Abstract
IMPORTANCE
Li-Fraumeni syndrome, usually characterized by germline TP53 mutations, is associated with markedly elevated lifetime risks of multiple cancers, and has been linked to an increased risk of early-onset colorectal cancer.
OBJECTIVE
To examine the frequency of germline TP53 alterations in patients with early-onset colorectal cancer.
DESIGN, SETTING, AND PARTICIPANTS
This was a multicenter cross-sectional cohort study of individuals recruited to the Colon Cancer Family Registry (CCFR) from 1998 through 2007 (genetic testing data updated as of January 2015). Both population-based and clinic-based patients in the United States, Canada, Australia, and New Zealand were recruited to the CCFR. Demographic information, clinical history, and family history data were obtained at enrollment. Biospecimens were collected from consenting probands and families, including microsatellite instability and DNA mismatch repair immunohistochemistry results. A total of a 510 individuals diagnosed as having colorectal cancer at age 40 years or younger and lacking a known hereditary cancer syndrome were identified from the CCFR as being potentially eligible. Fifty-three participants were excluded owing to subsequent identification of germline mutations in DNA mismatch repair genes (n = 47) or biallelic MUTYH mutations (n = 6).
INTERVENTIONS
Germline sequencing of the TP53 gene was performed. Identified TP53 alterations were assessed for pathogenicity using literature and international mutation database searches and in silico prediction models.
MAIN OUTCOMES AND MEASURES
Frequency of nonsynonymous germline TP53 alterations.
RESULTS
Among 457 eligible participants (314, population-based; 143, clinic-based; median age at diagnosis, 36 years [range, 15–40 years]), 6 (1.3%; 95%CI, 0.5%–2.8%) carried germline missense TP53 alterations, none of whom met clinical criteria for Li-Fraumeni syndrome. Four of the identified TP53 alterations have been previously described in the literature in probands with clinical features of Li-Fraumeni syndrome, and 2 were novel alterations.
CONCLUSIONS AND RELEVANCE
In a large cohort of patients with early-onset colorectal cancer, germline TP53 mutations were detected at a frequency comparable with the published prevalence of germline APC mutations in colorectal cancer. With the increasing use of multigene next-generation sequencing panels in hereditary cancer risk assessment, clinicians will be faced with the challenge of interpreting the biologic and clinical significance of germline TP53 mutations in families whose phenotypes are atypical for Li-Fraumeni syndrome.
More than 10% of all colon cancers and nearly one-fifth of all rectal cancer diagnoses occur in patients younger than 50 years, yet the minority of early-onset cases can be attributed to the 3 most common hereditary colorectal cancer (CRC) syndromes: Lynch syndrome, familial adenomatous polyposis (FAP), or MUTYH-associated polyposis (MAP).1–4 Other hereditary syndromes linked to early onset CRC include Peutz-Jeghers syndrome, Cowden syndrome, juvenile polyposis, and Li-Fraumeni syndrome (LFS), yet their prevalence is poorly understood.5,6
Li-Fraumeni syndrome is an inherited cancer syndrome, usually caused by germline TP53 mutations, in which patients classically develop early-onset cancers, including leukemias, brain tumors, sarcomas, breast carcinomas, and adrenocortical carcinomas.7–10 Beyond these so-called core cancers, data have shown that TP53 mutation carriers are also at increased risk for a wide array of other malignant neoplasms, including bronchoalveolar, pancreatic, gastric, ovarian, and colorectal cancers.6,10–12 Germline TP53 testing is recommended for individuals who meet strict clinical criteria, including classical LFS criteria8 or Chompret criteria,13,14 none of which include CRC as a component cancer.
With data linking germline TP53 mutations to early-onset CRC, however, TP53 testing is included on most multigene next-generation sequencing panels now commercially available for hereditary CRC risk assessment.6,15 As the availability of such panels grows, the number of patients undergoing TP53 mutation analysis will likely markedly increase.15 In order for clinicians to provide effective and appropriate counseling to patients undergoing TP53 testing as part of multigene risk assessment, an accurate understanding of this gene’s contribution to hereditary and early-onset CRC is needed. This study’s aim was to estimate the proportion of participants with early-onset CRC who carry germline TP53 mutations.
Methods
Colon Cancer Family Registry
The Colon Cancer Family Registry (CCFR) is an international consortium created in 1997 to facilitate collaboration for interdisciplinary studies in the genetic epidemiology of CRC.16 The CCFR consists of participants and families ascertained through both clinic-based and population-based recruitment in the United States, Canada, Australia, and New Zealand. All participants provided written informed consent for the use of blood samples and tumor tissue in cancer research and for inclusion in the CCFR through one of the following registry centers: Mayo Clinic (Rochester, Minnesota), Fred Hutchinson Cancer Research Center (Seattle, Washington), University of Southern California Consortium (Los Angeles), Lunenfeld Tanenbaum Research Institute (Toronto, Ontario, Canada), and University of Melbourne (Melbourne, Australia). Demographic information, clinical history, and family history data were obtained using standardized instruments, as previously described.16 Biospecimens, including tumor tissue, were collected from consented probands and families, along with data on tumor microsatellite instability (MSI) and DNA mismatch repair (MMR) immunohistochemistry (IHC) status. All protocols were approved by the appropriate institutional review boards. All samples and data have been anonymized. Patients were not compensated for their participation.
Study Population
Participants enrolled in the CCFR were potentially eligible for analysis if they were diagnosed as having CRC at age 40 years or younger and were not known to carry a germline mutation in any of the genes linked to Lynch syndrome (MLH1, MSH2, MSH6, PMS2, or EPCAM) or MAP. The CCFR has not routinely recruited individuals with FAP phenotypes and has not performed systematic germline APC analysis on enrolled participants. A few individuals with known germline APC mutations who had been enrolled at individual CCFR sites were included in the pool of potentially eligible participants for this study.
