Abstract
There are numerous examples of sex differences in brain and behavior and in susceptibility to a broad range of brain diseases. For example, gene expression is sexually dimorphic during brain development, adult life, and aging. These differences are orchestrated by the interplay between genetic, hormonal, and environmental influences. However, the molecular mechanisms that underpin these differences have not been fully elucidated. Because recent studies have highlighted the key roles played by epigenetic processes in regulating gene expression and mediating brain form and function, this chapter reviews emerging evidence that shows how epigenetic mechanisms including DNA methylation, histone modifications, and chromatin remodeling, and non-coding RNAs (ncRNAs) are responsible for promoting sexual dimorphism in the brain. Differential profiles of DNA methylation and histone modifications are found in dimorphic brain regions such as the hypothalamus as a result of sex hormone exposure during developmental critical periods. The elaboration of specific epigenetic marks is also linked with regulating sex hormone signaling pathways later in life. Furthermore, the expression and function of epigenetic factors such as the methyl-CpG-binding protein, MeCP2, and the histone-modifying enzymes, UTX and UTY, are sexually dimorphic in the brain. ncRNAs are also implicated in promoting sex differences. For example, X inactivation-specific transcript (XIST) is a long ncRNA that mediates X chromosome inactivation, a seminal developmental process that is particularly important in brain. These observations imply that understanding epigenetic mechanisms, which regulate dimorphic gene expression and function, is necessary for developing a more comprehensive view of sex differences in brain. These emerging findings also suggest that epigenetic mechanisms are, in part, responsible for the differential susceptibility between males and females that is characteristic of a spectrum of neurological and psychiatric disorders.
Keywords: DNA methylation, Epigenetics, Histone modifications, MicroRNAs, Non-coding RNAs, Sex differences, X chromosome inactivation
Introduction
Decades of research have focused on characterizing sexually dimorphic features in mammalian brain form and function. These range from differences in higher order cognitive and behavioral traits to those found at cellular and molecular levels. These include but are not limited to language abilities; social and reproductive behaviors; pain perception; responses to hormonal stimuli; overall and regional brain volumes; trajectories of brain development; patterns of neural network activation; region-specific neuronal numbers, morphology, and connectivity; and synaptic plasticity (Becker, 2008). Significant efforts have concentrated on elucidating the mechanisms responsible for promoting these sex-specific characteristics and revealed that sex steroid hormones and sex chromosomes play key roles in brain sexual differentiation during developmental critical periods and in brain sexual dimorphism throughout the life span (Becker, 2008). Sex differences in brain and behavior have been attributed to the effects of sex hormones including permanent (“organizational”) effects, such as those that promote masculinization and defeminization, and reversible (“activational”) effects. Brain sexual dimorphism is also mediated by the complement of genes encoded on the sex chromosomes, which are expressed in a sex-specific manner that is independent of the effects of sex hormones. In fact, in the genomic era, it has become increasingly clear that sex chromosome and autosomal gene expression and post-transcriptional RNA processing (e.g., alternative splicing) are sexually dimorphic in the brain in a region-, cell type-, and developmental stage-specific manner (Berchtold et al., 2008; Blekhman et al., 2010; Galfalvy et al., 2003; Lahr et al., 1995; Mayer et al., 1998; Reinius and Jazin, 2009; Reinius et al., 2008; Vawter et al., 2004; Weickert et al., 2009; Xu et al., 2002; Yang et al., 2006). However, the factors responsible for regulating sex-specific gene expression and function are not well characterized. Recent evidence has begun to unravel how epigenetic mechanisms, in a complex regulatory network that involves sex steroid hormone activity and sex chromosomes, promote sex differences in neural gene expression and function (Dunn et al., 2010; McCarthy et al., 2009).
The emerging field of epigenetics has already revolutionized our understanding of brain structure and function because it explains how specific genes and gene networks are dynamically regulated during development, homeostasis, and plasticity (Mehler, 2008). These epigenetic processes include DNA methylation, post-translational histone modifications, higher order chromatin remodeling, and non-coding RNA (ncRNA) regulation (Mehler, 2008). These epigenetic mechanisms are now being implicated in the molecular underpinnings of sex differences in the brain (Dunn et al., 2010; McCarthy et al., 2009). For example, sex differences have been noted in profiles of DNA methylation, histone modifications, and ncRNAs as well as in the expression of various epigenetic effector proteins (see below). The organizational effects of sex steroid hormone exposure during sexual differentiation are mediated, at least in part, by these epigenetic processes. Furthermore, X chromosome inactivation (XCI), the cellular mechanism by which female cells transcriptionally silence one X chromosome to compensate for having two genomic doses of X genes, is mediated by a series of epigenetic modifications that is initiated by an ncRNA (Brown et al., 1991). Epigenetic mechanisms are also responsible for governing gene-hormonal–environmental interactions, including the transgenerational programming that occurs in response to dietary influences and stress (Mehler, 2008). Furthermore, epidemiological studies have shown that the incidence of diverse neurological and psychiatric diseases is sex biased (Becker, 2008). These include a spectrum of neuroimmunological, neurodevelopmental, neurodegenerative, and psychiatric disorders. Because epigenetic factors play roles in promoting sex-specific neural gene expression and function, understanding these mechanisms may provide insights into the underpinnings of well-documented but often poorly explained sex differences in the susceptibility to these neurological and psychiatric disease states.
Sex differences in gene expression
Gene expression is sexually dimorphic in the brains of various species including humans and is important for mediating sex differences in brain and behavior. A number of studies have focused on characterizing the expression profiles for genes found on sex chromosomes as well as those on autosomal chromosomes during brain development, adult life, and aging (Berchtold et al., 2008; Galfalvy et al., 2003; Lahr et al., 1995; Mayer et al., 1998; Reinius and Jazin, 2009; Reinius et al., 2008; Vawter et al., 2004; Weickert et al., 2009; Xu et al., 2002; Yang et al., 2006). For example, an examination of whole brain preparations from adult mice showed that 612 genes are expressed in a sexually dimorphic manner, including 355 with female-biased and 257 with male-biased expression (Yang et al., 2006). Functional analysis of this entire set of genes revealed significant enrichment for RNA helicase activity, highlighting the potential roles of RNA metabolism in mediating sex differences in the brain (see below). Furthermore, genes encoding chemokine ligands, heat shock proteins, and histocompatibility antigens were overrepresented in male-biased genes, whereas genes involved in Janus Kinase-Signal Transducer and Activator of Transcription (JAK-STAT) signaling and lactation pathways were overrepresented in female-biased genes. A complementary study of the developing mouse brain revealed that 51 genes are expressed in a sexually dimorphic manner at embryonic day (E) 10.5, which is prior to the secretion of gonadal hormones (Dewing et al., 2003). These include genes with key roles in neural development, such as Gli3 and Wnt10b, which exhibit female- and male-enhanced expression, respectively. These factors are members of seminal signaling pathways that regulate brain patterning and cell fate specification within the E10.5 developmental time window (Hebert and Fishell, 2008). In fact, at E10.5, the volume of the ventral telencephalon is 50% greater in mice lacking functional Gli3 than in wild-type mice (Yu et al., 2009). The higher level of Gli3 expression in female brains during development is consistent with one of the well-characterized sex differences in the brain—female brains are typically smaller compared to their male counterparts.
