellness Promotion. Subjects were included who had a diagnosis of sarcoidosis as proven by tissue biopsy per guidelines (Costabel and Hunninghake, 1999) and have a negative PPD. TB subjects were included who had a positive TB culture and were HIV negative. Subjects were excluded, who were positive for HIV or were receiving high dose immune suppressive medication that was defined as prednisone more than 15 mg per day alone or in combination with immune modulatory medications. Subjects who had positive PPD or quantiferon test were excluded from the sarcoidosis group. All study subjects signed a written informed consent.
2.3. Bronchoalveolar Lavage
BAL cells were obtained, after informed consent, during diagnostic bronchoscopy from subjects with active sarcoidosis as previously described (Rastogi et al., 2011). BAL cells were suspended in 500 μL of RNAlater and stored at − 80 °C.
2.4. Collection of Total Leukocytes from Sarcoidosis Subjects
Leukocytes from 36 sarcoid subjects were isolated from whole blood using LeukoLOCK filters as previously described (Glatt et al., 2009).
2.5. Human Macrophage (EL-1) and Human Lung Embryonic Fibroblast (MRC-5) Cell Cultures
Both cell lines were obtained from ATCC and cultured as per ATTC recommendations. From each cell line 1–2 mg RNA was isolated to construct the cDNA library.
2.6. Serum Collection
Using standardized phlebotomy procedures blood samples were collected and allowed to clot and then centrifuged at 2500 rpm for 10 min. Supernatants were stored at − 80 °C.
2.7. Construction of T7 Phage Display cDNA Libraries
Total RNA was isolated using the RNeasy Midi kit (Qiagen, Valencia, CA). Integrity of the RNA samples was assessed using the Agilent 2100 bioanalyzer. Total RNA, in the amount of 1–2 mg, was subjected to two cycles of polyA purification to minimize ribosomal RNA contamination as suggested by the manufacturer (Qiagen, Valencia, CA). The construction of phage cDNA libraries was performed using Novagen's Orient Express cDNA Synthesis (Random Primer System) and Cloning system as per manufacturer's suggestions (EMD Biosciences-Novagen). Each library was cloned using modified linkers that allow us to distinguish the phage clones (Chatterjee et al., 2006). The number of clones in each of the 4 libraries was titrated by plaque assay as per manufacturer's instructions (EMD Biosciences-Novagen). Finally, the same number of phages from each BAL, WBC, EL-1 and MRC5 library were pooled to generate a complex sarcoid library (CSL).
2.8. Biopanning of T7 Phage Displayed cDNA Library with Human Sera
Differential biopanning for negative and positive selection was performed using sera from healthy controls to remove the non-specific IgG, and sarcoidosis sera for selective enrichment according to manufacturer's suggestions (T7 Select System, TB178; EMD Biosciences-Novagen). Protein G Plus-agarose beads (Santa Cruz Biotechnology) were used for serum IgG immobilization. Four rounds of biopannings were performed and the selected phage libraries were used for microarray immunoscreening. Each cycle of biopanning consisted of passing the entire phage library through protein G beads coated with IgG from pooled sera of healthy controls, then passing through beads coated with IgGs from individual serum of sarcoid subjects.
2.9. Microarray Construction and Immunoscreening
Informative phage clones were randomly picked and amplified after several rounds of biopannings and their lysates were arrayed in quintuplicates onto nitrocellulose FAST slides (Grace Biolabs, OR) using the ProSys 5510TL robot (Cartesian Technologies, CA). The nitrocellulose slides were then blocked with a solution of 1% BSA in PBS for 1 h at room temperature followed by another hour of incubation with serum at a dilution of 1:300 in 1 × PBS as primary antibodies, together with mouse anti-T7 capsid antibody (0.15 μg/mL) and BL21 Escherichia coli cell lysates (5 μg/mL). BL21 E. coli cell lysates were added to remove antibodies specific to E. coli from the serum. The microarrays were then washed three times at room temperature with a solution of PBS/0.1% Tween20 for 4 min. Secondary antibodies consisted of goat anti-human IgG Alexa Fluor 647 (red fluorescent dye) 1 μg/mL and goat anti-mouse IgG Alexa Fluor 532 (green fluorescent dye) 0.05 μg/mL. After 1 h of incubation in the dark, the microarrays were washed 3 times with a solution of PBS/0.1% Tween20 for 4 min at room temperature, and 2 times in PBS for 4 min at room temperature and then air dried.
2.10. Sequencing of Phage cDNA Clones
Individual phage clones were PCR amplified using T7 phage forward primer 5′ GTTCTATCCGCAACGTTATGG 3′ and reverse primer 5′ GGAGGAAAGTCGTTTTTTGGGG 3′ and sequenced by Genwiz (South Plainfield, NJ), using T7 phage sequence primer TGCTAAGGACAACGTTATCGG.
2.11. Data Acquisition and Pre-processing
Following the immunoreaction, the microarrays were scanned in an Axon Laboratories 4100 scanner (Palo Alto, CA) using 532 and 647 nm lasers to produce a red (Alexa Fluor 647) and green (Alexa Fluor 532) composite image. Using the ImaGene 6.0 (Biodiscovery) image analysis software, the binding of each sarcoid specific peptide with IgGs in each serum was then analyzed and expressed as a ratio of red-to-green fluorescent intensities. The microarray data were further read into the R environment v2.3.0 (Team RDC, 2004) and processed by a sequence of pre-processing, including background correction, omission of poor quality spots and log2 transformations. Within array loess normalization was performed for each spot and summarized by median of triplicates and followed by between array quantile normalization.
2.12. Statistical Analysis
We performed microarray analysis using sera from sarcoid patients and healthy controls in two independent sets of experiments. Technical and biological sources of variation were expected in the design of experiment. As opposed to pooling all datasets, one powerful and robust method is to integrate results from individual datasets. We expected to obtain a higher confidence list of markers than by using individual datasets. To detect differentially expressed antigens between sarcoidosis samples and healthy controls, an integrative analysis of two datasets was performed. Limma's empirical Bayes moderated t-test identified fold-changes in expression of antigens that differed significantly between sarcoidosis and controls for each dataset separately. Then an integrative-analysis method, an adaptively-weighted method with one-sided correction (AW-OC) (Li and Tseng, 2011), was performed to combine the statistics from both datasets. The integrative method was designed to test whether an antigen is consistently over- or under-expressed in sarcoidosis subjects in both datasets. False discovery rate (FDR) was estimated using the Benjamini–Hochberg method (Benjamini and Hochberg, 1995).