Germline Analysis
Archived genomic DNA from 510 potentially eligible participants was analyzed by a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory (Laboratory for Molecular Medicine, Cambridge, Massachusetts) using bidirectional Sanger sequencing of the 11 exons of the TP53 gene (NM_000546) (see eMethods in the Supplement).
PubMed searches and querying of the International Agency for Research on Cancer TP53 Mutation Database17,18 were used to identify previous literature reports of the specific TP53 alterations and assess their functional significance. Alamut mutation interpretation software (Interactive Biosoftware) was used to access PolyPhen-2 and SIFT for in silico pathogenicity analyses and to assess species conservation.19,20 Minor allele frequencies (MAFs) for all identified alterations were queried from the Exome Variant Server.21
Results
Fifty-three participants were excluded from analysis owing to subsequent testing that identified pathogenic germline mutations in MLH1 (n = 28 participants), MSH2 (n = 14), MSH6 (n = 2), PMS2 (n = 3), and biallelic MUTYH mutations (n = 6). The final study population consisted of 457 participants with a history of CRC at age 40 years or younger (median age at diagnosis, 36 years [range, 15–40 years]). Microsatellite instability and/or MMR IHC results were available on tumors from 326 participants (71%), 47 of whom had MSI-H and/or mismatch repair deficient (MMR-D) findings (Table 1). A total of 162 participants (35%) had prior negative or inconclusive germline testing of at least 1 MMR gene (MLH1, MSH2, MSH6, and PMS2). A total of 397 participants (87%) had prior MUTYH testing, 387 of whom had negative results, and 10 of whom carried monoallelic MUTYH mutations.
Table 1.
Characteristics of 457 Participants Diagnosed as Having Colorectal Cancer at Age 40 Years or Younger
| Characteristic | No. (%) |
|---|---|
| Sex | |
| Male | 204 (45) |
| Female | 253 (55) |
| Race | |
| White | 382 (84) |
| Black/African American | 11 (2) |
| Hispanic | 7 (2) |
| Asian | 16 (4) |
| Native American | 1 (<1) |
| Middle Eastern | 4 (1) |
| Other | 3 (1) |
| Missing/unknown | 33 (7) |
| Cancer history | |
| 1 CRC only | 389 (85) |
| 1 CRC + other cancer(s) | 34 (7) |
| Synchronous/metachronous CRCs only | 26 (6) |
| Synchronous/metachronous CRCs + other cancer(s) | 8 (2) |
| Site of colorectal cancer | |
| Right colon | 121 (26) |
| Left colon | 105 (23) |
| Rectum | 149 (33) |
| Appendix | 7 (2) |
| Missing/unknown | 75 (16) |
| MSI and MMR IHC statusa | |
| MSI-H and/or abnormal MMR IHC | 47 (10) |
| MSS/MSI-L and/or normal MMR IHC | 279 (61) |
| Missing tumor testing data | 131 (29) |
| Prior genetic testing results (Lynch genes and/or MUTYH)b | |
| Prior genetic testing of ≥1 Lynch genes | 162 (35) |
| Negative result for MLH1 testing | 142 (31) |
| MLH1 VUS detected | 12 (3) |
| Negative result for MSH2 testing | 129 (28) |
| MSH2 VUS detected | 2 (<1) |
| Negative result for MSH6 testing | 40 (9) |
| MSH6 VUS detected | 2 (<1) |
| Negative result for PMS2 testing | 22 (5) |
| PMS2 VUS detected | 1 (<1) |
| Negative result for MUTYH testing | 387 (85) |
| Monoallelic MUTYH mutation detected | 10 (2) |
| MUTYH VUS detected | 0 |
| Recruitment method | |
| Population-based | 314 (69) |
| Clinic-based | 143 (31) |
Abbreviations: CRC, colorectal cancer; MSI, microsatellite instability; MMR IHC, mismatch repair immunohistochemistry testing; MSI-H, high-level microsatellite instability; MSI-L, low-level microsatellite instability; MSS, microsatellite stable; VUS, variant of uncertain significance.
No participants had discordant MSI and MMR IHC results.
Rows are not mutually exclusive; numerous participants had prior genetic testing for more than 1 gene.
Six of the 457 participants (1.3%; 95%CI, 0.5%–2.8%) were found to carry germline TP53 missense alterations that have not been previously described as benign changes. Four were recruited through population-based ascertainment, and 2 through clinic-based ascertainment. Statistical significance was set at P = .05. There was no significant difference in the clinical and pathological characteristics of TP53 carriers vs non-carriers, except that 4 of the TP53 carriers’ CRCs were left-sided tumors (P = .01); the sites of the remaining 2 carriers’ tumors were undefined. Microsatellite instability status was available for 2 of the carriers’ tumors, both of which were microsatellite stable. Based on available clinical data, none of these 6 probands met either classic LFS criteria8 or Chompret criteria13,14 for germline TP53 analysis. Two of the TP53 probands were younger than 30 years at the time of CRC diagnosis. A Multiplex Ligation-dependent probe Amplification (MLPA) analysis of a randomly selected subset of 100 participants did not find any TP53 deletions.