Studies performed utilizing human brain tissues have revealed complementary dimorphic gene expression profiles, highlighting sex differences in genes that are encoded on sex and autosomal chromosomes. For example, in adult humans, SRY and ZFY were found in the hypothalamus and frontal cortex of the male but not female brain (Mayer et al., 1998); nine Y and two X chromosome-encoded genes displayed significant sex differences in prefrontal cortex (Galfalvy et al., 2003); and six genes, including X inactivation-specific transcript (XIST) (see below), were expressed in a sexually dimorphic manner in the dorsolateral prefrontal cortex, anterior cingulate cortex, and cerebellum (Vawter et al., 2004). A study of human brain development revealed that, at mid-gestation, some of the most significant sex differences in gene expression are found for genes encoded on the male-specific region of the Y chromosome (Reinius and Jazin, 2009). These genes included RPS4Y1, PCDH11Y, DDX3Y, USP9Y, NLGN4Y, EIF1AY, UTY, ZFY, TMSB4Y, CYorf15B, and PRKY, and the majority (10 out of 11) are expressed widely throughout the brain—in all 12 regions analyzed. Notably, these profiles overlap but are distinct from those present in rodents. For example, some of these human genes have rodent homologs, encoded on somatic chromosomes or on the X chromosome, that are not expressed dimorphically in the brain, or they do not have known rodent homologs. These observations suggest, not surprisingly, that sex differences in the human brain have evolved along with increasing complexity in brain form and function. Similarly, a recent analysis of gene expression in the human prefrontal cortex during development and adult life revealed sex differences, especially in infancy, for 130 transcripts (Weickert et al., 2009). These include zinc finger transcription factors, intracellular signaling molecules, and heat shock proteins. Among these, the Y-chromosome transcripts, PCHD11Y and NLGN4Y, are highly expressed in infant males and may have roles in promoting male-specific cortical network formation as these factors are implicated in mediating neuronal connectivity.
Furthermore, a study of 55 individuals cognitively intact and 20–99 years of age at the time of death revealed that changes in profiles of gene expression occurring during human brain aging are sexually dimorphic (Berchtold et al., 2008). Intriguingly, males exhibited expression changes for more than three times as many genes as females. The majority of these (66%) were down regulated and the most significant changes were found in the transition to the sixth and seventh decades of life. In contrast, females had an equivalent number of upregulated and downregulated genes and exhibited progressive expression changes with aging that were most significant in the eighth and ninth decades. Functional analysis of these gene sets revealed that downregulated genes in males are uniquely enriched (compared to females) for categories related to energy production, RNA metabolism, and protein synthesis/transport, suggesting a global decrease in metabolic capacity with aging. In contrast, down-regulated genes in females are uniquely enriched for categories related to cell–cell communication and neuronal morphogenesis. For upregulated genes, sex differences in enriched categories are less prominent but still present including, for example, RNA catabolism in males and integrin signaling in females. Moreover, this analysis was performed utilizing tissue from the hippocampus, entorhinal cortex, superior frontal gyrus, and postcentral gyrus and demonstrated region-specific sexually dimorphic patterns of aging. These observations suggest that sex differences in brain aging may play a role in the differential susceptibilities to the onset and progression of brain disorders.
Additional evidence suggests that sex differences in gene expression are evolutionarily conserved in the primate brain. A gene expression microarray study performed using cortex preparations from adult humans, macaques, and marmo-sets demonstrated that 85 genes exhibit common sex-biased expression in humans and macaques and that two genes, XIST and heat shock factor binding protein 1 (HSBP1), exhibit female-biased expression in all three species (Reinius et al., 2008). This study further demonstrated that the coding regions of genes with female-biased expression are more evolutionarily constrained compared to genes with male-biased and non-sex-biased expression (Reinius et al., 2008).
Sex determining region Y (SRY) is a high mobility group (HMG) box transcription factor encoded on the Y chromosome that is crucial for promoting differentiation of the bipotent gonadal primordium into the testis (Berta et al., 1990). In fact, it is referred to as the testis-determining factor because it is necessary and sufficient for male sex determination and because it leads to disorders of sexual development when mutated. Remarkably, sex differences in the brain can, at least in part, be attributed to the expression and function of SRY not only in the testis but also in the male brain. In adult male mice, SRY is found in the hypothalamus and midbrain (Lahr et al., 1995). Specifically, in adult male mice, SRY is expressed in tyrosine hydroxylase (TH)-expressing neurons within the locus coeruleus, substantia nigra, and ventral tegmental area where it may play a role in regulating TH expression and catecholamine biosynthesis (Milsted et al., 2004). When SRY is down regulated in male rat brains utilizing antisense oligonucleotides, TH expression decreases and motor functions are compromised though no effect on neuronal numbers is observed (Dewing et al., 2006). Similarly, in adult male humans, SRY is expressed in the hypothalamus and frontal and temporal cortex (Mayer et al., 1998). In a human male-derived neuroblastoma BE(2)C cell line, SRY serves as a transcriptional regulator of monoamine oxidase-A (MAO-A), an enzyme involved in catecholamine metabolism that is encoded on the X chromosome (Wu et al., 2009). These observations suggest that, through diverse mechanisms, SRY has effects on sexual dimorphism in adult brain function and potentially in neurological and psychiatric disorders associated with disruption of catecholaminergic pathways.
R-spondin 1 (RSPO1) is a more recently described factor encoded on an autosomal chromosome that acts as an active ovarian determinant (Parma et al., 2006). In addition to being expressed in the ovaries, it is also found transiently but at significant levels in regions of the developing murine central nervous system, including the roof plate of the neural tube with prominent expression in the forebrain (peak expression between E10 and E11) (Kamata et al., 2004). These findings imply that RSPO1 plays a role in mediating neural development, where it may be involved in Wnt and associated β-catenin signaling similar to its involvement in these pathways in the ovaries (Kamata et al., 2004). It is not clear whether this neural developmental expression is sexually dimorphic and whether RSPO1 plays a role in promoting sex differences in brain and behavior. However, it is intriguing to speculate that, because SRY represses the effects of RSPO1 signaling on gene expression in the developing gonads (Lau and Li, 2009), a similar mechanism of action may exist in neural cells where these factors are co-expressed.
Some genes found on sex chromosomes are expressed in specific patterns in the brain, implying that they are important for mediating particular brain functions. One hypothesis is that homologous gene pairs from the X and Y chromosomes are functionally equivalent. However, homologous genes may be differentially regulated in a temporal and spatial manner, suggesting that the two genes are not comparable (Xu et al., 2002). For example, Usp9x and Usp9y are homologous ubiquitin-specific proteases derived from the X and Y chromosomes, respectively. One study of sex differences in Usp9x expression in mice reported that Usp9x is clearly found in fetal, neonatal, and adult female brains, while Usp9y is expressed in fetal and neonatal males but only weakly in the adult (Xu et al., 2002). This sexually dimorphic expression profile for Usp9x in the adult mouse brain is related to the complement of sex chromosomes (Xu et al., 2005a). Further studies observed that in adult mice Usp9x is specifically expressed in neocortex, hippocampal subregions, cerebellar Purkinje cells, and the rostral migratory stream (Friocourt et al., 2005; Xu et al., 2005b), where it is implicated in diverse processes including synaptic development and plasticity (Xu et al., 2005b) and self-renewal, differentiation, and migration of neural progenitors (Friocourt et al., 2005; Jolly et al., 2009). These findings highlight how sexual dimorphism in the expression of homologous genes may be responsible for promoting sex differences in important neurobiological processes.