To identify a panel of markers that classify sarcoidosis samples and controls, we used a strategy of univariate marker selection followed by multivariate modeling. The top antigens differentially expressed in the two groups were selected using the above described AW-OC approach. We used the top genes that were consistently up- or down-regulated in both datasets. The top markers were then required by the supervised classification models to achieve the most sensitivity and specificity in differentiating sarcoid and controls. The multivariate classification models chosen for this study were K-nearest neighbors (KNN) and support vector machine (SVM). The cross-validation technique was used to prevent the overfitting of data analysis due to a large number of antigens used to discriminate between sarcoid and control subjects. The study was performed in two nested 10-fold cross-validation loops, an inner loop to select the optimal number of antigens and an outer loop to measure the optimized model performance with estimation of the area under the receiver operating characteristic (AUROC) sensitivity and specificity. The receiver operating characteristic curves were estimated through 10-fold cross-validation. A moderated t-test was carried out to identify the significant clones between healthy controls, sarcoidosis and tuberculosis.
3. Results
3.1. Generation of cDNA Libraries Representative of Sarcoidosis Antigens
Both PBMCs and alveolar macrophages (AMs) play an important role in initiation of sarcoidosis granuloma (Rastogi et al., 2011). It has been shown that extracts from sarcoidosis BAL cells and peripheral blood monocytes (PBMCs) are able to initiate a Kveim-like reaction (Siltzbach and Ehrlich, 1954, Holter et al., 1992). Therefore, we used total BAL cells and WBCs from patients with biopsy proven sarcoidosis to develop a cDNA antigen library. We used BAL cells and WBC as sources of antigens in order to increase the diversity of sarcoidosis antigens. To increase the chance of identifying sarcoidosis antigen(s), we isolated RNA from BAL samples obtained from 20 patients with active sarcoidosis to generate the BAL cDNA library. The patients' characteristics are shown in Table 1 (left panel). The LeukoLock system was used to isolate RNA from total WBC obtained from a different cohort of 36 sarcoidosis subjects to build the WBC cDNA library. The patients' characteristics are shown in Table 1 (right panel). Two other sources of cDNA, one from cultured human splenic monocytes (EL-1) and another from lung embryonic fibroblasts (MRC5) were used to generate two additional libraries. These sources were added to increase the chance of discovering potential sarcoidosis antigens. Each cDNA underwent two cycles of PolyA selection to minimize ribosomal contamination. These four libraries were developed as described in the Materials and Methods section. Each library was cloned using modified linkers; ECOR1/HindIII was used for BAL cDNA, ALA for WBC cDNA, LEU for MARC5 cDNA and THR for EL1 cDNA (S1). The use of these linkers enabled us to identify the original library for each antigen.
Table 1.
Subject demographics, chest X-ray stages, and organ involvements.
| BAL derived RNA | Leukocyte derived RNA | ||
|---|---|---|---|
| Age (Mean ± SEM) | 30 ± 8 | Age (Mean ± SEM) | 36 ± 11.2 |
| BMI (Mean ± SEM) | 27.7 ± 8.7 | BMI (Mean ± SEM) | 31 ± 5.4 |
| Gender, N (%) | Gender, N (%) | ||
| Male | 7 (33) | Male | 12 (33) |
| Female | 13 (67) | Female | 24 (67) |
| Race, N (%) | Race, N (%) | ||
| African American | 17 (87) | African American | 32 (88) |
| Caucasian | 3 (13) | Caucasian | 4 (12) |
| CXR stage, N (%) | CXR Stage, N (%) | ||
| 1 | 2 (6) | 1 | 1 (3) |
| 2 | 14 (67) | 2 | 13 (41) |
| 3 | 4 (27) | 3 | 12 (37) |
| 4 | 0 | 4 | 6 (19) |
| Lung | 18 | Lung | 33 |
| Extrapulmonary | 16 | Extrapulmonary | 31 |
| Neuro-ophthalmologic | 6 | Neuro-ophthalmologic | 11 |
| Skin | 6 | Skin | 13 |
| Liver | 2 | Liver | 4 |
| Heart | 1 | Heart | 2 |
| Prednisone | 1 | Prednisone | 3 |
| IMD | 0 | IMD | 14 |
| Smoking | Smoking | ||
| None | 12 | None | 26 |
Age, BMI and disease duration values are presented as means and variability in SD or range where indicated. N = number of patients and percent is shown in parentheses. IMD = Immunomodulatory drugs.
3.2. Differential Biopanning of Sarcoidosis Phage cDNA Display Libraries
The four phage cDNA display libraries (BAL, WBC, EL-1 and MARC5) were combined to generate a complex sarcoidosis library (CSL). To isolate a large panel of antigens, differential biopanning of the T7 phage cDNA display library was performed on the combined complex sarcoid library. A negative biopanning selection was done using 10 pooled sera from healthy controls to remove non-specific IgG, while 2 sarcoidosis sera were used for positive selective enrichment. One serum was obtained from a woman (P51) with systemic sarcoidosis who had uveitis and another serum was collected from a male subject (P197) who had active systemic sarcoidosis with renal involvement. Both patients had pulmonary involvements. Each clone was derived either from P51 or from P197. The titer of the complex library was assessed (Supplemental Fig. 2A) and individual phage clones were amplified by PCR (Supplemental Fig. 2B).
3.3. High-throughput Protein Microarray Immunoreaction to Select Sarcoidosis Specific Antigens
A total of 1152 potential antigens were randomly selected from the two highly enriched pools of T7 phage cDNA libraries (Fig. 1). These antigens were robotically spotted on nitrocellulose Fast slides and were hybridized with sera of sarcoidosis patients or healthy controls. The binding of each of the arrayed potential sarcoidosis-specific peptides with antibodies in sera was quantified with Alexa Fluor 647 (red-fluorescent dye)-labeled goat anti-human antibody. The amount of phage particles at each spot throughout the microarray was detected using a mouse monoclonal antibody to the T7 capsid protein and quantified using Alexa Fluor 532 (green-fluorescent dye)-labeled goat anti-mouse antibody (Fig. 1). To correct for any small variation in the amount of antibody binding in each spot that may be due to different amounts of phage spotted on the microarray, the ratio of intensity of Alexa Fluor 647 over Alexa Fluor 532 was calculated for each spot. Following immunoreaction, the microarray data were processed by a sequence of transformations and then analyzed. The intra-assay reproducibility was assessed by comparing the results among five replicates printed within the same chip for each clone.
Fig. 1.