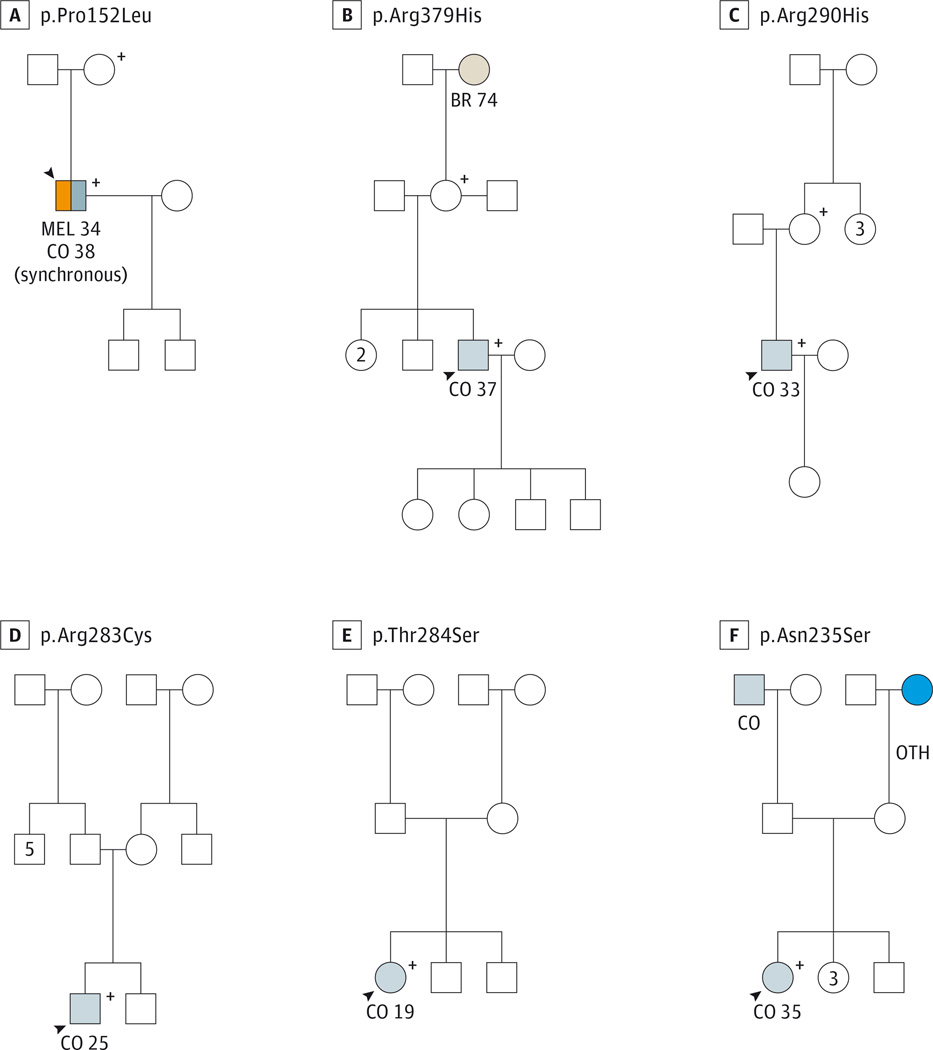
Participant 1 (of the 6 carrying the germline TP53 missense alterations) was a white man with a prior melanoma at age 34 years who was diagnosed as having synchronous sigmoid adenocarcinomas at age 38 years (Figure), and carried a c.445C>T TP53 alteration, resulting in the amino acid change p.Pro152Leu (Table 2). Germline TP53 p.Pro152Leu alterations have been reported numerous times in the literature in probands meeting Chompret criteria,14 including a proband with a pediatric choroid plexus carcinoma.22 The p.Pro152Leu TP53 alteration was also described in multiple reports of pediatric adrenocortical carcinoma probands, several of whom had relatives with histories of breast cancer and other malignant neoplasms, who carried the p.Pro152Leu alteration.23–25 Our prior report12 on gastric cancer in LFS included a family with the p.Pro152Leu TP53 alteration, in which 3 separate carriers were diagnosed as having gastric carcinoma at ages 45, 52, and 58 years.
Figure. Pedigrees of 6 Participants With Early-Onset Colorectal Cancer Found to Carry Germline TP53 Alterations.
A–F, Panels represent patients 1 to 6, respectively; see Results section. BR indicates breast cancer; CO, colorectal cancer; MEL, melanoma; OTH, other cancer. Squares represent male family members, and circles represent female family members. Numbers represent age in years at diagnosis. The numbers and letters at the top of each panel indicate the specific germline TP53 mutation carried by the family described in each panel. Plus signs indicate that the individual was confirmed to carry the germline TP53 alteration. Shading indicates that the individual was affected with cancer. The arrowheads indicate the specific study participant for that family.
Table 2.
Characteristics of TP53 Alterations Identified Among 457 Participants With Colorectal Cancer (CRC) at Age 40 Years or Youngera
| Patient No./ Sex/Ageb |
Family History | Nucleotide Change |
Genome Loc | Amino Acid Change |
Minor Allele Freq |
Species Conservation |
Exon |
In silico Pathogenicity Assessment Resultsc |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Chompret Crit |
Classic LFS Crit |
PolyPhen-2 | SIFT | |||||||
| 1/M/38 | No | No | c.455C>T | g.7578475 | p.Pro152Leu | Undefined | Highly conserved | 5 | Probably damaging | Not tolerated |
| 2/M/37 | No | No | c.1136G>A | g.7572973 | p.Arg379His | Undefined | Moderately conserved | 11 | Benign | Tolerated |
| 3/M/33 | No | No | c.869G>A rs55819519 |
g.7577069 | p.Arg290His | 0.0003EA 0.0002AA |
Moderately conserved | 8 | Benign | Tolerated |
| 4/M/25 | No | No | c.847C>T rs149633775 |
g.7577091 | p.Arg283Cys | 0.0005EA 0.0002AA |
Moderately conserved | 8 | Benign | Not tolerated |
| 5/F/19 | No | No | c.850A>T | g.7577088 | p.Thr284Ser | Undefined | Moderately conserved | 8 | Possibly damaging | Tolerated |
| 6/F/35 | No | No | c.704A>G rs114340710 |
g.7577577 | p.Asn235Ser | 0.0001EA 0.0002AA |
Moderately conserved | 7 | Benign | Tolerated |
Abbreviations: AA, African American; crit, criteria; EA, European American; freq, frequency; LFS, Li-Fraumeni syndrome; loc, location.