Sex differences in epigenetic regulatory mechanisms
DNA methylation
Recent evidence suggests that DNA methylation may play a key role in mediating sex differences in brain and behavior. DNA methylation refers to the covalent modification of cytosine dinucleo- tides in DNA that occurs in gene regulatory regions as well as in inter- and intragenic sequences (Fig. 1) (Mehler, 2008). It is an important mechanism for regulating genes associated with these regions and more global gene expression as well as for mediating additional epigenetic processes, such as XCI and imprinting (Mehler, 2008). Members of the DNA methyltransferase (DNMT) family of enzymes catalyze DNA methylation. MethylCpG-binding domain proteins (MBDs) bind to methylated DNA and mediate the effects of DNA methylation on gene transcription and other processes. The expression and function of these factors in neural cells is tightly regulated and is activity dependent (Sharma et al., 2008). Dynamic changes in DNA methylation are involved in modulating cell-, tissue-, and developmental stage-specific gene expression. Furthermore, these DNA methylation profiles are linked to a broad spectrum of processes including neural development, homeostasis, and plasticity (Feng et al., 2010; Mehler, 2008; Miller et al., 2010).
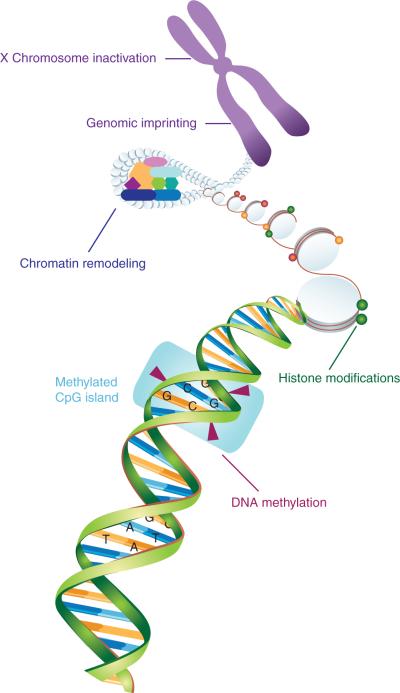
Fig. 1.
Epigenetic mechanisms. This diagram represents major classes of epigenetic mechanisms, including DNA methylation, histone modifications and chromatin remodeling, genomic imprinting, and X chromosome inactivation.
DNA methylation controls the expression of important sexually dimorphic genes, such as SRY, during gonadal development (Nishino et al., 2004). DNA methylation may be similarly relevant in the brain for regulating the expression of sexually dimorphic genes. For example, a recent study showed that Usp9x is expressed at higher levels in adults than in newborn mice. Correspondingly, the Usp9x promoter and gene body were less methylated in adults than in newborns (Xu, 2005). These observations suggest that decreased methylation is responsible for promoting increased expression of Usp9x. Similarly, another study found that, in the cortex of male and female mice, expression of the estrogen receptor-α (ERα) is high in early postnatal development and begins to decline at postnatal day (PN) 10, becoming absent in the adult (Westberry et al., 2010). This decrease in ERα expression was associated with significant hypermethylation of ERα gene regulatory regions for both sexes. Furthermore, this study found that methyl-CpG-binding protein 2 (MeCP2), an MBD, is recruited to the ERα promoter at PN10 and that MeCP2 mutants have increased ERα expression at subsequent postnatal time points. These observations suggest that DNA methylation regulates ERα expression in an MeCP2-dependent manner.
MeCP2 binds to methylated genomic loci and recruits additional epigenetic regulatory factors, which, in turn, modulates gene expression and local and long-range chromatin structural and functional dynamics. Interestingly, a recent study of rat brain development found sex differences in the expression of mecp2 in the amygdala and ventromedial hypothalamus (VMH) but not within the preoptic area (POA) at PN1, with males expressing significantly less mecp2 than females (Kurian et al., 2007). The sex differences in mecp2 expression were transient, largely disappearing by P10. However, they were found, at least in part, in sexually dimorphic brain regions during steroid-sensitive periods of brain development, suggesting that MeCP2-mediated epigenetic regulation could be important for promoting these sex differences in the brain. An additional study that focused on decreasing mecp2 expression within the developing amygdala supported this conclusion. It demonstrated that targeted disruption of mecp2 expression within the developing amygdala reduced juvenile social play behavior in males without altering juvenile sociability or adult anxiety-like behavior (Kurian et al., 2008). Juvenile social play behavior in females was not affected. These observations highlight the role of MeCP2 in organizing sex-specific behavior.
In addition, DNA methylation profiles are sexually dimorphic within the brain. A recent study found that male rats exhibit higher levels of ERα promoter methylation than females within the developing preoptic area, a sexually dimorphic brain region (Kurian et al., 2010). Also, these profiles of DNA methylation can be modulated by hormonal and other influences. Estradiol exposure altered ERα promoter methylation. Simulated maternal grooming of females, which represents a maternal interaction that is sexually dimorphic during the neonatal period, resulted in masculinization of ERα promoter methylation and gene expression.
Histone modifications and chromatin regulation
Recent evidence suggests that histone modifications and chromatin regulation may also play roles in mediating sex differences in brain and behavior. Chromatin refers to the packaging of DNA, and modulation of chromatin structure regulates the accessibility and activity of regulatory and functional DNA sequences, including their transcriptional activation (Fig. 1) (Mehler, 2008). Chromatin encompasses DNA and histone proteins that form a “beads on a string” structure, nucleosomes that serve as the basic units of chromatin, and higher order chromatin structures. Histone modifications, nucleosome repositioning, and chromatin remodeling are key epigenetic mechanisms for regulating specific genes and more extensive genomic regions. Specific classes of enzymes (e.g., histone acetytransferases [HATs] and histone deacetylases [HDACs]) mediate reversible histone modifications (Mehler, 2008). These modifications may include but are not limited to acetylation, methylation, phosphorylation, ubiquitylation, SUMOylation, and ADP-ribosylation. ATP-dependent chromatin remodeling enzymes play a critical role in modulating higher order chromatin structure. Specific histone modifications and chromatin remodeling enzymes and associated macromolecular complexes may promote diverse cellular processes including but not limited to activation or repression of transcription.
The expression of a number of histone and chromatin regulatory factors is sexually dimorphic within the brain. For example, SET nuclear oncogene (SET) is expressed in a sexually dimorphic manner in the brain (Reinius et al., 2008). It encodes a histone chaperone that is partly responsible for regulating the access of transcription machinery to chromatin (Gamble and Fisher, 2007). The cell cycle regulator and cell fate determinant, Geminin is expressed in a sexually dimorphic manner in the brain (Yang et al., 2006). It has been shown to control the timing of neurogenesis by blocking the activity of proneural basic helix-loop-helix (bHLH) proteins through inhibition of their interactions with Brg1, a member of the SWItch/Sucrose NonFermentable (SWI/SNF) family of chromatin remodeling proteins (Seo et al., 2005). H2A histone family member Z (H2A.Z), a histone variant, is implicated in diverse cellular processes including gene activation, chromosome segregation, heterochromatin silencing, and progression through the cell cycle. Furthermore, it serves as a key component of ERα signaling pathways. H2A.Z is incorporated into the promoter regions of ERα target genes when these are activated, and ERα directly associates with the H2A.Z promoter modulating its expression (Gevry et al., 2009; Svotelis et al., 2010)
Utx is a H3K27-specific demethylase (Hong et al., 2007), and a recent study reported that it has sex-specific regional expression profiles in mouse brain (Xu et al., 2008). Utx levels were higher in females than in males in all brain regions tested except the amygdala. This study also reported higher levels of expression of Utx compared with its paralog, Uty, in XY P19 embryonal carcinoma (EC) cell-derived neurons attributable to higher levels of H3K4Me2 within the Utx promoter and H4K16Ac in the gene body (Xu et al., 2008). Both of these histone marks are associated with transcriptional activation. These observations suggest that differences in the regional expression profiles of Utx and Uty in the male brain—Utx is expressed preferentially in the amygdala and Uty in the paraventricular nucleus (PVN) of the hypothalamus—may arise because of differential profiles of histone modifications.
