Abstract
Mismatches generated during eukaryotic nuclear DNA replication are removed by two evolutionarily conserved error correction mechanisms acting in series, proofreading and mismatch repair (MMR). Defects in both processes are associated with increased susceptibility to cancer. To better understand these processes, we have quantified base selectivity, proofreading and MMR during nuclear DNA replication in Saccharomyces cerevisiae. In the absence of proofreading and MMR, the primary leading and lagging strand replicases, polymerase ε and polymerase δ respectively, synthesize DNA in vivo with somewhat different error rates and specificity, and with apparent base selectivity that is more than 100 times higher than measured in vitro. Moreover, leading and lagging strand replication fidelity rely on a different balance between proofreading and MMR. On average, proofreading contributes more to replication fidelity than does MMR, but their relative contributions vary from nearly all proofreading of some mismatches to mostly MMR of other mismatches. Thus accurate replication of the two DNA strands results from a non-uniform and variable balance between error prevention, proofreading and MMR.
Keywords: Replication fidelity, DNA polymerase, Genome stability, Mutation rate
1. Introduction
In an unperturbed eukaryotic cell cycle, high nuclear DNA replication fidelity is achieved through the sequential operation of three processes: the selectivity of three replicative DNA polymerases (replicases) for inserting correct nucleotides into properly aligned DNA substrates, exonucleolytic proofreading of mismatches made during replication, and DNA mismatch repair (MMR) of rare errors that escape proofreading. In Escherichia coli, where DNA polymerase III is the primary replicase for both DNA strands, the relative contribution of these processes to replication fidelity has been determined by comparing spontaneous mutation rates in a wild type strain to rates in strains deficient in either proofreading by Pol III, in MMR, or in both error correction mechanisms [1]. A similar approach has been applied to replication of the Saccharomyces cerevisiae nuclear genome [2,3], but in a less comprehensive manner and at a time when the identities of the major leading and lagging strand replicases were uncertain. Now however, compelling evidence using base substitution patterns (see [4] and references therein) and more recently using strand-specific ribonucleotide incorporation [5–8] as biomarkers of replicase actions indicates that the leading strand is primarily replicated by DNA polymerase ε (Pol ε), the product of the yeast POL2 gene. Synthesis of the nascent lagging strand involves limited synthesis by Pol α (POL1), which is naturally deficient in 3′-exonucleolytic activity and cannot proofread mismatches, which it generates at a rate of about 10−4 in vitro [9]. Synthesis by Pol α is followed by extensive synthesis by Pol δ (the product of the yeast POL3 gene). When this knowledge of strand specific replicase activity is combined with use of a mutational reporter gene placed close to a well-studied replication origin, it is now possible to deduce the identity of the mismatches that are being generated, proofread or corrected by MMR during nuclear DNA replication in a yeast cell.
Unlike Pol α, the catalytic subunits of Pol ε [10] and Pol δ [11] have 3′-exonuclease activity for proofreading their own errors. Moreover, there is evidence to suggest that the exonuclease activity of Pol δ, but not that of Pol ε, likely proofreads errors made by Pol α during lagging strand replication [12], and more recent evidence that Pol δ can proofread errors made by Pol ε [13]. Not only is proofreading more complicated in yeast as compared to E. coli, but so too is eukaryotic MMR more complicated than in E. coli. This is because two complexes of eukaryotic MutS Homologs are present for MMR. Single base–base mismatches that can result in base substitutions are primarily repaired by Msh2–Msh6 (MutSα), with Msh2–Msh3 (MutSβ) having but a much smaller role [14,15]. However, MutSα and MutSβ can both participate in repairing insertion-deletion (indel) mutations containing one or two unpaired bases, and MutSβ has primary responsibility for repairing mismatches containing multiple unpaired bases [16].
In this study, we have examined the contribution of all three replication fidelity processes to genome stability in budding yeast. The complexity described above necessitates the use of several yeast strains to quantify the contributions of nucleotide selectivity, proofreading and MMR to leading and lagging strand DNA replication fidelity. Here we use the URA3 reporter gene that scores all types of substitutions in many different sequence contexts. We compare mutation rates in a wild type strain to rates in strains defective in proofreading by Pol δ (pol3-5DV, subsequently referred to as pol3-exo-) [17], or defective in proofreading by Pol ε (pol2-04, subsequently referred to as pol2-exo-) [10], in both cases resulting from mutations in their exonuclease active sites. Rates in MMR-proficient strains are compared to rates in msh6Δ strains that cannot repair the vast majority of single base-base mismatches due to inactivation of MutSα. However, in these strains, most indel mismatches are repaired by MutSβ, thus reducing the possibility of error catastrophe due to lethal indels in strains lacking both proofreading and MMR. Here we have determined mutation rates and mutational spectra in all strains, and used these data to calculate rates for each type of base substitution. Pairwise comparisons of rates among strains then allowed us to estimate the contributions of the three fidelity processes to different single base-base mismatches in different sequence contexts, and to do so for leading and lagging strand replication. The results indicate that on average, proofreading contributes more to replication fidelity than MMR, but the relative importance of proofreading and MMR for correcting replication errors varies widely. In the absence of both MMR and proofreading, Pols δ and ε generate errors in vivo at much lower rates than have been measured in vitro, suggesting that additional fidelity mechanisms may operate at the replication fork in vivo.
2. Methods
2.1. Strain construction
The identities and sources of the strains used in this study are listed in Supplementary Table 1. For this study, we used pol2-04 (Pol ε) and pol3-5DV (Pol δ) mutants that are subsequently referred to as pol2-exo- and pol3-exo-, respectively. In the pol2-04 mutant, alanines were substituted for D290 and E292 in the 3′-exonuclease active site, inactivating 3′-exonuclease activity but leaving polymerase activity similar to that of wild type Pol ε [10]. The homologous allele for Pol δ is pol3-01 (D321A, E323A [18]), but this allele is lethal in combination with msh6Δ [19]. We therefore used the pol3-5DV mutant, in which a valine was substituted for D520 in the exonuclease active site to inactivate 3′-exonuclease activity but leave polymerase activity similar to that of wild type Pol δ [20].
2.2. Mutation rate measurements and mutational spectra
Spontaneous mutation rates at the URA3 locus were measured by fluctuation analysis as described [21,22]. URA3 mutation spectra were obtained by sequencing the URA3 gene in collections of independent 5-fluoroorotic acid-resistant (5-FOAR) colonies. Each 5-FOAR colony from the double mutant strains was obtained from an independent spore. Genomic DNA was isolated from 5-FOAR colonies, the URA3 gene was PCR-amplified and the DNA product was sequenced. Rates for each type of mutation are calculated as the total number of each type of mutation divided by the total number of 5-FOAR mutants sequenced and then multiplied by the total mutation rate. For individual types of base substitutions, the substitution rate per base pair per generation was calculated by dividing the mutation rate by the number of sites in the URA3 gene where that event is known to result in 5-FOAR (Supplementary Fig. 1). The contributions of base selectivity, proofreading and mismatch repair to replication fidelity were calculated as described below.