Schematic diagram of discovery of sarcoidosis antigens. The process of combining phage-display technology, protein microarray and bioinformatic tools to select a panel of novel clones for the diagnosis of sarcoidosis. A cDNA library was constructed from a pool of total RNA isolated from 20 BAL samples and 36 WBC samples from sarcoid patients, and then combined with RNA extracts from cultured human monocytes and human embryonic fibroblasts. After digestion, the cDNA library was inserted into the T7 phage vector and packaged into T7 phages to generate a sarcoid cDNA–phage-display library. Several rounds of biopannings of the library were performed with pooled control sera for negative selection, and with sarcoid sera for positive enrichments. After four rounds of biopanning, enriched sarcoid specific peptide clones were cultured onto LB agar plates. A total of 1152 single colonies, including positive and negative clones, were randomly picked and propagated into 96-well plates. Phage–clone lysates were then printed robotically onto coated glass slides to create a sarcoid–phage–protein microarray. Cy5 (red fluorescent dye)-labeled antihuman antibody was used to detect IgGs in human serum that were reactive to peptide clones, and a Cy3 (green fluorescent dye)-labeled antibody was used to detect the phage capsid protein in order to normalize for spotting. Thus, if a phage clone carries a peptide that is non-reactive to human IgG, the spot will remain green suggesting an unreactive clone, whereas clones reactive to human IgG give raise to a red signal. A total of 115 sarcoid sera, 64 healthy control sera and 17 TB sera were tested on the 1152 phage peptide microarray. Bioinformatically analyzed data identified 259 antigens with the highest level of differentiation between sarcoidosis and healthy controls.
3.4. Selection of a Panel of Antigens and Estimation of Neural Network Classifier Performance in Sarcoidosis
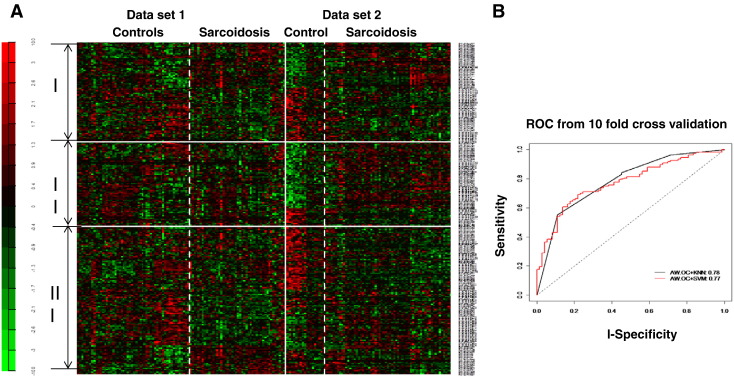
A novel aspect of our work was the integration of data from two independent trials of printing allowing the development of two datasets obtained from two independent cohorts of sarcoidosis patients and healthy controls utilized for hybridization. To generate the first dataset, sera from 54 sarcoidosis subjects and 45 healthy controls were immune-screened against 1152 sarcoidosis specific peptides. In the second dataset, sera from 19 healthy controls and 61 sarcoidosis subjects were immune-screened using the same platform of clones. Sera used in both datasets for hybridization had not been previously used for biopanning or selection of clones. Table 2 shows the clinical characteristics of sarcoidosis and healthy control subjects. Within array loess normalization was performed for each spot and summarized by median of triplicates and followed by between array quantile normalization. After preprocessing, 1101 antigens common in both datasets were used for further analysis. Univariate and multivariate analyses were performed. Limma's empirical Bayes moderated t-test was used to identify fold-changes in expression of antigens that differed significantly between sarcoidosis and controls for each dataset separately. Then both datasets were combined using an integrative-analysis method, an adaptively-weighted method with one-sided correction (AW-OC) (Li and Tseng, 2011). Out of the 1101 potential antigens, 259 showed a strong differentiation between sarcoidosis and healthy control subjects with adjusted p value (q value) of < 0.05 and FDR (false discovery rate) of < 0.05. Fig. 2A shows the heatmap of the 259 significant antigens that were differentially expressed in both datasets. Seventy eight markers out of 259 were consistently over- or under-expressed in sarcoidosis subjects. Fig. 2B shows the AUROC for this classifier. KNN method performed slightly better than SVM. Using the highly significant 32 antigens selected by AW-OC and KNN methods to classify sarcoidosis and healthy controls (AW-OC + KNN), the area under the curve (AUROC) was 0.78, with a sensitivity of 89% and a specificity of 83% estimated after 10-fold cross-validation (Fig. 2B).
Table 2.
Patient characteristics.
| Control subjects | TB subjects | ||
|---|---|---|---|
| Age | 29.7 ± 13.4 y | 33 ± 7.4 | 23 ± 14 |
| BMI | 29 ± 10.4 | 28 ± 3.6 | 21 ± 7.9 |
| Gender, N | |||
| Female | 87 (75) | 48 (75) | 7 (42) |
| Male | 28 (25) | 16 (25) | 10 (58) |
| Race, N | |||
| African American | 107 (89) | 44 (69) | 6 (36) |
| Caucasian | 8 (11) | 20 (31) | 11 (64) |
| CXR stage, N | |||
| 0 | 3 (2) | NA | NA |
| 1 | 18 (15) | NA | NA |
| 2 | 49 (43) | NA | NA |
| 3 | 45 (39) | NA | NA |
| Organ involvements, | |||
| Neuro-ophthalmologic | 33 (28) | NA | – |
| Lung | 109 (94) | NA | 17/17 |
| Skin | 50 (45) | NA | – |
| Multiorgan | 70 (52) | NA | – |
| PPDa | Negative | NA | – |
| TB cultureb | Negative | NA | Positive |
Some patients had multiple organ involvements.
NA = not applicable.
PPD = Mantoux test (purified protein derivative).
TB cultures obtained from BAL, tissue or sputum.
Fig. 2.
A) Heatmap generated by applying meta-analysis using microarray analysis of 2 separate datasets derived from 115 sarcoidosis patients and 64 healthy controls. Data reflecting 259 antigens expressed significantly differently between healthy controls and sarcoidosis subjects in immunoscreening using sera. The 259 antigens are further divided into three categories according to the AW-OC method. I: 78 antigens are consistently over- or under-expressed in sarcoidosis in both datasets; II: 115 antigens are over- or under-expressed in sarcoidosis in the second dataset only; III: 66 antigens are over- or under-expressed in sarcoidosis in the first dataset only. B) Receiver operating characteristics (ROC) curve demonstrating the performance of 32 classifiers to discriminate between healthy controls and sarcoidosis subjects.