One interesting hypothesis is that Utx and perhaps Uty may play a role in neural developmental processes. In fact, Utx is known to be involved in regulating HOX genes (Agger et al., 2007), which are important for brain development. Uty is also implicated in modulating cell fate. Furthermore, Utx and Uty are targeted in a cell type-specific manner in mouse forebrain-derived neural cells by the epigenetic regulators, Repressor element-1 silencing transcription factor (REST) and corepressor for element-1-silencing transcription factor (CoREST) (Abrajano et al., 2009a,b). These factors are, in turn, responsible for promoting neuronal and glial cell fate decisions. Utx has been associated with regulating neural gene families such as protocadherins and olfactory receptors (Wang et al., 2010).
Histone modification profiles are sexually dimorphic within the brain. For example, a recent study found sex differences in specific histone modifications that are associated with brain development in mice (Tsai et al., 2009). Specifically, males exhibited higher levels of H3K9/14Ac and H3K9Me3 in the cortex and hippocampus. Sex differences in acetylation were detected at E18 and PN0, whereas sex differences in methylation were observed at PN6. These modifications are associated with gene activation and repression, respectively. Furthermore, prenatal treatment of females with testosterone resulted in the masculinization of H3K9/14Ac profiles but did not affect H3K9Me3 levels linking sex steroid hormone activity with differential effects on the regional profiles of histone modifications during developmental critical periods.
Histone modifications and chromatin remodeling may control sexual differentiation in brain structures, such as the principal nucleus of the bed nucleus of the stria terminalis (BNSTp). A greater volume and number of cells are typical of the BNSTp in male mice compared to females as a result of developmental testosterone exposure. A recent study found that the histone deacetylase inhibitor (HDACi), valproic acid, specifically prevents masculinization of the BNSTp in males and in testosterone-treated females but does not affect females not treated with testosterone (Murray et al., 2009). These observations suggest that the effects of testosterone on the developing BNSTp are mediated by histone acetylation. Similarly, valproic acid treatment may also promote the masculinization of vasopressin expression in female mice (Murray et al., 2009).
The testis-determining factor, SRY, is a transcription factor that exerts its effects on target genes, at least in part, through interactions with chromatin remodeling complexes. SRY associates directly with the KRAB-O protein and recruits the KAP1 core-pressor machinery (KAP1-NuRD-SETDB1-HP1) to silence its target genes (Oh et al., 2005; Peng et al., 2009). Intriguingly, a recent study reported that KRAB/KAP1 recruitment is established by the long-range spreading of H3K9Me3 marks and HP1β, potentially implicating SRY in transcriptional repression through the spread of heterochromatin (Groner et al., 2010). One caveat is that SRY target genes in the brain are presently unknown. However, KAP1 is expressed at high levels in the mouse brain and is necessary for KRAB-mediated epigenetic regulation of gene expression in the hippocampus (Jakobsson et al., 2008).
Nuclear receptor function is mediated by the recruitment of various epigenetic coregulatory complexes to specific genomic loci. For example, sex steroid hormone receptor activity is associated with the nuclear receptor corepressor (NCoR) (Zhang et al., 1998). This coregulatory complex includes HDAC3 and interacts with MBDs, including MeCP2. One study showed that NCoR levels are sexually dimorphic and likely to be estradiol mediated, with females expressing higher levels in the developing amygdala and hypothalamus (Jessen et al., 2010). Furthermore, manipulations of NCoR levels in the amygdala during development suggest that it has organizational effects on juvenile social play and anxiety-like behavior (Jessen et al., 2010).
Short non-coding RNAs
Non-protein-coding DNA comprises the vast majority of mammalian genomes, including more than 98% of the human genome (Mehler and Mattick, 2006). Recent studies have demonstrated that these sequences are pervasively transcribed forming numerous classes of short ncRNAs including but not limited to short-interfering RNAs (siRNAs), microRNAs (miRNAs), P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and long ncRNAs (lncRNAs; i.e., transcripts longer than 200 nucleotides) (Fig. 2). These ncRNAs are expressed in distinct cell-, tissue-, and developmental stage-specific profiles and are involved in nearly every biological process (Amaral et al., 2008). The mechanisms of actions for these ncRNAs are still emerging, but they include regulating aspects of DNA methylation, chromatin architecture, transcription, post-transcriptional RNA processing, and translation (Mattick et al., 2009). Various classes of ncRNAs have roles in promoting the mammalian sexual phenotype (McFarlane and Wilhelm, 2009). For example, a number of studies have reported that hundreds of siRNAs, miRNAs, piRNAs, and snoRNAs are expressed in the mouse testes and ovaries (Ahn et al., 2010; Mishima et al., 2008). Similarly, genes with key roles in sex determination in the gonad, such as FOXL2 and WT1, are associated with antisense lncRNAs that may regulate their expression and function (Campbell et al., 1994; Cocquet et al., 2005). Also, a significant percentage of transcriptional units on the Y chromosome are thought to encode lncRNAs, including many that are expressed in the testis and potentially involved in spermatogenesis (Makrinou et al., 2001; Skaletsky et al., 2003).
Fig. 2.
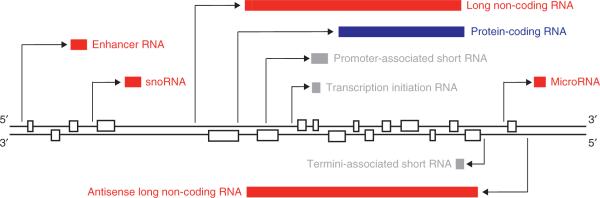
Non-coding RNA (ncRNA) transcription. This schematic shows how multiple interleaved and overlapping RNAs are encoded within the genome. These include protein-coding RNAs and various classes of ncRNAs, which are described in the text as well as others.
The roles of short ncRNAs in sexual dimorphism in the brain have not been studied in detail. One preliminary study revealed that miRNAs are expressed in a sexual dimorphic manner in various regions of the adult murine brain, including hippocampus, frontal cortex, and cerebellum (Koturbash et al., 2010). In females relative to males, three miRNAs (miR-181b, miR-34c, and miR-488*) were up regulated in the hippocampus and one miRNA (miR-130b) was up regulated in the cerebellum. By contrast, in males relative to females, four miRNAs (miR-206, miR-214, miR-329, and miR-124a) were up regulated in the hippocampus and two miRNAs (miR-182 and miR-183) were up regulated in the frontal cortex. These miRNAs are likely to be important for modulating the expression of sex-specific gene networks within these brain regions and may underlie sexual dimorphism in brain form and function. For example, miR-329 is essential for activity-dependent dendritic outgrowth of hippocampal neurons (Khudayberdiev et al., 2009). Sex differences in hippocampal dendritic spine morphology and plasticity are well documented, suggesting that miR-329 may play a role in mediating these differences. Indirect evidence also supports a role of miRNAs in promoting brain sexual dimorphism. miRNAs in other dimorphic tissues, such as liver, are subject to regulation by sex steroid hormones (Delic et al., 2010). Furthermore, in silico analysis reveals that genes expressed in the brain in a sexually dimorphic manner may be regulated by miRNAs. For example, the human SRY transcript is a predicted target of 10 miRNAs, including let-7a and let-7e, members of the let-7 family of miRNAs that are highly expressed in brain (Betel et al., 2008).