3. Results and discussion
3.1. Approach
We measured mutation rates in yeast strains that were wild type for proofreading and MMR, deficient in proofreading (pol2-exo- or pol3-exo-) only, deficient in MMR of base-base mismatches (msh6Δ), or deficient in proofreading and MMR (pol2-exo- msh6Δ or pol3-exo- msh6Δ). Because the double mutant strains are highly mutable and rapidly accumulates mutations that could modulate mutation rates, we minimized the number of generations used to measure rates by sporulating diploid strains that were heterozygous for proofreading. Spore viability for the POL2/pol2-exo-msh6Δ/MSH6 and POL3/pol3-exo- msh6Δ/MSH6 strains were 94% and 96%, respectively. The pol2-exo- msh6Δ mutants have normal colony size and the pol3-exo- msh6Δ mutants are smaller than normal (Supplementary Fig. 2). 60–100 independent, double mutant spore colonies were used to measure the rate of resistance to 5-FOA, which primarily scores mutations in the URA3 gene, including substitutions resulting from all 12 possible single base mismatches at numerous locations in the coding sequence (Supplementary Fig. 1). In the strains used here, URA3 is located about 2000 base pairs from ARS306, an early-firing and efficient replication origin on chromosome 3. The next closest origin (ARS307) is about 35,000 base pairs away, such that the replication fork originating at ARS306 will generate the majority of errors in URA3, an interpretation strongly supported by recent ribonucleotide mapping studies [5–8]. In this situation, the coding sequence depicted in Supplementary Fig. 1 is the template for lagging strand synthesis, and the complementary non-coding sequence (not shown) is the template for leading strand replication.
3.2. Mutation rates and specificity
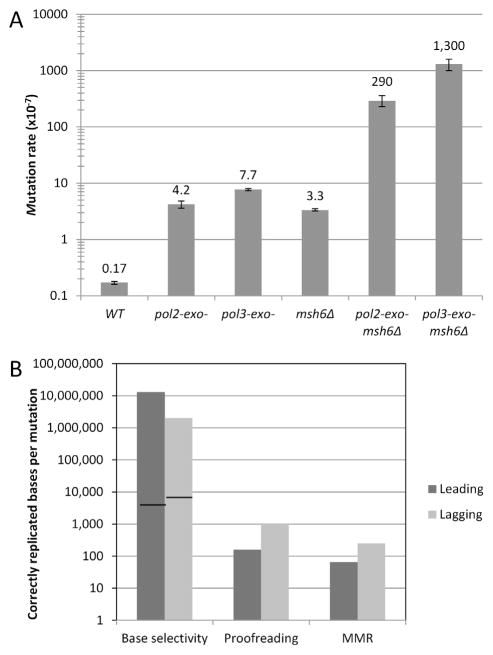
In the wild type strain, the base substitution rate for resistance to 5-FOA is low, and close to the unbiased substitution rate for the whole yeast genome [4]. This fact, and the observation that the vast majority of the 5-FOA-resistant mutants have mutations in URA3 (Table 1), underscores the utility and reliability of URA3 as a reporter for genome stability. Compared to the wild type strain, mutation rates in the pol2-exo-, pol3-exo- and msh6Δ single mutant strains are elevated by 19- to 45-fold (Fig. 1A). These increases are consistent with earlier studies in yeast demonstrating that defects in proofreading or MMR alone elevate mutation rates (see [23,24] and references therein). The mutation rates in the pol2-exo- msh6Δ and pol3-exo- msh6Δ double mutant strains are much higher than the sum of the single mutant strains (Fig. 1A, right). Again, the results are consistent with earlier studies [18,25] indicating that proofreading and MMR act in series to correct replication errors by Pols δ and ε.
Table 1.
Total and specific mutation rates in six yeast strains.
| WT | msh6Δ | pol2-exo- | pol2-exo- msh6Δ | pol3-exo- | pol3-exo- msh6Δ | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation rate (×10−7) | 0.17 | 3.3 | 4.2 | 290 | 7.7 | 1300 | |||||||||||||
| 95% Confidence intervals |
(0.16–0.18) | (3.2–3.5) | (3.6–4.8) | (230–260) | (7.3–8.0) | (1000–1600) | |||||||||||||
| # 5FOA Resistant clones sequenced |
211
|
691
|
170
|
178
|
123
|
361
|
|||||||||||||
| Mutation rates (×10−9) |
Mutation rates (×10−9) |
Mutation rates (×10−9) |
Mutation rates (×10−9) |
Mutation rates (×10−9) |
Mutation rates (×10−9) |
||||||||||||||
| Target sizea |
Total | Mutation typeb |
Per basec |
Total | Mutation typeb |
Per basec |
Total | Mutation typeb |
Per basec |
Total | Mutation typeb |
Per basec |
Total | Mutation typeb |
Per basec |
Total | Mutation typeb |
Per basec |
|
| Base substitutions | 285 | 122 | 9.8 | 0.034 | 292 | 140 | 0.49 | 139 | 340 | 1.2 | 137 | 22000 | 78 | 91 | 570 | 2 | 399 | 140000 | 500 |
| Transitions | 130 | 47 | 3.8 | 0.029 | 182 | 87 | 0.67 | 20 | 49 | 0.38 | 39 | 6400 | 49 | 64 | 400 | 3.1 | 363 | 130000 | 1000 |
| A to G | 18 | 4 | 0.32 | 0.018 | 2 | 0.96 | 0.053 | 1 | 2.5 | 0.14 | 2 | 330 | 18 | 0 | 6.3 | 0.35 | 2 | 720 | 40 |
| T to C | 27 | 7 | 0.56 | 0.021 | 20 | 9.6 | 0.35 | 3 | 7.4 | 0.27 | 2 | 330 | 12 | 37 | 230 | 8.6 | 154 | 55000 | 2100 |
| C to T | 36 | 11 | 0.89 | 0.025 | 6 | 2.9 | 0.08 | 4 | 9.9 | 0.27 | 7 | 1100 | 32 | 6 | 38 | 1 | 5 | 1800 | 50 |
| G to A | 49 | 25 | 2 | 0.041 | 154 | 74 | 1.5 | 12 | 30 | 0.61 | 28 | 4600 | 93 | 21 | 130 | 2.7 | 202 | 73000 | 1500 |
| Transversions | 225 | 75 | 6 | 0.027 | 110 | 53 | 0.23 | 119 | 290 | 1.3 | 98 | 16000 | 71 | 27 | 170 | 0.75 | 36 | 13000 | 58 |
| A to T | 39 | 9 | 0.73 | 0.019 | 3 | 1.4 | 0.037 | 91 | 220 | 5.8 | 8 | 1300 | 33 | 5 | 31 | 0.8 | 3 | 1100 | 28 |
| T to A | 51 | 7 | 0.56 | 0.011 | 3 | 1.4 | 0.028 | 8 | 20 | 0.39 | 5 | 810 | 16 | 3 | 19 | 0.37 | 3 | 1100 | 21 |
| T to G | 42 | 8 | 0.64 | 0.015 | 3 | 1.4 | 0.034 | 0 | 2.5d | 0.059 | 0 | 160 | 3.9 | 11 | 69 | 1.6 | 11 | 4000 | 94 |
| A to C | 25 | 5 | 0.4 | 0.016 | 6 | 2.9 | 0.11 | 6 | 15 | 0.59 | 2 | 330 | 13 | 0 | 6.3 | 0.25 | 0 | 360 | 14 |
| C to A | 38 | 11 | 0.89 | 0.023 | 48 | 23 | 0.6 | 1 | 2.5 | 0.065 | 1 | 160 | 4.3 | 5 | 31 | 0.82 | 16 | 5800 | 150 |
| G to T | 58 | 19 | 1.5 | 0.026 | 40 | 19 | 0.33 | 11 | 27 | 0.47 | 82 | 13000 | 230 | 2 | 13 | 0.22 | 2 | 720 | 12 |
| C to G | 20 | 6 | 0.48 | 0.024 | 1 | 0.48 | 0.024 | 1 | 2.5 | 0.12 | 0 | 160 | 8.1 | 0 | 6.3 | 0.31 | 0 | 360 | 18 |
| G to C | 33 | 10 | 0.81 | 0.024 | 6 | 2.9 | 0.087 | 1 | 2.5 | 0.075 | 0 | 160 | 4.9 | 1 | 6.3 | 0.19 | 1 | 360 | 11 |
Number of base pairs in URA3 where this substitution confers 5-FOA resistance, from Supplementary Fig. 1.