3.5. Characterization of 10 Most Significant Sarcoidosis Antigens
Based on the results of AW-OC integrative-analysis, we identified the top 10 high performance antigens that predict sarcoidosis. To further characterize the performance of each clone, we next calculated the AUROC, and sensitivity and specificity given the optimal cutoff of the clones. Fig. 3 depicts the ROC curves for individual sarcoid antigens and their adjusted p value (q value). As shown, each antigen has a different specificity and sensitivity as well as ROC to predict the presence of sarcoidosis. ROC for these antigens ranged from the highest of 0.84 to the lowest of 0.7. Nine of 10 antigens were clearly increased in sarcoidosis versus healthy controls. To further characterize the identified antigens, we have sequenced these 10 highest ranked clones. After obtaining the sequences of clones, the Expasy program was used to translate the cDNA sequences to protein sequences. Protein blast using Blastn and tblastn algorithms of the BLAST program were applied to identify the highest homology to our identified proteins or peptides. Additionally, we compared these results with corresponding nucleotide sequences using nucleotide blast. We also determined the predicted amino acid in frame with phage T7 gene 10 capsid proteins. Five antigens (RNAPII, SAMDHI, DNAJC1, TPT1 and SH3YL1) among the top 10 clones fit the definition of an epitope containing known gene products in the reading frame of known genes (Wang et al., 2005). The other five contained peptides coded by the inserted gene fragments leading to out of frame peptides, which meets the definition of mimotopes (Wang et al., 2005). Full length of proteins and genes of 10 sarcoidosis clones are shown in Supplementary Table 1. As sarcoidosis sera reacted to these out of frame peptides, it is likely that these clones represent sarcoidosis antigens produced as a result of altered reading frames or alternative splicing. Interestingly, when a similar technique was applied to discovery of cancer antigens, numerous out of frame peptides were discovered (Lin et al., 2007, Wang et al., 2005). Table 3 shows the 10 most significant sarcoidosis antigens, gene names and q-values.
Fig. 3.
ROC for top 10 sarcoidosis clones.
Table 3.
Top 10 most significant sarcoidosis clones, clone name, gene names, adjusted p values (q-values), AUC, sensitivity and specificity, confidence interval (CI).
| Clone | Increased in sarcoidosis vs healthy controls | Gene name | q value | AUC | Sensitivity %, 95% CI | Specificity %, 95% CI |
|---|---|---|---|---|---|---|
| P51_BP3_287 (MRC5) | Small inducible cytokine A21 precursor | CCL21 | 1.9 × 10− 20 | 0.84 | 78 | 82 |
| P51_BP3_281 (BAL) | Methionine aminopeptidase 1 | Metap 1 | 1.0 × 10− 20 | 0.78 | 70 | 82 |
| P51_BP4_388 (EL-1) | Activated RNA polymerase II transcription cofactor variant 4 | RNAP II | 0.00045 | 0.75 | 70 | 74 |
| P51_BP4_596 (WBC) | RNA methyltransferase | CLI_3190 | 0.00045 | 0.72 | 72 | 74 |
| P51_BP4_566 (WBC) | Tumor necrosis factor receptor superfamily member 21 precursor. Also known as death receptor 6 (DR6) | TNFRSF21 | 0.0009 | 0.74 | 70 | 71 |
| P51_BP3_283 (WBC) | Monocyte differentiation antigen CD14 | CD14 | 0.0009 | 0.74 | 68 | 65 |
| P51_BP3_47 (EL-1) | DnaJ (Hsp40) homologue subfamily C member 1 precursor | DNAJC1 | 0.002 | 0.72 | 60 | 82 |
| P197_BP4_885 (BAL) | Amyloid β A4 precursor protein-binding family B member 1-interacting protein | APBB1 | 0.007 | 0.79 | 75 | 82 |
| P51_BP4_577 (BAL) | Fibroblast growth factor binding protein 2 precursor | FGFBP-2 | 0.009 | 0.70 | 64 | 68 |
| Clone | Decreased in sarcoidosis vs healthy controls | |||||
| P197_BP4_755 (BAL) | SH3 domain-containing YSC84 like protein 1 | SH3YL1 | 1.0 × 10− 20 | 0.77 | 65 | 82 |
3.6. Complex Sarcoidosis Library Detects Unique Antigens in the Sera of Tuberculosis Patients
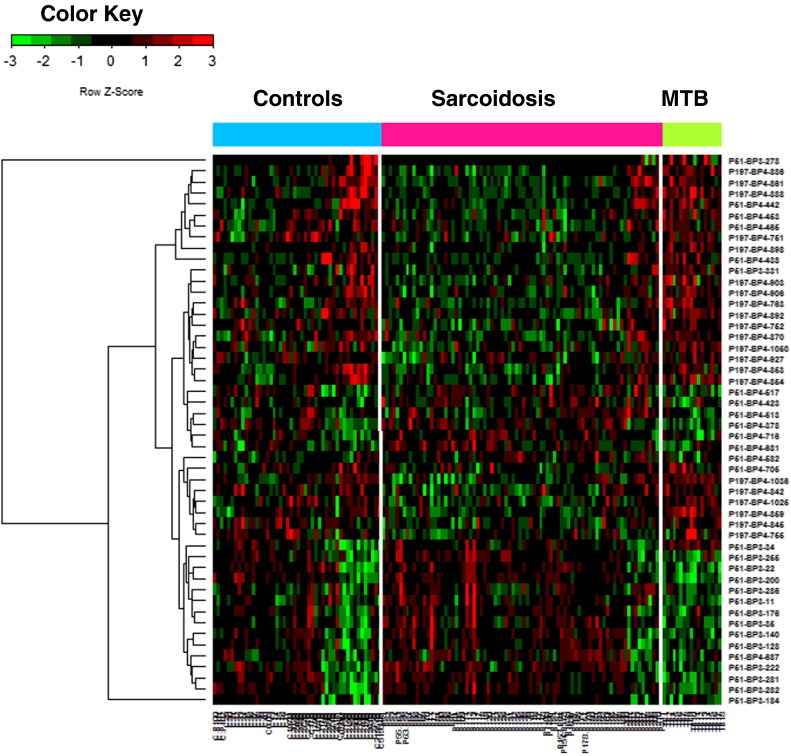
In view of the clinical and pathological similarities between MTB and sarcoidosis, a most useful clinical antigen(s) should discriminate between these two conditions. To this end, using the antigens identified by biopanning the CSL library, a microarray was constructed and then this construct was interrogated with sera from 17 culture positive MTB subjects. Using a moderate t-test and a q value of < 0.05 in this system, we identified 238 clones differentially expressed between TB and healthy controls and 380 clones differentially expressed between TB and sarcoidosis. Fig. 4 shows a Venn diagram depicting the overlap between 259 sarcoidosis markers, 238 TB vs. control and 380 TB vs. sarcoidosis markers. Clearly, 47 clones differentiate both sarcoidosis and TB from healthy controls, while 5 of them cannot differentiate sarcoidosis from TB. From these clones, 164 were found to be TB specific, and different from both healthy controls and sarcoidosis clones. Fig. 5 shows the heatmap of 50 significant clones differentially expressed in all three groups. Similarly to the sarcoidosis antigens, we have analyzed the specificity and sensitivity of TB clones to predict the presence of TB (Table 4). Finally, we sequenced 10 TB antigens, and sequence homologies were searched using the same algorithm as previously described. Table 4 shows the 10 TB-specific antigens as compared to healthy controls as well as sarcoidosis. After sequence analysis and homology search, we found one identical sequence between TB and sarcoidosis clone. Although the identified clone's name was different: P51_BP3_287 versus P51_BP3_174, and they performed differently in sarcoidosis versus TB as indicated in q value (compare Table 3, Table 4). However, using NCBI blast databases (mycobacterium toxoid and the universal blast) on the same sequence, two different proteins could be identified. Supplementary Table 2 shows the full length of protein and genes of 10 TB antigens. Surprisingly, TB clones show much higher sensitivity and specificity; similarly the AUROC was larger for the majority of TB antigens (Table 4).