None of the 6 TP53 probands carried a known germline variant of uncertain significance in MLH1, MSH2, MSH6, or PMS2; none carried a monoallelic MUTYH mutation.
Age in years at CRC diagnosis.
Interactive Biosoftware.
Participant 2 was a white man with sigmoid adenocarcinoma at age 37 years, who carried a c.1136G>A (p.Arg379His) TP53 alteration. To our knowledge, germline p.Arg379His TP53 alterations have not been previously described in the literature. A somatic p.Arg379His TP53 alteration has been described in a patient with an astrocytoma; however, that patient’s tumor had another somatic TP53 alteration (p.Val218Gly)with a conservative amino acid change as well as 2 silent somatic TP53 alterations, prompting the authors of the case report to label the p.Arg379His alteration “possibly noncausative.”26
Participant 3 was a white man with CRC at age 33 years, who carried a c.869G>A (p.Arg290His) TP53 alteration. Germline p.Arg290His TP53 alterations were reported in a Portuguese family27 meeting Chompret criteria13 in which 2 relatives carrying the p.Arg290His TP53 alteration were diagnosed as having astrocytomas at ages 29 and 31 years.27 Another report28 included a French Canadian woman with breast cancer at age 44 years who carried the p.Arg290His TP53 alteration; although there were multiple other cancers in this family, they did not meet Chompret criteria.14,28 One report29 included a male proband with rhabdomyosarcoma at age 2 years and a brain tumor at 10 years of age who carried the p.Arg290His alteration and 2 other germline TP53 variants (p.Arg156His and p.Arg267Gln). This patient’s mother, who had had metachronous breast cancers at 35 and 43 years of age, was confirmed to carry the p.Arg156His and p.Arg267Gln alterations, and the maternal family history met Chompret14 criteria. The patient’s cancer-free father carried the p.Arg290His alteration.29 The p.Arg290His TP53 alteration was also described in a female proband29 with a brain tumor at age 9 years whose maternal grandfather (mutation status unknown) died of a brain tumor at age 40 years and whose paternal first cousin (mutation status unknown) died of rhabdomyosarcoma at age 4 years.
Participant 4 was a white man with sigmoid CRC at age 25 years, who carried a c.847C>T (p.Arg283Cys) TP53 alteration. Prior reports of germline p.Arg283Cys TP53 alterations include a proband with metachronous breast cancers and a subsequent leiomyosarcoma who carried both the p.Arg283Cys TP53 alteration and a pathogenic p.Arg2394X alteration in BRCA2.30 This proband’s family history included a sister with ovarian cancer at age 39 years, a mother with breast and ovarian cancers at ages 60 and 65 years, respectively, and a daughter with glioblastoma at age 41 years, none of whom had had testing for the TP53 or BRCA2 mutations.30 Another report31 described a female proband with diffuse-type gastric carcinoma at age 52 years who tested negative for a germline CDH1 mutation but carried a germline TP53 p.Arg283Cys alteration. Her family history included a mother and maternal uncle with gastric cancer (ages unknown), a sister with leukemia (age 17 years), and another sister with liver carcinoma (age 34 years), none of whom had been tested for the TP53 p.Arg283Cys alteration.31 Other reports of germline p.Arg283Cys TP53 alteration carriers include a woman32 with HER2/neu-negative breast cancer (age unknown), for whom family history data were unknown, and a 25-year-old proband33 with a desmoplastic small round cell tumor and no family history of cancer.
Participant 5 was a woman (race unknown) with descending colon adenocarcinoma at age 19 years, who carried a c.850A>T (p.Thr284Ser) TP53 alteration. To our knowledge, germline p.Thr284Ser TP53 alterations have not been previously described in the literature, although a somatic p.Thr284Ser TP53 alteration in a patient with B-cell chronic lymphocytic leukemia has been reported.34
Participant 6 was a white woman with CRC at age 35 years, who carried a c.704A>G (p.Asn235Ser) TP53 alteration. Prior reports of germline p.Asn235Ser TP53 alterations include that of a proband with an embryonal rhabdomyosarcoma of the vagina at age 19 years, with no family history of cancer.35 Another report36 included a female proband with breast cancer at age 26 years, found to carry the germline p.Asn235Ser TP53 alteration, whose mother had ovarian cancer and whose sister had breast cancer (age and mutation status were unknown for both). The germline p.Asn235Ser TP53 alteration was found in a Finnish woman37 with bilateral breast cancer at age 57 years and her nephew with an ependymoma at age 19 years; other members of this family with unknown germline TP53 status included 3 women with breast cancer at unknown ages and 3 cases of gastric cancer at unknown ages.37 One large family38 who met classic LFS criteria8 was found to carry the p.Asn235Ser alteration as well as an intronic splice site (IVS5-1G>A) TP53 mutation. The pathogenicity of the p.Asn235Ser alteration was questioned by the report’s authors because it did not segregate with cancer phenotype, whereas the IVS5-1G>A mutation did.38
Discussion
Overall, 1.3% of this large cohort of patients with early-onset CRC was found to carry germline TP53 alterations. None of these probands had clinical histories meeting the criteria for TP53 testing, and 3 of these alterations were confirmed to be carried by the participants’ cancer-free parents. Four of these alterations have been previously reported in the literature as germline TP53 alterations, and at least some of the probands described in such reports had clinical histories consistent with LFS.
Prior studies examining the rates of cancer susceptibility gene mutations in early-onset CRC have typically focused on testing for Lynch syndrome, FAP, and MAP, which are presumed to be more common causes of early-onset CRC than LFS. In another study3 of population-based CCFR patients, 5.6% of a random sample of participants with CRC younger than 50 years carried germline mutations in MLH1, MSH2, or MSH6. A retrospective analysis of a cohort of Spanish patients with CRC at 50 years oldor younger found that, after excluding those with polyposis phenotypes, 7.8% carried germline MLH1, MSH2, or MSH6 mutations and an additional 2.8% carried biallelic MUTYH mutations.4 Similarly, a retrospective analysis39 of an American cohort of patients with CRC diagnosed before age 36 years found that, after excluding those with more than 10 colorectal adenomas, 29% carried MLH1, MSH2, or MSH6 mutations, although virtually all of these carriers had a personal and/or family history of Lynch-associated cancer. Notably, they also found that 1 of the 96 participants in their cohort carried a germline TP53 mutation, although systematic TP53 testing was not otherwise performed.39 Our work adds to this literature in that it is the first study, to our knowledge, to systematically test patients with early-onset CRC for germline TP53 mutations. Although at first glance, the fraction of participants (1.3%) found to carry germline TP53 alterations in our study is small, it is comparable with the proportion of inherited CRC thought to be attributable to germline APC mutations.5
Another key finding of our study is that none of the TP53 alteration carriers had personal and/or family histories that met clinical criteria for LFS. Prior studies40,41 have found a 4% to 5% prevalence of germline TP53 mutations in population-based cohorts of women with early-onset breast cancer and, similar to our data, found that most mutation carriers do not meet clinical criteria for LFS. Such findings raise the fundamental question as to whether carriage of a germline TP53 mutation should be pathognomonic for diagnosing LFS. If a substantial fraction of TP53 mutation carriers indeed fail to meet clinical criteria for LFS, this calls into doubt the assumption that all germline TP53 alterations confer the 73% to 100% life-time risks of cancer associated with classic LFS10 and raises important issues regarding the impact of ascertainment on counseling of families found to carry germline alterations.
With the advent of multigene panels, exome sequencing, and other comprehensive strategies for hereditary cancer risk assessment, a growing number of patients will likely undergo germline analysis of TP53 and other cancer susceptibility genes, even in the absence of classic phenotypic features.15,42 A predominant concern about such approaches is that they will reveal a large number of patients with germline variant of uncertain significance, most of which will be missense mutations; this can be anxiety-provoking and potentially misinterpreted by both patients and clinicians.15,42
All 6 of the TP53 alterations identified in this study were missense mutations, thus raising questions regarding their pathogenicity. Three features of TP53 mutations in LFS make missense variant of uncertain significance (VUS) assessment particularly challenging. First, de novo alterations are thought to account for as many as 20% of pathogenic TP53 mutations, which thus prevents reassurance when a TP53 alteration carrier lacks a family history of cancer.43 Second, a disproportionate majority of pathogenic TP53 mutations are missense mutations, rather than nonsense mutations, splice site mutations, or insertion/deletions.10,44,45 Third, there are compelling data to suggest that missense TP53 mutations may actually be more oncogenic than other types of loss-of-function TP53 mutations, thereby highlighting the importance of appropriate pathogenicity assessment for any TP53 missense alterations.10,44 Notably, 5 of the TP53 alterations identified in our study were in the gene’s DNA-binding domain (codons 94–292), which is where most LFS-causing mutations are found.10
Our study’s primary strength is its use of a large, multicenter cohort of patients with CRC recruited through both clinic-based and population-based means, which serves to limit potential ascertainment bias. The availability of detailed family history data collected in a uniform fashion by cancer genetics researchers, allowed assessment of whether the families met clinical criteria for various hereditary CRC syndromes. The use of centralized TP53 mutation analysis, so as to minimize procedural errors, is another strength.
We acknowledge that our study has limitations. We are unable to verify the completeness and accuracy of reported family histories since the initiation of the study, and it is unknown if family members may have subsequently developed LFS-associated tumors, which is particularly important owing to the young age of the probands studied. Furthermore, because participants with known hereditary CRC syndromes were excluded, we are unable to claim that our study defines the true prevalence of TP53 alterations in patients with early-onset CRC. Because many participants in our study had not previously undergone Lynch syndrome testing, it is possible that this cohort included patients with undiagnosed Lynch syndrome. If indeed a fraction of our cohort has unrecognized Lynch syndrome, then the true prevalence of TP53 mutations in patients with early-onset CRC without Lynch syndrome would actually be higher than the 1.3% rate observed in this study. The true prevalence of most hereditary CRC syndromes remains undefined, because most prior large studies have involved some sort of clinical preselection, rather than population-based testing. Thus, to precisely define the prevalence of patients with CRC with cancer susceptibility gene mutations, future studies using multiplex panel testing, whole genome sequencing, or some other form of comprehensive germline analysis will be needed.
Our in silico assessments and literature searches are imperfect tools to interpret the clinical and biologic significance of identified TP53 alterations, and we are thus unable to precisely classify their pathogenicity or determine whether they are truly the etiologic basis of the observed early-onset CRC. Although this is admittedly another limitation of our study, it also highlights a prominent real-world challenge that clinicians will face with more widespread TP53 testing in patients lacking classic LFS phenotypes. The typical management for TP53 mutation carriers involves aggressive and early radiographic, laboratory, endoscopic, and sometimes even surgical risk-reduction strategies, based on the notion that TP53 mutations confer a LFS phenotype with a near-100% lifetime risk of malignant disease.10,46,47 Our study was unable to define the penetrance of our participants’ TP53 alterations, although these findings raise the hypothesis that some pathogenic TP53 mutations do not confer a classic LFS phenotype.