Rates are the total mutation rate times the proportion of the observed mutations of that type (left-most column) among the total 5-FOAR clones sequenced.
Mutation rate per phenotypically detectable base pair.
Italicized values in bold are “less than or equal to” rates for events not seen, calculated based on the maximum mutation rate had one event been observed.
Fig. 1.
Mutation rates and fidelity factors for base selectivity, proofreading, and MMR on both the leading and lagging strands. (A) Mutation rates were measured at the agp1::URA3-OR1 locus in WT, pol2-exo-, pol3-exo-, msh6Δ, pol2-exo- msh6Δ, and pol3-exo- msh6Δ strains. Please note that the y-axis is on a logarithmic scale. (B) The apparent fidelity factors for base selectivity, proofreading, and MMR are depicted for the leading strand (dark gray) and the lagging strand (light gray). The y-axis values are number of correctly replicated or repaired bases per mutation and is on a logarithmic scale. The lines within the bars for base selectivity indicate the base selectivity of Pol ε[46] and Pol δ [45] measured in vitro.
Also, the mutation rate in the pol3-exo- msh6Δ strain is higher than that in the pol2-exo- msh6Δ strain. Given evidence that the exonuclease activity of Pol δ, but not that of Pol ε, proofreads errors made by Pol α[12], and evidence that Pol δcan also proofread errors made by Pol ε [13], the higher mutation rate in the pol3-exo- msh6Δ strain could be due to loss Pol δ proofreading of errors made by any of the three replicases, whereas loss Pol ε may only proofread its own errors. The different mutation rates in the pol3-exo- msh6Δ and pol2-exo- msh6Δ strains could also be related to differences in activation of the S phase checkpoint in proofreading-deficient strains (see [26] and references therein).
To determine rates for individual substitutions, mutational spectra were obtained by sequencing the URA3 gene in collections of independent 5-FOAR colonies. The spectra for wild type versus msh6Δ, for pol2-exo- versus pol2-exo- msh6Δ, and for pol3-exo-versus pol3-exo- msh6Δ strains are shown in Figs. 2, 3 and 4, respectively. Table 1 lists the number of occurrences and the rate per generation for each type of substitution, both before and after correcting the rate for the known number of detectable sites in URA3 (from Supplementary Fig. 1). From these rates, and knowing which strand acts as the template for the majority of leading or lagging strand replication, we calculated the contributions of base selectivity, proofreading and MMR for total substitutions (Fig. 1B) and for substitutions resulting from formation of each of the 12 possible single base mismatches generated during leading and lagging strand replication (Table 2). The contribution of MMR was calculated by dividing the total base substitution rate in the exo- msh6Δ strains by the corresponding base substitution rates in the exo-strains. The contribution of proofreading to replication fidelity was determined by dividing mutation rates in the exo- msh6Δ strains by the corresponding rates in the msh6Δ strain. Apparent base selectivity was calculated as the inverse of the mutation rate for the two exo- msh6Δ strains that lack both proofreading and MMR. We also examined the effects of DNA sequence context by calculating rates and fidelity factors for substitutions at specific base pairs in URA3 (Fig. 5). The latter examples focus on mismatches generated when dTTP, the dNTP present at the highest concentration in yeast [27], is misinserted opposite template G, C or T.
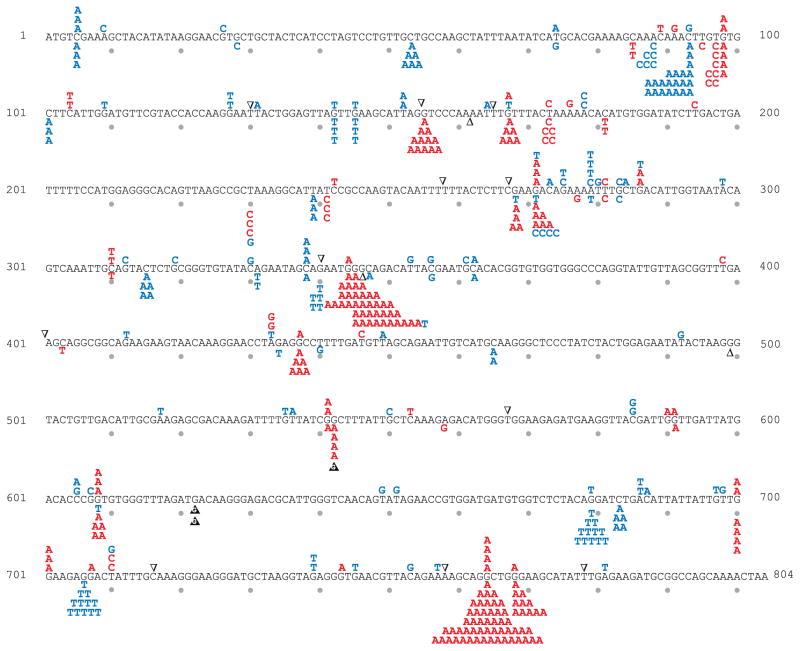
Fig. 2.