Fig. 4.
Venn diagram depicts differential phage clone significances among sarcoidosis, TB and healthy controls (q < 0.01). Venn diagram shows the overlap between 259 sarcoidosis clones and 238 TB clones as compared to healthy controls, as well as 380 TB clones versus sarcoidosis. Forty seven clones could differentiate both sarcoidosis and TB from healthy controls. Five clones could not discriminate between TB and sarcoidosis.
Fig. 5.
Heatmap generated from the microarray analysis using 3 datasets derived from 115 sarcoidosis patients, 64 control subjects and 17 TB patients. Fifty antigens showed significant differential expression among the three groups.
Table 4.
Top 10 most significant tuberculosis clones, clone name, gene names, adjusted p values (q-values), AUC, sensitivity and specificity, confidence interval (CI).
| Clone | Increased in TB vs sarcoidosis subjects | Gene name | q value | AUC | Sensitivity %, 95% CI | Specificity %, 95% CI |
|---|---|---|---|---|---|---|
| P51_BP3_174 (MRC5) | Ferredoxin (Mycobacterium tuberculosis) | Fed A | 4.9 × 10− 15 | 0.87 | 88 | 83 |
| P51_BP4_610 (BAL) | WDFY3 protein (Homo sapiens) | WDFY3 | 4.1 × 10− 12 | 0.92 | 88 | 84 |
| P51_BP3_266 (EL-1) | Membrane protein (Mycobacterium tuberculosis) | MFS | 6.7 × 10− 10 | 0.9 | 82 | 93 |
| P51_BP3-166 (BAL) | Leucine rich PPR-motif containing protein (Homo sapiens) | LRPPRC | 1.3 × 10− 9 | 0.81 | 71 | 90 |
| P51_BP4_704 (BAL) | HLA-DR alpha (Homo sapiens) | HLA-DR | 1.1 × 10− 8 | 0.89 | 94 | 83 |
| P197_BP4_763 (BAL) | Transketolase (Mycobacterium tuberculosis) | TKT | 2.7 × 10− 6 | 0.86 | 82 | 76 |
| P51-BP4_563 (BAL) | Dihydroxy acid dehydratase (Mycobacterium tuberculosis) | Rv0189C | 1.04 × 10− 6 | 0.85 | 76 | 86 |
| Clone | Decreased in TB vs sarcoidosis subjects | |||||
| P51_BP3_113 (BAL) | Chain A Mycobacterium tuberculosis | BfrA | 1.2 × 10− 10 | 0.9 | 88 | 85 |
| P51_BP3_200 (BAL) | Disabled homologue 2 isoform 2 (Homo sapiens) | DAB2 | 1.5 × 10− 9 | 0.92 | 82 | 91 |
| P51_BP4_622 (BAL) | Transcription elongation factor B polypeptide 2 isoform (Homo sapiens) | TCEB2 | 6.9 × 10− 7 | 0.89 | 82 | 89 |
4. Discussion
Our work was inspired by the classic observation that the intradermal injection of a suspension of granulomatous splenic tissue (Kveim–Siltzbach test) induces granuloma formation weeks later in patients with sarcoidosis, suggesting the presence of antigen(s) in granuloma tissue and host immunoreactivity to those antigen(s) (Iannuzzi et al., 2007, Hajizadeh et al., 2007, Hiramatsu et al., 2003, Dubaniewicz et al., 2006). Kveim-like effects have also been observed using non-viable BAL cell extracts or PBMCs derived from sarcoidosis subjects (Munro and Mitchell, 1987, Siltzbach and Ehrlich, 1954, Holter et al., 1992, Kataria and Holter, 1996). Several studies have attempted to identify specific antigens that can discriminate sarcoidosis from healthy subjects or from patients with other granulomatous diseases such as TB (Hajizadeh et al., 2007, Chen and Moller, 2007). Most of these studies used limited proteomics or genomics to search for tissue antigens (Hajizadeh et al., 2007, Richter et al., 1999, Song et al., 2005).
We tested the hypothesis that antigen presenting cells, especially AMs might harbor elusive sarcoidosis antigen (s) in their phagosomes. By applying novel high throughput technology, here, we attempted to overcome the current gap by constructing phage–protein microarrays in which peptides derived from a unique sarcoidosis cDNA library were expressed as a phage fusion protein. The phage–protein microarrays were screened to identify phage–peptide clones that bind antibodies in serum samples from patients with sarcoidosis but not in those from controls. Importantly, we immune-screened the same microarray constructs using sera of culture positive TB patients.
