Highlights
-
•
Coronavirus spike proteins can be cleaved by a multitude of host cell proteases.
-
•
Proteolytic activation of spike is a crucial step to activate its fusogenicity.
-
•
The spike protein can be cleaved at multiple sites.
-
•
Modulation of spike cleavage can have profound effects on tropism and pathogenesis.
Keywords: Cleavage activation, Coronavirus, Pathogenesis, Protease, Spike protein, Tropism
Abstract
Coronaviruses are a large group of enveloped, single-stranded positive-sense RNA viruses that infect a wide range of avian and mammalian species, including humans. The emergence of deadly human coronaviruses, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV) have bolstered research in these viral and often zoonotic pathogens. While coronavirus cell and tissue tropism, host range, and pathogenesis are initially controlled by interactions between the spike envelope glycoprotein and host cell receptor, it is becoming increasingly apparent that proteolytic activation of spike by host cell proteases also plays a critical role. Coronavirus spike proteins are the main determinant of entry as they possess both receptor binding and fusion functions. Whereas binding to the host cell receptor is an essential first step in establishing infection, the proteolytic activation step is often critical for the fusion function of spike, as it allows for controlled release of the fusion peptide into target cellular membranes. Coronaviruses have evolved multiple strategies for proteolytic activation of spike, and a large number of host proteases have been shown to proteolytically process the spike protein. These include, but are not limited to, endosomal cathepsins, cell surface transmembrane protease/serine (TMPRSS) proteases, furin, and trypsin. This review focuses on the diversity of strategies coronaviruses have evolved to proteolytically activate their fusion protein during spike protein biosynthesis and the critical entry step of their life cycle, and highlights important findings on how proteolytic activation of coronavirus spike influences tissue and cell tropism, host range and pathogenicity.
1. Introduction
Coronaviruses are a wide-ranging family of viruses that infect many species of birds and mammals, including humans (Woo et al., 2009). They possess a remarkable ability for interspecies transmission as exemplified by the emergence of the deadly human virus severe acute respiratory syndrome coronavirus (SARS-CoV) (Drosten et al., 2003, Peiris et al., 2003, Peiris et al., 2004), and more recently, Middle East respiratory syndrome coronavirus (MERS-CoV) (van Boheemen et al., 2012, Zaki et al., 2012), both of which are thought to have originated in bats (Li et al., 2005b, Wang et al., 2014), followed by an intermediate host stage (civet cats and camels, respectively) (Alagaili et al., 2014, Haagmans et al., 2013, Hemida et al., 2014, Wang and Eaton, 2007), before crossing into the human population (Drexler et al., 2014). Such zoonotic potential is of particular concern, especially since global trade, deforestation, massive urbanization and high density farming practices increase the likelihood of sparking new and severe zoonotic outbreaks (Cutler et al., 2010).
The success of coronaviruses in their ability to jump between species may be attributed, in part, to the diverse array of virus entry strategies they deploy to infect target cells (Belouzard et al., 2012, Bosch and Rottier, 2008). Coronavirus entry is largely controlled by the spike surface envelope glycoprotein (S) since it bears both receptor binding and membrane fusion capabilities (Masters and Perlman, 2013). As such, the S glycoprotein is a crucial determinant of tissue and cell tropism as well as host range. Coronaviruses are notable because at each step of virus entry, which includes receptor binding, activation of fusion, and internalization, a multitude of mechanisms and strategies have evolved (Belouzard et al., 2012). For example, depending on the coronavirus species, the S protein can mediate binding to a proteinaceous receptor or to carbohydrate moieties.
Coronaviruses can enter cells via fusion either directly at the cell surface or can be internalized through the endosomal compartment. The mouse hepatitis virus (MHV) is a prime example of the flexibility in entry mechanisms used. A variant of the MHV-4 strain was shown to be able to fuse directly at the cell surface at neutral pH and also enter cells through an endocytic route (Nash and Buchmeier, 1997), whilst the MHV-2 strain enters cells through endocytosis by a clathrin-dependent mechanism (Pu and Zhang, 2008, Qiu et al., 2006).
Importantly, coronaviruses employ a diversity of cues, such as receptor binding, low pH and proteolytic activation, to activate the S protein, allowing a timely release of the fusion peptide into target membranes (Bosch and Rottier, 2008). This enables a spatio-temporally controlled orchestration of viral entry steps.
Along with binding to the host cell receptor, fusion of the viral envelope with host cell membranes is a critical step in establishing successful infection for enveloped viruses such as coronaviruses. The coronavirus S envelope glycoprotein is a class I viral fusion protein (Bosch et al., 2003). The S protein is often activated for fusion by means of proteolytic processing by host cell proteases, an activation process that is typical of class I viral fusion proteins (White et al., 2008). Remarkably, for some coronaviruses, such as SARS-CoV, cleavage of S can occur at two distinct sites (Belouzard et al., 2009). A variety of proteases have been shown to mediate coronavirus activation (Belouzard et al., 2012).
Notably, certain viruses harboring class I viral fusion proteins, like influenza virus and Newcastle disease virus, display characteristically expanded or modified cell and tissue tropism, and altered viral pathogenesis following mutation of the cleavage site that results in a change in proteolytic activation (Klenk and Garten, 1994b). This is very well exemplified by the hemagglutinin (HA) protein of highly pathogenic avian influenza (HPAI) virus strains, where transition from a monobasic site, typically cleaved by trypsin-like proteases, to a polybasic site, allows cleavage by ubiquitously expressed furin-like proteases, enabling systemic spread of the virus within an infected host. Thus, small mutational changes in amino acid composition at cleavage sites can have a drastic impact on tissue and cell tropism, host range, and pathogenesis (Klenk and Garten, 1994b, Nagai, 1993).
Here, we put into perspective the wide variety of entry activation mechanisms employed by coronaviruses and present the wide diversity of host cell proteases known to activate coronavirus S proteins. We review what is known about coronavirus proteolytic processing of the S protein and its link with pathogenicity, cell and tissue tropism and host range. We also analyze and compare the amino acid sequence composition of two identified coronavirus S cleavage sites, S1/S2 and S2′, in a wide range of coronavirus species encompassing all four coronavirus genera. Finally, we propose using protease sequence recognition motifs on coronavirus S protein as a novel marker to assess pathogenicity and host range, as well as forming the basis for effective therapeutic intervention.
2. Coronavirus S protein
Because they possess an envelope, coronavirus entry into host target cells requires the successful completion of two critical steps. The first is binding to the cell surface by means of attachment to a host cell receptor. The second is fusion of the viral envelope with cellular membranes allowing release of the virus genome into the host cell's cytoplasm, enabling viral replication to ensue. Both steps are controlled by the S envelope protein (Bosch and Rottier, 2008). As S is a class I viral fusion protein, we will first introduce important basic features of the prototypical class I viral fusion protein, influenza virus HA.
2.1. A prototypical class I fusion protein: Influenza virus hemagglutinin (HA)
There are three classes of virus fusion proteins known, which are classified according to their structural features (Harrison, 2013). While there is a wide degree of variability in the structure and mechanisms involved in the fusion process among these classes, all virus fusion proteins undergo major structural transitions, and ultimately form a final, compact and low-energy trimeric structure, the so-called trimer of hairpins. During the conformational changes, viral and cellular membranes are brought into close proximity. This in turn induces hemifusion, followed by complete fusion of viral and cellular membranes and formation of a pore that can expand, allowing for viral genetic material access into the cell (White et al., 2008).
The very well characterized class I fusion protein influenza virus HA is useful in introducing key structural and mechanistic concepts shared with coronavirus S. Structurally, the salient features of influenza HA are that it assembles in homotrimers orientated perpendicular to the virion membrane surface, has two functional subunits, HA1 which binds to sialic acid receptors and HA2 which contains the fusion machinery, featuring mainly alpha-helical secondary structures (Skehel and Wiley, 2000, White et al., 2008). HA2 contains two heptad repeats, which are structural motifs consisting of chains of seven amino acids that are critical for the fusion function and are another characteristic feature of class I fusion proteins (Chambers et al., 1990). HA is synthesized as an uncleaved precursor named HA0. Fusion activation occurs thanks to endoproteolysis by host cell proteases, such as trypsin-like proteases (Lazarowitz and Choppin, 1975, Lazarowitz et al., 1973a), a cleavage event that processes HA0 into HA1 and HA2 fragments, with both fragments held together by disulfide bonds (White et al., 2008). A fusion peptide, consisting mainly of apolar residues, is found at the N-terminus of HA2 buried at the subunit interface, and is exposed upon cleavage of HA0 and conformational changes of HA.
A key initial step in the HA fusion process is its priming by proteolytic cleavage that separates HA into HA1 and HA2 fragments. At this stage, the HA is in a metastable “spring-loaded” conformation. The receptor-binding subunit HA1 binds to sialic receptors found at the surface of target cells. This triggers uptake and internalization of the virion via the endocytic pathway. During maturation of the endosome, the virion becomes exposed to an increasingly acidic environment (note that this can also occur outside the cell in acidic tissue fluids). This drop in pH is the crucial trigger that provokes further conformational changes of HA allowing for full exposure of the hydrophobic fusion peptide on HA2 (Carr and Kim, 1993): the fusion peptide extends to the tip of the molecule allowing for insertion into the target endosomal membrane, forming an intermediate structure called the prehairpin (White et al., 2008). Further structural rearrangements of several prehairpins occur in which the alpha-helical heptad repeats assemble and bundle up into a compact coiled-coil structure, the six-helix bundle (6HB) with the C-terminal heptad repeats wrapping around the N-terminal heptad repeats (Chen et al., 1999, White et al., 2008). During this dramatic change in structure, viral and target endosomal membranes come into closer proximity. This allows for hemifusion and then full fusion to occur, generating an expanding fusion pore and ultimately allowing release of viral genetic material into the host cell. It is after fusion has occurred, that the HA adopts a structurally stable conformation called the trimer of hairpin.
2.2. The coronavirus spike (S) protein
The coronavirus S protein is a type I transmembrane protein located at the surface of the virion, with a large ectodomain and very short endodomain (Fig. 1 ) (Masters and Perlman, 2013). As a class I viral fusion protein, it shares many structural and mechanistic features of influenza virus HA (Bosch et al., 2003, Masters and Perlman, 2013). It is the largest of the coronavirus structural proteins with an overall length ranging between ∼1200 and 1400 amino acids, and is often heavily glycosylated, with between 21 and 35 N-glycosylation sites found within S monomers. Individual monomers of S proteins assemble into trimers giving rise to club-shaped protrusions forming characteristic corona-shaped structures around the virion (Fig. 1A; Belouzard et al., 2012). The S protein ectodomain can be divided into two functional domains. The S1 domain holds the receptor binding properties while the S2 domain harbors the fusion machinery (Fig. 1B). Within the S1 domain, two subdomains have been characterized. The N-terminal domain (NTD) is found at the S1 N-terminal half, while the C-domain is located at the S1 C-terminal half. The NTD of some coronaviruses can bind carbohydrate moieties such as 9-O-acetylated neuraminic acid (bovine coronavirus, BCoV, and human coronavirus HCoV-OC43), while the C-domain binds to proteinaceous receptors. Due to structural similarities between coronavirus S NTD and host cell proteins, it has been suggested that the NTD emerged from acquisition of a host sugar-binding galectin-like domain in an ancestral coronavirus (Peng et al., 2011). The NTD is dispensable for some coronaviruses, such as human coronavirus 229E (HCoV-229E) and porcine respiratory coronavirus (PRCoV), a naturally occurring variant of transmissible gastroenteritis virus (TGEV), as their NTD appears to have been deleted from their spike protein. Notably in the latter case, the deletion of the NTD leads to a shift of tissue tropism, from enteric tract for TGEV, to the respiratory tract for PRCoV. Depending on the coronavirus species, the receptor binding domain (RBD) can be found in the NTD or C-domain, or both. A structural study on murine hepatitis virus (MHV) S has demonstrated that its NTD, which has evolved to bind to the proteinaceous receptor mouse CEACAM1 and no longer carbohydrates, still adopts a galectin fold structure (Peng et al., 2011). Other crystal structure studies have shown that the coronavirus C-domain can adopt either a beta-sandwich fold, as is the case for the human alphacoronavirus HCoV-NL63 S (Wu et al., 2009), or a beta-sheet fold, as shown for the human betacoronavirus SARS-CoV (Li et al., 2005a). It is noteworthy that while both HCoV-NL63 and SARS-CoV S bind the same receptor, ACE2, their C-domains are structurally distinct and the modality of their respective binding to the receptor differs (Hofmann et al., 2006).
Fig. 1.
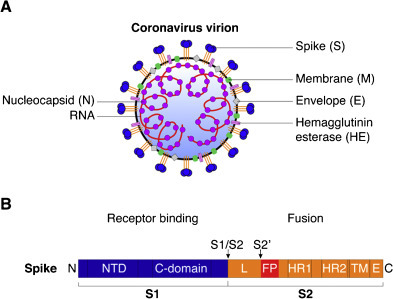
Structural features of coronavirus spike (S) envelope glycoprotein. (A) Diagram of a coronavirus virion with the main structural proteins depicted. The S protein assembles in trimers and projects outward from the virion to form a crown-like structure. The hemagglutinin esterase protein (HE) is found only in lineage A betacoronaviruses. (B) Diagram of coronavirus S protein with the two well-defined cleavage sites, S1/S2 and S2′ (arrows). The S protein is composed of two subunits, the S1 receptor-binding subunit, and the S2 fusion subunit. NTD: N-terminal domain of S1, C-domain: C-terminal domain of S1, L: linker region between S1/S2 and S2′ sites, FP: putative fusion peptide, HR1: heptad repeat 1, HR2: heptad repeat 2, TM: transmembrane domain, E: endodomain. Not drawn to scale.
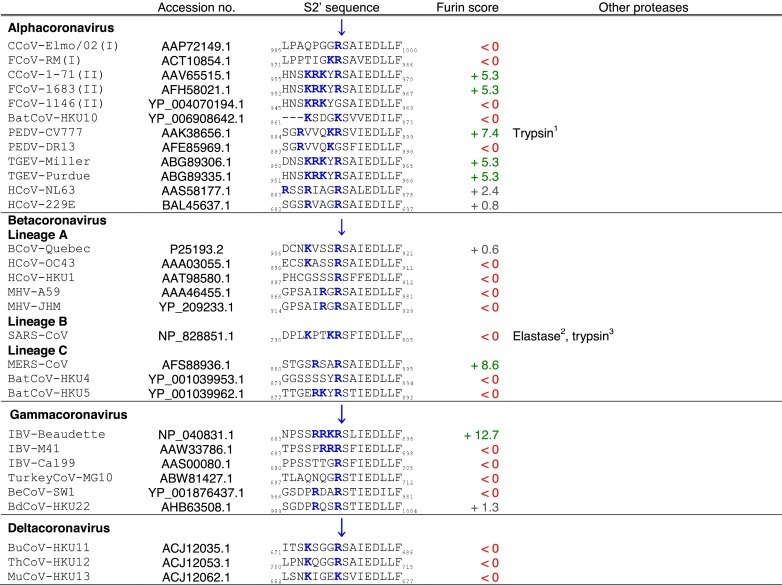
The S2 domain contains two heptad repeats (HR), which are composed of a chain of seven amino acid patterns, abcdefg, where a and d are hydrophobic residues, allowing formation of alpha-helical structures. The heptad repeats represent a defining structure of class I viral fusion proteins (Fig. 1B). These participate in the formation of coiled-coil structure during membrane fusion. Initially, the precise location of the coronavirus fusion peptide was unclear (Belouzard et al., 2012), however, it is becoming increasingly apparent that the fusion peptide is most likely located adjacent to one of the two cleavage sites, S2′ (Burkard et al., 2014, Madu et al., 2009, Masters and Perlman, 2013). This region contains a remarkably conserved motif, I-E-D-L-L-F, which is present in coronavirus S proteins from all four genera (Table 2). Similarly to the influenza HA fusion peptide (Cross et al., 2009), the proposed coronavirus fusion peptide contains mostly hydrophobic residue (I: isoleucine, L: leucine, and F: phenylalanine) with some negatively charged residues (E: glutamic acid, and D: aspartic acid).
Table 2.
Coronavirus spike (S) S2′ cleavage sites.
The transmembrane domain of the S protein is found in the region C-terminal of the S2 domain (Fig. 1B). A short cytoplasmic tail or endodomain is located at the C-terminal end of S. It contains conserved stretches of cysteine residues, which can be palmitoylated. Palmitoylation of cysteine residues within the endodomain was found to be important for regulating fusogenicity of S (Petit et al., 2007, Shulla and Gallagher, 2009).
Cryo-EM studies on single particles of the SARS-CoV have shed light on the overall architecture of S protein trimers and the conformational changes they undergo during fusion (Beniac et al., 2006, Beniac et al., 2007). While individual structures of key domains of S, such as the receptor binding domain (Li et al., 2005a, Peng et al., 2011, Wu et al., 2009) or the six-helix bundle (Xu et al., 2004), have been extensively studied, it is important to note that, to date, a complete crystal structure determination of the S ectodomain for any coronavirus is still lacking. Such a structure would be very useful to explain accessibility of sites to proteases within S, the precise location of N-linked glycans, and to compare it with other class I viral fusion proteins with known structures, such as influenza virus HA.
3. Coronavirus spike (S) proteolytic activation
3.1. Coronavirus spike (S) can be cleaved at different sites
A peculiar feature of the coronavirus S protein is that it can contain more than one proteolytic cleavage site (Fig. 1B) (Masters and Perlman, 2013). The first identified cleavage site was found to be located at the S1/S2 boundary (Table 1 ), and another more recently characterized site was found to be within S2 (Table 2 ), upstream of the putative fusion peptide, and called S2′ (Belouzard et al., 2009). S can also be cleaved at other less well-characterized sites. Depending on the coronavirus and which host cell it infects, cleavage at these different sites can occur at different stages of the virus life cycle, such as during S protein biosynthesis in producer cells and during virus entry into target cells. Cleavage events occurring during biosynthesis or virus entry can be critical factors in modulating pathogenicity, cell and tissue tropism as well as host range. An important distinction from influenza HA is that after cleavage of coronavirus S, the S1 and S2 domains are not held by disulfide bonds, but remain associated non-covalently. Since the two domains are not held covalently, the S1 domain may be shed from the S2 stalk domain of the protein.
Table 1.
Coronavirus spike (S) S1/S2 cleavage sites.
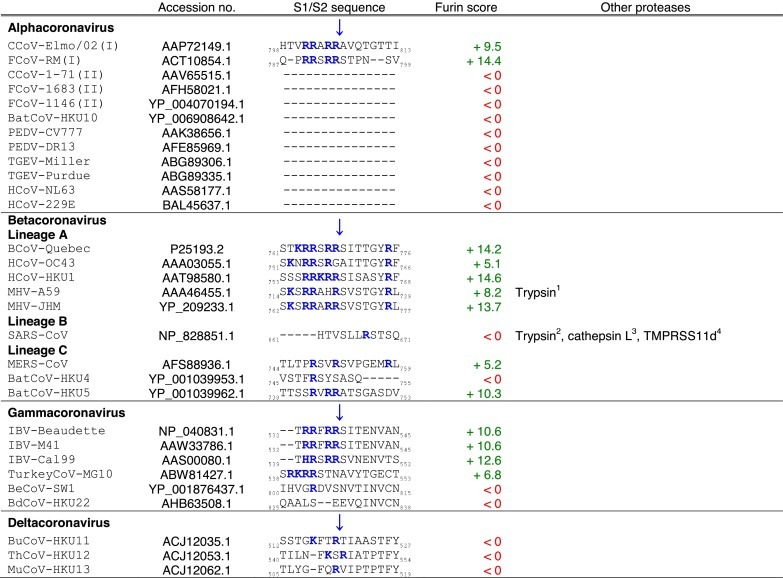
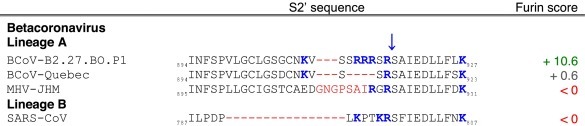
Tables. Analysis of coronavirus spike (S) cleavage sites.Table 1. Coronavirus S S1/S2 cleavage sites. The amino acid sequences of coronavirus S S1/S2 sites from the four coronavirus genera were aligned using ClustalX. Table 2. Coronavirus S S2′ cleavage sites. The amino acid sequences of coronavirus S S2′ sites from the four coronavirus genera were aligned using ClustalX. For Table 1, Table 2, the serotype of CCoV and FCoV are denoted in brackets (I or II). Table 3. Furin score and spike (S) amino acid sequence alignment of the S2′ region of passaged BCoV-B2.27.BO.P1 and other betacoronaviruses. Amino acid sequence alignment of S2′ region of S proteins of passaged BCoV-B2.27.BO.P1, BCoV-Quebec, MHV-JHM and SARS-CoV, based on sequence information by Borucki and collaborators (Borucki et al., 2013). Residues colored in red indicate positions with insertions/deletions. To generate furin scores in Table 1, Table 2, Table 3, sequences were queried into the PiTou 2.0 furin prediction algorithm that gives a score, with positive numbers (green) indicating predicted furin cleavage, while negative numbers (red) denote no predicted cleavage by furin. Furin scores that are borderline (<3) are denoted in grey. For comparison, the avian influenza strain A/muscovy duck/VietNam/209/2005(H5), which harbors the following polybasic cleavage site, R-R-R-K-R, has a +9.1 furin score. Other proteases, known to cleave coronavirus S1/S2 or S2′ sites are shown in the “Other proteases” column. Note that some proteases are known to cleave coronavirus S proteins, however, because the precise location of their cleavage site(s) has not been determined, they are not shown in Table 1, Table 2. Blue arrows denote the position of potential sites of cleavage. Basic arginine (R) and lysine (K) residues are highlighted in blue and bold font.
Proteases recognize specific amino acid sequences found in their substrates, with the general residue designation: P6–P6′ (Fig. 2A). The cleavage site or scissile bond is located between the residue positions P1 and P1′.
Fig. 2.
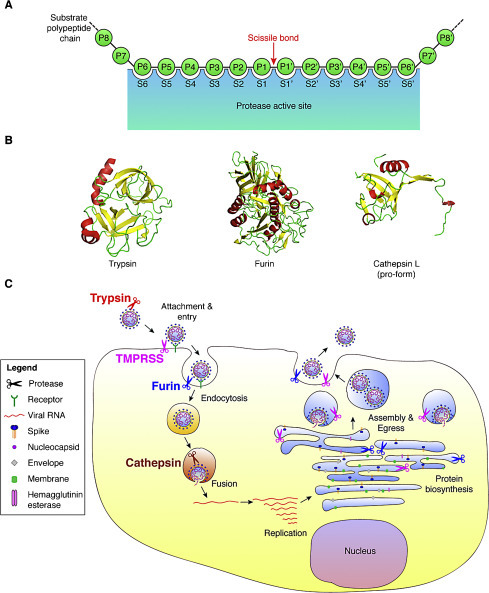
Host cell proteases involved in activating the coronavirus spike (S) protein. (A) Schematic of a protease cleavage site and substrate binding pocket. The sites within the protease that accommodate substrate residues are designated with the letter S. The residues of the substrate protein involved in recognition and proteolytic processing are denoted with the letter P. The scissile bond is cleaved by the protease and the residues involved in this bond are denoted P1–P1′. (B) Structures of three common host cell proteases known to activate coronavirus S: crystal structures of trypsin (PDB: 2PTN), furin (PDB: 1P8J), and the pro-form of cathepsin L (PDB: 1CJL). (C) Diagram of a coronavirus life cycle and the various host cell proteases known to cleave and activate some coronavirus S proteins. Note that for certain coronaviruses, fusion can occur directly at the plasma membrane.
3.2. Cellular proteases involved in coronavirus activation
3.2.1. Trypsin
Trypsin (Fig. 2B) is a prototype serine endopeptidase and has been studied extensively in the context of virus glycoprotein cleavage-activation. Trypsin cleaves at neutral pH, with an optimum of approximately 8.0. Biochemically, trypsin strongly prefers to cleave at arginine (R) or lysine (K) residues, although almost all viral substrates get cleaved at arginines. Other flanking residues are in general non-discriminatory, however basic or hydrophobic residues at P2 decreases the catalytic activity, as does a proline (P) residue at P3 (Halfon et al., 2004). Overall the requirement for cleavage is for a single basic residue at P1. As trypsin is not very selective in terms of substrate recognition, there are potentially many sites within coronavirus S proteins that may be cleaved by it. However, within the native trimeric conformation of S, only a few of these sites, if any, are accessible to trypsin cleavage. Trypsin is expressed in the respiratory tract, where its activity is inhibited by alpha-1 antitrypsin. Trypsin is principally a digestive enzyme, with active trypsin found in the small intestine; as such trypsin is likely to be able to directly cleave the S proteins of many enteric coronaviruses. Probably the best example of this is porcine epidemic diarrhea virus (PEDV), where most strains are highly trypsin-dependent. In the case of PEDV produced in Vero cells, trypsin cleavage only occurs after receptor binding, and not on free particles (Park et al., 2011a). For those coronaviruses with respiratory tissue tropism, cell-culture based studies using trypsin likely act as a surrogate for more biologically relevant proteases such as members of the TMPRSS family, which have similar substrate specificities. A good example of this is SARS-CoV produced in VeroE6 cells where trypsin can override the need for cathepsin-mediated cleavage, and shifts the virus to a low-pH independent route of entry, possibly at the plasma membrane. As with PEDV, prior binding to the viral receptor is a pre-requisite for SARS-CoV fusion activation (Matsuyama et al., 2005). In other cases, such as with feline coronavirus (FCoV), trypsin can activate the virus for fusion without the need for prior engagement with the viral receptor (Costello et al., 2013).
3.2.2. Furin and the proprotein convertase (PC) family
A large number of coronaviruses, from different genera, possess at their S1/S2 boundary site a furin cleavage site (Table 1). Furin (Fig. 2B) and furin-like proteases cleave at paired basic residues, and belong to the group of proteases called proprotein convertases.
The PC family is comprised of nine serine proteases that can cleave a vast number of cellular and microbial protein substrates. Furin belongs to the subset of PCs that includes proprotein convertase 1 (PC1), PC2, PC4, PC5, paired basic amino acid cleaving enzyme 4 (PACE4) and PC7, which generally cleave at single or paired basic residues within the following motif R/K-(X)0,2,4,6-R/K (X: any amino acid) (Seidah and Prat, 2012). The other two PCs, subtilisin/kexin-isozyme-1 (SKI-1) and proprotein convertase subtilisin/kexin type 9 (PCSK9) cleave at non-basic residue sites. SKI-1 recognizes R-X(-L/V/I)-X motifs (L: leucine, V: valine, I: isoleucine) and PCSK9 only autoproteolytically cleaves itself at the V–F–A–Q motif (F: phenylalanine, A: alanine, Q: glutamine). PCs are crucial regulatory proteases as they can activate and sometimes inactivate a very wide variety of substrates, such as enzymes and other proteases, cell adhesion factors, growth factors and hormones. The critical role of PCs is further demonstrated by knockout mouse experimental models, where individual knockout of furin, PC5, PACE4 or SKI-1 are lethal at different stages of development (Seidah and Prat, 2012).
PCs are known to be subverted by pathogenic microorganisms as they are recruited by viruses to process viral proteins, such as HIV gp160, respiratory syncytial virus (RSV) F protein, human papilloma virus minor capsid protein L2, dengue virus prM, and Chikungunya virus E3E2 (McCune et al., 1988, Ozden et al., 2008, Richards et al., 2006, Rodenhuis-Zybert et al., 2010, Zimmer et al., 2001). They are also implicated in the cleavage of bacterial toxins, e.g. Pseudomonas exotoxin and Shiga toxin (Molloy and Thomas, 2001).
Because furin can be viewed as a representative PC (Thomas, 2002), and because it is the most frequently characterized PC to be involved in processing and activating coronavirus S proteins, we will focus on this protease. However, it is important to recognize that there is a degree of redundancy between substrate recognition motifs by the different PCs, especially between furin, PC5 and PACE4. In line with this, a study has shown that overexpression of PACE4 and PC5 could enhance proteolytic cleavage of SARS-CoV S, along with furin (Bergeron et al., 2005).
Like all PCs, furin is a serine protease, containing a critical serine residue in its catalytic triad. Also, like all PCs, it is related to the bacterial subtilisin family of protease, in which the prototypical yeast protease kexin is found. PCs require Ca2+ ions to be active. While most PCs, including furin require neutral pH for activity, a study has shown that self-activation of furin via protonation of a regulatory histidine residue (His-69) requires a slightly acidic environment (Williamson et al., 2013). The minimal substrate recognition motif for furin is R-X-X-R, with the R-X-R/K-R motif being highly favorable. The fairly strict requirement for basic arginines at P1 and P4 positions as well as lysine at P2 is due to furin's binding pocket containing complementary charged residues (Henrich et al., 2003). Also, flanking serine (S) residues in the vicinity of the cleavage site appear to be highly favored by furin.
Several computational tools, such as ProP 1.0 and PiTou 2.0, have been made available for users to query amino acid sequences and predict whether or not they can be cleaved by furin (Duckert et al., 2004, Tian et al., 2012). The PiTou 2.0 tool offers the advantage of analyzing the 20 residue recognition motif used by furin (Tian, 2009, Tian et al., 2011), and takes into account the binding strength and solvent accessibility of substrate residues, allowing for high sensitivity and specificity predictions (Tian et al., 2012).
Furin is widely expressed and is often considered as a ubiquitous protease. However it should be noted that in most cases, the levels of furin expression are low (Shapiro et al., 1997). Furin is produced in the ER, and traffics along the secretory pathway, where it can process coronavirus S proteins in infected cells. Furin is a membrane bound protease that can be shed in the extracellular space, is constitutively secreted, and is mainly found in the trans-golgi network (TGN) where it is activated and can traffic to the plasma membrane. Furin can also recycle back from the plasma membrane to the TGN through an endocytic pathway (Seidah and Prat, 2012).
It is noteworthy that while most coronaviruses that are cleaved by furin or furin-like proteases are cleaved at only one site, the S1/S2 site, there are examples of coronaviruses, such as IBV and MERS-CoV that can be additionally cleaved at the putative fusion peptide-proximal site S2′ (Table 2), as described in Section 2.4 Proteolytic activation mechanisms of coronaviruses.
3.2.3. Cathepsins
The cathepsins comprise a group of cysteine, serine, and aspartyl proteases with both endo- and exo-peptidase activity that are typically found in endosomes and lysosomes and function as degradative enzymes, as well as in antigen processing.
Cathepsins may also be secreted however, especially in cancer cells (Mohamed and Sloane, 2006). As a general rule, the substrate specificity of cathepsins is quite broad, and analysis of the residues recognized by these enzymes seems to differ depending on the analyses used. Biochemical analysis of peptide substrates indicates that arginine (R) residues are preferred at the P1 position, often with an aromatic residue at P2 (Choe et al., 2006, Polgár, 1989, Rawlings and Barrett, 2004). However, databases of natural cathepsin substrates (e.g. MEROPS) show a preference for non-polar residues such as glycine (G), alanine (A), serine (S), threonine (T), and glutamine (Q) at the P1 position, along with a bulky hydrophobic residue at P2. Given their predominant endosomal/lysosomal location there is a distinct requirement of low pH for enzymatic activity. Cathepsin L (Fig. 2B) is most commonly associated with activation of viral glycoproteins, and has a pH optimum of 3.0 to 6.5. Cathepsin L is known to process SARS-CoV S, as well as a range of other coronaviruses, including MERS-CoV, HCoV-229E, and MHV-2 (Kawase et al., 2009, Qiu et al., 2006, Shirato et al., 2013, Simmons et al., 2005). Cathepsin B can also be involved in coronavirus entry, and has some distinct properties compared to cathepsin L and other cathepsins; it has a generally higher pH optimum and, while preferring an aromatic P2 residue, can process di-basic substrates, in contrast to the other cathepsins (Choe et al., 2006, Mort, 2004, Polgár, 1989). Coronaviruses using cathepsin B for entry include type II feline coronavirus (FCoV) and MHV-2 (Qiu et al., 2006, Regan et al., 2008).
3.2.4. Other proteases: Transmembrane protease/serine (TMPRSS) proteases, elastase, plasmin
Members of the TMPRSS family are membrane-bound tryspin-like serine proteases that are found on the cell surface or in the secretory pathway of cells (Antalis et al., 2011, Simmons et al., 2013). TMPRSS proteases are part of the larger family of proteases, the type II transmembrane serine proteases or TTSP (Bugge et al., 2009). TTSPs are subdivided into four groups: the HAT/DESC, Hepsin/TMPRSS, Matriptase, and Corin subfamilies. TMPRSS proteases are type II transmembrane proteins. They are widely expressed in the respiratory tract and are involved in the activation of many respiratory viruses. Both MERS-CoV S and SARS-CoV S can be activated by TMPRSS family members, as well as HCoV-229E S (Bertram et al., 2013, Gierer et al., 2013, Glowacka et al., 2011, Shirato et al., 2013). Intriguingly, TMPRSS2 was shown to be important for the late stages of PEDV life cycle (Shirato et al., 2011). While the substrate specificity of TMPRSS proteases is not well defined, it is generally thought to be similar to that of trypsin.
Recently, the TTSPs mosaic serine protease large form (MSPL), and differentially expressed in squamous cell carcinoma 1 (DESC1) were shown to activate MERS-CoV and SARS-CoV S proteins for cell–cell and virus–cell fusion (Zmora et al., 2014). MSPL and DESC1 were also shown to activate the HA proteins of influenza virus. The authors demonstrated by quantitative RT-PCR that while MSPL was readily expressed in human lung tissue, DESC1 was only expressed at low levels. Remarkably, TMPRSS13, a splice variant form of MSPL, was shown to be sensitive to inhibition by the naturally occurring Kunitz-type serine protease inhibitor, hepatocyte growth factor activator inhibitor type 1 (HAI-1) (Hashimoto et al., 2010). HAI-1 was shown earlier to be involved in the regulation and inhibition of TTSPs such as hepsin, matriptase and prostatin (Fan et al., 2005, Kirchhofer et al., 2005, Lin et al., 1999). A related protease inhibitor, HAI-2 was shown to inhibit influenza virus infection in vitro and its administration protected animals in a mouse model for influenza infection (Hamilton et al., 2014). It would be of interest to test such naturally occurring inhibitors for their effects on coronavirus entry and infection.
Elastase has been shown to activate SARS-CoV S. While the biological relevance of this has focused on elastase produced by neutrophils during the inflammatory response, most experimental studies have used pancreatic elastase. Like trypsin, pancreatic elastase can shift SARS-CoV entry to a low pH-independent route (Belouzard et al., 2010, Matsuyama et al., 2005), although the amino acid preferences for elastase are very different to trypsin, with alanine (A), serine (S), glycine (G) and valine (V) residues being preferred at the P1 position (Bieth, 2004).
Plasmin is a key enzyme in blood clot lysis and its major natural substrates are fibrinogen and fibrin (Castellino, 2004, Castellino and Ploplis, 2003). It is produced from its precursor plasminogen. Plasmin is also involved in a variety of other cellular processes in various tissues. Like trypsin, plasmin cleaves at K- and R-bonds, but is much less efficient and cleaves only a subset of such bonds. In part, this is due to differences in the P2 residues, where bulky hydrophobic residues are preferred (Backes et al., 2000, Hervio et al., 2000). Thus plasmin is a much more selective enzyme than trypsin. Plasmin is known to activate certain influenza virus HAs (Lazarowitz et al., 1973b, Sun et al., 2010, Tse et al., 2013). Although plasmin has been shown to activate SARS-CoV S (Kam et al., 2009), there remains little direct biological evidence for a role of plasmin in activating SARS-CoV or other coronaviruses.
Another possible source of proteases that may impact coronavirus pathogenesis is proteases produced by co-infecting bacteria. Many bacterial species secrete proteases as virulence factors, or elicit secretion of host proteases, and these proteases are likely to have the capacity to activate virus envelope glycoproteins under certain co-infection conditions. Such a situation has been explored for influenza virus (Böttcher-Friebertshauser et al., 2013), but has not generally been evaluated in coronaviruses, except for infection of mice with SARS-CoV and Pasteurella pneumotropica, a low-pathogenicity bacterium that elicits elastase production in the lungs (Ami et al., 2008).
3.3. Spatio-temporal regulation of coronavirus fusion
One important component of S protein activation is that protease action is integrated into the life cycle of the virus, with cleavage able to occur during biosynthesis of the S protein during virus assembly, in the extracellular space on released particles, or on the cell surface and in endosomal compartments during virus entry (Fig. 2C). As a general rule, cleavage during biosynthesis is likely to be carried out by furin and related PCs that have steady-state localization to the ER/Golgi and trans-Golgi compartments. Cleavage by trypsin and TMPRSS family members would be expected to occur in the extracellular space and cell surface, respectively. Cleavage during virus entry is expected to be mediated by cathepsins. However, it is important to note that intracellular proteases cycle between membrane compartments, and many proteases can shed catalytically active ectodomains from the cell surface. For instance, TMPRSS proteases can be active in the Golgi, furin can be present on the cell surface or in endosomes, and some cathepsins can be active in the secretory pathway or outside the cell. Such spatio-temporal protease cleavage events can be modulated extensively in different cell types and by the physiological status of a given cell type.
3.4. Proteolytic activation mechanisms of coronaviruses
3.4.1. Severe acute respiratory syndrome coronavirus (SARS-CoV)
The emergence of SARS-CoV in the human population reinforced research in coronavirus biology. In particular, the entry pathways and activation of SARS-CoV, a betacoronavirus, have become the subject of intense study. The proteolytic activation mechanisms of SARS-CoV S protein are perhaps amongst the best characterized so far and provide a good model to investigate coronavirus S proteolytic processing because many details of the proteases involved and cleavage sites processed are known. SARS-CoV also represents an example of how virus envelope glycoproteins can be activated by a diverse array of mechanisms.
Initially, a study employing retroviral pseudovirions produced in HEK-293T cells and harboring SARS-CoV S has found that SARS-CoV entry was sensitive to lysosomotropic agents, indicating a role for endosomal low pH (Simmons et al., 2004). Using similar HEK-293T produced SARS-CoV S pseudovirions, it was later shown that the dependency of virus entry on low-pH was because SARS-CoV S can be activated by endosomal cathepsin L, which requires low pH for its enzymatic activity (Simmons et al., 2005). Surprisingly, SARS-CoV seems to have evolved a multi-pronged approach regarding the activation of its S as many other proteases were shown to enable S-mediated entry and fusion, most of which act on the virus outside of the cell. Cathepsin L was shown to cleave SARS-CoV S at residue T678 in the S1/S2 boundary (Table 1) (Bosch et al., 2008). Trypsin was shown to activate S-induced syncytia formation, without the need to acidify the extracellular milieu (Simmons et al., 2004), indicating that the low pH requirement observed for SARS-CoV entry is not due to S requiring low pH for fusion competency. Trypsin was found to cleave SARS-CoV S at R667 in the S1/S2 site (Table 1) (Li et al., 2006). Furthermore, using native SARS-CoV infection in VeroE6 cells, it was shown that along with trypsin, thermolysin and elastase were also able to activate SARS-CoV fusion and entry (Matsuyama et al., 2005). The authors showed that cleavage activation by trypsin and thermolysin on virions bound on the cell surface allowed a much more efficient entry (>100-fold enhancement of infectivity) than endosomal cathepsin-mediated entry. Notably, prior treatment of virions before cell attachment decreased infectivity. This result suggests that fusion-active cleaved SARS-CoV S proteins are unstable, and the conformational changes induced by the cleavage event are transient and irreversible.
The location of the cleavage sites within SARS-CoV S has been investigated. Using wild type and mutated SARS-CoV S pseudovirions produced in HEK-293T cells, it was found that S could be cleaved by trypsin at two distinct sites, one located at the boundary of S1 and S2, the “classical” S1/S2 site (R667 P1 residue), and another newly identified site, S2′ (R797 P1 residue), found upstream of the putative fusion peptide and within the S2 domain (Table 1, Table 2) allowing for activation of fusion and entry (Belouzard et al., 2009). Importantly, trypsin cleavage of SARS-CoV S is thought to be sequential, with the S1/S2 cleavage occurring first and enhancing subsequent cleavage at S2′. It is the second cleavage event, at S2′, that is believed to be crucial for fusion activation of S. The S1/S2 cleavage appears dispensable for syncytia formation and virus–cell fusion.
Elastase-mediated cleavage was shown to occur at the S2′ site of SARS-CoV S on pseudovirions produced in HEK-293T cells, with the P1 residue found to be a threonine, T795 (Table 2) (Belouzard et al., 2010). Cleavage at this site shifts the starting position of the putative fusion peptide, which modulates but does not abrogate fusogenicity of S. A study has shown that plasmin, a known protease activator of influenza HA, could, along with trypsin, cleave SARS-CoV S expressed in BHK-21 cells (Kam et al., 2009).
The respiratory-tract resident and cell surface-expressed TMPRSS2 and TMPRSS11a proteases were shown to cleave and mediate activation of SARS-CoV entry in studies using HEK-293T-produced SARS-CoV S pseudoviruses (Glowacka et al., 2011, Kam et al., 2009). This is a feature shared with influenza HA, which is known to be activated by TMPRSS proteases (Böttcher et al., 2006). Remarkably, it was found that TMPRSS2 associates with the SARS-CoV receptor, ACE2, forming a receptor-protease complex allowing for efficient entry of the virus, directly at the cell surface (Shulla et al., 2011). Another TMPRSS protease, TMPRSS11d also known as human airway trypsin-like protease (HAT), was also shown to proteolytically activate SARS-CoV S in a HEK-293T expression system (Bertram et al., 2011). Using mass spectrometry, the authors demonstrated that TMPRSS11d cleaved S at R667, in the S1/S2 cleavage site (Table 1). While TMPRSS2 cleavage appears to occur at different sites within S, TMPRSS11d cleavage occurs mostly at the S1/S2 site (R667), which may explain why TMPRSS11d activation appears sufficient for cell–cell fusion but not virus–cell fusion. After TMPRSS2 cleavage, shed fragments of S, including a large 150 kDa fragment, are detected in the supernatant of cells and were shown to act as decoys to anti-S neutralizing antibodies (Glowacka et al., 2011).
Factor Xa is yet another protease that was shown to activate SARS-CoV S for entry into host cells (Du et al., 2007). The authors showed that the S protein of SARS-CoV retroviral pseudovirions produced in HEK-293T cells could be cleaved by factor Xa at the S1/S2 boundary. This cleavage event occurred when the pseudovirions were incubated with susceptible cells that were shown to express the protease.
Overall, the study of SARS-CoV S was instrumental in defining cleavage sites and putting forward the notion that the coronavirus S protein can be cleaved by a wide variety of proteases, suggestive of a relatively high degree of plasticity in cleavage mechanisms employed.
3.4.2. Murine hepatitis virus (MHV)
The betacoronavirus MHV has been extensively studied and represents a good model for coronavirus biology and in particular for studying coronavirus entry mechanisms. In groundbreaking work on MHV strain A59 (MHV-A59), the S protein found on virions (then known as E2), was shown to be cleaved during its biosynthesis when produced in four infected murine cell lines: 17 Cl 1, Sac-, DBT and L2 (Frana et al., 1985). Pulse-chase experiments demonstrated that the cleavage event occurred before virion release from producer cells. The authors showed that the degree of cleavage varied depending on which cell type the S protein was expressed in. A study from the same group showed that the 180 kDa precursor S protein of MHV-A59 virions produced in the 17 Cl 1 mouse cell line was cleaved approximately in two halves, with equal amounts of uncleaved and cleaved S and yielding two ∼90 kDa cleavage products (Sturman et al., 1985). The proteolytic processing of S was found to be potentiated by trypsin treatment of virions produced in murine 17 Cl 1 cells. The site where trypsin cleaved MHV-A59 S was mapped to the S1/S2 site (Table 1) (Luytjes et al., 1987). It was later shown that MHV-A59 S is cleaved by furin during biosynthesis and trafficking of S through the trans-Golgi network in murine LR-7 cells (de Haan et al., 2004). MHV-A59 S contains a minimal furin cleavage motif (R-R-A-H-R-S), containing at least P1 and P4 arginine residues (R-X-X-R) at the S1/S2 boundary, a feature that is shared by some alphacoronaviruses and many other coronaviruses from the beta- and gamma-genera (Table 1). Mutating the S1/S2 site was shown to greatly affect the degree of cleavage of the S protein (de Haan et al., 2004). Another important concept that comes out from MHV studies is that the degree of cleavage at S1/S2 found when S is expressed and matures in different cell lines reflects the varying abundance of protease expression.
The MHV-2 strain was found to have lost this cleavage site and its S is uncleaved on virions exiting murine DBT producer cells (Yamada et al., 1997). When expressed in cells, MHV-2 S fails to induce syncytia at neutral pH. MHV-2 S-induced syncytia can form if exogenous protease treatment is applied. It appears that MHV-2 still requires proteolytic processing for fusion competency. Indeed, MHV-2 S-mediated entry of virions produced in 17 Cl 1 murine cells is sensitive to inhibitors of the endosomal compartment resident proteases cathepsin B or L in murine fibroblast L2 target cells (Qiu et al., 2006). This suggests that for MHV-2, the timing and nature of proteases involved has been switched from furin-like proteolytic processing during virus maturation in producer cells to endosomal cathepsins during entry in target cells.
Recent work reported that the S protein of MHV-A59, in addition to being cleaved at S1/S2 during protein biosynthesis in mouse LR7 producer cells, is cleaved a second time at a site near the putative fusion peptide, named S2*, during the viral entry process (Wicht et al., 2014a). This site is analogous to the S2′ site in SARS-CoV S (Belouzard et al., 2009). The authors devised a clever experimental setup using conditional biontinylation assay that allows specific labeling and detection of MHV S proteins that have undergone the fusion process. This allows the analysis of any cleavage activation that may have occurred before fusion takes place. A new ∼80 kDa band was detected by Western blot analysis, which the authors conclude to be the fusion subunit. A comprehensive study, using siRNA screenings and a replication-independent fusion assay, has shown that MHV-A59 entry necessitates clathrin-mediated endocytosis and that the virions fuse with lysosomal membranes (Burkard et al., 2014). The study also conclusively demonstrates that MHV-A59 requires cleavage of S, at the S2′ site. Inhibition of MHV-A59 entry could be obtained by using a pan-lysosomal protease inhibitor. The identity of the specific protease involved awaits elucidation.
3.4.3. Bovine coronavirus (BCoV)
A recent study on the betacoronavirus BCoV has uncovered important features of coronavirus population adaptation during host, tissue, and cell tropism shifts (Borucki et al., 2013). Intriguingly, the authors found a critical role for a genotype containing a S variation at the S2′ site that rapidly expands and dominates the population during the course of passaging in cell culture (Table 3 ). Using a deep-sequencing approach the authors looked at the effect of passaging field strains of BCoV obtained in nasal washes on the genotype composition of the population. The viruses were passaged in human or bovine macrophage cell lines (THP-1 and BOMAC) as well as in a human rectal cell line (HRT-18). Surprisingly, they found that a variant genotype, a minority present in unpassaged samples, and which harbored a mutated S containing a multibasic insert composed of three arginines at S2′, was independently selected during passaging of three different nasal wash samples in either BOMAC, THP-1 or HRT-18 cell lines (for example, the B2.27.BO.P1 BOMAC passaged virus, Table 3). Surprisingly, the study showed that just a single passage in BOMAC cells was sufficient to expand the minority variant to become dominant in the viral population. It is notable that the insertion of the three basic arginines (R-R-R) is very similar to what is observed in the HA polybasic sites of highly pathogenic avian influenza virus subtypes H5 and H7 (Kawaoka and Webster, 1988, Klenk and Garten, 1994a, Lee et al., 2006). The work on BCoV demonstrates the important role of cleavage sites in shifting coronavirus genetic populations depending on the proteolytic environment.
Table 3.
Furin score and spike (S) amino acid sequence alignment of the S2′ region of passaged BCoV-B2.27.BO.P1 and other betacoronaviruses.
3.4.4. Infectious bronchitis virus (IBV)
The avian gammacoronavirus IBV was the first described coronavirus. It provokes extensive impairment of respiratory and urogenital tracts of chickens, and has been well studied. Many strains of IBV have been characterized, most of which have a restricted cell and tissue tropism, often limited to primary chicken cells. All IBV strains harbor a well-defined furin cleavage site at the S1/S2 site (R-R-F-R-R-S, Table 1) that is cleaved during protein biosynthesis. Strikingly, a lab adapted strain of IBV, the Beaudette strain, which has been heavily egg- and cell-culture adapted has acquired a mutation at its S2′ site, giving rise to the following motif S-R-R-R/K-R-S. Such a site is deemed ideal for furin recognition and cleavage as it harbors the critical P1 and P4 arginines (R), along with a highly favorable basic P2 lysine (K) or arginine (R) as well as flanking serines (S) at P1′ and P6 (Yamada and Liu, 2009). Notably, the IBV-Beaudette S2′ site is also cleaved during protein biosynthesis in S-transfected Huh-7 cells and in IBV-Beaudette-infected Vero cells, and this has been shown to be important for syncytia formation as well as entry of the virus (Belouzard et al., 2009, Yamada and Liu, 2009). Furthermore, IBV-Beaudette has a characteristically widened tissue and cell tropism and can productively infect many different cell types in vitro, including human cells. Importantly, high levels of furin expression, such as that observed in Huh-7 cells, was found to correlate tightly with highly productive infection by IBV-Beaudette produced in Vero cells (Tay et al., 2012). The wide cell and tissue tropism is in sharp contrast with the very restricted cell and tissue tropism observed for the other strains of IBV, which do not harbor a S2′ furin cleavage site. Similarly to the concept found for avian influenza, modification of its S2′ cleavage site may have allowed IBV-Beaudette to be cleaved by ubiquitously expressed furin, allowing for this expansion in tissue and cell tropism. This unusual feature is shared with MERS-CoV, as described in paragraph 2.4.7. Middle East respiratory syndrome coronavirus (MERS-CoV).
3.4.5. Porcine epidemic diarrhea virus (PEDV)
An alphacoronavirus, PEDV causes severe outbreaks of diarrheal disease in pigs mostly centering in East Asia (Park et al., 2011b). The virus recently emerged in the United States and is of particular concern because of the high mortality rates associated with piglet infections. In an early study, trypsin was found to activate PEDV for infection in Vero cells (Hofmann and Wyler, 1988). The S protein of Vero cell-produced PEDV was shown to be cleaved by trypsin when the virus is engaged with its receptor, but not on free virions (Park et al., 2011a). This suggests that PEDV S protein requires receptor-binding-induced conformational changes in order for trypsin cleavage to occur. TMPRSS2 was found to play a role in releasing PEDV particles from Vero producer cells stably expressing TMPRSS2 (Shirato et al., 2011). PEDV produced in Vero cells was also shown to enter cells via a clathrin-dependent endocytic pathway, also involving low pH and the activity of serine proteases (Park et al., 2014). In a recent study, notable findings about proteolytic activation of PEDV S have been uncovered (Wicht et al., 2014b). The authors focused on analyzing and comparing two strains of PEDV propagated in Vero cells, the first, CV777 which is an isolate that, like most PEDV isolates, requires trypsin activation for infection in cell culture, and the other, DR13, is a cell culture adapted strain, and has evolved to become trypsin-independent. The authors were able to demonstrate that CV777 S harbored a critical arginine (R) residue at the S2′ site that is cleaved by trypsin (Table 2). Notably, they found that in the cell-culture adapted strain DR13, an R to G mutation occurred at the S2′ site and explains why the strain has evolved to become trypsin-independent. As most coronavirus S proteins contain an arginine (R) at the S2′ site (Table 2), it would be interesting to study whether the findings of Wicht et al. can be generalized.
3.4.6. Feline coronavirus (FCoV)
Studies on FCoV offer deep insights into the role of S and in particular its proteolytic processing and fusion activation in coronavirus pathogenesis and host cell tropism. The FCoV is an alphacoronavirus and there are two known serotypes, based on differential S antigenicity (Pedersen, 2009). While serotype II viruses are the most extensively studied, notably due to their propensity to grow well in vitro, such as in CrFK feline kidney cells, they are not the most clinically prevalent. The serotype I is the most prevalent but is difficult to culture and so has yet to be extensively studied, although some strains, such as Black (TN-406) and UCD1, can be grown in feline AK-D cells and feline macrophage-like cell line Fcwf-4, respectively (Neuman et al., 2006, Pedersen, 2009). In both serotypes I and II, FCoV exists as two biotypes or pathotypes that display extremely contrasting pathogenicity. The low pathogenicity biotype, feline enteric coronavirus (FECV) is a highly prevalent virus that transmits easily through the fecal-oral route and generally causes only mild and self-limiting enteritis. In contrast, the highly pathogenic biotype, feline infectious peritonitis or FIPV, causes an acute, systemic and immune-mediated disease that is invariably fatal and represents a leading cause of infectious disease deaths in cats. The current understanding on FIP pathogenesis, the so-called “internal mutation theory”, is that it arises from an initial FECV infection in which the virus acquires mutations within the infected animal that leads to its conversion from the low pathogenic form to the virulent FIPV form (Vennema et al., 1998). A key difference between FECV and FIPV is their tissue and cell tropism: while FECV is generally restricted to the epithelial cells of the gastrointestinal tract, FIPV has the ability to readily infect monocytes and macrophages (Stoddart and Scott, 1989). It is this change in cell and tissue tropism, from gut epithelial cells to motile immune cells, that is thought to be the crucial step toward systemic spread of the virus. Several studies have mapped mutations that are linked to the transition between the biotypes, located in the accessory 3c and ORF 7 genes (Herrewegh et al., 1995, Pedersen et al., 2012), but there is a growing body of evidence indicating that mutations within the S protein are key to the development of FIP (Chang et al., 2012, Licitra et al., 2013). In the following section we will describe the current understanding on the S mutations that are thought to be linked with FIP.
3.4.6.1. Type I feline coronavirus (FCoV)
Type I FCoV have S proteins harboring two putative cleavage sites: S1/S2 and S2′ (Table 1, Table 2). A study on serotype I strains of FCoV has demonstrated that alphacoronaviruses could harbor a cleaved S protein (de Haan et al., 2008). In particular, serotype I UCD strain, a FECV obtained from the feces of experimentally infected cats, was shown to contain a S1/S2 site (R-R-S-R-R-S) that is cleaved by furin. In contrast, the authors show that UCD1, a FIPV adapted for growth in Fcwf cells, has acquired a mutation at the P1 position (R-R-S-R-G-S) abrogating cleavage by furin. It was also shown that because the S1/S2 site in UCD1 S was left uncleaved, it retained an intact heparan sulfate binding motif (B-B-X-B, B: basic residue), allowing the virus to bind the polysaccharide, a property that is not shared with its parental strain UCD. Since UCD1 arose from cell-culture adaptation, it is an important example of how a change in S cleavage site can lead to acquisition of new characteristics, such as heparan sulfate binding, that can lead to modulation of cell tropism and pathogenicity.
Licitra and colleagues have studied S S1/S2 cleavage site sequence diversity found in circulating field strains of type I FECV and compared them with those found in FIPV (Licitra et al., 2013). Remarkably, FECV S sequences harbored a highly conserved furin cleavage motif (R-R-S/A-R-R-S), an unusual property for an alphacoronavirus, which is shared with type I canine coronavirus (Table 1). However, a strong correlation was found between FIPV S sequences and presence of one or more non-silent mutations at the S1/S2 cleavage site, introducing a range of hydrophobic or non-polar residues. Biochemical assays employing fluorogenic peptide mimetics of the mutated cleavage sites confirmed that the substitutions found in FIPV modulated furin cleavage. A more recent study, using an extended sample set of type I FCoV, and which compares S1/S2, including the P6 position, and the S2′ cleavage sites of FECV and FIPV, confirmed the strong association between FIPV samples and mutations at the S1/S2 and S2′ cleavage sites (Licitra et al., manuscript in preparation). It is hypothesized that mutation at the S1/S2 and S2′ cleavage sites may switch S activating protease requirements, allowing for a change in tissue and cell tropism. Further research is required to investigate this hypothesis. A practical outcome of this discovery would be to use the type I S1/S2 and S2′ sites as biotype markers to discriminate between type I FECV and FIPV infections.
3.4.6.2. Type II feline coronavirus (FCoV)
The type II FCoV has a S protein that is distinct from type I FCoV as it was demonstrated to have arisen from recombination with canine coronavirus (CCoV) S (Herrewegh et al., 1998). Although the precise mechanisms involved differ from type I FCoV, a study on the prototypical FECV (1683) and FIPV (1146) type II FCoV strains, produced in canine A-72 cells, has shown that there is also a link between modified proteolytic processing requirements and cell tropism (Regan et al., 2008). Whereas FECV-1683 entry was shown to be highly dependent on endosomal cathepsin B and L activity and low pH, FIPV-1146 was shown to be dependent on cathepsin B activity but not cathepsin L activity. Importantly, while FECV-1683 S was demonstrated to be cleaved by both cathepsin B and L, FIPV-1146 could only be cleaved by cathepsin B. This difference in proteolytic processing may influence which cell types each virus infects, as individual protease expression profiles differ from cell type to cell type. It would be important to investigate further whether this change in proteolytic activation requirement is due to a mutation found within S, particularly at the S2′ site of FIPV-1146 S, as the R to G mutation found at that site (Table 2) is reminiscent of the R to G mutation found in PEDV S2′ (Wicht et al., 2014b).
3.4.7. Middle East respiratory syndrome coronavirus (MERS-CoV)
MERS-CoV, a recently emerged virus from the Middle East that is associated with severe pneumonia with a high case fatality rate, is another example of a coronavirus whose S protein can be activated by a multitude of proteases. Since its isolation and discovery in 2012, the complete viral genome was swiftly sequenced (van Boheemen et al., 2012), and its receptor, dipeptidyl peptidase 4 (DPP4 or CD26), rapidly identified (Raj et al., 2013).
In a report using pseudovirions produced in HEK-293T cells as well as protease inhibitors, it was first shown that TMPRSS2 and endosomal cathepsins could activate MERS-CoV virus–cell fusion (Gierer et al., 2013). The authors also reported the sensitivity of MERS-CoV entry to the lysosomotropic weak base NH4Cl, a result attributed to a decreasing of activity of endosomal cathepsins, which require low pH.
The role of cell surface-expressed TMPRSS2 in the activation of MERS-CoV was confirmed using native MERS-CoV virions produced in Vero cells, and it was estimated that the entry of the virus in Vero cells overexpressing the protease was increased 100-fold, compared to control cells (Shirato et al., 2013). The authors described how TMPRSS2 could greatly potentiate MERS-CoV infection-induced cell–cell fusion in Vero cells stably expressing the protease. The study also corroborated that endosomal cathepsin L could activate MERS-CoV S. Notably, the study showed that depending on the target cell type, MERS-CoV could use different activation pathways.
Along with TMPRSS2, TMPRSS4 was also shown to activate MERS-CoV S-mediated cell–cell fusion in a co-transfection HEK-293T cell expression system (Qian et al., 2013).
More recently, a study has revealed that MERS-CoV S from native virions produced in Vero cells could be cleaved by furin at two distinct sites, S1/S2 and S2′, with the former cleavage occurring during S biosynthesis in the Vero producer cells, and the latter, during virus entry into target cells (Millet and Whittaker, 2014). Similarly to the situation with IBV-Beaudette, it was also shown that expression of high levels of furin, such as in Huh-7 cells, along with the receptor DPP4, is associated with increased susceptibility to MERS-CoV infection. The role of furin during entry of MERS-CoV was confirmed by a recent study by Burkard and colleagues (Burkard et al., 2014). Intriguingly, a similar two-step activation mechanism for entry by furin was demonstrated for the paramyxovirus respiratory syncytial virus (RSV) (Gonzalez-Reyes et al., 2001, Krzyzaniak et al., 2013, Zimmer et al., 2001). It was shown for bovine RSV that the dual cleavage events generated a small peptide fragment, the virokinin, which is released upon activation and entry and has bioactive properties implicated in exacerbating pathogenesis (Valarcher et al., 2006, Zimmer et al., 2003).
4. Discussion: Spike (S) cleavage motifs as coronavirus cell and tissue tropism, host range, and virulence markers
In recent years, it has become more appreciated that the protease-mediated activation of membrane fusion is a versatile process that can be exploited by coronaviruses not only for the basic requirement of entry into host cells, but also to modulate this entry process and allow changes in cell, tissue and species tropism. In concert with changes in the receptor binding site(s) of S, the ability to modulate infectivity via cleavage site changes makes coronaviruses of particular concern as emerging pathogens. In particular, a recent study on MERS-CoV has strengthened the concept that along with the virus receptor, the repertoire of proteases a given cell type expresses can be a determining factor for infectivity (Barlan et al., 2014).
Our present understanding is that probably all coronavirus S are cleaved at some point during infection, in line with their designation as class I viral fusion proteins. In many cases, cleavage occurs at the S1/S2 position; a location that (based on the situation with other class I viral fusion proteins such an influenza HA) is at the expected junction between the S1 (receptor binding) and S2 (fusion) domains. We believe that in most cases, this cleavage will occur during the process of S protein maturation and virus assembly. S1/S2 cleavage may not be obligatory, however. While in several cases, coronaviruses appear to be released from cells with uncleaved S in many other cases the S1/S2 site is cleaved, often by furin. Previously, it has been suggested that alphacoronaviruses have uncleaved S, betacoronavirus S may or not be cleaved, with gammacoronavirus S having obligatory cleavage. However this now appears to be an oversimplification; for instance, S proteins of alphacoronaviruses in the feline and canine serotype I lineage have strong furin cleavage sites and have been shown to be furin cleaved, and based on bioinformatics (Table 1), S proteins of gammacoronaviruses in aquatic mammalian hosts appear not to be cleaved by furin. It appears that different coronaviruses have different requirements for S1/S2 cleavage.
The more recently identified cleavage site within the fusion domain (S2′) is more directly connected to the proposed fusion peptide, and so may function in a more analogous manner to the influenza HA cleavage site. As such, we believe that S2′ cleavage is an obligatory event for coronavirus entry (Burkard et al., 2014), with the possibility of cleavage at the S1/S2 site priming subsequent cleavage at the second (S2′) site (Belouzard et al., 2009, Millet and Whittaker, 2014). There is accumulating evidence that S2′ cleavage mainly occurs during virus entry (either at the cell surface or in endosomal compartments), and may be a relatively transient event. The transient nature of the cleavage means that in many cases it is difficult to observe this cleavage event based on conventional techniques (e.g. Western blots). While the nature of the proteases cleaving at S2′ remain unclear, it is likely that cathepsins would be involved, as well as potentially cell-surface TMPRSS2-like proteases. Bioinformatic analysis of S2′ indicates that a strong furin motif is present on laboratory-selected viruses (IBV-Beaudette and BCoV-B2.27.BO.P1, Table 2, Table 3). The presence of a robust furin cleavage site on MERS-CoV S at S2′ may represent a novel, if not unique, situation for a naturally occurring coronavirus. However, bioinformatics also predicts that some alphacoronavirus S (PEDV, TGEV, type II FCoV and type II CCoV) also have pronounced furin cleavage motifs at S2′. It will be important to test this possibility experimentally.
Another parallel to point out between cleavage sites of coronaviruses and influenza viruses is that it appears that the presence of a furin cleavage site adjacent to the putative fusion peptide can arise by either insertion of a polybasic site, as is the case for HPAI viruses of the H5 and H7 subtypes (Kawaoka and Webster, 1988, Lee et al., 2006) and the bovine coronavirus B2.27.BO.P1 (Table 3) (Borucki et al., 2013), or by incremental substitutions such as in avian influenza virus H9 subtype (Tse et al., 2014), MERS-CoV (Millet and Whittaker, 2014), or IBV-Beaudette (Tay et al., 2012; Table 2). Notably, MHV S contains what appears to be an insertion of a stretch of residues upstream of the cleavage site (Table 3), which are clearly linked to fusion activity (Taguchi and Shimazaki, 2000). In this regard, there are also parallels with influenza virus, where peptide insertions upstream of the cleavage site/fusion peptide modulate the fusion activity of H7 influenza viruses (Hamilton et al., 2012, Perdue, 2008). In addition, work on influenza virus HA have shown that glycosylations can have profound effects on cleavage activation, as bulky sugar moieties can create steric hindrance around the cleavage site, restricting access for proteases (Kawaoka et al., 1984, Tse et al., 2014). The role of glycosylations in modulating proteolytic activation of the coronavirus S protein, which is known to be heavily glycosylated, has been relatively unexplored and awaits further investigation.
Inhibitors of host cell proteases, including TMPRSS2, PCs and furin, have been proposed as potential antiviral therapeutics for coronavirus infections, in particular for the treatment of SARS and MERS infections (Bergeron et al., 2005, Gierer et al., 2013, Millet and Whittaker, 2014, Simmons et al., 2013). Targeting host cell proteases in the context of a virus infection presents the advantage of minimizing the development of viral drug resistance observed when targeting virus proteins. The list of cellular protease inhibitors approved for clinical use for various diseases, such as trypsin-like, factor X, and neutrophil elastase inhibitors, is growing (Turk, 2006). However, it is important to consider that such treatments increase the risk of higher toxicity and side effects as host cell proteases, such as furin, are essential for a great number of cellular processes (Seidah and Prat, 2012, Turk, 2006). Clearly, targeting host cell proteases will require more research efforts before actual therapeutics can be safely and effectively used against coronavirus infections.
Many open questions remain, including the specific proteases involved, the compartment and pH at which cleavage occurs, what proportion of S need to get cleaved within a single virus particle/S trimer and how many virus particle in a population get cleaved. New technologies such as single particle studies and selective biotinylation assays will hopefully provide answers to these questions in the future (Costello et al., 2013, Wicht et al., 2014a). In addition to the S1/S2 and S2′ sites, it is also possible that the S protein gets cleaved in other locations, giving rise to the concept that S undergoes a progressive “destabilization”, based on a combination of proteolysis (sometimes at multiple sites), low pH, and receptor binding, as part of its fusion activation (Gierer et al., 2014, Simmons et al., 2011). In this regard coronaviruses may show more similarity to Ebola virus, another virus with a class I fusion protein, than it does to influenza virus (White et al., 2008). It is also important to note that many coronavirus receptors are membrane ectopeptidases (i.e. ACE2, APN, DPP4). To date however, there is no evidence for proteolytic involvement of these receptors in virus entry (Bosch et al., 2014).
Bioinformatic analysis of the coronavirus S cleavage sites suggests the possibility of using this data within the framework of virulence or bio-markers that can differentiate different coronaviruses. For type I FCoV S, the S1/S2 and S2′ cleavage sites are likely to be a biotype markers differentiating FECV from FIPV, and for type II FCoV S a similar situation may also occur for their S2′ site. Notably, the amino acid sequence within the two cleavage sites are often highly conserved within groups of viruses, and so sequencing information of the cleavage sites may be useful in the future for virus typing, rapid identification of new viruses and as a “virulence marker”. This proposed virus typing can be applied to so-called polytropic coronaviruses, in particular IBV-Beaudette and MERS-CoV. These studies indicate that a second furin cleavage positioned at the S2′ site may be viewed as a marker of expanded cell and tissue tropism, host range and perhaps pathogenicity (Millet and Whittaker, 2014, Tay et al., 2012). At present, modulation of the S2′ site seems to be correlated most strongly to changes in tissue and cell tropism, host range and pathogenesis (e.g. IBV, MERS-CoV, PEDV). However, in other cases changes in the S1/S2 site are also strongly correlated (e.g. type I FCoV) with modification of cell tropism and pathogenesis. Overall, it seems likely that modulation of either of two protease cleavage sites by coronaviruses can have a profound impact on disease outcome, depending on the individual coronavirus.
The coronavirus field has gained tremendously thanks to the study of S proteolytic processing by host cell proteases. As shown in this review, such investigations are crucial for the development of the study of coronavirus entry, cell and tissue tropism, host range, and pathogenesis. While many questions remain, we look forward to future breakthroughs and discoveries in this active and lively area of investigation.
Acknowledgements
We thank Weishan Huang for critical reading of this manuscript. We are grateful to all members of the Whittaker laboratory for helpful discussions. This work was supported by National Institutes of Health Grant R21 AI111085.
References
- Alagaili A.N., Briese T., Mishra N., Kapoor V., Sameroff S.C., de Wit E., Munster V.J., Hensley L.E., Zalmout I.S., Kapoor A., Epstein J.H., Karesh W.B., Daszak P., Mohammed O.B., Lipkin W.I. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio. 2014;5(2):e00884-14. doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ami Y., Nagata N., Shirato K., Watanabe R., Iwata N., Nakagaki K., Fukushi S., Saijo M., Morikawa S., Taguchi F. Co-infection of respiratory bacterium with severe acute respiratory syndrome coronavirus induces an exacerbated pneumonia in mice. Microbiol. Immunol. 2008;52(2):118–127. doi: 10.1111/j.1348-0421.2008.00011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antalis T.M., Bugge T.H., Wu Q. Membrane-anchored serine proteases in health and disease. Prog. Mol. Biol. Transl. Sci. 2011;99:1–50. doi: 10.1016/B978-0-12-385504-6.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes B.J., Harris J.L., Leonetti F., Craik C.S., Ellman J.A. Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat. Biotechnol. 2000;18(2):187–193. doi: 10.1038/72642. [DOI] [PubMed] [Google Scholar]
- Barlan A., Zhao J., Sarkar M.K., Li K., McCray P.B., Jr., Perlman S., Gallagher T. Receptor variation and susceptibility to MERS coronavirus infection. J. Virol. 2014;88(9):4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U.S.A. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Madu I., Whittaker G.R. Elastase-mediated activation of the SARS coronavirus spike protein at discrete sites within the S2 domain. J. Biol. Chem. 2010;285(30):22758–22763. doi: 10.1074/jbc.M110.103275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac D.R., Andonov A., Grudeski E., Booth T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13(8):751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac D.R., deVarennes S.L., Andonov A., He R., Booth T.F. Conformational reorganization of the SARS coronavirus spike following receptor binding: implications for membrane fusion. PLoS One. 2007;2(10):e1082. doi: 10.1371/journal.pone.0001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron E., Vincent M.J., Wickham L., Hamelin J., Basak A., Nichol S.T., Chretien M., Seidah N.G. Implication of proprotein convertases in the processing and spread of severe acute respiratory syndrome coronavirus. Biochem. Biophys. Res. Commun. 2005;326(3):554–563. doi: 10.1016/j.bbrc.2004.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H., Thiel V., Pöhlmann S. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87(11):6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Muller M.A., Lavender H., Gnirss K., Nehlmeier I., Niemeyer D., He Y., Simmons G., Drosten C., Soilleux E.J., Jahn O., Steffen I., Pöhlmann S. Cleavage and activation of the SARS-coronavirus spike-protein by human airway trypsin-like protease. J. Virol. 2011;85(24):13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieth . Leukocyte elastase. In: Barrett, Rawlings, Woessner, editors. second ed. vol. 2. 2004. pp. 1517–1523. (Handbook of Proteolytic Enzymes). (2 vols) [Google Scholar]
- Borucki M.K., Allen J.E., Chen-Harris H., Zemla A., Vanier G., Mabery S., Torres C., Hullinger P., Slezak T. The role of viral population diversity in adaptation of bovine coronavirus to new host environments. PLoS One. 2013;8(1):e52752. doi: 10.1371/journal.pone.0052752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Bartelink W., Rottier P.J.M. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82(17):8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., Rottier P. Nidovirus entry into cells. In: Perlman, Gallagher, Snijder, editors. Nidoviruses. ASM Press; Washington, D.C.: 2008. pp. 157–178. [Google Scholar]
- Bosch B.J., Smits S.L., Haagmans B.L. Membrane ectopeptidases targeted by human coronaviruses. Curr. Opin. Virol. 2014;6C:55–60. doi: 10.1016/j.coviro.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher E., Matrosovich T., Beyerle M., Klenk H.-D., Garten W., Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006;80(19):9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher-Friebertshauser E., Klenk H.D., Garten W. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog. Dis. 2013;69(2):87–100. doi: 10.1111/2049-632X.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge T.H., Antalis T.M., Wu Q. Type II transmembrane serine proteases. J. Biol. Chem. 2009;284(35):23177–23181. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier P.J.M., Bosch B.J., de Haan C.A.M. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10(11):e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr C.M., Kim P.S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73(4):823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- Castellino . Plasmin. In: Barrett, Rawlings, Woessner, editors. second ed. vol 2. 2004. pp. 1692–1699. (Handbook of Proteolytic Enzymes). (2 vols) [Google Scholar]
- Castellino Ploplis. Kluwer Academic; New York, NY: 2003. Human Plasminogen: Structure, Activation, and Function, Plasminogen Structure, Activation, and Regulation; pp. 3–17. [Google Scholar]
- Chambers P., Pringle C.R., Easton A.J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- Chang H.W., Egberink H.F., Halpin R., Spiro D.J., Rottier P.J. Spike protein fusion Peptide and feline coronavirus virulence. Emerg. Infect. Dis. 2012;18(7):1089–1095. doi: 10.3201/eid1807.120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Skehel J.J., Wiley D.C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. U.S.A. 1999;96(16):8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe Y., Leonetti F., Greenbaum D.C., Lecaille F., Bogyo M., Bromme D., Ellman J.A., Craik C.S. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J. Biol. Chem. 2006;281(18):12824–12832. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- Costello D.A., Millet J.K., Hsia C.-Y., Whittaker G.R., Daniel S. Single particle assay of coronavirus membrane fusion with proteinaceous receptor-embedded supported bilayers. Biomaterials. 2013;34(32):7895–7904. doi: 10.1016/j.biomaterials.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross K.J., Langley W.A., Russell R.J., Skehel J.J., Steinhauer D.A. Composition and functions of the influenza fusion peptide. Protein Pept. Lett. 2009;16(7):766–778. doi: 10.2174/092986609788681715. [DOI] [PubMed] [Google Scholar]
- Cutler S.J., Fooks A.R., van der Poel W.H. Public health threat of new, reemerging, and neglected zoonoses in the industrialized world. Emerg. Infect. Dis. 2010;16(1):1–7. doi: 10.3201/eid1601.081467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Haijema B.J., Schellen P., Schreur P.W., te Lintelo E., Vennema H., Rottier P.J.M. Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J. Virol. 2008;82(12):6078–6083. doi: 10.1128/JVI.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Stadler K., Godeke G.-J., Bosch B.J., Rottier P.J.M. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell–cell but not virus–cell fusion. J. Virol. 2004;78(11):6048–6054. doi: 10.1128/JVI.78.11.6048-6054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Du L., Kao R.Y., Zhou Y., He Y., Zhao G., Wong C., Jiang S., Yuen K.-Y., Jin D.-Y., Zheng B.-J. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem. Biophys. Res. Commun. 2007;359(1):174–179. doi: 10.1016/j.bbrc.2007.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckert P., Brunak S.r., Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004;17(1):107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- Fan B., Wu T.D., Li W., Kirchhofer D. Identification of hepatocyte growth factor activator inhibitor-1B as a potential physiological inhibitor of prostasin. J. Biol. Chem. 2005;280(41):34513–34520. doi: 10.1074/jbc.M502119200. [DOI] [PubMed] [Google Scholar]
- Frana M.F., Behnke J.N., Sturman L.S., Holmes K.V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 1985;56(3):912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Kramer-Kuhl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., von Hahn T., Simmons G., Hofmann H., Pöhlmann S. The spike-protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2 and is targeted by neutralizing antibodies. J. Virol. 2013;87(10):5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Muller M.A., Heurich A., Ritz D., Springstein B., Karsten C.B., Schendzielorz A., Gnirss K., Drosten C., Pöhlmann S. Inhibition of proprotein convertases abrogates processing of the MERS-coronavirus spike protein in infected cells but does not reduce viral infectivity. J. Infect. Dis. 2014 doi: 10.1093/infdis/jiu407. PII: jiu407, [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pöhlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes L., Ruiz-Arguello M.B., Garcia-Barreno B., Calder L., Lopez J.A., Albar J.P., Skehel J.J., Wiley D.C., Melero J.A. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 2001;98(17):9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Al Dhahiry S.H.S., Reusken C.B.E.M., Raj V.S., Galiano M., Myers R., Godeke G.-J., Jonges M., Farag E., Diab A., Ghobashy H., Alhajri F., Al-Thani M., Al-Marri S.A., Al Romaihi H.E., Al Khal A., Bermingham A., Osterhaus A.D.M.E., AlHajri M.M., Koopmans M.P.G. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2013;14(2):140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon S., Baird T., Craik C. Trypsin. In: Barrett, Rawlings, Woessner, editors. second ed. vol. 2. Elsevier; Amsterdam: 2004. pp. 1483–1488. (Handbook of Proteolytic Enzymes). (2 vols) [Google Scholar]
- Hamilton B.S., Chung C., Cyphers S.Y., Rinaldi V.D., Marcano V.C., Whittaker G.R. Inhibition of influenza virus infection and hemagglutinin cleavage by the protease inhibitor HAI-2. Biochem. Biophys. Res. Commun. 2014;450(2):1070–1075. doi: 10.1016/j.bbrc.2014.06.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B.S., Sun X., Chung C., Whittaker G.R. Acquisition of a novel eleven amino acid insertion directly N-terminal to a tetrabasic cleavage site confers intracellular cleavage of an H7N7 influenza virus hemagglutinin. Virology. 2012;434(1):88–95. doi: 10.1016/j.virol.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Principles of virus structure. In: Knipe D.M., Howley P.M., editors. sixth ed. vol. 1. Lippincot Williams & Wilkins; Philadelphia, PA: 2013. pp. 52–86. (Fields Virology). (2 vols) [Google Scholar]
- Hashimoto T., Kato M., Shimomura T., Kitamura N. TMPRSS13, a type II transmembrane serine protease, is inhibited by hepatocyte growth factor activator inhibitor type 1 and activates pro-hepatocyte growth factor. FEBS J. 2010;277(23):4888–4900. doi: 10.1111/j.1742-4658.2010.07894.x. [DOI] [PubMed] [Google Scholar]
- Hemida M.G., Chu D.K., Poon L.L., Perera R.A., Alhammadi M.A., Ng H., Siu L.Y., Guan Y., Alnaeem A., Peiris M. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis. 2014;20(7):1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich S., Cameron A., Bourenkov G.P., Kiefersauer R., Huber R., Lindberg I., Bode W., Than M.E. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat. Struct. Mol. Biol. 2003;10(7):520–526. doi: 10.1038/nsb941. [DOI] [PubMed] [Google Scholar]
- Herrewegh A.A., Smeenk I., Horzinek M.C., Rottier P.J., de Groot R.J. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72(5):4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., Vennema H., Horzinek M.C., Rottier P.J.M., de Groot R.J. The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes. Virology. 1995;212(2):622–631. doi: 10.1006/viro.1995.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervio L.S., Coombs G.S., Bergstrom R.C., Trivedi K., Corey D.R., Madison E.L. Negative selectivity and the evolution of protease cascades: the specificity of plasmin for peptide and protein substrates. Chem. Biol. 2000;7(6):443–453. doi: 10.1016/s1074-5521(00)00125-3. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Simmons G., Rennekamp A.J., Chaipan C., Gramberg T., Heck E., Geier M., Wegele A., Marzi A., Bates P., Pöhlmann S. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 2006;80(17):8639–8652. doi: 10.1128/JVI.00560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26(11):2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.-W., Okumura Y., Kido H., Ng L.F.P., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One. 2009;4(11):e7870. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y., Naeve C.W., Webster R.G. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology. 1984;139(2):303–316. doi: 10.1016/0042-6822(84)90376-3. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Webster R.G. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1988;85(2):324–328. doi: 10.1073/pnas.85.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., Matsuyama S., Taguchi F. Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 2009;83(2):712–721. doi: 10.1128/JVI.01933-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhofer D., Peek M., Lipari M.T., Billeci K., Fan B., Moran P. Hepsin activates pro-hepatocyte growth factor and is inhibited by hepatocyte growth factor activator inhibitor-1B (HAI-1B) and HAI-2. FEBS Lett. 2005;579(9):1945–1950. doi: 10.1016/j.febslet.2005.01.085. [DOI] [PubMed] [Google Scholar]
- Klenk H.D., Garten W. Activation cleavage of viral spike proteins by host proteases. In: Wimmer E., editor. Cellular Receptors for Animal Viruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1994. pp. 241–280. [Google Scholar]
- Klenk H.D., Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2(2):39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- Krzyzaniak M.A., Zumstein M.T., Gerez J.A., Picotti P., Helenius A. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the f protein. PLoS Pathog. 2013;9(4):e1003309. doi: 10.1371/journal.ppat.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz S.G., Choppin P.W. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975;68(2):440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- Lazarowitz S.G., Compans R.W., Choppin P.W. Proteolytic cleavage of the hemagglutinin polypeptide of influenza virus. Function of the uncleaved polypeptide HA. Virology. 1973;52(1):199–212. doi: 10.1016/0042-6822(73)90409-1. [DOI] [PubMed] [Google Scholar]
- Lazarowitz S.G., Goldberg A.R., Choppin P.W. Proteolytic cleavage by plasmin of the HA polypeptide of influenza virus: host cell activation of serum plasminogen. Virology. 1973;56(1):172–180. doi: 10.1016/0042-6822(73)90296-1. [DOI] [PubMed] [Google Scholar]
- Lee C.-W., Lee Y.-J., Senne D.A., Suarez D.L. Pathogenic potential of North American H7N2 avian influenza virus: a mutagenesis study using reverse genetics. Virology. 2006;353(2):388–395. doi: 10.1016/j.virol.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Li F., Berardi M., Li W., Farzan M., Dormitzer P.R., Harrison S.C. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 2006;80(14):6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.-F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Licitra B.N., Millet J.K., Regan A.D., Hamilton B.S., Rinaldi V.D., Duhamel G.E., Whittaker G.R. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg. Infect. Dis. 2013;19(7):1066–1073. doi: 10.3201/eid1907.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-Y., Anders J., Johnson M., Dickson R.B. Purification and characterization of a complex containing matriptase and a Kunitz-type serine protease inhibitor from human milk. J. Biol. Chem. 1999;274(26):18237–18242. doi: 10.1074/jbc.274.26.18237. [DOI] [PubMed] [Google Scholar]
- Luytjes W., Sturman L.S., Bredenbeek P.J., Charite J., van der Zeijst B.A., Horzinek M.C., Spaan W.J. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology. 1987;161(2):479–487. doi: 10.1016/0042-6822(87)90142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83(15):7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters, Perlman . Coronaviridae. In: Knipe D.M., Howley P.M., editors. sixth ed. vol. 1. Lippincot Williams & Wilkins; Philadelphia, PA: 2013. pp. 825–858. (Fields Virology). (2 vols) [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. U.S.A. 2005;102(35):12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune J.M., Rabin L.B., Feinberg M.B., Lieberman M., Kosek J.C., Reyes G.R., Weissman I.L. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53(1):55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed M.M., Sloane B.F. Cysteine cathepsins: multifunctional enzymes in cancer. Nat. Rev. Cancer. 2006;6(10):764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- Molloy S.S., Thomas G. Furin. In: Rose E.D., David S.S., editors. Vol. 22. Academic Press; San Diego, CA: 2001. pp. 199–235. (The Enzymes). [Google Scholar]
- Mort . Cathepsin B. In: Barrett, Rawlings, Woessner, editors. second ed. vol. 2. Elsevier; 2004. pp. 1079–1086. (Handbook of Proteolytic Enzymes). (2 vols) [Google Scholar]
- Nagai Y. Protease-dependent virus tropism and pathogenicity. Trends Microbiol. 1993;1(3):81–87. doi: 10.1016/0966-842X(93)90112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash T.C., Buchmeier M.J. Entry of mouse hepatitis virus into cells by endosomal and nonendosomal pathways. Virology. 1997;233(1):1–8. doi: 10.1006/viro.1997.8609. [DOI] [PubMed] [Google Scholar]
- Neuman B., Adair B., Yoshioka C., Quispe J., Milligan R., Yeager M., Buchmeier M. Ultrastructure of SARS-CoV, FIPV, and MHV revealed by electron cryomicroscopy. In: Perlman S., Holmes K., editors. vol. 581. Springer; US: 2006. pp. 181–185. (The Nidoviruses). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden S., Lucas-Hourani M., Ceccaldi P.-E., Basak A., Valentine M., Benjannet S., Hamelin J., Jacob Y., Mamchaoui K., Mouly V., Desprès P., Gessain A., Butler-Browne G., Chrétien M., Tangy F., Vidalain P.-O., Seidah N.G. Inhibition of chikungunya virus infection in cultured human muscle cells by furin inhibitors. J. Biol. Chem. 2008;283(32):21899–21908. doi: 10.1074/jbc.M802444200. [DOI] [PubMed] [Google Scholar]
- Park J.E., Cruz D.J., Shin H.J. Receptor-bound porcine epidemic diarrhea virus spike protein cleaved by trypsin induces membrane fusion. Arch. Virol. 2011;156(10):1749–1756. doi: 10.1007/s00705-011-1044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Cruz D.J., Shin H.J. Clathrin- and serine proteases-dependent uptake of porcine epidemic diarrhea virus into Vero cells. Virus Res. 2014;191C:21–29. doi: 10.1016/j.virusres.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Kim H.K., Song D.S., Moon H.J., Park B.K. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field isolates in Korea. Arch. Virol. 2011;156(4):577–585. doi: 10.1007/s00705-010-0892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C. A review of feline infectious peritonitis virus infection: 1963–2008. J. Feline Med. Surg. 2009;11(4):225–258. doi: 10.1016/j.jfms.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Liu H., Scarlett J., Leutenegger C.M., Golovko L., Kennedy H., Kamal F.M. Feline infectious peritonitis: role of the feline coronavirus 3c gene in intestinal tropism and pathogenicity based upon isolates from resident and adopted shelter cats. Virus Res. 2012;165(1):17–28. doi: 10.1016/j.virusres.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10(12):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Sun D., Rajashankar K.R., Qian Z., Holmes K.V., Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. U.S.A. 2011;108(26):10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdue M.L. Molecular determinants of pathogenicity for avian influenza viruses. In: Swayne D.E., editor. Avian Influenza. Blackwell Publishing; Ames, IA: 2008. pp. 23–41. [Google Scholar]
- Petit C.M., Chouljenko V.N., Iyer A., Colgrove R., Farzan M., Knipe D.M., Kousoulas K.G. Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion. Virology. 2007;360(2):264–274. doi: 10.1016/j.virol.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgár L. CRC Press; Boca Raton, FL: 1989. Cysteine Proteases B. Cathepsins, Mechanisms of Protease Action; pp. 125–128. [Google Scholar]
- Pu Y., Zhang X. Mouse hepatitis virus type 2 enters cells through a clathrin-mediated endocytic pathway independent of Eps15. J. Virol. 2008;82(16):8112–8123. doi: 10.1128/JVI.00837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Dominguez S.R., Holmes K.V. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One. 2013;8(10):e76469. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Hingley S.T., Simmons G., Yu C., Das Sarma J., Bates P., Weiss S.R. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006;80(12):5768–5776. doi: 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings Barrett. Introduction: the clans and families of cysteine peptidases. In: Barrett, Rawlings, Woessner, editors. second ed. vol. 2. Elsevier; Amsterdam: 2004. pp. 1051–1071. (Handbook of Proteolytic Enzymes). (2 vols) [Google Scholar]
- Regan A.D., Shraybman R., Cohen R.D., Whittaker G.R. Differential role for low pH and cathepsin-mediated cleavage of the viral spike protein during entry of serotype II feline coronaviruses. Vet. Microbiol. 2008;132(3–4):235–248. doi: 10.1016/j.vetmic.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards R.M., Lowy D.R., Schiller J.T., Day P.M. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc. Natl. Acad. Sci. U.S.A. 2006;103(5):1522–1527. doi: 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenhuis-Zybert I.A., van der Schaar H.M., da Silva Voorham J.M., van der Ende-Metselaar H., Lei H.Y., Wilschut J., Smit J.M. Immature dengue virus: a veiled pathogen? PLoS Pathog. 2010;6(1):e1000718. doi: 10.1371/journal.ppat.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah N.G., Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discovery. 2012;11(5):367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- Shapiro J., Sciaky N., Lee J., Bosshart H., Angeletti R.H., Bonifacino J.S. Localization of endogenous furin in cultured cell lines. J. Histochem. Cytochem. 1997;45(1):3–12. doi: 10.1177/002215549704500102. [DOI] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Middle East respiratory syndrome coronavirus (MERS-CoV) infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013;87(23):12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Matsuyama S., Ujike M., Taguchi F. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J. Virol. 2011;85(15):7872–7880. doi: 10.1128/JVI.00464-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Gallagher T. Role of spike protein endodomains in regulating coronavirus entry. J. Biol. Chem. 2009;284:32725–32734. doi: 10.1074/jbc.M109.043547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85(2):873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Bertram S., Glowacka I., Steffen I., Chaipan C., Agudelo J., Lu K., Rennekamp A.J., Hofmann H., Bates P., Pöhlmann S. Different host cell proteases activate the SARS-coronavirus spike-protein for cell–cell and virus–cell fusion. Virology. 2011;413(2):265–274. doi: 10.1016/j.virol.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 2004;101(12):4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100(3):605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Stoddart C.A., Scott F.W. Intrinsic resistance of feline peritoneal macrophages to coronavirus infection correlates with in vivo virulence. J. Virol. 1989;63(1):436–440. doi: 10.1128/jvi.63.1.436-440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman L.S., Ricard C.S., Holmes K.V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90 K cleavage fragments. J. Virol. 1985;56(3):904–911. doi: 10.1128/jvi.56.3.904-911.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Tse L.V., Ferguson A.D., Whittaker G.R. Modifications to the hemagglutinin cleavage site control the virulence of a neurotropic H1N1 influenza virus. J. Virol. 2010;84(17):8683–8690. doi: 10.1128/JVI.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Shimazaki Y.K. Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: involvement in fusion activity. J. Gen. Virol. 2000;81:2867–2871. doi: 10.1099/0022-1317-81-12-2867. [DOI] [PubMed] [Google Scholar]
- Tay F.P.L., Huang M., Wang L., Yamada Y., Xiang Liu D. Characterization of cellular furin content as a potential factor determining the susceptibility of cultured human and animal cells to coronavirus infectious bronchitis virus infection. Virology. 2012;433(2):421–430. doi: 10.1016/j.virol.2012.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002;3(10):753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S. A 20 residues motif delineates the furin cleavage site and its physical properties may influence viral fusion. Biochem. Insights. 2009;2(BCI-2-Tian):9–20. [Google Scholar]
- Tian S., Huajun W., Wu J. Computational prediction of furin cleavage sites by a hybrid method and understanding mechanism underlying diseases. Sci. Rep. 2012;2:261. doi: 10.1038/srep00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Huang Q., Fang Y., Wu J. FurinDB: a database of 20-residue furin cleavage site motifs, substrates and their associated drugs. Int. J. Mol. Sci. 2011;12(2):1060–1065. doi: 10.3390/ijms12021060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse L.V., Hamilton A.M., Friling T., Whittaker G.R. A novel activation mechanism of avian influenza virus H9N2 by furin. J. Virol. 2014;88(3):1673–1683. doi: 10.1128/JVI.02648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse L.V., Marcano V.C., Huang W., Pocwierz M.S., Whittaker G.R. Plasmin-mediated activation of pandemic H1N1 influenza virus hemagglutinin is independent of the viral neuraminidase. J. Virol. 2013;87(9):5161–5169. doi: 10.1128/JVI.00210-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B. Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Discovery. 2006;5(9):785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- Valarcher J.F., Furze J., Wyld S.G., Cook R., Zimmer G., Herrler G., Taylor G. Bovine respiratory syncytial virus lacking the virokinin or with a mutation in furin cleavage site RA(R/K)R109 induces less pulmonary inflammation without impeding the induction of protective immunity in calves. J. Gen. Virol. 2006;87(6):1659–1667. doi: 10.1099/vir.0.81755-0. [DOI] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6) doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Poland A., Foley J., Pedersen N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243(1):150–157. doi: 10.1006/viro.1998.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.F., Eaton B.T. Bats, civets and the emergence of SARS. In: Childs J., Mackenzie J., Richt J., editors. vol. 315. Springer; Heidelberg: 2007. pp. 325–344. (Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission). [Google Scholar]
- Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y., Ji W., Li Y., Wu Y., Wang J., Iwamoto A., Woo P.C., Yuen K.Y., Yan J., Lu G., Gao G.F. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16(3):328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicht O., Burkard C., de Haan C.A., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Identification and characterization of a proteolytically primed form of the murine coronavirus spike proteins after fusion with the target cell. J. Virol. 2014;88(9):4943–4952. doi: 10.1128/JVI.03451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicht O., Li W., Willems L., Meuleman T.J., Wubbolts R.W., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014;88(14):7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D.M., Elferich J., Ramakrishnan P., Thomas G., Shinde U. The mechanism by which a propeptide-encoded ph sensor regulates spatiotemporal activation of furin. J. Biol. Chem. 2013;288(26):19154–19165. doi: 10.1074/jbc.M112.442681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009;234(10):1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Wu K., Li W., Peng G., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. U.S.A. 2009;106(47):19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F., Rao Z. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279(47):49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Liu D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009;83(17):8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y.K., Takimoto K., Yabe M., Taguchi F. Acquired fusion activity of a murine coronavirus MHV-2 variant with mutations in the proteolytic cleavage site and the signal sequence of the S protein. Virology. 1997;227(1):215–219. doi: 10.1006/viro.1996.8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zimmer G., Budz L., Herrler G. Proteolytic activation of respiratory syncytial virus fusion protein: cleavage at two furin consensus sequences. J. Biol. Chem. 2001;276(34):31642–31650. doi: 10.1074/jbc.M102633200. [DOI] [PubMed] [Google Scholar]
- Zimmer G., Rohn M., McGregor G.P., Schemann M., Conzelmann K.-K., Herrler G. Virokinin, a bioactive peptide of the tachykinin family, is released from the fusion protein of bovine respiratory syncytial virus. J. Biol. Chem. 2003;278(47):46854–46861. doi: 10.1074/jbc.M306949200. [DOI] [PubMed] [Google Scholar]
- Zmora P., Blazejewska P., Moldenhauer A.-S., Welsch K., Nehlmeier I., Wu Q., Schneider H., Pöhlmann S., Bertram S. DESC1 and MSPL activate influenza A viruses and emerging coronaviruses for host cell entry. J. Virol. 2014;88(20):12087–12097. doi: 10.1128/JVI.01427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