URA3 mutation spectra for the wild type and msh6Δ strains. Mutations observed in the wild type and msh6Δ strains are shown above and below the URA3 coding sequence, respectively. Red letters are transitions, blue letters are transversions, open black triangles are single base pair deletions and filled triangles with a letter are single base pair insertions.
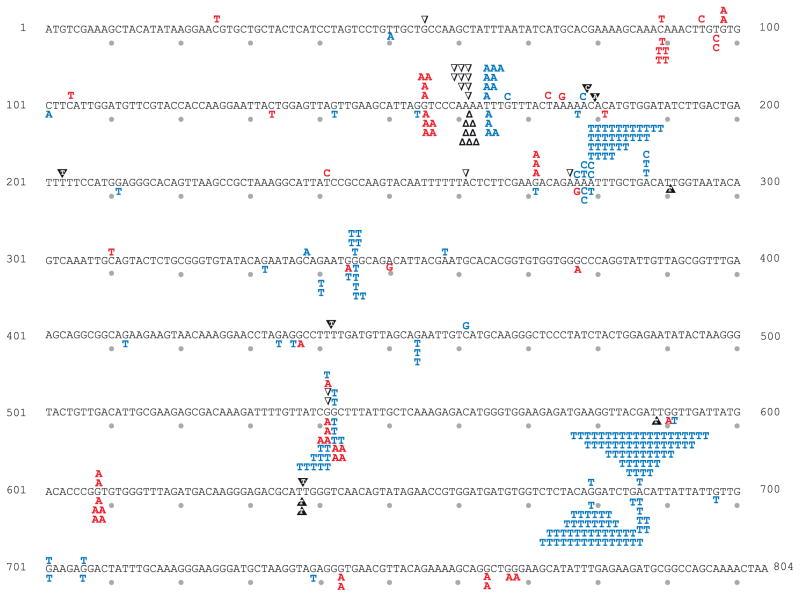
Fig. 3.
URA3 mutation spectra for pol2-exo- and pol2-exo- msh6Δ strains. As in Fig. 2.
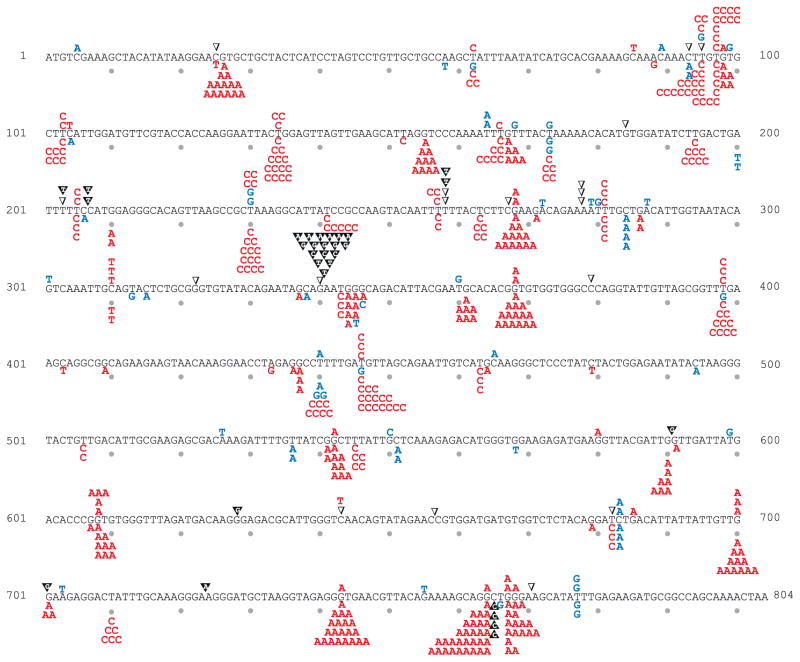
Fig. 4.
URA3 mutation spectra for pol3-exo- and pol3-exo- msh6Δ strains. As in Fig. 2.
Table 2.
Contributions of selectivity, proofreading and MMR to replication fidelity.
| Leading strand
|
Lagging strand
|
|||||
|---|---|---|---|---|---|---|
| Base selectivity | Proofreading | MMR | Base selectivity | Proofreading | MMR | |
| 12000000 | 160 | 65 | 2000000 | 1000 | 250 | |
| Transitions | 21000000 | 73 | 130 | 1000000 | 1500 | 330 |
| T-dG | 55000000 | 340 | 130 | 490000 | 5800 | 240 |
| A-dC | 83000000 | 34 | 44 | 25000000 | 750 | >=120 |
| G-dT | 32000000 | 400 | 120 | 670000 | 990 | 550 |
| C-dA | 11000000 | 62 | 150 | 20000000 | 630 | 48 |
| Transversions | 14000000 | 300 | 54 | 17000000 | 250 | 77 |
| T-dT | 30000000 | 910 | 5.8 | 47000000 | 750 | 58 |
| A-dA | 63000000 | 570 | 41 | 36000000 | 750 | 35 |
| A-dG | >=260000000 | =<110 | a | >=69000000 | =<130 | a |
| T-dC | 77000000 | 110 | 22 | 11000000 | 2800 | 58 |
| G-dA | 230000000 | 7.1 | 66 | 81000000 | 38 | 58 |
| C-dT | 4300000 | 700 | 490 | 6600000 | 250 | 180 |
| G-dG | >=120000000 | =<340 | =<66 | 92000000 | 130 | 58 |
| C-dC | >=200000000 | =<57 | =<66 | >=56000000 | =<750 | b |
Too few events detected to quantify a contribution.
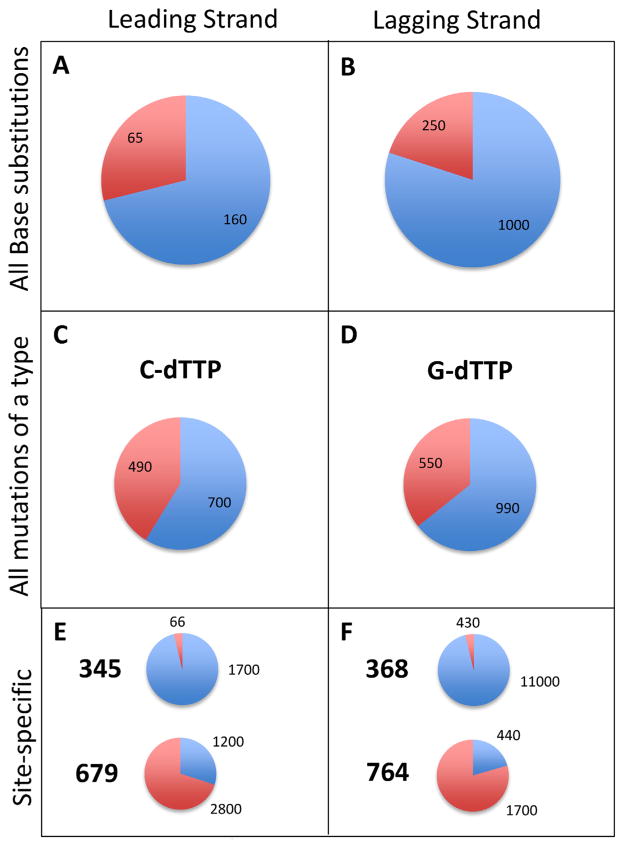
Fig. 5.