The average length of identified peptides for sarcoidosis antigens was between 9 and 130 amino acids (AA), while the average peptide length for TB antigens was 9–209 AA. Among 10 sarcoidosis specific phage peptides, we identified 5 expression sequence tags with in frame epitopes. Five other reactive antigens were relatively short out of frame peptides meeting the criteria to be considered as mimotopes (mimetic sequence of a true epitope) (Wang et al., 2005). Similarly, among 10 sequenced TB specific phage peptides, we identified 5 in frame epitopes with full length in frame proteins with homology to known human sequences. Five other sequences were relatively short peptides with homology to various known MTB proteins (Table 4). Interestingly, TB antigens had much higher specificity and sensitivity as compared to antigens selective to sarcoidosis as indicated by higher AUCs (Table 4). Although the significance of mimotopes is not clear, it has been shown that some out of frame peptides are immunogenic and can activate MHC class I molecules (Schirmbeck et al., 2005). Due to smaller peptide sequences of mimotopes, they may have homology with diverse proteins. Prior studies using similar techniques in various cancers had similarly identified out of frame peptides (Lin et al., 2007, Chatterjee et al., 2006, Wang et al., 2005). Detection of mimotopes in this method may be due to out of frame peptide synthesis secondary to altered ribosomal function, or may correspond to open reading frames or generation of displayed peptides due to competition for binding during phage selection during phage insertion.
Although our primary goal in this study was to identify the immune signature in sarcoidosis, we have identified a panel of antigens differentially expressed in sarcoidosis and tuberculosis as compared to healthy subjects. Table 3, Table 4 summarize the 10 most significant clones we identified in sarcoidosis and tuberculosis respectively. Further studies with larger sample sizes need to verify current data.
In recent years several groups have attempted to identify specific signatures to distinguish between tuberculosis and sarcoidosis using transcriptomics or gene expression profilings (Koth et al., 2011, Maertzdorf et al., 2012, Berry et al., 2010). Yet, most of these methods led to the discovery of a series of markers or expression signatures that failed to discriminate between these two diseases (Koth et al., 2011, Stone et al., 2013). This is partly due to the fact that several inflammatory or infectious diseases such as CD, lupus, sarcoidosis and tuberculosis may respond to various antigens with activation of similar pathways leading to similar transcriptomes and/or inflammatory gene expression profiles (Koth et al., 2011, Maertzdorf et al., 2012, Berry et al., 2010). For instance, Maertzdorf et al. found more similarity in the activated pathways than differences between sarcoidosis and MTB (Maertzdorf et al., 2012). Their results in sarcoidosis were similar to those results by Berry et al. indicating the importance of the interferon pathway (IFN) signature in MTB (Maertzdorf et al., 2012, Berry et al., 2010). In addition, considerable pathway overlap was identified between lupus, sarcoidosis and TB (Maertzdorf et al., 2012). However, despite similar genetic or transcriptomic signatures, these diseases are clinically entirely different and require different therapy. Tuberculosis, a global infectious disease caused by the intracellular bacterium Mycobacterium tuberculosis remains a worldwide health problem (http://www.who.int). One barrier for eradication of tuberculosis besides the lack of effective vaccination is the lack of reliable antigens to evaluate the activity of the disease and its response to treatment (Nahid et al., 2011). Standard methods to diagnose TB and to monitor response to treatment rely on sputum microscopy and culture. The current CDC/NIH roadmap emphasizes the need for development of new TB antigens as alternative methods (Nahid et al., 2011). In view of this background, perhaps surprisingly, our microarray platform could discriminate tuberculosis from sarcoidosis and healthy controls. In addition to antigens for sarcoidosis, we have detected more than 300 clones specifically for tuberculosis. Interestingly, a considerable number of these clones were TB specific and related to bacterial growth of M. tuberculosis, and its metabolism (Table 4). Recently a tremendous effort has been put toward elucidating the antibody response to MTB antigens, which has implications for the development of new antigens to diagnose and monitor successful treatment, as well as to develop effective vaccination (Kunnath-Velayudhan et al., 2010). Yet, a consistent immune response to MTB has not been found. Most other studies searching for antigens in TB have identified unspecific markers primarily involving host response such as C-reactive protein, serum amyloid A and others, but not MTB specific antigens (Agranoff et al., 2006, De Groote et al., 2013). MTB has the ability to survive within host macrophages, largely escaping immune surveillance and maintaining its ability for replication and person to person transmission (Meena and Rajni, 2010). The primary goal of our project was to discover antigens related to sarcoidosis. Yet, in addition we have detected specific antigens for TB. These results are surprising to us, as the question remains, how can the sarcoidosis library detect TB specific antigens? Lungs are environmentally highly exposed to numerous bacteria, and our library is predominantly derived from BAL cells that contain all types of immune cells, including macrophages that might have integrated messages from MTB. Hence, we speculate that this could be the reason why the CSL is able to detect TB specific antigens. Still, the major question is why BAL cells of patients with sarcoidosis can harbor MTB messages, yet respond to PPD skin testing negatively, since all of our donors with sarcoidosis were PPD negative.
Similar to gene-expression profiling and the pattern-recognition approaches utilizing serum proteomics, our method may have the limitations of background signals, and sample-selection bias. To minimize these problems, we have applied an integrative-analysis method, an adaptively-weighted statistical method on two sets of data acquired in two independent experiments. The discriminatory power of antibody signatures was validated by analyzing data from two completely different cohorts of patients.
In summary, we have developed a novel T7 phage display library derived from macrophages from BAL, monocytes from blood leukocytes of patients with sarcoidosis that may display a significant segment of the universe of potential sarcoidosis and MTB antigens that can be specially recognized by high IgG antibodies in sarcoidosis and MTB sera. Our results support the hypothesis that sarcoidosis sera can recognize antigens presented in sarcoidosis materials. Current study of the antibody response can advance how proteomics can be used to harness immunity to identify and treat diseases, because it investigates antibody–antigen interactions and also evaluates the effects on antibody responses of pathogen and host characteristics. Further studies with a larger cohort group of patients and/or longitudinal studies are needed to investigate the role of these antigens in sarcoidosis and its organ involvement.
Contributors
Harvinder Talwar contributed to the study design, sample processing, conducted the analysis, and interpreted the data. Rita Rosati developed the T7 phage library and participated in the study design. Jia Li contributed to data preprocessing and statistical analysis. Dana Kissner provided access to patients with TB. Samiran Ghosh contributed to data processing and statistical analysis. Félix Fernández-Madrid contributed to study design and data analysis. Lobelia Samavati conceived and designed the study, participated in all areas of the research such as patient selection and oversaw patient enrollment, data analysis and writing of the manuscript.
Conflicts of Interest
The authors declare that no conflict of interest exists. Wayne State University has filed for comprehensive world-wide patent protection related to this technology through the United States Patent and Trademark Office (USPTO).
Acknowledgments
We thank all patients and healthy volunteers for their participation in this study. This project has been funded by NIH grant R21HL104481-01A1 awarded to L.S. and with the support of the Department of Medicine, Wayne State University. We would like to thank Dr. Michael A. Tainsky for his invaluable assistance in completing this study.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.03.007.