Other classes of short ncRNAs may be dimorphically expressed and play roles in sex differences in brain function but these remain to be elucidated (Fig. 2). One interesting example may be the recently characterized enhancer RNAs (eRNAs) that are transcribed from neuronal enhancers (Kim et al., 2010). These eRNAs are expressed in an activity-dependent manner and their levels correlate with the levels of mRNA synthesis at nearby genes. They have also been linked to the function of nuclear receptors (Kim et al., 2010). Therefore, it may be possible that eRNAs mediate the relatively well-recognized effects of sex steroid hormones on neurotransmission and synaptic plasticity (Foy et al., 2010).
Long non-coding RNAs and X chromosome inactivation
In contrast to short ncRNAs, it is not known whether many lncRNAs are expressed in the brain in a sexual dimorphic manner. It seems likely that they are because of extensive experimental and bioinformatic evidence for thousands of lncRNA genes in mammalian genomes, including many with seminal roles in epigenetic regulation in the brain (Qureshi et al., 2010). Sex differences in the expression of one lncRNA, XIST, have been well characterized in the brain (see above). XIST is transcribed from the X inactivation center (Xic) on the X chromosome to be inactivated (Xi) and plays a key role in XCI (Chow and Heard, 2009). XIST initiates XCI by coating and silencing the inactive X-chromosome in cis. TSIX, an lncRNA encoded within the Xic that is transcribed antisense to XIST, plays a crucial role in XCI as well. It represses XIST activity thereby designating which of the X-chromosomes will remain active.
In general, XCI is thought to occur randomly with respect to the parental origin of the X chromosome (Chow and Heard, 2009); however, a recent study challenges this hypothesis (Gregg et al., 2010a,b). This high-resolution transcriptomic analysis in mice identified preferential expression from the maternally inherited X chromosome in various neuronal subpopulations, including glutamatergic neurons of the female cortex (Gregg et al., 2010a). In addition, while the majority of genes on the Xi are transcriptionally silenced, some genes escape XCI and are expressed from the Xi where they may contribute to dimorphic gene expression, including in brain. For example, PCDH11X expression levels in brain are sexually dimorphic because of its inactivation status (Lopes et al., 2006). One study of human females found that 15% of X-linked genes escape XCI and an additional 10% have variable patterns (Carrel and Willard, 2005). Another study performed in mice demonstrated that genes escaping XCI lack H3K27Me3, a developmentally regulated histone modification associated with XCI (Yang et al., 2010). Humans and mice exhibit a great deal of variability in genes escaping XCI in somatic tissues. Genes escaping XCI are subject to stronger purifying selection than inactivated genes, particularly those with Y chromosome homologs (Park et al., 2010a). These genes may be important contributors to sexually dimorphic features. Intriguingly, ncRNAs may be subject to escape from XCI as well (Song et al., 2009) though this has not been well established in the brain.
Genomic imprinting
Genomic imprinting is an epigenetic mechanism for gene silencing that is mediated by DNA methylation, histone modifications, and ncRNAs (Bartolomei, 2009). Imprinted genes are monoallelically expressed in a parent-of-origin-dependent manner and implicated in various biological processes, including development. Imprinting is particularly relevant within the brain, as evidenced by the aberrant neurobiological phenotypes associated with perturbations in imprinting, linkage analyses showing associations sensitive to the parental origin of the region of interest, and preferential transmission of neurological and psychiatric disorders from a single parent (Davies et al., 2008). Furthermore, the profiles of imprinting are highly complex within the brain and exhibit temporal, spatial, cell type-specific, sex-specific, and inter-individual variability (Davies et al., 2005b, 2008; Gregg et al., 2010a,b). Notably, studies of Turner syndrome (45, XO) have found X-linked parent-of-origin effects on brain development and cognitive functioning, suggesting that imprinted genes on the X chromosome may be important in mediating these effects. Studies in mice have found that Xlr3b, Xlr4b, and Xlr4c are maternally expressed X-linked genes imprinted in the brain and that Xlr3b, a factor putatively involved in DNA repair and/or chromatin remodeling, may play roles in behavioral phenotypes associated with a mouse model of Turner syndrome (Davies et al., 2005a, 2006; Kopsida et al., 2010). Intriguingly, FAM9B is a human ortholog of Xlr3b, whose normal function is unknown (see below) (Kopsida et al., 2010; Thomas et al., 1999). Furthermore, a recent genome-wide study of adult mouse cortex and hypothalamus found sex-specific parent-of-origin allelic effects for 347 autosomal genes (Gregg et al., 2010a,b). For example, females had 150 genes and males had 48 genes exhibiting sex-specific imprinted features in the POA, a sexually dimorphic region. These observations suggest that it is important to consider how imprinted genes, including those encoded on the X chromosome and on autosomes, may contribute to sex differences in brain and behavior.
Clinical correlations
There are many examples of complex diseases with sex-specific features ranging from asthma and diabetes to multiple sclerosis (MS), autism, and depression. The interplay between genetic, hormonal, and environmental factors is responsible for these sex differences and is likely to be mediated by epigenetic alterations that, in turn, dynamically regulate sexually dimorphic gene expression and function (Kaminsky et al., 2006). Herein, we call attention to diverse neurological and psychiatric disorders whose incidence and natural history are different between men and women and highlight emerging evidence that suggests how epigenetic mechanisms are responsible for the sex-specific pathophysiology of these disorders.
Neuroimmunological disorders
MS is an immune-mediated, demyelinating, and neurodegenerative disease that is caused by genetic and environmental factors (Mechelli et al., 2010). Genetic epidemiology suggests that the major histocompatibility complex (MHC) class II genes account for a significant percentage of MS genetic risk. However, the concordance rate for monozygotic twins is only 30%, highlighting the importance of environmental factors in disease pathogenesis. Epigenetic mechanisms have been implicated in MS, consistent with their roles in a range of other autoimmune disorders (AIDs) (Brooks et al., 2010). For example, the peptidyl argininedeiminase 2 (PAD2) gene promoter is selectively hypomethylated in MS brains (Mastronardi et al., 2007) and is associated with abnormal expression of this enzyme, which compromises the integrity of myelin. Similarly, deregulation of miRNA expression may play a role in the pathophysiology of MS, possibly through effects on T-cell differentiation and maturation (Du et al., 2009; Keller et al., 2009). The HDACi, Trichostatin A (TSA), has been shown to ameliorate experimental autoimmune encephalomyelitis, an animal model of MS (Camelo et al., 2005). The entire MHC class II gene family and the adjacent histone cluster are strongly induced by the application of TSA (Gialitakis et al., 2006). Furthermore, environmental factors associated with MS risk such as vitamin D, Epstein–Barr virus, stress, diet, and smoking are all associated with effects on the epigenome.
In terms of sex differences, MS is approximately three times more common in women than in men, and current data reveal that this ratio is increasing in many populations (Koch-Henriksen and Sorensen, 2010). Male sex is a prognostic factor for a more severe disease course. Furthermore, MS susceptibility is subject to maternal parent-of-origin effects and increased penetrance in females. Recent studies implicate epigenetic modifications at MHC gene loci in mediating some of these sex differences in MS (Chao et al., 2009, 2010). MS pathophysiology is also sensitive to hormonal factors and is characterized by a decrease in disease activity during pregnancy and worsening of symptoms in the post-partum period (Niino et al., 2009). These effects are linked to estrogens and may be related to expression of ERα, which is epigenetically regulated (Imamura, 2010).