The contributions of proofreading and MMR to correcting base-base mismatches. Results on the left are for the pol2-exo- strains, and results on the right are for the pol3-exo- strains. The contributions of proofreading and MMR to correcting: (A) and (B) total base-base mismatches, (C) total C-dTTP mismatches (D) total G-dTTP mismatches, (E) individual C-dTTP mismatches at URA3 base pairs 345 and 679, and (F) individual G-dTTP mismatches at URA3 base pairs 368 and 764. The numbers superimposed within or above the pie slices indicate the error correction factors, either for proofreading (in blue) or MMR (in red).
3.3. Contribution of MMR to replication fidelity
Dividing the total base substitution rate in the pol2-exo- msh6Δ strain (78 × 10−9, Table 1) by the corresponding base substitution rate in the pol2-exo- strain (1.2 × 10−9) estimates the contribution of MMR to the fidelity of replication of the nascent leading strand. This comparison indicates that on average, MMR corrects 64 of every 65 single base-base mismatches generated by Pol ε in the absence of its intrinsic exonuclease activity (Fig. 1B, right-most dark gray bar, and Table 2). The corresponding comparison of the pol3-exo- msh6Δ strain to the pol3-exo- strain indicates even greater MMR efficiency for lagging strand errors, where 249 of every 250 mismatches are corrected. These mismatches are generated during lagging strand replication by Pol δ lacking its intrinsic exonuclease activity, as well as by Pol α, which lacks an intrinsic proofreading activity but whose mistakes may sometimes be edited by Pol δ [12]. These results confirm that MMR efficiently corrects replication errors in both nascent strands. They further imply that MMR is somewhat more efficient at correcting lagging strand mismatches as compared to leading strand mismatches. The latter interpretation is similar to that derived from previous studies examining mismatches containing modified bases [28,29], and from studies of mutator replicases rendered inaccurate by replacing an amino acid in the polymerase active site while leaving the proofreading exonuclease active site unperturbed (see [4,21,22,30–32] and references therein). An important difference between the present study and the earlier reports is that here we monitor mismatches involving unmodified bases in cells where a proofreading exonuclease has been inactivated but the polymerase active site and nucleotide selectivity remain wild type. In all these studies, MMR appears to be more efficient at correcting lagging strand errors as compared to leading strand errors, a fact that we previously suggested may reflect a higher density of signals for MMR in the lagging strand, possibly including PCNA [33] and the 5′-ends of Okazaki fragments in the discontinuously replicated lagging strand [32]. The idea that MMR is somewhat more efficient at correcting lagging strand replication errors due to a higher density of 5D́NA ends is further supported by studies showing that Exonuclease 1 (Exo1), a 5′-exonuclease, contributes more to MMR of lagging as compared to leading strand [34,35], and that Exo1 makes a larger contribution to MMR of errors made by Pol α as compared to errors made by Pol δ, which are always more distal to 5′-DNA ends of Okazaki fragments [35].
On average, transition mismatches are repaired more efficiently than are transversion mismatches (Table 2). This result is consistent with similar observations on MMR in E. coli [1]. Nonetheless, a general conclusion that transition mismatches are repaired more efficiently than transversion mismatches in yeast is unwarranted, because apparent MMR efficiencies vary widely among the 12 mismatches. For example, in the pol2-exo- background, one of the four transversion mismatches (template C-incoming dTTP, 490-fold MMR correction factor, Table 2) is repaired more efficiently than any of the four purine–pyrimidine mismatches that result in transitions. Moreover, this same pyrimidine–pyrimidine mismatch (C-dT, 490-fold) is repaired much more efficiently than another pyrimidine–pyrimidine mismatch (T-dT, 5.8-fold). MMR efficiency also varies in the pol3-exo- background, but less so (range of 16-fold, Table 2). In some cases, the efficiency of repairing a mismatch containing the same two bases varies depending on which base is in the template and which base is the incoming nucleotide. The largest such difference is in the pol2-exo- background, where MMR repairs 489 of 490 template C–dT mismatches but only 21 of 22 T–dC mismatches (Table 2). Similar but smaller MMR biases are also observed in the pol3-exo- background (Table 2). Theoretically, these variations may reflect the binding affinity of yeast MutSα for mismatches of different composition, which vary more than 10-fold [36], or they may reflect MMR events downstream of MutSα binding.
3.4. Contribution of proofreading to replication fidelity
The contribution of proofreading to replication fidelity was determined by dividing mutation rates in the pol2-exo- msh6Δ or pol3-exo- msh6Δ strain by the corresponding rates in the msh6Δ strain. When all substitutions are considered, the average contribution of proofreading to replication fidelity in vivo is 160-fold for Pol ε and 1000-fold for Pol δ (Fig. 2, middle, and Table 2). These results establish that, on average, (i) proofreading contributes several-fold more than MMR to both leading and lagging strand replication fidelity in vivo, and (ii) proofreading of lagging strand mismatches is more efficient than proofreading of leading strand mismatches (Fig. 1B). Just as for MMR, proofreading efficiency also varies widely (Table 2). In the pol2-exo- background, transversion mismatches are proofread four times more efficiently than are transition mismatches (300-fold versus 73-fold), whereas the opposite is true in the pol3-exo- background (250 versus 1500). In the pol2-exo- background, proofreading efficiency among the mismatches varies by more than 100-fold, from 910-fold for T–dT to only 7-fold for G–dA in the pol2-exo- strain, and from 5800-fold for T–dG to 28-fold for G–dA. Mismatch symmetry also matters, as exemplified by the 11-fold difference in proofreading efficiency between a C–dT and T–dC mismatch in the pol3-exo- background (250 versus 2800).