Appendix A. Supplementary Data
Supplementary Fig. 1. Modified linkers distinguishing between origins of each library. Each cDNA library was tagged with a modified linker: ECOR1/HindIII was used for BAL cDNA, ALA for WBC cDNA, LEU for MARC5 cDNA and THR for EL1 cDNA.
Supplementary Fig. 2A. Graphical representation of the output eluent phage titers as a function of biopanning (BP) showing exponential enrichment of the output eluent phage titers after the completion of each cycle of biopannings.
Supplementary Fig. 2B. PCR amplification of the phage clones picked up from biopannings 3 & 4 (BP3 & BP4) showing retention of diversity in the pool of immunoreactive phage.
Supplementary Table 1. Full length of sequence analysis of the top 10 sarcoid phage clones using NCBI BLAST.
Supplementary Table 2. Full length of sequence analysis of the top 10 TB phage clones using NCBI BLAST.
References
- Agranoff D., Fernandez-Reyes D., Papadopoulos M.C. Identification of diagnostic markers for tuberculosis by proteomic fingerprinting of serum. Lancet. 2006;368(9540):1012–1021. doi: 10.1016/S0140-6736(06)69342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple hypothesis testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Berry M.P., Graham C.M., McNab F.W. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bons J.A., Drent M., Bouwman F.G., Mariman E.C., van Dieijen-Visser M.P., Wodzig W.K. Potential biomarkers for diagnosis of sarcoidosis using proteomics in serum. Respir. Med. 2007;101(8):1687–1695. doi: 10.1016/j.rmed.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Chatterjee M., Mohapatra S., Ionan A. Diagnostic markers of ovarian cancer by high-throughput antigen cloning and detection on arrays. Cancer Res. 2006;66(2):1181–1190. doi: 10.1158/0008-5472.CAN-04-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.S., Moller D.R. Expression profiling in granulomatous lung disease. Proc. Am. Thorac. Soc. 2007;4(1):101–107. doi: 10.1513/pats.200607-140JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.S., Wahlstrom J., Song Z. T cell responses to mycobacterial catalase-peroxidase profile a pathogenic antigen in systemic sarcoidosis. J. Immunol. 2008;181(12):8784–8796. doi: 10.4049/jimmunol.181.12.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.S., Song Z., Willett M.H. Serum amyloid A regulates granulomatous inflammation in sarcoidosis through Toll-like receptor-2. Am. J. Respir. Crit. Care Med. 2010;181(4):360–373. doi: 10.1164/rccm.200905-0696OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costabel U. Sarcoidosis: clinical update. Eur. Respir. J. Suppl. 2001;32:56s–68s. [PubMed] [Google Scholar]
- Costabel U., Hunninghake G.W. ATS/ERS/WASOG statement on sarcoidosis. Sarcoidosis Statement Committee. American Thoracic Society. European Respiratory Society. World Association for Sarcoidosis and Other Granulomatous Disorders. Eur. Respir. J. 1999;14(4):735–737. doi: 10.1034/j.1399-3003.1999.14d02.x. [DOI] [PubMed] [Google Scholar]
- De Groote M.A., Nahid P., Jarlsberg L. Elucidating novel serum biomarkers associated with pulmonary tuberculosis treatment. PLoS One. 2013;8(4):e61002. doi: 10.1371/journal.pone.0061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubaniewicz A. Mycobacterium tuberculosis heat shock proteins and autoimmunity in sarcoidosis. Autoimmun. Rev. 2010;9(6):419–424. doi: 10.1016/j.autrev.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Dubaniewicz A., Dubaniewicz-Wybieralska M., Sternau A. Mycobacterium tuberculosis complex and mycobacterial heat shock proteins in lymph node tissue from patients with pulmonary sarcoidosis. J. Clin. Microbiol. 2006;44(9):3448–3451. doi: 10.1128/JCM.01433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eishi Y., Suga M., Ishige I. Quantitative analysis of mycobacterial and propionibacterial DNA in lymph nodes of Japanese and European patients with sarcoidosis. J. Clin. Microbiol. 2002;40(1):198–204. doi: 10.1128/JCM.40.1.198-204.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Madrid F., VandeVord P.J., Yang X. Antinuclear antibodies as potential markers of lung cancer. Clin. Cancer Res. 1999;5(6):1393–1400. [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Madrid F., Tang N., Alansari H. Autoantibodies to annexin XI-A and other autoantigens in the diagnosis of breast cancer. Cancer Res. 2004;64(15):5089–5096. doi: 10.1158/0008-5472.CAN-03-0932. [DOI] [PubMed] [Google Scholar]
- Fox J.L., Berman B., Teirstein A.S., France D.S., Reed M.L. Quantitation of cutaneous Langerhans cells of sarcoidosis patients. J. Invest. Dermatol. 1983;80(6):472–475. doi: 10.1111/1523-1747.ep12534905. [DOI] [PubMed] [Google Scholar]
- Glatt S.J., Chandler S.D., Bousman C.A. Alternatively spliced genes as biomarkers for schizophrenia, bipolar disorder and psychosis: a blood-based spliceome-profiling exploratory study. Curr. Pharmacogenomics Person. Med. 2009;7(3):164–188. doi: 10.2174/1875692110907030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D., Agarwal R., Aggarwal A.N., Jindal S.K. Molecular evidence for the role of mycobacteria in sarcoidosis: a meta-analysis. Eur. Respir. J. 2007;30(3):508–516. doi: 10.1183/09031936.00002607. [DOI] [PubMed] [Google Scholar]
- Hajizadeh R., Sato H., Carlisle J. Mycobacterium tuberculosis antigen 85A induces Th-1 immune responses in systemic sarcoidosis. J. Clin. Immunol. 2007;27(4):445–454. doi: 10.1007/s10875-007-9080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu J., Kataoka M., Nakata Y. Propionibacterium acnes DNA detected in bronchoalveolar lavage cells from patients with sarcoidosis. Sarcoidosis Vasc. Diffuse Lung Dis. 2003;20(3):197–203. [PubMed] [Google Scholar]
- Holter J.F., Park H.K., Sjoerdsma K.W., Kataria Y.P. Nonviable autologous bronchoalveolar lavage cell preparations induce intradermal epithelioid cell granulomas in sarcoidosis patients. Am. Rev. Respir. Dis. 1992;145(4 Pt 1):864–871. doi: 10.1164/ajrccm/145.4_Pt_1.864. [DOI] [PubMed] [Google Scholar]