While skewing of XCI has been found in other AIDs, one study of XCI in patients with MS revealed no significant differences in the frequency of skewing in affected females compared to controls (Knudsen et al., 2007). However, skewing of XCI was more common in progressive MS rather than relapsing remitting forms of the disease, suggesting that skewing of XCI is associated with more severe or advanced disease. One caveat is that this analysis was performed utilizing blood, and the degree of correlation between XCI in blood and neural tissues is not well characterized, particularly in the context of disease. As discussed above, there is potential variability in genes escaping XCI in somatic tissues. This is particularly relevant because regions of the X chromosome, particularly Xp22.1, that contain genes in polyamine pathways have the potential to become deregulated, and these have been linked to various AIDs. It is well established that polyamines, such as spermine, are ubiquitous nuclear components with functions in chromatin compaction, maintenance of DNA structure, RNA processing, and translation (Childs et al., 2003). Endogenous retroviral elements found in patients with MS may also be implicated in epigenetic regulation and sex differences in MS. For example, human endogenous retroviral family W (HERV-W) RNA is present in circulating viral particles that may be responsible for activating pro-inflammatory and autoimmune cascades. HERV-W RNA has been linked to MS pathogenesis, and intriguingly, HERV-W copies are present on the X chromosome (Perron et al., 2009). Furthermore, sex-specific imprinting in the brain has been observed for interleukin-18 (Il18) (Gregg et al., 2010a), a pleiotropic cytokine that is implicated in mediating neuroinflammatory and neurodegenerative processes in MS (Alboni et al., 2010).
Neurodevelopmental disorders
Various epigenetic regulatory factors are encoded on the X chromosome and lead to neurodevelopmental disorders when they are mutated. For example, α-thalassemia mental retardation X-linked protein (ATRX) is an SWI/SNF chromatin remodeling protein that plays a role in brain development, globin regulation, and sexual development (Tang et al., 2004). ATRX mutations lead to X-linked α-thalassemia, mental retardation, and gonadal and urogenital abnormalities. Similarly, the histone demethylase, JARID1C/SMCX, has been linked to mental retardation and to autism spectrum disorders (ASDs) (Abidi et al., 2008; Adegbola et al., 2008; Claes et al., 2000; Jensen et al., 2005; Santos et al., 2006). ASDs exhibit a strong male preponderance. Some theories consider ASDs to be the result of differential manifestations of male behavioral traits. The pathogenesis of ASDs is unknown but has been linked to epigenetic mechanisms that may play sex-specific roles. For example, factors that are expressed in the brain in a sexually dimorphic manner, such as RORA, may be differentially regulated in the autistic brain and subject to epigenetic regulation (Dewing et al., 2003; Nguyen et al., 2010). Imprinted and X-linked genes have been associated with ASDs. Notably, patients with Turner's syndrome only exhibit ASDs when the maternal X chromosome is present. In addition, ASDs are associated with micro-deletions that encompass FAM9B, the human homolog of the mouse X imprinted Xlr3b gene (Thomas et al., 1999). A deletion of the ANKRD15 gene that exhibits imprinting-like inheritance is similarly linked with congenital cerebral palsy (Lerer et al., 2005). Affected individuals harbor a paternal deletion and a normal maternal allele that is repressed. The only difference between the alleles is differential methylation in the CpG island flanking the DMRT gene located 3′ of the ANKRD15 gene. Interestingly, DMRT is involved in sex determination and its Drosophila homolog is required for the development of male-specific neurons and sexual behavior (Rideout et al., 2010). Additional studies have linked sex chromosome aneuploidy with neurodevelopmental phenotypes (Lenroot et al., 2009).
Other neurological and psychiatric disorders
Sex differences have been noted in the incidence of neurodegenerative disorders such as Alzheimer's and Parkinson's diseases (AD and PD) and may be influenced by underlying epigenetic mechanisms. The risk of AD is higher in women than in men (Musicco, 2009). Various studies suggest a role for genomic imprinting and parent-of-origin effects in AD pathogenesis (Bassett et al., 2002). For example, selective patterns of hypometabolism have been noted in maternally inherited AD (Mosconi et al., 2010). Furthermore, amyloid precursor protein (APP) is a gene that causes certain forms of early onset AD. APP promoter methylation is higher in females and differentially regulated by sex steroids in mouse cerebral cortex (Mani and Thakur, 2006). Also, APP has been linked to male sexual behavior (Park et al., 2010b). In contrast to AD, men are about 1.5 times more likely to develop PD than women. Interestingly, a study performed utilizing nigral dopaminergic neurons from postmortem brains of sporadic PD patients revealed that major cellular signaling pathways involved in PD pathogenesis have distinct patterns of deregulation between males and females (Simunovic et al., 2010). This sex-specific molecular pathophysiology may be linked to the effects of SRY on catecholaminergic pathways in the basal ganglia and cortex (see above).
Moreover, sex differences have been found in the incidence of brain cancers (Deorah et al., 2006) and in sex-specific toxicity profiles and therapeutic outcomes (Borgmann et al., 2009). These, too, may have epigenetic underpinnings. For example, mutations of the histone H3K27 demethylase, UTX, which is expressed in neurons in a sexually dimorphic manner, have been found in human cancers including glioblastoma multiforme (GBM) (van Haaften et al., 2009). Sex-specific responses to brain radiation exposure have also been noted in the expression of miRNAs in the hippocampus, cerebellum, and frontal cortex of mice (Koturbash et al., 2010). Moreover, alterations in multiple components of the epigenome have been implicated in the pathogenesis of a spectrum of primary nervous system tumors and in signaling pathways mediating CNS cancer initiation, progression, and responses to therapy (Mehler, 2008). These observations suggest that sexually dimorphic epigenetic regulatory mechanisms may represent promising avenues for better understanding and treating brain cancers.
In addition to neurological diseases, a number of psychiatric disorders are also characterized by sex differences that may be influenced by epigenetic factors. Sex differences are commonly found in the incidence of psychiatric diseases, and sex-specific epigenetic interactions have been noted across the life span in association with these disorders (Vigod and Stewart, 2009). Furthermore, the heritability of psychiatric diseases, such as major depression, is greater in women than in men (Kendler et al., 2006). Potential epigenetic mechanisms explaining these observations include, for example, that imprinted genes mediate susceptibility to psychiatric disorders (Kopsida et al., 2010). There is an ever-expanding body of evidence for a continuum of epigenetic mechanisms and for sex differences in neuropsychiatric diseases. Future studies will be necessary to delineate the complex interrelationships between these pathogenic factors and their contributions to disease susceptibility, onset, progression, and response to treatment.
Acknowledgments
M.F.M. is supported by grants from the National Institutes of Health (MH66290, NS38902, NS071571), as well as by the F.M. Kirby, Alpern Family, Mildred and Bernard H. Kayden and Roslyn and Leslie Goldstein Foundations.