3.5. Variations in the relative importance of proofreading and MMR
Replication errors in the URA3 open reading frame are non-uniformly distributed (Figs. 2–4). For example, among 178 independent 5-FOAR colonies from the pol2-exo- msh6Δ strain (Table 1), 43 colonies contained a G to T substitution at base pair 679 (Fig. 3). By comparison, a combined total of only 39 G to T substitutions was observed at all 56 other G-C base pairs in URA3 where this substitution results in resistance to 5-FOA (Supplementary Fig. 1). These substitutions reflect uncorrected misinsertion of dTTP opposite template C during leading strand replication by Pol ε. The existence of this and other such hotspots provides the opportunity to quantify the relative contributions of proofreading and MMR to correcting individual mismatches of the same base composition and symmetry. In Fig. 5, average proofreading and MMR correction factors for all substitutions (panels A and B) are compared to average correction factors for two mismatches (panels C and D), and to site-specific examples of these same mismatches (panels E and F). All of these mismatches involve the misinsertion of a dTTP, the dNTP present at the highest concentration in yeast [27], and whose misincorporation is responsible for the greatest proportion of substitutions. In the pol2-exo- strain, misinsertion of dTTP opposite template C at base pair 345 is corrected more efficiently by proofreading than by MMR, whereas the opposite is true for the same mismatch but in a different sequence context at base pair 679 (Fig. 5E). Similarly in the pol3-exo- strain, misinsertion of dTTP opposite template G at base pair 368 is corrected more efficiently by proofreading than by MMR, but the opposite is true for the same error but at base pair 764 (Fig. 5F). These examples clearly illustrate that even when mismatches of the same composition and symmetry are considered, the relative contributions of proofreading and MMR to genome stability strongly depend on the local sequence environment. Possible explanations for these observations come from studies showing that proofreading depends on local base-base stacking interactions and on the concentrations of correct dNTPs to be incorporated after misinsertion (reviewed in [24]). Both parameters affect partitioning between polymerization to embed the mismatch in the DNA duplex and thus protect it from removal, versus fraying of the primer terminus to permit movement of the error to the exonuclease active site for removal. Compared to proofreading, these parameters may be less relevant to MMR, whose efficiency may depend more on the affinity of MutSε to bind a mismatch, the flexibility of a mismatched DNA duplex, and/or steps further downstream in the MMR pathway. The observations showing that the relative contributions of proofreading and MMR to replication fidelity vary by mismatch composition, sequence environment, replicase and DNA strand (Table 2, Fig. 5) illustrate the difficulty in predicting (i) the consequences of defects in these two processes on specific genes or other functional non-coding sequences in the genome, and (ii) the relative importance of proofreading versus MMR in preventing the mutations that drive tissue-specific tumorigenesis [23,37–42].
3.6. Contribution of base selectivity to replication fidelity
Base selectivity was calculated here as the inverse of the mutation rate for the pol2-exo- msh6Δ or pol3-exo- msh6Δ strains that lack proofreading and MMR. Note that mismatches in the pol3-exo-msh6Δ strain can be caused by either Pol δ or Pol α, such that the calculated selectivity values are estimates of “apparent” base selectivity, which could be different in vivo (discussed further below). The calculated selectivity values show that on average, Pol ε and Pol δ create only one mismatch that results in a base substitution for every 1.3 × 107 or 2.0 × 106 correct bases incorporated, respectively (Fig. 1B). Thus base selectivity appears to be by far the largest contributor to the fidelity of both leading and lagging strand replication. Interestingly, while Pol ε discriminates about equally well against transition and transversion mismatches (fidelity factors of 2.1 × 107 and 1.4 × 107 correctly replicated bases per mismatch, respectively, Table 2), Pol δ discriminates against transversions (blue events in Fig. 4) better than transitions (red events in Fig. 4) by 17-fold (Table 2, 17 × 106 versus 1 × 106). Among different mismatches generated in the pol2-exo- background, selectivity against A–dG mismatches (≥2.6 × 108) is 60-fold higher than against C–dT mismatches (4.3 × 106), whereas in the pol3-exo- background, selectivity against G–dG mismatches (9.2 × 107) is 140-fold higher than against G–dT mismatches (6.7 × 105). These results imply that, during nuclear DNA replication in vivo, (i) apparent base selectivity is higher for leading than lagging strand replication, and (ii) the two major nuclear replicases have substantially different misincorporation specificities regarding mismatch composition (Table 2) and sequence context (different hotspots in Figs. 3 and 4).
When the dNTPs responsible for the substitutions in Table 1 are totaled irrespective of the template base, dTTP is misincorporated more often than the other three dNTPs, and during replication of both strands. This fact is interesting because the intracellular concentration of dTTP in yeast is about 2 to 3-fold higher than the concentrations of the other three dNTPs [27]. This suggests that this natural dNTP pool imbalance influences replication error rate and error specificity by Pols ε and α, which has implications in mammalian cells where dNTP pools are also naturally imbalanced [43,44].
The rank order of selectivity against certain mismatches is somewhat unexpected. Previous studies in a variety of systems led to the idea that the most commonly formed base–base mismatch is G–T, whereas pyrimidine–pyrimidine mismatches, especially C–dC, are rarely formed. This generally appears to be the case for lagging strand replication, where selectivity against G–dT and T–dG is at least 10-fold lower than for the other 10 mismatches, and where selectivity against C–C and the other pyrimidine–pyrimidine mismatches is very high (Table 2). However, this does not appear to be the case for leading strand replication, where the lowest selectivity is for the C–dT mismatch, and selectivity against this and two of the other three pyrimidine–pyrimidine mismatches (T–dC and T–dT) is similar to selectivity against G–dT and T–dG mismatches. Differences in the error specificity of Pol ε and Pol δ are still apparent, albeit reduced, even after subtracting the events at the hottest of the substitution hot spots in the spectra (e.g., transversions at base pairs 279, 679 and 686 in Fig. 3). These data suggest that the active sites of the two major nuclear replicases impose different structural, kinetic and/or thermodynamic constraints on formation of mismatches of the same composition.
3.7. In vivo influences promoting increased replication fidelity
Proofreading-deficient Pol ε and Pol δ generate only one error for every 12,000,000 or 2,000,000 correct bases replicated in vivo, respectively (Fig. 1B). Both values are much higher than the base selectivity of proofreading-deficient yeast Pol ε and Pol δ measured during DNA synthesis in vitro (black lines in bars in Fig. 1B), where one error is observed for every 4.200 and 7700 correct bases incorporated, respectively [45,46]. These differences are large, 2900-fold for Pol ε and 280-fold for Pol α. There are several non-exclusive possibilities for these differences. One fact that may be relevant is that the DNA synthesis reaction mixtures in vitro contain all four dNTPs at a concentration of 100 μM, whereas dNTP concentrations in yeast are lower and slightly imbalanced [27,47]. Other reaction parameters in vitro may be less than optimal for high selectivity, including the pH and magnesium concentration, both of which are known to affect the fidelity of DNA synthesis in vitro [48–50]. Replication accessory proteins not yet examined in vitro could also improve fidelity in vivo, although studies to date have not revealed large increases in nucleotide selectivity by accessory proteins.
A perhaps more interesting possibility for the higher “apparent” base selectivity in vivo is extrinsic proofreading. Biochemical [51] and genetic evidence [2,12] suggest that errors made by naturally exonuclease-deficient Pol α, whose primary role is in lagging strand replication, can be proofread by Pol δ. Thus it is possible that in the absence of only one of the two proofreading activities intrinsic to Pol ε and Pol δ, the exonuclease that remains intact in one replicase can proofread mismatches generated by the other major, but proofreading-deficient replicase [2,12]. Indeed, a recent study [13] provides evidence that Pol δ proofreads errors made by Pol ε. Theoretically, extrinsic proofreading may also be catalyzed by other 3′-exonucleases (e.g., see [52]). To the extent that extrinsic proofreading may occur during replication in yeast, this implies that (i) the actual base selectivity of the replicases in vivo could be substantially lower than calculated here, and therefore more in line with the estimates from studies in vitro, and (ii) the base selectivity calculated here for lagging strand replication may be a mixture of the base selectivity of Pol α plus Pol ε, because both polymerases contribute to the mature lagging strand (see [21,53], and more recently [5,8]).
Also of interest is genetic evidence suggesting that Pol ε does not efficiently proofread errors made by Pol α [12] or errors made by proofreading-deficient Pol ε [13]. Given the participation of Pol δ in several other DNA transactions in cells, it may be that Pol δ is generally more efficient at extrinsic proofreading than is Pol ε. This could account for the 10-fold greater difference in apparent base selectivity in vivo versus in vitro mentioned above (2900-fold for the leading strand, 280-fold for the lagging strand). Possibly relevant here is evidence that Pol ε has a strong interaction with the CMG (Cdc45, Mcm2-7, and GINS) complex, while Pol δ has a very weak interaction with the same complex [54]. In contrast, Pol δ has a strong interaction with PCNA, which stimulates its processivity. It is possible that if mismatches created by Pol ε during leading strand replication cannot be proofread by its intrinsic exonuclease, Pol ε can transiently dissociate from the terminal mismatch to allow Pol δ proofreading, and then Pol ε may re-engage and continue leading strand replication [54,55]. Still other possibilities may lie in the identity of yet-to-be disclosed suppressors of mutability in MMR-deficient pol2-4 strains, leading to the suggestion that factors in addition to proofreading and MMR influence leading-strand DNA replication fidelity [56].
3.8. Complementarity among the three DNA replication fidelity processes
The observation that the two major nuclear replicases have substantially different base selectivity against different mismatches means that the two downstream error correction processes must meet different challenges in order to achieve high fidelity replication of both DNA strands. That proofreading and MMR have evolved to effectively meet these different challenges is illustrated by the following observations. (i) Average base selectivity is lower during lagging strand replication, but average proofreading and MMR efficiencies are higher, whereas the opposite holds for leading strand replication (Fig. 1B). (ii) On average, transitions are generated at a 20-fold higher rate in the pol3-exo- msh6Δ strain (1300 × 10−7, Table 1) as compared to the pol2-exo- msh6Δ strain (64 × 10−7), and this difference is counterbalanced by the fact that proofreading of transition mismatches is 20-fold more efficient on the lagging strand (Table 2, correction factor 1500-fold, as compared to the leading strand, 73-fold). (iii) As compared to the above example, average selectivity against transversion mismatches is similar during leading and lagging strand replication, and so too are proofreading and MMR efficiencies (Table 2). (iv) Selection against a C–dT mismatch is relatively low in the pol2-exo- background, and highly efficient proofreading and MMR counterbalance this low selectivity. (v) Conversely, high selectivity against certain mismatches, such as C–dA, A–dC, G–dA, is associated with lesser contributions of proofreading and MMR. These observations, in combination with earlier studies of engineered variants of replicases harboring polymerases active site point mutations (see [4] and references therein), support the idea that the eukaryotic replicases, their attendant 3′-exonucleases, and MMR have coevolved to most efficiently correct replication errors made at the highest rates and in the most risky sequence contexts, in order to accurately replicate both DNA strands of the nuclear genome. This idea, and the fact that the contribution to replication fidelity of base selectivity, proofreading and MMR all strongly depend on mismatch composition and symmetry, the replicase, and the strand and sequence context in which the mismatch resides, likely contribute to defining the composition of nuclear genomes. The balancing act among the three replicases may partly underlie differences in life span and tissue-specific tumorigenesis in mice encoding proofreading deficient alleles of Pols ε and δ [57]. These differences may also be relevant to the nature and number of mutations that are thought to drive tissue-specific tumor development in humans harboring mutations in the exonuclease motifs of Pols ε and δ [23,37–42].
Supplementary Material
Acknowledgments
The authors thank Dmitry Gordenin and members of the Genome Integrity and Structural Biology Laboratory at the NIEHS for helpful discussions on experiments, and Kin Chan and Deepa Singh for valuable comments on the manuscript. This work was supported by the Division of Intramural Research of the National Institutes of Health, National Institute of Environmental Health Sciences, by Projects Z01 ES065070 and Z01 ES065089 to TAK.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.dnarep.2015.04.006
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest with this work.
References
- 1.Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 2.Morrison A, Sugino A. The 3′ → 5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 3.Greene CN, Jinks-Robertson S. Spontaneous frameshift mutations in Saccharomyces cerevisiae: accumulation during DNA replication and removal by proofreading and mismatch repair activities. Genetics. 2001;159:65–75. doi: 10.1093/genetics/159.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lujan SA, Clausen AR, Clark AB, MacAlpine HK, MacAlpine DM, Malc EP, Burkholder AB, Fargo DC, Gordenin DA, Kunkel TA. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;24:1751–1764. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clausen AR, Lujan SA, Burkholder AB, Orebaugh CD, Williams JS, Clausen MF, Malc EP, Mieczkowski PA, Fargo DC, Smith DJ, Kunkel TA. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol. 2015;22:185–191. doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh KD, Balachander S, Hesselberth JR, Storici F. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nat Methods. 2015;12:251–257. doi: 10.1038/nmeth.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daigaku Y, Keszthelyi A, Muller CA, Miyabe I, Brooks T, Retkute R, Hubank M, Nieduszynski CA, Carr AM. A global profile of replicative polymerase usage. Nat Struct Mol Biol. 2015;22:192–198. doi: 10.1038/nsmb.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reijns MA, Kemp H, Ding J, de Proce SM, Jackson AP, Taylor MS. Lagging-strand replication shapes the mutational landscape of the genome. Nature. 2015;518:502–506. doi: 10.1038/nature14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunkel TA, Hamatake RK, Motto-Fox J, Fitzgerald MP, Sugino A. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:4447–4458. doi: 10.1128/mcb.9.10.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′–5′ exonuclease activity. Proc Natl Acad Sci U S A. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon M, Giot L, Faye G. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10:2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Flood CL, Rodriguez GP, Bao G, Shockley AH, Kow YW, Crouse GF. Replicative DNA polymerase delta but not epsilon proofreads errors in cis and in trans. PLoS Genet. 2015;11:e1005049. doi: 10.1371/journal.pgen.1005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaab WE, Risinger JI, Umar A, Kunkel TA, Barrett JC, Tindall KR. Characterization of distinct human endometrial carcinoma cell lines deficient in mismatch repair that originated from a single tumor. J Biol Chem. 1998;273:26662–26669. doi: 10.1074/jbc.273.41.26662. [DOI] [PubMed] [Google Scholar]