- Hunninghake G.W., Costabel U., Ando M. ATS/ERS/WASOG statement on sarcoidosis. American Thoracic Society/European Respiratory Society/World Association of Sarcoidosis and other Granulomatous Disorders. Sarcoidosis Vasc. Diffuse Lung Dis. 1999;16(2):149–173. [PubMed] [Google Scholar]
- Iannuzzi M.C., Rybicki B.A., Teirstein A.S. Sarcoidosis. N. Engl. J. Med. 2007;357(21):2153–2165. doi: 10.1056/NEJMra071714. [DOI] [PubMed] [Google Scholar]
- Kataria Y.P., Holter J.F. Sarcoidosis: a model of granulomatous inflammation of unknown etiology associated with a hyperactive immune system. Methods. 1996;9(2):268–294. doi: 10.1006/meth.1996.0033. [DOI] [PubMed] [Google Scholar]
- Koth L.L., Solberg O.D., Peng J.C., Bhakta N.R., Nguyen C.P., Woodruff P.G. Sarcoidosis blood transcriptome reflects lung inflammation and overlaps with tuberculosis. Am. J. Respir. Crit. Care Med. 2011;184(10):1153–1163. doi: 10.1164/rccm.201106-1143OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnath-Velayudhan S., Salamon H., Wang H.Y. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc. Natl. Acad. Sci. U. S. A. 2010;107(33):14703–14708. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Tseng G.C. An adaptively weighted statistic for detecting differential gene expression when combining multiple transcriptomic studies. Ann. Appl. Stat. 2011;5(2A):994–1019. [Google Scholar]
- Lin H.S., Talwar H.S., Tarca A.L. Autoantibody approach for serum-based detection of head and neck cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16(11):2396–2405. doi: 10.1158/1055-9965.EPI-07-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J., Weiner J., III, Mollenkopf H.J. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc. Natl. Acad. Sci. U. S. A. 2012;109(20):7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena L.S., Rajni Survival mechanisms of pathogenic Mycobacterium tuberculosis H37Rv. FEBS J. 2010;277(11):2416–2427. doi: 10.1111/j.1742-4658.2010.07666.x. [DOI] [PubMed] [Google Scholar]
- Munro C.S., Mitchell D.N. The Kveim response: still useful, still a puzzle. Thorax. 1987;42(5):321–331. doi: 10.1136/thx.42.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid P., Saukkonen J., Mac Kenzie W.R. CDC/NIH workshop. Tuberculosis biomarker and surrogate endpoint research roadmap. Am. J. Respir. Crit. Care Med. 2011;184(8):972–979. doi: 10.1164/rccm.201105-0827WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi M., Takemura T., Guzman J. Localization of Propionibacterium acnes in granulomas supports a possible etiologic link between sarcoidosis and the bacterium. Mod. Pathol. 2012;25(9):1284–1297. doi: 10.1038/modpathol.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald-Richter K.A., Drake W.P. The etiologic role of infectious antigens in sarcoidosis pathogenesis. Semin. Respir. Crit. Care Med. 2010;31(4):375–379. doi: 10.1055/s-0030-1262205. [DOI] [PubMed] [Google Scholar]
- Prince J.E., Kheradmand F., Corry D.B. 16. Immunologic lung disease. J. Allergy Clin. Immunol. 2003;111(2 Suppl.):S613–S623. doi: 10.1067/mai.2003.124. [DOI] [PubMed] [Google Scholar]
- Rastogi R., Du W., Ju D. Dysregulation of p38 and MKP-1 in response to NOD1/TLR4 stimulation in sarcoid bronchoalveolar cells. Am. J. Respir. Crit. Care Med. 2011;183(4):500–510. doi: 10.1164/rccm.201005-0792OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter E., Kataria Y.P., Zissel G., Homolka J., Schlaak M., Muller-Quernheim J. Analysis of the Kveim–Siltzbach test reagent for bacterial DNA. Am. J. Respir. Crit. Care Med. 1999;159(6):1981–1984. doi: 10.1164/ajrccm.159.6.9701038. [DOI] [PubMed] [Google Scholar]
- Schirmbeck R., Riedl P., Fissolo N., Lemonnier F.A., Bertoletti A., Reimann J. Translation from cryptic reading frames of DNA vaccines generates an extended repertoire of immunogenic, MHC class I-restricted epitopes. J. Immunol. 2005;174(8):4647–4656. doi: 10.4049/jimmunol.174.8.4647. [DOI] [PubMed] [Google Scholar]
- Siltzbach L.E., Ehrlich J.C. The Nickerson–Kveim reaction in sarcoidosis. Am. J. Med. 1954;16(6):790–803. doi: 10.1016/0002-9343(54)90443-x. [DOI] [PubMed] [Google Scholar]
- Song Z., Marzilli L., Greenlee B.M. Mycobacterial catalase-peroxidase is a tissue antigen and target of the adaptive immune response in systemic sarcoidosis. J. Exp. Med. 2005;201(5):755–767. doi: 10.1084/jem.20040429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone R.C., Du P., Feng D. RNA-Seq for enrichment and analysis of IRF5 transcript expression in SLE. PLoS One. 2013;8(1):e54487. doi: 10.1371/journal.pone.0054487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RDC . R Foundation for Statistical Computing; Vienna (Austria): 2004. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Wang X., Yu J., Sreekumar A. Autoantibody signatures in prostate cancer. N. Engl. J. Med. 2005;353(12):1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen X., Hu Y. Preliminary characterizations of a serum biomarker for sarcoidosis by comparative proteomic approach with tandem-mass spectrometry in ethnic Han Chinese patients. Respir. Res. 2013;14:18. doi: 10.1186/1465-9921-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Modified linkers distinguishing between origins of each library. Each cDNA library was tagged with a modified linker: ECOR1/HindIII was used for BAL cDNA, ALA for WBC cDNA, LEU for MARC5 cDNA and THR for EL1 cDNA.
Supplementary Fig. 2A. Graphical representation of the output eluent phage titers as a function of biopanning (BP) showing exponential enrichment of the output eluent phage titers after the completion of each cycle of biopannings.
Supplementary Fig. 2B. PCR amplification of the phage clones picked up from biopannings 3 & 4 (BP3 & BP4) showing retention of diversity in the pool of immunoreactive phage.
Supplementary Table 1. Full length of sequence analysis of the top 10 sarcoid phage clones using NCBI BLAST.
Supplementary Table 2. Full length of sequence analysis of the top 10 TB phage clones using NCBI BLAST.