References
- Abidi FE, Holloway L, Moore CA, Weaver DD, Simensen RJ, Stevenson RE, et al. Mutations in JARID1C are associated with X-linked mental retardation, short stature and hyperreflexia. Journal of Medical Genetics. 2008;45:787–793. doi: 10.1136/jmg.2008.058990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. Differential deployment of REST and corest promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS One. 2009a;4:e7665. doi: 10.1371/journal.pone.0007665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. REST and corest modulate neuronal subtype specification, maturation and maintenance. PLoS One. 2009b;4:e7936. doi: 10.1371/journal.pone.0007936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adegbola A, Gao H, Sommer S, Browning M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD). American Journal of Medical Genetics. Part A. 2008;146A:505–511. doi: 10.1002/ajmg.a.32142. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Ahn HW, Morin RD, Zhao H, Harris RA, Coarfa C, Chen ZJ, et al. Microrna transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Molecular Human Reproduction. 2010;16:463–471. doi: 10.1093/molehr/gaq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboni S, Cervia D, Sugama S, Conti B. Interleukin 18 in the CNS. Journal of Neuroinflammation. 2010;7:9. doi: 10.1186/1742-2094-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS. Genomic imprinting: Employing and avoiding epigenetic processes. Genes and Development. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett SS, Avramopoulos D, Fallin D. Evidence for parent of origin effect in late-onset alzheimer disease. American Journal of Medical Genetics. 2002;114:679–686. doi: 10.1002/ajmg.10648. [DOI] [PubMed] [Google Scholar]
- Becker JB. Sex differences in the brain: From genes to behavior. Oxford University Press; Oxford; New York: 2008. [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN, et al. Genetic evidence equating SRY and the testis-determining factor. Nature. 1990;348:448–450. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microrna.Org resource: Targets and expression. Nucleic Acids Research. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Research. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgmann K, Dikomey E, Petersen C, Feyer P, Hoeller U. Sex-specific aspects of tumor therapy. Radiation and Environmental Biophysics. 2009;48:115–124. doi: 10.1007/s00411-009-0216-1. [DOI] [PubMed] [Google Scholar]
- Brooks WH, Le Dantec C, Pers JO, Youinou P, Renaudineau Y. Epigenetics and autoimmunity. Journal of Autoimmunity. 2010;34:J207–J219. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Camelo S, Iglesias AH, Hwang D, Due B, Ryu H, Smith K, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin a ameliorates experimental autoimmune encephalomyelitis. Journal of Neuroimmunology. 2005;164:10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Campbell CE, Huang A, Gurney AL, Kessler PM, Hewitt JA, Williams BR. Antisense transcripts and protein binding motifs within the wilms tumour (WT1) locus. Oncogene. 1994;9:583–595. [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Chao MJ, Herrera BM, Ramagopalan SV, Deluca G, Handunetthi L, Orton SM, et al. Parent-of-origin effects at the major histocompatibility complex in multiple sclerosis. Human Molecular Genetics. 2010;19:3679–3689. doi: 10.1093/hmg/ddq282. [DOI] [PubMed] [Google Scholar]
- Chao MJ, Ramagopalan SV, Herrera BM, Lincoln MR, Dyment DA, Sadovnick AD, et al. Epigenetics in multiple sclerosis susceptibility: Difference in transgenerational risk localizes to the major histocompatibility complex. Human Molecular Genetics. 2009;18:261–266. doi: 10.1093/hmg/ddn353. [DOI] [PubMed] [Google Scholar]
- Childs AC, Mehta DJ, Gerner EW. Polyamine-dependent gene expression. Cellular and Molecular Life Sciences. 2003;60:1394–1406. doi: 10.1007/s00018-003-2332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Current Opinion in Cell Biology. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Claes S, Devriendt K, Van Goethem G, Roelen L, Meireleire J, Raeymaekers P, et al. Novel syndromic form of X-linked complicated spastic paraplegia. American Journal of Medical Genetics. 2000;94:1–4. doi: 10.1002/1096-8628(20000904)94:1<1::aid-ajmg1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Cocquet J, Pannetier M, Fellous M, Veitia RA. Sense and antisense Foxl2 transcripts in mouse. Genomics. 2005;85:531–541. doi: 10.1016/j.ygeno.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Davies W, Isles AR, Burgoyne PS, Wilkinson LS. X-linked imprinting: Effects on brain and behaviour. Bioessays. 2006;28:35–44. doi: 10.1002/bies.20341. [DOI] [PubMed] [Google Scholar]
- Davies W, Isles AR, Humby T, Wilkinson LS. What are imprinted genes doing in the brain? Advances in Experimental Medicine and Biology. 2008;626:62–70. doi: 10.1007/978-0-387-77576-0_5. [DOI] [PubMed] [Google Scholar]
- Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, et al. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nature Genetics. 2005a;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- Davies W, Isles AR, Wilkinson LS. Imprinted gene expression in the brain. Neuroscience and Biobehavioral Reviews. 2005b;29:421–430. doi: 10.1016/j.neubiorev.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Delic D, Grosser C, Dkhil M, Al-Quraishy S, Wunderlich F. Testosterone-induced upregulation of miRNAs in the female mouse liver. Steroids. 2010;75:998–1004. doi: 10.1016/j.steroids.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Deorah S, Lynch CF, Sibenaller ZA, Ryken TC. Trends in brain cancer incidence and survival in the united states: Surveillance, epidemiology, and end results program, 1973 to 2001. Neurosurgical Focus. 2006;20:E1. doi: 10.3171/foc.2006.20.4.E1. [DOI] [PubMed] [Google Scholar]
- Dewing P, Chiang CW, Sinchak K, Sim H, Fernagut PO, Kelly S, et al. Direct regulation of adult brain function by the male-specific factor SRY. Current Biology. 2006;16:415–420. doi: 10.1016/j.cub.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Dewing P, Shi T, Horvath S, Vilain E. Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Brain Research. Molecular Brain Research. 2003;118:82–90. doi: 10.1016/s0169-328x(03)00339-5. [DOI] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nature Immunology. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- Dunn GA, Morgan CP, Bale TL. Sex-specificity in transgenerational epigenetic programming. Hormones and Behavior. 2010 May 17; doi: 10.1016/j.yhbeh.2010.05.004. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nature Neuroscience. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy MR, Baudry M, Akopian GK, Thompson RF. Regulation of hippocampal synaptic plasticity by estrogen and progesterone. Vitamins and Hormones. 2010;82:219–239. doi: 10.1016/S0083-6729(10)82012-6. [DOI] [PubMed] [Google Scholar]
- Friocourt G, Kappeler C, Saillour Y, Fauchereau F, Rodriguez MS, Bahi N, et al. Doublecortin interacts with the ubiquitin protease DFFRX, which associates with microtubules in neuronal processes. Molecular and Cellular Neurosciences. 2005;28:153–164. doi: 10.1016/j.mcn.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Galfalvy HC, Erraji-Benchekroun L, Smyrniotopoulos P, Pavlidis P, Ellis SP, Mann JJ, et al. Sex genes for genomic analysis in human brain: Internal controls for comparison of probe level data extraction. BMC Bioinformatics. 2003;4:37. doi: 10.1186/1471-2105-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nature Structural and Molecular Biology. 2007;14:548–555. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- Gevry N, Hardy S, Jacques PE, Laflamme L, Svotelis A, Robert F, et al. Histone H2A.Z is essential for estrogen receptor signaling. Genes and Development. 2009;23:1522–1533. doi: 10.1101/gad.1787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gialitakis M, Kretsovali A, Spilianakis C, Kravariti L, Mages J, Hoffmann R, et al. Coordinated changes of histone modifications and HDAC mobilization regulate the induction of MHC class II genes by trichostatin A. Nucleic Acids Research. 2006;34:765–772. doi: 10.1093/nar/gkj462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, et al. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, et al. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genetics. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert JM, Fishell G. The genetics of early telencephalon patterning: Some assembly required. Nature Reviews. Neuroscience. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T. Epigenetic setting for long-term expression of estrogen receptor alpha and androgen receptor in cells. Hormones and Behavior. 2010 Jul 7; doi: 10.1016/j.yhbeh.2010.05.018. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Jakobsson J, Cordero MI, Bisaz R, Groner AC, Busskamp V, Bensadoun JC, et al. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60:818–831. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. American Journal of Human Genetics. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen HM, Kolodkin MH, Bychowski ME, Auger CJ, Auger AP. The nuclear receptor corepressor has organizational effects within the developing amygdala on juvenile social play and anxiety-like behavior. Endocrinology. 2010;151:1212–1220. doi: 10.1210/en.2009-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly LA, Taylor V, Wood SA. USP9X enhances the polarity and self-renewal of embryonic stem cell-derived neural progenitors. Molecular Biology of the Cell. 2009;20:2015–2029. doi: 10.1091/mbc.E08-06-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata T, Katsube K, Michikawa M, Yamada M, Takada S, Mizusawa H. R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochimica et Biophysica Acta. 2004;1676:51–62. doi: 10.1016/j.bbaexp.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Kaminsky Z, Wang SC, Petronis A. Complex disease, gender and epigenetics. Annals of Medicine. 2006;38:530–544. doi: 10.1080/07853890600989211. [DOI] [PubMed] [Google Scholar]
- Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;4:e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Gatz M, Gardner CO, Pedersen NL. A swedish national twin study of lifetime major depression. American Journal of Psychiatry. 2006;163:109–114. doi: 10.1176/appi.ajp.163.1.109. [DOI] [PubMed] [Google Scholar]
- Khudayberdiev S, Fiore R, Schratt G. MicroRNA as modulators of neuronal responses. Communicative and Integrative Biology. 2009;2:411–413. doi: 10.4161/cib.2.5.8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen GP, Harbo HF, Smestad C, Celius EG, Akesson E, Oturai A, et al. X chromosome inactivation in females with multiple sclerosis. European Journal of Neurology. 2007;14:1392–1396. doi: 10.1111/j.1468-1331.2007.01987.x. [DOI] [PubMed] [Google Scholar]
- Koch-Henriksen N, Sorensen PS. The changing demographic pattern of multiple sclerosis epidemiology. Lancet Neurology. 2010;9:520–532. doi: 10.1016/S1474-4422(10)70064-8. [DOI] [PubMed] [Google Scholar]
- Kopsida E, Mikaelsson MA, Davies W. The role of imprinted genes in mediating susceptibility to neuropsychiatric disorders. Hormones and Behavior. 2010 Apr 18; doi: 10.1016/j.yhbeh.2010.04.005. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Koturbash I, Zemp F, Kolb B, Kovalchuk O. Sex-specific radiation-induced microRNAome responses in the hippocampus, cerebellum and frontal cortex in a mouse model. Mutation Research. 2010 May 15; doi: 10.1016/j.mrgentox.2010.05.007. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Kurian JR, Bychowski ME, Forbes-Lorman RM, Auger CJ, Auger AP. Mecp2 organizes juvenile social behavior in a sex-specific manner. Journal of Neuroscience. 2008;28:7137–7142. doi: 10.1523/JNEUROSCI.1345-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian JR, Forbes-Lorman RM, Auger AP. Sex difference in mecp2 expression during a critical period of rat brain development. Epigenetics. 2007;2:173–178. doi: 10.4161/epi.2.3.4841. [DOI] [PubMed] [Google Scholar]
- Kurian JR, Olesen KM, Auger AP. Sex differences in epigenetic regulation of the estrogen receptor-alpha promoter within the developing preoptic area. Endocrinology. 2010;151:2297–2305. doi: 10.1210/en.2009-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr G, Maxson SC, Mayer A, Just W, Pilgrim C, Reisert I. Transcription of the Y chromosomal gene, Sry, in adult mouse brain. Brain Research. Molecular Brain Research. 1995;33:179–182. doi: 10.1016/0169-328x(95)00136-g. [DOI] [PubMed] [Google Scholar]
- Lau YF, Li Y. The human and mouse sex-determining SRY genes repress the Rspol/beta-catenin signaling. Journal of Genetics and Genomics. 2009;36:193–202. doi: 10.1016/S1673-8527(08)60107-1. [DOI] [PubMed] [Google Scholar]
- Lenroot RK, Lee NR, Giedd JN. Effects of sex chromosome aneuploidies on brain development: Evidence from neuroimaging studies. Devevelopmental Disabilities Research Reviews. 2009;15:318–327. doi: 10.1002/ddrr.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerer I, Sagi M, Meiner V, Cohen T, Zlotogora J, Abeliovich D. Deletion of the ANKRD15 gene at 9p24.3 causes parent-of-origin-dependent inheritance of familial cerebral palsy. Human Molecular Genetics. 2005;14:3911–3920. doi: 10.1093/hmg/ddi415. [DOI] [PubMed] [Google Scholar]
- Lopes AM, Ross N, Close J, Dagnall A, Amorim A, Crow TJ. Inactivation status of PCDH11X: Sexual dimorphisms in gene expression levels in brain. Human Genetics. 2006;119:267–275. doi: 10.1007/s00439-006-0134-0. [DOI] [PubMed] [Google Scholar]
- Makrinou E, Fox M, Lovett M, Haworth K, Cameron JM, Taylor K, et al. TTY2: A multicopy Y-linked gene family. Genome Research. 2001;11:935–945. doi: 10.1101/gr.175901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani ST, Thakur MK. In the cerebral cortex of female and male mice, amyloid precursor protein (APP) promoter methylation is higher in females and differentially regulated by sex steroids. Brain Research. 2006;1067:43–47. doi: 10.1016/j.brainres.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Mastronardi FG, Noor A, Wood DD, Paton T, Moscarello MA. Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. Journal of Neuroscience Research. 2007;85:2006–2016. doi: 10.1002/jnr.21329. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- Mayer A, Lahr G, Swaab DF, Pilgrim C, Reisert I. The Y-chromosomal genes SRY and ZFY are transcribed in adult human brain. Neurogenetics. 1998;1:281–288. doi: 10.1007/s100480050042. [DOI] [PubMed] [Google Scholar]
- McCarthy MM, Auger AP, Bale TL, De Vries GJ, Dunn GA, Forger NG, et al. The epigenetics of sex differences in the brain. Journal of Neuroscience. 2009;29:12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane L, Wilhelm D. Non-coding RNAs in mammalian sexual development. Sexual Development. 2009;3:302–316. doi: 10.1159/000284688. [DOI] [PubMed] [Google Scholar]
- Mechelli R, Annibali V, Ristori G, Vittori D, Coarelli G, Salvetti M. Multiple sclerosis etiology: Beyond genes and environment. Expert Reviews of Clinical Immunology. 2010;6:481–490. doi: 10.1586/eci.10.11. [DOI] [PubMed] [Google Scholar]
- Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Progress in Neurobiology. 2008;86:305–341. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Non-coding RNAs in the nervous system. Journal of Physiology. 2006;575:333–341. doi: 10.1113/jphysiol.2006.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, et al. Cortical DNA methylation maintains remote memory. Nature Neuroscience. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milsted A, Serova L, Sabban EL, Dunphy G, Turner ME, