Abstract
Glutamylation, the most prevalent tubulin posttranslational modification, marks stable microtubules and regulates recruitment and activity of microtubule- interacting proteins. Nine enzymes of the tubulin tyrosine ligase-like (TTLL) family catalyze glutamylation. TTLL7, the most abundant neuronal glutamylase, adds glutamates preferentially to the β–tubulin tail. Coupled with ensemble and single- molecule biochemistry, our hybrid X-ray and cryo-electron microscopy structure of TTLL7 bound to the microtubule delineates a tripartite microtubule recognition strategy. The enzyme uses its core to engage the disordered anionic tails of α– and β–tubulin, and a flexible cationic domain to bind the microtubule and position itself for β–tail modification. Furthermore, we demonstrate that all single-chain TTLLs with known glutamylase activity utilize a cationic microtubule-binding domain analogous to that of TTLL7. Therefore, our work reveals the combined use of folded and intrinsically disordered substrate recognition elements as the molecular basis for specificity among the enzymes primarily responsible for chemically diversifying cellular microtubules.
INTRODUCTION
The αβ-tubulin heterodimer, the basic building block of the microtubule polymer, consists of a globular core and negatively charged, diversely posttranslationally modified, and intrinsically disordered C-terminal tails that decorate the microtubule exterior. Most of the chemical complexity of tubulin observed in cells arises from the differential action of tubulin tyrosine ligase (TTL) and the homologous tubulin tyrosine ligase-like (TTLL) enzymes. The latter are phylogenetically widespread and functionally diversified proteins that carry out either glutamylation or glycylation of tubulin tails. These tails are in the vicinity of known binding sites for motors and microtubule associated proteins (MAPs) where they tune their interactions. Their chemically diverse posttranslational modifications may thus constitute a “tubulin code” read by cytoskeletal effectors (reviewed in Verhey and Gaertig, 2007), analogous to the histone code (Jenuwein and Allis, 2001). Understanding the molecular underpinnings for the biochemical characteristics of TTLLs is essential to elucidate the tubulin code, since it is the combinatorial action of these enzymes that generates the complex microtubule modification patterns observed in cells that differentiate microtubules for distinct functions in specific tissues or subcellular structures.
Glutamylation, the most abundant tubulin posttranslational modification in the adult mammalian brain (Audebert et al., 1994), is the ATP-dependent addition of glutamates, either singly or sequentially in chains, to conserved internal glutamate residues of the α- or β-tubulin tails. Glutamylation is conserved in all metazoans as well as protists, where it is key to the assembly and function of cilia and flagella (reviewed in Verhey and Gaertig, 2007; Garnham and Roll-Mecak, 2012). Mass spectrometry of mammalian brain tubulin revealed one to six glutamates added to both α- and β-tails at distinct conserved locations (Alexander et al., 1991; Eddé et al., 1990; Redeker et al., 1992; Rudiger et al., 1992). The longest glutamate chains are on axonemal microtubules (Geimer et al., 1997; Schneider et al., 1998) and centrioles (Bobinnec et al., 1998), where tubulin tails contain as many as 21 glutamates (Schneider et al., 1998). Humans have nine glutamylases of the TTLL family (van Dijk et al., 2007; reviewed in Garnham and Roll-Mecak, 2012). Of these, TTLL7, which is conserved from the acorn worm to primates, is the most abundantly expressed in the mammalian nervous system where it is critical for neurite outgrowth and localization of dendritic MAPs (Ikegami et al., 2006). It is primarily responsible for the dramatic increase in tubulin glutamylation during postnatal neuronal maturation (Ikegami et al., 2006).
Misregulation of tubulin glutamylation leads to several physiological abnormalities. Hyperglutamylation in Purkinje cell degeneration (pcd) mice leads to neurodegeneration (Rogowski et al., 2010), while depressed glutamylation impairs cilia-facilitated mucous flow, inhibits neurite outgrowth and compromises synaptic function (Ikegami et al., 2007; Ikegami et al., 2006; Ikegami et al., 2010; Janke et al., 2005). Moreover, mutations in TTLL glutamylases or proteins required for their localization have been implicated in several degenerative disorders including retinal dystrophy (Sergouniotis et al., 2014) and Joubert syndrome (Lee et al., 2012). At the molecular level, glutamylation is enriched on stable microtubules and regulates the interaction between microtubules and molecular motors or MAPs. Glutamylation enhances the processivity of kinesin-1 and 2 (O'Hagan et al., 2011; Sirajuddin et al., 2014), regulates inner arm dynein motility in flagella (Kubo et al., 2010; Suryavanshi et al., 2010) and creates microtubule “fast tracks” for efficient motor based vesicle transport from the Golgi to the plasma membrane (Bulinski et al., 1997; Spiliotis et al., 2008). It also modulates the binding of MAPs tau, MAP1A and B, and MAP2 (Bonnet et al., 2001; Boucher et al., 1994) and enhances the activity of the microtubule severing enzymes katanin and spastin (Lacroix et al., 2010; Roll-Mecak and Vale, 2008; Sharma et al., 2007).
Tubulin tyrosine ligase (TTL) serves as the structural blueprint for the TTLL family of enzymes (Janke et al., 2005; Szyk et al., 2011; van Dijk et al., 2007). TTL selectively binds to the isolated tubulin dimer (Raybin and Flavin, 1975; Szyk et al., 2011). The preference of TTL for the free tubulin dimer has the cellular consequence that newly polymerized, dynamic microtubules are enriched in tyrosination. In contrast, and despite conservation of their structural core with that of TTL (Szyk et al., 2011), TTLLs preferentially modify the microtubule polymer (Mukai et al., 2009; van Dijk et al., 2007) and glutamylation is enriched on stable, long-lived microtubules. Different TTLL glutamylases exhibit a preference for either the α- or β-tubulin tails (van Dijk et al., 2007) and cellular motors respond differentially to α- or β-tail glutamylation (Sirajuddin et al., 2014). To provide the molecular basis for understanding substrate specificity within this family of structurally homologous enzymes that are key players in establishing the tubulin code, we have now combined X-ray crystallography, cryo-electron microscopy (cryo-EM), and structure-based ensemble and single-molecule biochemistry to determine how TTLL7 recognizes and glutamylates microtubules. The first structural characterization of TTLL7 and its complex with its cognate substrate delineates a tripartite mode of microtubule recognition mediated by strong ionic interactions between both α- and β-tubulin C-terminal tails and the TTLL7 core, as well as engagement of the tubulin body by a flexible cationic microtubule binding domain (c-MTBD) critical for glutamylation activity. Moreover, we discovered analogous cationic MTBDs required for function in all TTLL family glutamylases with autonomous activity. Thus, our work provides a detailed molecular framework to understand microtubule glutamylation, an essential posttranslational modification conserved from ciliates to primates.
RESULTS
X-ray and EM Analysis of TTLL7 and its Microtubule Complex
To elucidate how TTLL7 recognizes the microtubule and modifies the β-tubulin tail, we employed a hybrid structural approach combining X-ray crystallography and cryo-EM to determine the three-dimensional structure of TTLL7 in isolation (Figure 1) as well as in complex with its cognate substrate, the microtubule (Figure 2). Homo sapiens TTLL7 constructs (residues 1-518 and residues 36-518, used for cryo-EM and X-ray crystallographic structure determination, respectively; Experimental Procedures) are active in glutamylating microtubules, resemble a naturally occurring form of TTLL7 lacking a C-terminal domain required for ciliary localization (Ikegami et al., 2006; van Dijk et al., 2007), and like the longer variant, exhibit a preference for modifying βtubulin (Ikegami et al., 2006; Mukai et al., 2009; van Dijk et al., 2007). The TTLL7 structure is the first reported for a TTLL family member. The TTLL7-ADP structure was refined to 2.55 Å resolution with a 26.9% Rfree factor (Table S1, Experimental Procedures). Although we co-crystallized TTLL7 with ADP, ATP or a slowly hydrolyzable ATP analog (Table S1 and Experimental Procedures), there are no significant structural differences between the resulting structures [root mean-square deviation (rmsd) < 0.3 Å over all atoms]; the γ-phosphate appears not to be engaged in either the ATP or AMPPNP structures, possibly because of the absence of the tubulin substrate (see below).
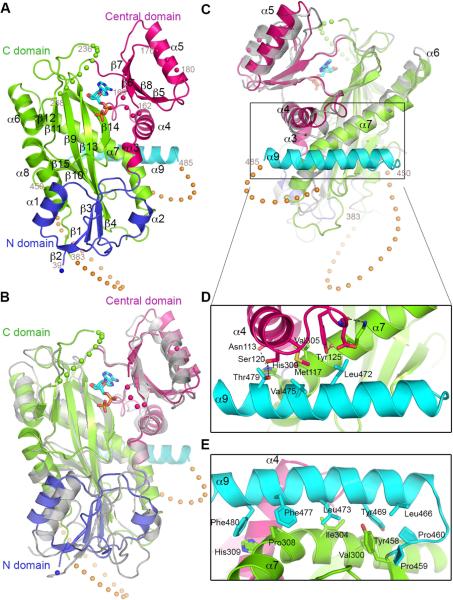
Figure 1. Crystal structure of the Homo sapiens TTLL7 core.
(A) Ribbon representation of the TTLL7 core bound to ADP. Nucleotide shown as a stick model. Spheres represent disordered polypeptide chain regions. c-MTBD residues, orange spheres.
(B,C) Superposition of TTLL7 and TTL (PDB ID 3TII; grey) crystal structures; (B), nucleotide binding view, (C), side-view, 90° rotated from (B).
(D, E) Close-up views showing residues important for helix α9 packing with the TTL-homologous core. See also Table S1 and Figure S1.
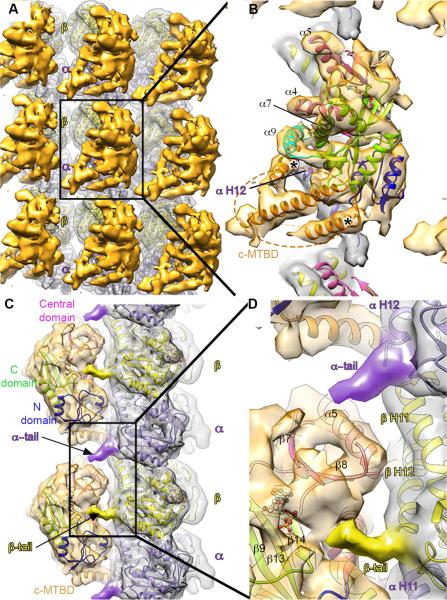
Figure 2. Cryo-EM 3D reconstruction of TTTL7 bound to the microtubule.
(A) Surface rendering of the TTLL7-bound microtubule. TTLL7 (gold) spans the αβ-tubulin dimer (silver). The transparent microtubule surface shows the docked atomic model of tubulin (PDB ID 1JFF, Lowe et al., 2001). α- and β-tubulin, purple and yellow, respectively.
(B) Boxed area in (A) expanded to show the TTLL7 crystal structure (colored as in Figure 1A) docked into the cryo-EM density, as well as the modeled helices in the c-MTBD (orange); tubulin colored as in (A). Asterisks lie above putative interactions between the c-MTBD and helix H12 of α-tubulin. Dashed orange ellipse indicates that the c-MTBD density extends beyond the partial α-helical model.
(C) Side view of a protofilament with docked tubulin and TTLL7 atomic models showing the interactions between the TTLL7 core and C-terminal tubulin tails of neighboring tubulin dimers. α- and β-tubulin tails purple and yellow, respectively. TTLL7 colored as in Figure 1A.
(D) Boxed area in (C) expanded to show the interactions between the α- and β-tubulin tails and the TTLL7 core. See also Table S2 and Figure S2.
To investigate the TTLL7-microtubule interaction, we solved the structure of the enzyme bound to both, heterogeneous bovine brain tubulin, and human tubulin with no posttranslational modifications by cryo-EM (Figure S2 and Experimental Procedures). Even though TTLL7 displays ~1.8-fold higher affinity for brain microtubules (Figure S3), the unmodified microtubules yielded more regular decoration, improving the resolution of the 3D maps from 10.6 to 7.95Å (Fourier shell correlation criterion at 0.5; Figures 2, S2, Table S2 and Experimental Procedures). EM reconstructions reported to date are of heterogeneous microtubules generated by randomly repolymerizing tubulin isolated from brain tissue, which contains at least 22 different charge variants, mostly due to isoform heterogeneity and abundant and variable glutamylation of tubulin tails (Zambito et al., 2002).
TTLL7 Architecture
Our X-ray structure reveals an elongated TTLL7 core (~40 ×60 ×45 Å3; Figure 1A) consisting of three domains (N-terminal, central and C-terminal, residues 36-100, 101-193 and 198-484, respectively). The TTLL7 core is structurally homologous to TTL (Szyk et al., 2011; Figures 1B and C; all-Cα rmsd = 3.5 Å) and comprises a central ten-stranded β-sheet of mixed polarity with strands contributed by both the N- and C-domains, surrounded by helices from all three domains (Figures 1A and S1). The TTLL7 core comprises a C-terminal helix (α9) absent from TTL (Figure 1). This helix packs against the face of the protein opposite to the active site and is stabilized by forming a four-helix bundle with helices α3, α4 and α7. It makes extensive apolar knobs-into-holes interactions with α7 of the C-domain and has a more polar interface with α4 of the central domain (Figures 1D and E). α3, α4 and α7 are also present in TTL; however, their surface residues are not conserved with TTLL7, consistent with the absence of α9 in TTL.
In our EM reconstruction, the microtubule-bound TTLL7 was easily identifiable as a large density spanning the αβ-tubulin heterodimer into which the TTLL7 X-ray structure could be fitted with high confidence (UCSF Chimera correlation coefficient 0.85; Figures 2A, 2B and Supplementary Movie S1). The cryo-EM TTLL7 density revealed a feature equivalent to ~10 kDa closely apposed to the microtubule shaft. This structural element extends from the ligase core toward the inter-protofilament groove, and likely corresponds to a ~65 amino acid, cationic insertion (residues 384-450) in the TTLL7 C-domain as well as residues immediately C-terminal to helix α9 (residues 484-518), neither of which are resolved in the crystal structure of the isolated enzyme (Figures 1A, 2B, S1 and S2G). This newly defined cationic microtubule-binding domain (c-MTBD) comprises residues highly conserved in TTLL7 orthologs, but absent from TTL (Figures S1 and 1C). Secondary structure predictions indicate that the c-MTBD is α-helix rich (Figure S2J). At sub-nanometer resolution, α-helices adopt a characteristic sausage-like shape. Such features are clearly discernible in the density corresponding to the c-MTBD. Based on secondary structure predictions and the docked TTLL7 crystal structure, helices were modeled into these features of the cryo-EM density that account for part of the c-MTBD (Figures 2B, S2G, Supplementary Movie S1 and Extended Experimental Procedures). Our hybrid structural analysis suggests that the c-MTBD is anchored to the TTLL7 core through helix α9 (Figure 2B).
A Tripartite TTLL7 Interface with the Microtubule
TTLL7 binds to helices H11 and H12 of both α- and β-tubulin (Figures 2B and C), at a site that partially overlaps with the microtubule surface used by the motors kinesin and dynein (Gigant et al., 2013; Redwine et al., 2012; Rice et al., 1999; Sindelar and Downing, 2010). Our EM map shows a limited interface between the TTLL7 core (excluding the c-MTBD) and the tubulin body that comprises two main contact points: the loop preceding TTLL7 helix α5 that interacts with helix H12 of β-tubulin, and the C-terminal half of helix α7 that is positioned between H11 and H12 of α-tubulin. TTLL7 helix α9 lies almost perpendicular to the microtubule interface, with its C-terminus pointing towards the inter-protomer groove at the center of the tubulin dimer (Figure 2B).
Strikingly, density attributable to both α- and β-tubulin C-terminal tails is reproducibly visible in our cryo-EM maps (Figure 2C). The anionic β-tubulin tail contacts TTLL7 at the N- and central domain junction with EM density attributable to the β-tail extending towards the TTLL7 active site, likely guided through interactions with conserved cationic residues lining the groove between these domains (Figure 2D). In the complex, the TTLL7 active site is more compact and appears to clamp down around the β-tail, compared to our crystal structure of the isolated enzyme in which the active site is open and partially disordered. Nucleotide (either ATP or ADP) is required for high-affinity binding to the microtubule as the apo enzyme displays five-fold lower affinity (Figure S3G), mostly due to a faster off-rate. Together, these results suggest a concerted remodeling of the active site that requires both the nucleotide and tubulin substrates. A similar disorder-to-order transition is found in TTL, which has an open, partially disordered active site in the absence of the α-tail (Szyk et al., 2011), but compacts and closes when engaging both tubulin and ATP (Prota et al., 2013). In our TTLL7-microtubule complex structure, the TTLL7 central domain is contacted in trans by the α-tail of the adjacent tubulin dimer on the same protofilament (Figures 2C and D). The density for the β-tail is better defined than that for the α-tail, suggesting the latter makes a weaker interaction with TTLL7.
TTLL7 makes a tripartite interaction with the microtubule. The flexible α- and β-tails from longitudinally adjacent tubulin dimers associate with the core of the enzyme, while the TTLL7 c-MTBD interacts primarily with the α-tubulin protomer of the αβ-tubulin dimer through two main contacts (Figure 2). The N-terminal region of the c-MTBD likely recognizes the C-terminal half of helix H12 of α-tubulin, while the C-terminal region interfaces with the N-terminal portion of α-tubulin helix H12 (Figure 2B). This multipronged microtubule recognition by TTLL7 ensures that the flexible β-tubulin tail is selectively guided into the active site. Consistent with this, TTLL7 modifies equally well peptides corresponding either to α or β-tubulin C-terminal sequences in isolation, but it preferentially modifies the β-tubulin tail when presented in the context of the microtubule (Figures S3A and B). Thus, our hybrid crystallographic and EM analysis reveals the structural basis for TTLL7 recognition of microtubules as well as its preferential modification of the β-tubulin tail.
Tubulin Tails Are Essential for TTLL7 Microtubule Recognition
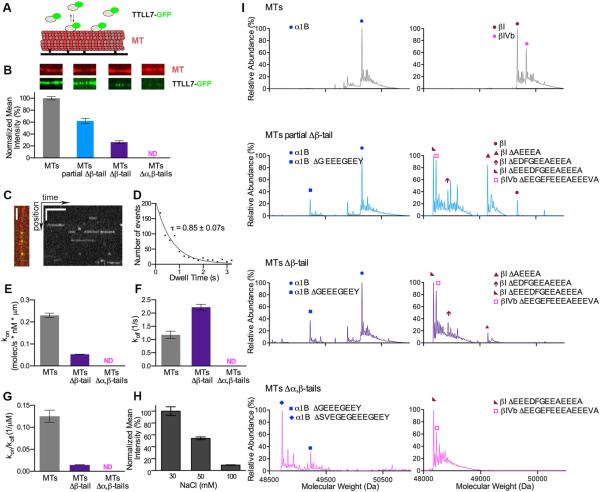
To evaluate the functional significance of the interactions between the two tubulin tails and TTLL7 revealed by our hybrid analysis, we used total internal reflection fluorescence (TIRF) microscopy and analyzed the association of GFP-tagged TTLL7 (the activity of GFP-tagged TTLL7 is similar to that of the untagged enzyme; Figures 3 and S3C) with intact microtubules as well as microtubules lacking either their β-tails, or both α- and β-tails. We find that the tubulin tails contribute significantly to the binding energy of this interaction. While TTLL7-GFP shows robust binding to microtubules, proteolytic removal of ~30% of the β-tubulin tails leads to a 40% decrease in binding (Figures 3B and 3I). Complete removal of the β-tails leads to a 74% reduction in binding, while removal of both α- and β-tails leads to total loss of binding (Figure 3B). Single-molecule kinetic measurements indicate that β-tail loss affects both the kon and koff of the enzyme, with the former affected disproportionally (78% reduction; Figures 3C, D, E, F and G). The affinity of TTLL7 for microtubules derived from our single-molecule kon and koff measurements closely matches that determined from bulk experiments (Kd ~ 1.9 μM versus 3 μM with brain microtubules; single-molecule and ensemble assays, respectively; Figures S3E and S3G). Binding of TTLL7 to microtubules missing both C-terminal tails was undetectable at concentrations as high as 60 μM, over 20-fold above the Kd of the enzyme for intact microtubules (Figure S3H). Consistent with the importance of electrostatic interactions, binding between the highly charged TTLL7 and microtubules is strongly modulated by ionic strength (Figure 3H).
Figure 3. Contribution of tubulin C-terminal tails to TTLL7 microtubule binding.
(A) Schematic of the microtubule-binding assay. Surface immobilized microtubules and GFP-tagged TTLL7 were imaged by TIRF microscopy.
(B) GFP-tagged TTLL7 was incubated with microtubules (MTs), microtubules missing ~30% of their β-tubulin tails (MTs partial Δβ-tail), microtubules missing all their β-tubulin tails (MTs Δβ-tail) and microtubules missing both α- and β-tubulin tails (MTs Δα,β-tails) in the presence of 1 mM ATP. TTLL7 normalized fluorescence intensity ± S.E.M. is shown (N = 45, 49, 44 microtubules for MTs, MTs partial Δβ-tail and MTs Δβ-tail, respectively). Representative microtubules and the corresponding TTLL7 channel are shown. The width of the panels corresponds to 6 µm. Mass spectra for the various microtubules are in (I).
(C) Single-molecule fluorescence analysis. Taxol-stabilized microtubules decorated with single TTLL7-GFP molecules (left) and corresponding kymograph (right). Scale bars, horizontal, 5 s; vertical, 2 μm.
(D) Distribution of durations of TTLL7-GFP interactions with the microtubule. A single-exponential fit to the histogram (corrected for photobleaching) yields a mean lifetime of interaction (τ) of 0.85 ± 0.07s (R2 = 0.9528, N = 807)
(E - G) kon (E), koff (F) and Ka (G) of TTLL7 with various types of microtubules. Error bars indicate S.E.M. N=807 for MTs and N=790 for MTs Δβ-tails obtained from four flow cells imaged on different days (Extended Experimental Procedures).
(H) TTLL7 binding to microtubules at various ionic strengths.
(I) Mass spectra of the types of microtubules used in assays shown in (B) – (G). See also Figure S3.
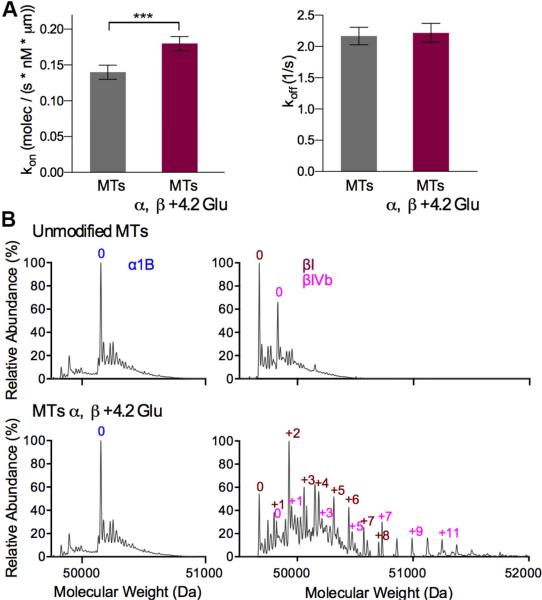
TTLL7 has two distinct but closely related activities. The enzyme initiates a branched chain from a genetically encoded internal glutamate of the tubulin tail, and also elongates this branch by addition of subsequent glutamates (Mukai et al., 2009). We examined how the presence of posttranslationally added glutamates on microtubules affects TTLL7 binding using single-molecule TIRF microscopy (Experimental Procedures). Glutamylation of β-tubulin produces a modest but statistically significant increase in the kon (28.5%) without affecting the koff (Figure 4A). The effect on the kon plateaus when the average number of glutamates on β-tubulin exceeds four (Figures S4A and S4B). Since favorable electrostatic interactions with the tails are determinant for TTLL7-microtubule association (Figure 3H), this falloff suggests that additional glutamates produce steric hindrance that offsets the favorable electrostatics conferred by the increased negative charge.
Figure 4. Kinetic analysis of TTLL7 binding to unmodified and glutamylated microtubules.
(A) kon and koff of the TTLL7 interaction with unmodified and modified microtubules. Error bars indicate S.E.M. N=1009 for unmodified MTs; N=1137 for MTs α, β+4.2 Glu obtained from at least four flow cells imaged on different days (Extended Experimental Procedures).
(B) Mass spectra of the unmodified and glutamylated microtubules used in (A). Number of glutamates on α- and β-tubulin were calculated from the weighted average of the mass spectra peak values for the different glutamylated species. See also Figure S4.
On microtubules with a larger number of glutamates on their β-tails (~ 8 Glu), TTLL7 exhibits a new behavior. A fraction (16.7%) of the molecules rather than associating statically with the microtubule, undergo one-dimensional diffusion. On hyperglutamylated microtubules (~ 28 Glu) this fraction increases to 36.4%. These diffusing molecules scan an average microtubule length of 0.38 ± 0.04 μm and have a mean microtubule interaction time of 2.6 ± 0.6 s (Figures S4C, S4D and S4E). The observed diffusive binding is independent of the elemental catalysis step as a catalytic dead mutant also undergoes 1D diffusion on modified microtubules (Figure S4H). This observation is consistent with our interpretation that long glutamate chains sterically hinder stable TTLL7 association with tubulin tails. We speculate that the observed 1D diffusion could provide negative feedback for enzyme activity. It could also be a mechanism for the enzyme to more effectively find tubulin tails that are not modified yet. TTLL7 has lower activity on glutamylated microtubules (Mukai et al., 2009).
Molecular Determinants of Tubulin Tail Recognition
TTLL7 modifies both α- and β-tubulin tail peptides in isolation (Figure S3A), but shows preference for the β-tubulin tail when presented in the context of the microtubule (Figure S3B). This suggests that the stereospecificity of the TTLL7 active site is limited and that the observed in vitro and in vivo selectivity for the β-tubulin tail must arise from interactions with the microtubule polymer mediated by the c-MTBD and the TTLL7 core. Such reliance on the contacts with the tubulin body may in turn provide TTLL7 the flexibility to attach multiple glutamates at multiple positions within the β-tubulin tail (Mukai et al., 2009).
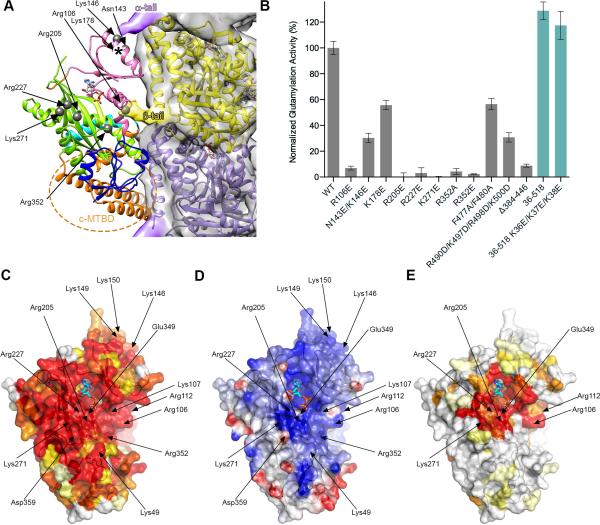
The TTLL7 molecular surface shows a high degree of phylogenetic conservation and pronounced cationic character, consistent with its interaction with the anionic tails and microtubule surface (Figure 5). The α-tubulin tail makes limited interactions with TTLL7, likely contacting residues in a disordered element connecting strands β7 and β8, and helix α5 (Figure 2D). Accordingly, mutation of invariant Lys178 reduces TTLL7 activity by 44%, while mutation of Asn143 and Lys146 in α5 by 70% (Figures 5A and B). The β-tubulin tail makes more extensive interactions with the TTLL7 core, consistent with its appearance in the EM reconstruction and dominant contribution to microtubule binding affinity (Figure 3). The molecular surface around the TTLL7 active site as well as the groove between the N- and central domains that contacts the β-tubulin tail is lined with conserved lysine and arginine residues (Figures 5C and D). Invariant Arg106 and Arg352 line the entrance to this groove and contact the N-terminal residues of the β-tail (Figures 5A and C). Mutation of either to glutamate leads to 93 and 96% loss in activity, respectively (Figure 5B). Consistent with their importance in engaging the β-tubulin tail, neither of these mutants can modify isolated β-tubulin peptides, unlike the wild-type (Figures S5C and S5F). Interestingly, while Arg106 is conserved across all TTLs and TTLLs, Arg352 is conserved only in TTLL7 sequences (Figure 5E), suggesting a specific role for this residue in engaging the β-tail.
Figure 5. Molecular determinants of microtubule glutamylation.
(A) Structure of the TTLL7:microtubule complex with conserved TTLL7 residues important for glutamylation denoted by grey spheres. EM surface is shown only for the microtubule for clarity (compare with Figure 2C). TTLL7 and tubulin colored as in Figure 1A and 2C, respectively. Dashed orange ellipse indicates that the c-MTBD extends beyond the partial α-helical model shown.
(B) Glutamylation activity of purified structure-guided TTTL7 mutants with taxol-stabilized microtubules. All mutations are in the H. sapiens TTLL7 construct (residues 1-518). Constructs with other boundaries are in teal. Error bars indicate S.E.M. (N ≥ 3)
(C) TTLL7 molecular surface in the same orientation as in (A) color-coded for conservation on a gradient from white (40% identity) to red (100% identity).
(D) TTLL7 molecular surface in the same orientation as in (A) color-coded for electrostatic potential (red, negative; blue, positive, ranging from −7 kBT to 7 kBT).
(E) TTLL7 molecular surface in the same orientation as in (A) color-coded for sequence conservation with TTLL family glutamylases (TTLL1, 4, 5, 6, 7, 9, and 13) on a gradient from white (40% identity) to red (100% identity). See also Figure S5.
The nucleotide is coordinated by residues from the central and C-domains (Figure 1A) conserved between TTL and all TTLL enzymes (Figures 5C and E), consistent with their common overall reaction mechanism, ATP-dependent peptide bond formation to a tubulin tail carboxylate (C-terminal α-carboxylate and internal γ-carboxylate for TTL and TTLLs, respectively). Invariant Arg202 in TTL coordinates the tubulin tail glutamate (Prota et al., 2013; Szyk et al., 2011). Mutation of the corresponding Arg205 in TTLL7 reduces activity to background both with microtubules and β-tail peptide (Figures 5 and S5D). Arg227 likely neutralizes the transition state during phosphate transfer and could also stabilize the incoming glutamate. Its mutation reduces TTLL7 activity by 97% (Figure 5B). The equivalent residue is invariant in all TTLLs and TTL (Szyk et al., 2011), indicative of its essential role. Invariant Lys271 flanks the active site cleft opposite from the β-tubulin tail entrance channel (Figures 5A, 5C and 5D). Its mutation to glutamate reduces activity by 99% (Figures 5B and S5E). Unlike Arg227 that is conserved across all TTLLs and TTLs, Lys271 is conserved only in glutamylases. This residue is a glutamate in TTL and does not participate in α-tail recognition (Prota et al., 2013). The more extended tail interaction site might reflect the fact that TTLL glutamylases ligate a glutamate to an internal position in the tubulin tail, requiring the tail to be engaged on both sides of the targeted glutamate, as opposed to TTL that ligates tyrosine to the α-tail carboxy terminus.
A Conserved Cationic Domain Important for Microtubule Recognition by TTLL7
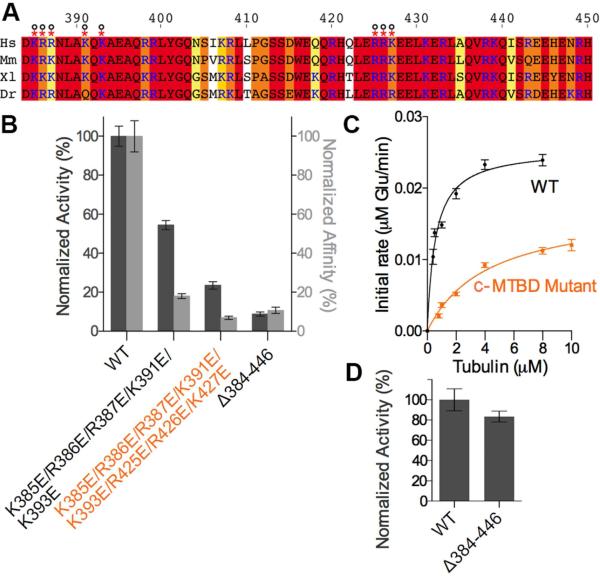
The c-MTBD binds the α-tubulin body mainly through contacts with the electronegative surface of helix H12 and the short helix preceding it. Helix α9 in TTLL7 anchors the C-terminal region of the c-MTBD domain (Figure 2). Destabilization of α9 through mutation of invariant Phe477 and Phe480 (engaged in hydrophobic interactions with conserved residues in TTLL7 helix α7; Figures 1D and E), reduces activity by 40% (Figure 5B). Deletion of c-MTBD residues 384-446 reduces glutamylation and microtubule binding by 89% and 81%, respectively (Figures 6A, 6B and S6). Conserved lysine and arginine residues constitute 28% of the c-MTBD sequence. Charge reversal of the first cluster of positive residues decreases binding by 45%. Charge-reversal mutations of additional positive residues clustered towards the C-terminus of the c-MTBD further reduces binding by 76 % (Figures 6A and 6B). This latter charge reversal mutant also displays a 7.5-fold weaker km with microtubules than the wild-type, but only a minor decrease in catalytic rate (0.1 ± 0.01 versus 0.09 ± 0.01 min−1; Figure 6C), indicating that the main function of this domain is microtubule binding. Consistent with the role of the c-MTBD in engaging the microtubule body, a mutant missing this domain modifies isolated β-tubulin peptides at a rate similar to the wild-type (Figure 6D). Similarly, charge reversal mutations of c-MTBD positively charged residues located C-terminal to helix α9 (R490D/K497D/R498D/K500D) reduces activity by 69% (Figure 5B). In contrast, deletion and charge reversal mutations of positive residues at the N-terminus of TTLL7 have no effect (Figure S1 and 5B). Our in vitro results are recapitulated by studies in living cells: while TTLL7-GFP shows co-localization with microtubules and strong glutamylation activity when overexpressed in human osteosarcoma cells (U2OS line), the c-MTBD deletion mutant shows diffuse cytoplasmic localization and no detectable glutamylation activity (Figures 7A and 7B).
Figure 6. A conserved microtubule-binding domain critical for TTLL7 function.
(A) Sequence alignment of the c-MTBD insertion in the TTLL7 C-domain from H.sapiens (Hs), Mus musculus (Mm), Xenopus laevis (Xl), and Danio rerio (Dr). Residues colored according to conservation as in Figure S1. H. sapiens TTLL7 residue numbers shown. Cationic residues in blue.
(B) Microtubule glutamylation (dark grey) and binding (light grey). Glutamylation assays performed as described in Experimental Procedures (N ≥ 3). Relative microtubule binding affinities were determined from fluorescence mean intensities (N = 57, 39, 46 and 44 microtubules for wild-type TTLL7 and the c-MTBD mutants). Error bars indicate S.E.M.
(C) Michaelis-Menten fit of microtubule glutamylation data for TTLL7 wild-type and TTLL7 c-MTBD charge-reversal mutant. Error bars represent standard error of the fit (N=4). The Km and kcat for TTLL7 and c-MTBD charge-reversal mutant are 0.6 ± 0.1 μM, 0.1 ± 0.01 min−1 and 4.2±0.7 μM, 0.09 +0.01 min−1, respectively.
(D) Glutamylation activity with isolated βIVb-tubulin peptide for TTLL7 and c-MTBD deletion mutant (N=4). Error bars indicate S.E.M. See also Figure S6.
Figure 7. A cationic microtubule-binding region critical for glutamylase activity.
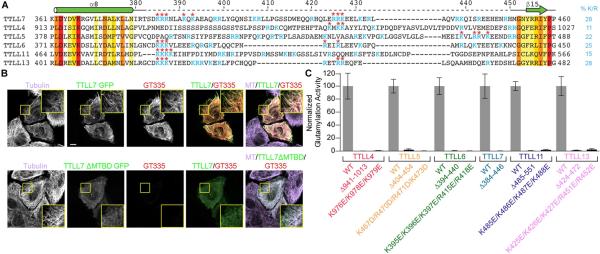
(A) Sequence alignment of the proposed microtubule binding C-domain insertions of M. musculus glutamylases TTLL4, 5, 6, 11 and 13. TTLL7 c-MTBD and adjacent secondary structure elements shown at the top. Residues colored according to conservation as in Figure S1. Percentage of cationic residues in the insertion indicated.
(B) Cellular distribution (green channel) and glutamylation activity (red channel) of H. sapiens wild-type TTLL7 and a c-MTBD deletion mutant. Scale bar, 10 μm.
(C) Glutamylation activity of GFP-tagged wild-type and mutant constructs of TTLL4, 5, 6, 7, 11, and 13 glutamylases as determined by Western blot using antibodies to GFP and glutamylation (N ≥ 3; Experimental Procedures). Error bars indicate S.E.M. See also Figure S7.
A Conserved Cationic Region Important for Glutamylation Across TTLL Family Glutamylases
Unlike TTL that tyrosinates monomeric tubulin, TTLL family members prefer microtubules (Mukai et al., 2009; van Dijk et al., 2007). To generalize our studies of TTLL7, we carried out structure-based sequence analyses of all TTLL glutamylases. This revealed the existence of an insertion in their C-domains at a position similar to that of the TTLL7 c-MTBD (Figure 7A and S7). The length of this analogous segment ranges between 50 and 80 amino acids and its composition is heavily biased towards positively charged residues (Figure 7A). To establish whether this cationic region important for substrate binding by TTLL7, contributes to the activity of other glutamylases of the TTLL family, we expressed internal deletion mutants of this region of six TTLL glutamylases in human tsa201 cells and determined their glutamylation activities (Figures 7A, 7C and Experimental Procedures). While wild-type TTLL enzymes exhibited robust glutamylation activity when overexpressed, the internal deletion constructs had either severely reduced or background levels of activity (Figure 7C). To ensure that this was not due to folding defects caused by the internal deletions (expression of full-length protein was confirmed for both wild-type and mutants), we also created several charge reversal mutations in this cationic region. As for the deletions, the charge reversal mutations also reduced the activity of all enzymes to near-background. Thus, our analysis has uncovered a functionally conserved cationic region that plays a key role in the function of all TTLL family glutamylases with autonomous glutamylation activity. We did not find an analogous cationic segment in TTLL1, 2 and 9. TTLL1 functions as part of a five-subunit complex and does not have glutamylation activity in isolation (Janke et al., 2005). Similarly, TTLL2 and 9 are not active in isolation and are thought to function as part of a complex (Kubo et al., 2014; van Dijk et al., 2007). We did not find an analogous domain in sequences of the glycylases TTLL3 and 8. However, we found that these enzymes contain a charged insertion in their N-terminal domains that may have an equivalent function.
DISCUSSION
We report the first atomic structure of any enzyme of the TTLL family. We also present the first structure of any microtubule posttranslational modification enzyme in complex with its physiological substrate, the microtubule. Because TTL and TTLL enzymes share a homologous core, it has been a puzzle since their initial discovery (Janke et al., 2005) how functional specialization arises. This specialization is what articulates the tubulin code. Despite their common fold, TTL modifies monomeric tubulin, while TTLL7 prefers microtubules. TTL preferentially acts on unpolymerized tubulin by recognizing an α-tubulin interface masked in the microtubule polymer. It binds at the edge of the αβ-tubulin dimer and makes contacts mostly with α-tubulin, guiding the flexible α-tail into its active site (Prota et al., 2013; Szyk et al., 2011). Because TTL binding occludes an α-tubulin polymerization interface, it inhibits microtubule assembly (Szyk et al., 2011; Szyk et al., 2013). Our hybrid structural analysis reveals that TTLL7 augments its TTL homologous core with a newly defined cationic microtubule-binding domain (Figures 1 and 2) and recognizes the microtubule polymer through multivalent interactions that involve the c-MTBD (absent in TTL), as well as both α- and β-tubulin tails, the former being provided in trans by the neighboring dimer on the same protofilament (Figure 2).
TTL displays exquisite specificity for the α-tubulin tail, ligating a tyrosine only to its C-terminal glutamate (Rüdiger et al., 1994). In contrast, TTLL7 catalyzes the addition of glutamates at multiple internal positions in the β-tail, and also that of subsequent glutamates to existing branched glutamates (Mukai et al., 2009). While for TTL the interaction with the α-tail is low affinity but high-specificity and additional binding energy is obtained from engaging the tubulin body (Szyk et al., 2011), for TTLL7 the interaction with the tails contributes predominantly to the binding affinity (Figure 3) but is more permissive stereochemically. Specificity for the β-tubulin tail is achieved through additional contacts with the microtubule (mainly through the c-MBTD) that position TTLL7 to selectively channel the β-tubulin tail into the active site. Consistent with the activity of TTLL7 on β-tail-internal glutamates, the TTLL7 active site is more open and residues involved in substrate recognition span a greater distance from the nucleotide-binding site than those of TTL. Thus, the TTLL7 active site has evolved a higher degree of plasticity to accommodate multiple biochemical activities (addition of branch-point glutamates at multiple positions, as well as glutamate chain elongation), while the TTL active site has evolved to stringently recognize the α-tail and add a single tyrosine to its terminal glutamate. Different TTLL family glutamylases have different substrate preferences (α- or β-tubulin tail). Additional structural studies of TTLL members with different substrate specificities will establish whether this strategy of conferring specificity through interactions with the microtubule body rather than stringent active site regioselectivity is a general strategy employed by TTLL family enzymes.
Our analyses show that TTLL7 achieves functional versatility by employing a three-pronged strategy to recognize the microtubule: it engages both α- and β-tubulin tails with its TTL homologous core, and uses the flexible c-MTBD to bind the tubulin body. All three interactions are negotiated between flexible, charged sequence elements (the anionic C-terminal tails; the cationic c-MTBD) and stably folded structural elements (the TTLL7 core; the tubulin body). Association through intrinsically disordered regions provides specificity, but entails high entropic cost, thereby producing accurate, but low affinity interactions (Dyson and Wright, 2002; reviewed in Roll-Mecak, 2015). The combined use of folded and intrinsically disordered substrate recognition elements could be a general mechanism employed by enzymes that use a single active site to catalyze multiple stereochemically distinct but chemically analogous reactions. The flexibility of the tubulin tails also likely facilitates a productive encounter of the enzyme with the rigid microtubule shaft, consistent with the predominant decrease in the kon when they are removed (Figure 3).
The c-MTBD employed by TTLL7 is present in other TTLL glutamylases with autonomous activity (TTLL4, 5, 6, 11, 13) where it is critical for function (Figure 7). Among these glutamylases, TTLL4 also modifies nucleosome-associated proteins (NAPs) 1 and 2 (van Dijk et al., 2008). It remains to be established whether the c-MTBD also serves to recognize those non-tubulin substrates. Intriguingly, the presence of a disordered glutamate-rich region in the C-terminus of NAPs is not sufficient for glutamylation by TTLL4, and additional regions far removed from the modification sites are required (van Dijk et al., 2008), suggesting the involvement of a c-MTBD-like engagement with the globular core of these NAPs. The equivalent cationic region is absent in glutamylases TTLL1, 2 and 9, which are thought to function as part of multi-subunit complexes and lack autonomous glutamylation activity. In these cases, an element analogous to the c-MTBD might be supplied in trans by their binding partners.
The tripartite recognition mechanism of TTLL7 has notable parallels with the “collaborative templating” mechanism of the class I CCA-adding enzyme (Shi et al., 1998). That enzyme uses non-catalytic domains to position itself on the upper helix and elbow of tRNA, while its active site unfolds and refolds around the acceptor end of tRNA for each sequential addition of CTP and ATP. This strategy allows the enzyme to hold the tRNA rigidly in place while its active site sequentially engages and repositions the 3’-terminal acceptor end (Tomita and Yamashita, 2014). TTLL7 uses interactions with the rigid microtubule shaft to position itself while its active site can fold and refold to enable both initiation at different sites in the tubulin tail and elongation of the branches thus formed. This could be a general mechanism used by enzymes that require distributive selectivity for their activities. The ability of TTLL7 to initiate and then elongate substrate chains is also reminiscent of poly-(ADP ribose) polymerases (PARPs). Those enzymes use common catalytic residues to both initiate and then elongate ADP ribose chains of varying lengths and branching structures (Gibson and Kraus, 2012). The two reactions are thought to be supported by differentially positioning the substrates within the active site, a mechanism that we propose is also operative in TTLL7. A comprehensive understanding of the kinetics and substrate recognition strategies of the large family of TTLL enzymes will enable us to understand the underlying principles that give rise to the complex microtubule modification patterns observed in cells as well as derive general design principles of enzyme active sites with distributive specificities.
EXPERIMENTAL PROCEDURES
Protein production, Crystallization and Structure Determination
Details regarding protein purification are in Extended Experimental Procedures. For crystallization studies TTLL7 (36-518) yielded crystals that were suitable for structure determination. Crystals of apo, ADP, ATP or AMPPNP bound TTLL7 grew at room temperature by hanging drop vapor diffusion in 0.1 M MES 6.0, 10% PEG 20K, 5 mM MgCl2, 0.5 mM tris(2-chloroethyl) phosphate (TCEP) and 0.5 mM nucleotide with symmetry of space group C2221 with one TTLL7 copy in the asymmetric unit. Data from SeMet and ethyl mercury phosphate (EMP) soaked TTLL7-AMMPNP crystals were collected at the Advanced Photon Source beamline 24-ID-E. Detailed procedures for structure determination and refinement can be found in Table S1 and Extended Experimental Procedures. Native data sets for ADP and TTLL7-AMPPNP were collected at Advanced Light Source beamlines 5.0.1 and 5.0.2, respectively. The structure of ADP-bound TTLL7 was solved by molecular replacement. The current TTLL7-ADP crystallographic model at 2.55 Å resolution has Rwork and Rfree of 22.2 and 26.9%, respectively (Table S1), with no unfavorable (ϕ,ψ) combinations. The current TTLL7-AMPPNP crystallographic model at 2.60 Å resolution has Rwork and Rfree of 21.0 and 24.6%, respectively (Table S1), with no unfavorable (ϕ,ψ) combinations.
Cryo-EM 3D Reconstruction
Undecorated 14 and 15 protofilament microtubule densities (Sui and Downing, 2010) were used as initial models for all preliminary reconstructions. The IHRSR (Egelman, 2007) procedure was used for multi-model projection matching of microtubule specimens with varying numbers of protofilaments (Alushin et al., 2014). A final refinement of the microtubule segment alignment parameters was performed in FREALIGN (Grigorieff, 2007) without further refinement of helical parameters. Fourier Shell Correlation (FSC) curves for microtubule reconstructions were used to estimate resolutions of each reconstruction using a cutoff of 0.5 (Figure S2E). In order to more accurately estimate the resolution of each region of the reconstructed density, a local resolution calculation was performed using the “blocres” and “blocfilt” functions in the Bsoft processing package (Heymann et al 2007). This analysis revealed that the majority of the tubulin density is in the range of 7-8Å, while the ligase portion ranges from 9-12Å resolution. Details regarding cryo-EM data collection, processing and 3D reconstruction are in Extended Experimental Procedures, Table S2 and Figure S2.
Glutamylation assays
Glutamylation activity was measured through [3H]-glutamate incorporation. Assays are described in Extended Experimental Procedures.
Mass Spectrometry Based Glutamylation Assays
Mass spectrometric analysis of glutamylation reactions is described in detail in Extended Experimental Procedures. Samples were analyzed by electrospray mass spectrometry as described in (Vemu et al., 2014) and Extended Experimental Procedures.
Single Molecule TIRF Analysis
Chambers were assembled as in (Szyk et al., 2014). For single molecule kinetic measurements TTLL7-GFP was perfused in assay buffer supplemented with oxygen scavengers prepared as in (Ziolkowska and Roll-Mecak, 2013). A complete description of data collection and analyses can be found in Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
-
1)
TTLL7 engages both the α- and β-tails of tubulin dimers
-
2)
TTL7 binds microtubules through a cationic domain that is functionally required for TTLL glutamylases
-
3)
Mutual disorder-to-order transitions mediate TTLL7-microtubule recognition
-
4)
TTLL7 substrate recognition is reminiscent of that of CCA-adding enzymes and PARPs
ACKNOWLEDGEMENTS
We thank staff at beamlines 24-ID (Advanced Photon Source) and 5.0.1 and 5.0.2. (Advanced Light Source) for support, D.-Y. Lee for access to mass spectrometers, C. Janke (Institute Curie) for TTLL expression plasmids, and A. Ferre-D’Amare, E. Giniger, S. Gottesman for critical reading of the manuscript. TEM studies were conducted at the National Resource for Automated Molecular Microscopy, supported by NIGMS (P41 GM103310).C.P.G. is supported by an NINDS postdoctoral fellowship. G.C.L. is supported by the Damon Runyon Cancer Research Foundation (DFS-#07-13), the Pew Scholars program, the Searle Scholars program, and NIH (DP2 EB020402). R.A.M. is supported by NIH (GM-052468). E.M.W-K is partly supported by NIH (GM095573). A.R.-M. is supported by the Searle Scholar program and the intramural programs of the National Institute of Neurological Disorders and Stroke and National Heart, Lung and Blood Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Protein Data Bank accession numbers for atomic coordinates and structure factor amplitudes are ### and ###. The EMDB accession code for the TTLL7:microtubule complex structure is ###. To be provided at proof stage.
AUTHOR CONTRIBUTIONS
C.P.G. did glutamylation assays, immunofluorescence, crystallized TTLL7, collected diffraction data and solved crystal structures with help from A. R.-M. A.V. did single molecule and microtubule-binding assays, prepared all TTLL7 and unmodified tubulin for EM. E.M.W.-K. collected EM data and processed it with help from G.C.L. E.M.W.-K., G.C.L. and R.A.M. interpreted EM data. I.Y. prepared TTLL7 mutants. A.S. did activity assays. A.R.-M. conceived and directed the project, designed experiments and wrote the manuscript with input from all authors. All authors reviewed the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, seven figures, two tables and a movie.
REFERENCES
- Alexander JE, Hunt DF, Lee MK, Shabanowitz J, Michel H, Berlin SC, MacDonald TL, Sundberg RJ, Rebhun LI, Frankfurter A. Characterization of posttranslational modifications in neuron-specific class III beta-tubulin by mass spectrometry. Proc Natl Acad Sci U S A. 1991;88:4685–4689. doi: 10.1073/pnas.88.11.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. High-resolution microtubule structures reveal the structural transitions in alphabeta-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert S, Koulakoff A, Berwald-Netter Y, Gros F, Denoulet P, Edde B. Developmental regulation of polyglutamylated alpha- and beta-tubulin in mouse brain neurons. J Cell Sci. 1994;107(Pt 8):2313–2322. doi: 10.1242/jcs.107.8.2313. [DOI] [PubMed] [Google Scholar]
- Bobinnec Y, Moudjou M, Fouquet JP, Desbruyeres E, Edde B, Bornens M. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil Cytoskeleton. 1998;39:223–232. doi: 10.1002/(SICI)1097-0169(1998)39:3<223::AID-CM5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Bonnet C, Boucher D, Lazereg S, Pedrotti B, Islam K, Denoulet P, Larcher JC. Differential binding regulation of microtubule-associated proteins MAP1A, MAP1B, and MAP2 by tubulin polyglutamylation. J Biol Chem. 2001;276:12839–12848. doi: 10.1074/jbc.M011380200. [DOI] [PubMed] [Google Scholar]
- Boucher D, Larcher JC, Gros F, Denoulet P. Polyglutamylation of tubulin as a progressive regulator of in vitro interactions between the microtubule-associated protein Tau and tubulin. Biochemistry. 1994;33:12471–12477. doi: 10.1021/bi00207a014. [DOI] [PubMed] [Google Scholar]
- Bulinski JC, McGraw TE, Gruber D, Nguyen HL, Sheetz MP. Overexpression of MAP4 inhibits organelle motility and trafficking in vivo. J Cell Sci. 1997;110(Pt 24):3055–3064. doi: 10.1242/jcs.110.24.3055. [DOI] [PubMed] [Google Scholar]
- Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. Structure and functional role of dynein's microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Eddé B, Rossier J, Le Caer JP, Desbruyères E, Gros F, Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Garnham CP, Roll-Mecak A. The chemical complexity of cellular microtubules: tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton. 2012;69:442–463. doi: 10.1002/cm.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimer S, Teltenkotter A, Plessmann U, Weber K, Lechtreck KF. Purification and characterization of basal apparatuses from a flagellate green alga. Cell Motil Cytoskeleton. 1997;37:72–85. doi: 10.1002/(SICI)1097-0169(1997)37:1<72::AID-CM7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Gigant B, Wang W, Dreier B, Jiang Q, Pecqueur L, Pluckthun A, Wang C, Knossow M. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat Struct Mol Biol. 2013;20:1001–1007. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Ikegami K, Heier RL, Taruishi M, Takagi H, Mukai M, Shimma S, Taira S, Hatanaka K, Morone N, Yao I, et al. Loss of alpha-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc Natl Acad Sci USA. 2007;104:3213–3218. doi: 10.1073/pnas.0611547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Mukai M, Tsuchida J.-i., Heier RL, Macgregor GR, Setou M. TTLL7 is a mammalian beta-tubulin polyglutamylase required for growth of MAP2-positive neurites. J Biol Chem. 2006;281:30707–30716. doi: 10.1074/jbc.M603984200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Sato S, Nakamura K, Ostrowski LE, Setou M. Tubulin polyglutamylation is essential for airway ciliary function through the regulation of beating asymmetry. Proc Natl Acad Sci U S A. 2010;107:10490–10495. doi: 10.1073/pnas.1002128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub J-M, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Yanagisawa HA, Liu Z, Shibuya R, Hirono M, Kamiya R. A conserved flagella-associated protein in Chlamydomonas, FAP234, is essential for axonemal localization of tubulin polyglutamylase TTLL9. Mol Biol Cell. 2014;25:107–117. doi: 10.1091/mbc.E13-07-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Yanagisawa HA, Yagi T, Hirono M, Kamiya R. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr Biol. 2010;20:441–445. doi: 10.1016/j.cub.2009.12.058. [DOI] [PubMed] [Google Scholar]
- Lacroix B, van Dijk J, Gold ND, Guizetti J, Aldrian-Herrada G, Rogowski K, Gerlich DW, Janke C. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J Cell Biol. 2010;189:945–954. doi: 10.1083/jcb.201001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Silhavy JL, Zaki MS, Schroth J, Bielas SL, Marsh SE, Olvera J, Brancati F, Iannicelli M, Ikegami K, et al. CEP41 is mutated in Joubert syndrome and is required for tubulin glutamylation at the cilium. Nat Genet. 2012;44:193–199. doi: 10.1038/ng.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- Mukai M, Ikegami K, Sugiura Y, Takeshita K, Nakagawa A, Setou M. Recombinant mammalian tubulin polyglutamylase TTLL7 performs both initiation and elongation of polyglutamylation on beta-tubulin through a random sequential pathway. Biochemistry. 2009;48:1084–1093. doi: 10.1021/bi802047y. [DOI] [PubMed] [Google Scholar]
- O'Hagan R, Piasecki BP, Silva M, Phirke P, Nguyen KC, Hall DH, Swoboda P, Barr MM. The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans. Curr Biol. 2011;21:1685–1694. doi: 10.1016/j.cub.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prota AE, Magiera MM, Kuijpers M, Bargsten K, Frey D, Wieser M, Jaussi R, Hoogenraad CC, Kammerer RA, Janke C, et al. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J Cell Biol. 2013;200:259–270. doi: 10.1083/jcb.201211017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybin D, Flavin M. An enzyme tyrosylating alpha-tubulin and its role in microtubule assembly. Biochem Biophys Res Commun. 1975;65:1088–1095. doi: 10.1016/s0006-291x(75)80497-9. [DOI] [PubMed] [Google Scholar]
- Redeker V, Melki R, Prome D, Le Caer JP, Rossier J. Structure of tubulin C-terminal domain obtained by subtilisin treatment. The major alpha and beta tubulin isotypes from pig brain are glutamylated. FEBS Lett. 1992;313:185–192. doi: 10.1016/0014-5793(92)81441-n. [DOI] [PubMed] [Google Scholar]
- Redwine WB, Hernandez-Lopez R, Zou S, Huang J, Reck-Peterson SL, Leschziner AE. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–1536. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme JC, Bosson A, Peris L, Gold ND, Lacroix B, Bosch Grau M, et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 2010;143:564–578. doi: 10.1016/j.cell.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A. Intrinsically disordered tubulin tails: complex tuners of microtubule functions? Semin Cell Dev Bio. 2015;37C:11–19. doi: 10.1016/j.semcdb.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451:363–367. doi: 10.1038/nature06482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger M, Plessman U, Kloppel KD, Wehland J, Weber K. Class II tubulin, the major brain beta tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett. 1992;308:101–105. doi: 10.1016/0014-5793(92)81061-p. [DOI] [PubMed] [Google Scholar]
- Rüdiger M, Wehland J, Weber K. The carboxy-terminal peptide of detyrosinated alpha tubulin provides a minimal system to study the substrate specificity of tubulin-tyrosine ligase. Eur J Biochem. 1994;220:309–320. doi: 10.1111/j.1432-1033.1994.tb18627.x. [DOI] [PubMed] [Google Scholar]
- Schneider A, Plessmann U, Felleisen R, Weber K. Posttranslational modifications of trichomonad tubulins; identification of multiple glutamylation sites. FEBS Lett. 1998;429:399–402. doi: 10.1016/s0014-5793(98)00644-9. [DOI] [PubMed] [Google Scholar]
- Sergouniotis PI, Chakarova C, Murphy C, Becker M, Lenassi E, Arno G, Lek M, MacArthur DG, Consortium UC-E, Bhattacharya SS, et al. Biallelic variants in TTLL5, encoding a tubulin glutamylase, cause retinal dystrophy. Am J Hum Genet. 2014;94:760–769. doi: 10.1016/j.ajhg.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Bryant J, Wloga D, Donaldson R, Davis RC, Jerka-Dziadosz M, Gaertig J. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J Cell Biol. 2007;178:1065–1079. doi: 10.1083/jcb.200704021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, Maizels N, Weiner AM. CCA addition by tRNA nucleotidyltransferase: polymerization without translocation? EMBO J. 1998;17:3197–3206. doi: 10.1093/emboj/17.11.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar CV, Downing KH. An atomic-level mechanism for activation of the kinesin molecular motors. Proc Natl Acad Sci U S A. 2010;107:4111–4116. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis ET, Hunt SJ, Hu Q, Kinoshita M, Nelson WJ. Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. J Cell Biol. 2008;180:295–303. doi: 10.1083/jcb.200710039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H, Downing KH. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure. 2010;18:1022–1031. doi: 10.1016/j.str.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi S, Edde B, Fox LA, Guerrero S, Hard R, Hennessey T, Kabi A, Malison D, Pennock D, Sale WS, et al. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr Biol. 2010;20:435–440. doi: 10.1016/j.cub.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Deaconescu AM, Piszczek G, Roll-Mecak A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat Struct Mol Biol. 2011;18:1250–1258. doi: 10.1038/nsmb.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Deaconescu AM, Spector J, Goodman B, Valenstein ML, Ziolkowska NE, Kormendi V, Grigorieff N, Roll-Mecak A. Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell. 2014;157:1405–1415. doi: 10.1016/j.cell.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Piszczek G, Roll-Mecak A. Tubulin tyrosine ligase and stathmin compete for tubulin binding in vitro. J Mol Biol. 2013;425:2412–2414. doi: 10.1016/j.jmb.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Yamashita S. Molecular mechanisms of template-independent RNA polymerization by tRNA nucleotidyltransferases. Frontiers in genetics. 2014;5:36. doi: 10.3389/fgene.2014.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk J, Miro J, Strub JM, Lacroix B, van Dorsselaer A, Edde B, Janke C. Polyglutamylation is a post-translational modification with a broad range of substrates. J Biol Chem. 2008;283:3915–3922. doi: 10.1074/jbc.M705813200. [DOI] [PubMed] [Google Scholar]
- van Dijk J, Rogowski K, Miro J, Lacroix B, Edde B, Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell. 2007;26:437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Vemu A, Garnham CP, Lee DY, Roll-Mecak A. Generation of differentially modified microtubules using in vitro enzymatic approaches. Methods Enzymol. 2014;540:149–166. doi: 10.1016/B978-0-12-397924-7.00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6:2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- Zambito AM, Knipling L, Wolff J. Charge variants of tubulin, tubulin S, membrane-bound and palmitoylated tubulin from brain and pheochromocytoma cells. Biochim Biophys Acta. 2002;1601:200–207. doi: 10.1016/s1570-9639(02)00472-7. [DOI] [PubMed] [Google Scholar]
- Ziolkowska NE, Roll-Mecak A. In vitro microtubule severing assays. Methods Mol Biol. 2013;1046:323–334. doi: 10.1007/978-1-62703-538-5_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.