- 15.Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2–Msh3 acts in repair of base-base mispairs. Mol Cell Biol. 2007;27:6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin YH, Garg P, Stith CM, Al-Refai H, Sterling JF, Murray LJ, Kunkel TA, Resnick MA, Burgers PM, Gordenin DA. The multiple biological roles of the 3′ → 5′ exonuclease of Saccharomyces cerevisiae DNA polymerase delta require switching between the polymerase and exonuclease domains. Mol Cell Biol. 2005;25:461–471. doi: 10.1128/MCB.25.1.461-471.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herr AJ, Ogawa M, Lawrence NA, Williams LN, Eggington JM, Singh M, Smith RA, Preston BD. Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 2011;7:e1002282. doi: 10.1371/journal.pgen.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. The 3′ → 5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc Natl Acad Sci U S A. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase epsilon mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014;74:1895–1901. doi: 10.1158/0008-5472.CAN-13-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reha-Krantz LJ. DNA polymerase proofreading: multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804:1049–1063. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Tran HT, Gordenin DA, Resnick MA. The 3′ → 5′ exonucleases of DNA polymerases delta and epsilon and the 5′ → 3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams LL, Marjavaara L, Knowels GM, Schultz EM, Fox EJ, Chabes A, Herr AJ. dNTP pool levels modulate mutator ohenotypes of error-prone DNA polymerase epsilon variants. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1422948112. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- 29.Kow YW, Bao G, Reeves JW, Jinks-Robertson S, Crouse GF. Oligonucleotide transformation of yeast reveals mismatch repair complexes to be differentially active on DNA replication strands. Proc Natl Acad Sci U S A. 2007;104:11352–11357. doi: 10.1073/pnas.0704695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larrea AA, Lujan SA, Nick McElhinny SA, Mieczkowski PA, Resnick MA, Gordenin DA, Kunkel TA. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci U S A. 2010;107:17674–17679. doi: 10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lujan SA, Williams JS, Pursell ZF, Abdulovic-Cui AA, Clark AB, Nick McElhinny SA, Kunkel TA. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012;8:e1003016. doi: 10.1371/journal.pgen.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta} Proc Natl Acad Sci U S A. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavlov YI, Newlon CS, Kunkel TA. Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- 34.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liberti SE, Larrea AA, Kunkel TA. Exonuclease 1 preferentially repairs mismatches generated by DNA polymerase alpha. DNA Repair (Amst) 2013;12:92–96. doi: 10.1016/j.dnarep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2–MSH6 complex and mispaired bases in DNA. J Biol Chem. 1999;274:26668–26682. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Church DN, Briggs SE, Palles C, Domingo E, Kearsey SJ, Grimes JM, Gorman M, Martin L, Howarth KM, Hodgson SV, Kaur K, Taylor J, Tomlinson IP The NSECG Collaborators. DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Hum Mol Genet. 2013;22:2820–2828. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Humphray S, Consortium C, Consortium WGS, Lucassen A, Holmes CC, Bentley D, Donnelly P, Taylor J, Petridis C, Roylance R, Sawyer EJ, Kerr DJ, Clark S, Grimes J, Kearsey SE, Thomas HJ, McVean G, Houlston RS, Tomlinson I. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinbrot E, Henninger EE, Weinhold N, Covington KR, Goksenin AY, Schultz N, Chao H, Doddapaneni H, Muzny DM, Gibbs RA, Sander C, Pursell ZF, Wheeler DA. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24:1740–1750. doi: 10.1101/gr.174789.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida R, Miyashita K, Inoue M, Shimamoto A, Yan Z, Egashira A, Oki E, Kakeji Y, Oda S, Maehara Y. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur J Hum Genet. 2011;19:320–325. doi: 10.1038/ejhg.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shlien A, Campbell BB, de Borja R, Alexandrov LB, Merico D, Wedge D, Van Loo P, Tarpey PS, Coupland P, Behjati S, Pollett A, Lipman T, Heidari A, Deshmukh S, Avitzur N, Meier B, Gerstung M, Hong Y, Merino DM, Ramakrishna M, Remke M, Arnold R, Panigrahi GB, Thakkar NP, Hodel KP, Henninger EE, Goksenin AY, Bakry D, Charames GS, Druker H, Lerner-Ellis J, Mistry M, Dvir R, Grant R, Elhasid R, Farah R, Taylor GP, Nathan PC, Alexander S, Ben-Shachar S, Ling SC, Gallinger S, Constantini S, Dirks P, Huang A, Scherer SW, Grundy RG, Durno C, Aronson M, Gartner A, Meyn MS, Taylor MD, Pursell ZF, Pearson CE, Malkin D, Futreal PA, Stratton MR, Bouffet E, Hawkins C, Campbell PJ, Tabori U. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47:257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 43.Kunz BA, Kohalmi SE, Kunkel TA, Mathews CK, McIntosh EM, Reidy JA International Commission for Protection Against Environmental Mutagens and Carcinogens. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat Res. 1994;318:1–64. doi: 10.1016/0165-1110(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 44.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 45.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 46.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J Biol Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 47.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 48.Eckert KA, Kunkel TA. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990;18:3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eckert KA, Kunkel TA. Fidelity of DNA synthesis catalyzed by human DNA polymerase alpha and HIV-1 reverse transcriptase: effect of reaction pH. Nucleic Acids Res. 1993;21:5212–5220. doi: 10.1093/nar/21.22.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eckert KA, Kunkel TA. Effect of reaction pH on the fidelity and processivity of exonuclease-deficient Klenow polymerase. J Biol Chem. 1993;268:13462–13471. [PubMed] [Google Scholar]
- 51.Perrino FW, Loeb LA. Hydrolysis of 3′-terminal mispairs in vitro by the 3′–5′ exonuclease of DNA polymerase delta permits subsequent extension by DNA polymerase alpha. Biochemistry. 1990;29:5226–5231. doi: 10.1021/bi00474a002. [DOI] [PubMed] [Google Scholar]
- 52.Nick McElhinny SA, Pavlov YI, Kunkel TA. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle. 2006;5:958–962. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niimi A, Limsirichaikul S, Yoshida S, Iwai S, Masutani C, Hanaoka F, Kool ET, Nishiyama Y, Suzuki M. Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity. Mol Cell Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Georgescu RE, Langston L, Yao NY, Yurieva O, Zhang D, Finkelstein J, Agarwal T, O’Donnell ME. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kunkel TA, Burgers PM. Delivering nonidentical twins. Nat Struct Mol Biol. 2014;21:649–651. doi: 10.1038/nsmb.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams LN, Herr AJ, Preston BD. Emergence of DNA polymerase epsilon antimutators that escape error-induced extinction in yeast. Genetics. 2013;193:751–770. doi: 10.1534/genetics.112.146910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, Treuting PM, Heddle JA, Goldsby RE, Preston BD. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci U S A. 2009;106:17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.