Conclusions
To our knowledge, this report represents the largest study to date examining germline TP53 alterations in individuals with early-onset CRC. Given that none of the TP53 probands in our study met clinical criteria for LFS, our data raise the question as to whether LFS should be defined by the presence of pathogenic germline TP53 mutations. This reflects a growing quandary in the field of cancer genetics, which, in recent years, has shifted toward using genotypic data (eg, carriage of a germline MMR mutation in Lynch syndrome) to define and diagnose specific hereditary cancer syndromes rather than the historical practice of using phenotypic information (eg, fulfillment of Amsterdam criteria in Lynch syndrome). Newer studies consistently show, however, that a subset of patients with germline mutations in MMR genes, APC, MUTYH, and now TP53, have particularly attenuated clinical histories, calling into question whether management recommendations should take both genotype and phenotype into account.48,49
For patients found to carry TP53 mutations in the setting of early-onset CRC but no other clinical features of LFS, our data suggest that clinicians may be able to reassure such probands that the lifetime risk of classic LFS cancers may not be as high as the 73% to 100% risk typically quoted, although confirmatory studies are certainly needed. These findings highlight the inevitable challenges raised by comprehensive approaches to hereditary cancer risk assessment, namely, the interpretation of missense alterations and VUS in cancer susceptibility genes, as well as the difficulties in estimating future cancer risks in families with atypical phenotypes. With modern techniques for comprehensively genotyping cancer patients, interpreting such germline results will undoubtedly be a prominent challenge in the counseling and management of at-risk individuals.
Supplementary Material
At a Glance.
Individuals with Li-Fraumeni syndrome (LFS) caused by germline TP53 mutations are estimated to have a 73% to 100% lifetime risk of cancer, including colorectal cancer (CRC).
The purpose of this study was to determine the frequency of germline TP53 alterations in individuals with early-onset CRC.
Of 457 participants diagnosed as having CRC at age 40 years or younger, 1.3% carried germline TP53 alterations.
None of the TP53 probands in this study had a personal or family cancer history that fulfilled clinical LFS criteria.
Cancer risk in TP53 mutation carriers may be different in patients presenting with early-onset CRC compared with those who present with classic LFS family histories.
Acknowledgments
Funding/Support: This study was funded by the National Institutes of Health (NIH) (National Cancer Institute [NCI]) K24CA113433 (Dr Syngal) and UM1CA167551 (Dr Haile), as well as an American Gastroenterological Association (AGA) Translational Research Award (Drs Syngal and Kucherlapati). The Colon Cancer Family Registry (CCFR) was supported by the NCI, NIH, of Health under RFA No. CA-95-011 and through cooperative agreements with members of the CCFR and Principal Investigators. Collaborating centers include the Australasian Colorectal Cancer Family Registry, Colon Cancer Family Registry: USC Consortium, Mayo Clinic Cooperative Family Registry for Colon Cancer Studies, Ontario Registry for Studies of Familial Colorectal Cancer, and Seattle Colorectal Cancer Family Registry.
Role of Funder/Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
We thank all staff and study participants of the CCFR for their invaluable contributions to this project.
Footnotes
Author Contributions: Drs Yurgelun and Syngal had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Drs Yurgelun and Masciari contributed equally to the study.
Study concept and design: Joshi, Hopper, Potter, Kucherlapati, Syngal.
Acquisition, analysis, or interpretation of data: Yurgelun, Masciari, Joshi. Mercado, Lindor, Gallinger, Hopper, Jenkins, Buchanan, Newcomb, Potter, Haile, Kucherlapati, Syngal.
Drafting of the manuscript: Yurgelun, Masciari, Joshi, Mercado, Haile, Kucherlapati.
Critical revision of the manuscript for important intellectual content: Yurgelun, Joshi, Lindor, Gallinger, Hopper, Jenkins, Buchanan, Newcomb, Potter, Kucherlapati, Syngal.
Statistical analysis: Mercado.
Obtained funding: Gallinger, Hopper, Jenkins, Newcomb, Kucherlapati, Syngal.
Administrative, technical, or material support: Masciari, Joshi, Mercado, Hopper, Buchanan, Newcomb, Potter, Haile, Kucherlapati, Syngal.
Study supervision: Hopper, Newcomb, Haile, Kucherlapati, Syngal.
Conflict of Interest Disclosures: Dr Yurgelun reports receiving research funding from Myriad Genetic Laboratories. Dr Syngal does not receive research funding from Myriad; however, she is Dr Yurgelun’s mentor and collaborator on the projects that will be supported by Myriad. No other disclosures are reported.
Group Information: The CCFR principal investigators are Robert W. Haile, PhD, Stanford Cancer Institute, Stanford, California; Mark A. Jenkins, PhD, University of Melbourne, Carlton, Victoria, Australia; and Noralane M. Lindor, MD, Mayo Clinic, Scottsdale, Arizona.
Disclaimer: The content of this article does not necessarily reflect the views or policies of the NCI or any of the collaborating centers in the CCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the CCFR.
Previous Presentations: Preliminary data from this study were presented as an abstract at the annual Digestive Disease Week; May 2–5, 2010; New Orleans, Louisiana; and as an oral abstract at the Annual Meeting of the Collaborative Group of the Americas on Inherited Colorectal Cancer; September 15–16, 2014; New Orleans, Louisiana.
REFERENCES
- 1.Ahnen DJ, Wade SW, Jones WF, et al. The increasing incidence of young-onset colorectal cancer: a call to action. Mayo Clin Proc. 2014;89(2):216–224. doi: 10.1016/j.mayocp.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Chang DT, Pai RK, Rybicki LA, et al. Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: an adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Mod Pathol. 2012;25(8):1128–1139. doi: 10.1038/modpathol.2012.61. [DOI] [PubMed] [Google Scholar]
- 3.Limburg PJ, Harmsen WS, Chen HH, et al. Prevalence of alterations in DNA mismatch repair genes in patients with young-onset colorectal cancer. Clin Gastroenterol Hepatol. 2011;9(6):497–502. doi: 10.1016/j.cgh.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giráldez MD, Balaguer F, Bujanda L, et al. MSH6 and MUTYH deficiency is a frequent event in early-onset colorectal cancer. Clin Cancer Res. 2010;16(22):5402–5413. doi: 10.1158/1078-0432.CCR-10-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoffel EM, Kastrinos F. Familial colorectal cancer, beyond Lynch syndrome. Clin Gastroenterol Hepatol. 2014;12(7):1059–1068. doi: 10.1016/j.cgh.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong P, Verselis SJ, Garber JE, et al. Prevalence of early onset colorectal cancer in 397 patients with classic Li-Fraumeni syndrome. Gastroenterology. 2006;130(1):73–79. doi: 10.1053/j.gastro.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms: a familial syndrome? Ann Intern Med. 1969;71(4):747–752. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 8.Li FP, Fraumeni JF, Jr, Mulvihill JJ, et al. A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988;48(18):5358–5362. [PubMed] [Google Scholar]
- 9.Garber JE, Goldstein AM, Kantor AF, Dreyfus MG, Fraumeni JF, Jr, Li FP. Follow-up study of twenty-four families with Li-Fraumeni syndrome. Cancer Res. 1991;51(22):6094–6097. [PubMed] [Google Scholar]
- 10.Malkin D. Li-fraumeni syndrome. Genes Cancer. 2011;2(4):475–484. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez KD, Noltner KA, Buzin CH, et al. Beyond Li Fraumeni syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol. 2009;27(8):1250–1256. doi: 10.1200/JCO.2008.16.6959. [DOI] [PubMed] [Google Scholar]
- 12.Masciari S, Dewanwala A, Stoffel EM, et al. Gastric cancer in individuals with Li-Fraumeni syndrome. Genet Med. 2011;13(7):651–657. doi: 10.1097/GIM.0b013e31821628b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chompret A, Abel A, Stoppa-Lyonnet D, et al. Sensitivity and predictive value of criteria for p53 germline mutation screening. J Med Genet. 2001;38(1):43–47. doi: 10.1136/jmg.38.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tinat J, Bougeard G, Baert-Desurmont S, et al. 2009 version of the Chompret criteria for Li Fraumeni syndrome. J Clin Oncol. 2009;27(26):e108–e109. doi: 10.1200/JCO.2009.22.7967. [DOI] [PubMed] [Google Scholar]
- 15.Domchek SM, Bradbury A, Garber JE, Offit K, Robson ME. Multiplex genetic testing for cancer susceptibility: out on the high wire without a net? J Clin Oncol. 2013;31(10):1267–1270. doi: 10.1200/JCO.2012.46.9403. [DOI] [PubMed] [Google Scholar]
- 16.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 17.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 18.Cleary SP, Cotterchio M, Jenkins MA, et al. Germline MutY human homologue mutations and colorectal cancer: a multisite case-control study. Gastroenterology. 2009;136(4):1251–1260. doi: 10.1053/j.gastro.2008.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzhubei IA, Schmidt S, Peshkin L, et al. Amethod and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Exome Variant Server NGESPE, Seattle WA. [Accessed May 2, 2014]; http://evs.gs.washington.edu/EVS/. [Google Scholar]
- 22.Dickens DS, Dothage JA, Heideman RL, Ballard ET, Jubinsky PT. Successful treatment of an unresectable choroid plexus carcinoma in a patient with Li-Fraumeni syndrome. J Pediatr Hematol Oncol. 2005;27(1):46–49. doi: 10.1097/01.mph.0000152569.60694.1f. [DOI] [PubMed] [Google Scholar]
- 23.Tabori U, Nanda S, Druker H, Lees J, Malkin D. Younger age of cancer initiation is associated with shorter telomere length in Li-Fraumeni syndrome. Cancer Res. 2007;67(4):1415–1418. doi: 10.1158/0008-5472.CAN-06-3682. [DOI] [PubMed] [Google Scholar]
- 24.Varley JM, McGown G, Thorncroft M, et al. Are there low-penetrance TP53 alleles? evidence from childhood adrenocortical tumors. Am J Hum Genet. 1999;65(4):995–1006. doi: 10.1086/302575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner J, Portwine C, Rabin K, Leclerc JM, Narod SA, Malkin D. High frequency of germline p53 mutations in childhood adrenocortical cancer. J Natl Cancer Inst. 1994;86(22):1707–1710. doi: 10.1093/jnci/86.22.1707. [DOI] [PubMed] [Google Scholar]
- 26.Hayes VM, Dirven CM, Dam A, et al. High frequency of TP53 mutations in juvenile pilocytic astrocytomas indicates role of TP53 in the development of these tumors. Brain Pathol. 1999;9(3):463–467. doi: 10.1111/j.1750-3639.1999.tb00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto C, Veiga I, Pinheiro M, et al. TP53 germline mutations in Portugal and genetic modifiers of age at cancer onset. Fam Cancer. 2009;8(4):383–390. doi: 10.1007/s10689-009-9251-y. [DOI] [PubMed] [Google Scholar]
- 28.Arcand SL, Maugard CM, Ghadirian P, et al. Germline TP53 mutations in BRCA1 and BRCA2 mutation-negative French Canadian breast cancer families. Breast Cancer Res Treat. 2008;108(3):399–408. doi: 10.1007/s10549-007-9608-6. [DOI] [PubMed] [Google Scholar]
- 29.Quesnel S, Verselis S, Portwine C, et al. p53 compound heterozygosity in a severely affected child with Li-Fraumeni syndrome. Oncogene. 1999;18(27):3970–3978. doi: 10.1038/sj.onc.1202783. [DOI] [PubMed] [Google Scholar]
- 30.Manoukian S, Peissel B, Pensotti V, et al. Germline mutations of TP53 and BRCA2 genes in breast cancer/sarcoma families. Eur J Cancer. 2007;43(3):601–606. doi: 10.1016/j.ejca.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 31.Keller G, Vogelsang H, Becker I, et al. Germline mutations of the E-cadherin (CDH1) and TP53 genes, rather than of RUNX3 and HPP1, contribute to genetic predisposition in German gastric cancer patients. J Med Genet. 2004;41(6):e89. doi: 10.1136/jmg.2003.015594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melhem-Bertrandt A, Bojadzieva J, Ready KJ, et al. Early onset HER2-positive breast cancer is associated with germline TP53 mutations. Cancer. 2012;118(4):908–913. doi: 10.1002/cncr.26377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell G, Ballinger ML, Wong S, et al. International Sarcoma Kindred Study. High frequency of germline TP53 mutations in a prospective adult-onset sarcoma cohort. PLoS One. 2013;8(7):e69026. doi: 10.1371/journal.pone.0069026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pekova S, Mazal O, Cmejla R, et al. A comprehensive study of TP53 mutations in chronic lymphocytic leukemia: analysis of 1287 diagnostic and 1148 follow-up CLL samples. Leuk Res. 2011;35(7):889–898. doi: 10.1016/j.leukres.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 35.Diller L, Sexsmith E, Gottlieb A, Li FP, Malkin D. Germline p53 mutations are frequently detected in young children with rhabdomyosarcoma. J Clin Invest. 1995;95(4):1606–1611. doi: 10.1172/JCI117834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornelis RS, van Vliet M, van de Vijver MJ, et al. Three germline mutations in the TP53 gene. Hum Mutat. 1997;9(2):157–163. doi: 10.1002/(SICI)1098-1004(1997)9:2<157::AID-HUMU8>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 37.Huusko P, Castrén K, Launonen V, et al. Germ-line TP53 mutations in Finnish cancer families exhibiting features of the Li-Fraumeni syndrome and negative for BRCA1 and BRCA2. Cancer Genet Cytogenet. 1999;112(1):9–14. doi: 10.1016/s0165-4608(98)00258-1. [DOI] [PubMed] [Google Scholar]
- 38.van Hest LP, Ruijs MW, Wagner A, et al. Two TP53 germline mutations in a classical Li-Fraumeni syndrome family. Fam Cancer. 2007;6(3):311–316. doi: 10.1007/s10689-006-9115-7. [DOI] [PubMed] [Google Scholar]
- 39.Jasperson KW, Vu TM, Schwab AL, et al. Evaluating Lynch syndrome in very early onset colorectal cancer probands without apparent polyposis. Fam Cancer. 2010;9(2):99–107. doi: 10.1007/s10689-009-9290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouchawar J, Korch C, Byers T, et al. Population-based estimate of the contribution of TP53 mutations to subgroups of early-onset breast cancer: Australian Breast Cancer Family Study. Cancer Res. 2010;70(12):4795–4800. doi: 10.1158/0008-5472.CAN-09-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lalloo F, Varley J, Moran A, et al. BRCA1, BRCA2 and TP53 mutations in very early-onset breast cancer with associated risks to relatives. Eur J Cancer. 2006;42(8):1143–1150. doi: 10.1016/j.ejca.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 42.Stadler ZK, Schrader KA, Vijai J, Robson ME, Offit K. Cancer genomics and inherited risk. J Clin Oncol. 2014;32(7):687–698. doi: 10.1200/JCO.2013.49.7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez KD, Buzin CH, Noltner KA, et al. High frequency of de novo mutations in Li-Fraumeni syndrome. J Med Genet. 2009;46(10):689–693. doi: 10.1136/jmg.2008.058958. [DOI] [PubMed] [Google Scholar]
- 44.Bougeard G, Sesboüé R, Baert-Desurmont S, et al. French LFS Working Group. Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families. J Med Genet. 2008;45(8):535–538. doi: 10.1136/jmg.2008.057570. [DOI] [PubMed] [Google Scholar]
- 45.Olivier M, Goldgar DE, Sodha N, et al. Li-Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Res. 2003;63(20):6643–6650. [PubMed] [Google Scholar]
- 46.Masciari S, Van den Abbeele AD, Diller LR, et al. F18-fluorodeoxyglucose-positron emission tomography/computed tomography screening in Li-Fraumeni syndrome. JAMA. 2008;299(11):1315–1319. doi: 10.1001/jama.299.11.1315. [DOI] [PubMed] [Google Scholar]
- 47.Wijnen J, Khan PM, Vasen H, et al. Majority of hMLH1 mutations responsible for hereditary nonpolyposis colorectal cancer cluster at the exonic region 15–16. Am J Hum Genet. 1996;58(2):300–307. [PMC free article] [PubMed] [Google Scholar]
- 48.Grover S, Kastrinos F, Steyerberg EW, et al. Prevalence and phenotypes of APC and MUTYH mutations in patients with multiple colorectal adenomas. JAMA. 2012;308(5):485–492. doi: 10.1001/jama.2012.8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreira L, Balaguer F, Lindor N, et al. EPICOLON Consortium. Identification of Lynch syndrome among patients with colorectal cancer. JAMA. 2012;308(15):1555–1565. doi: 10.1001/jama.2012.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.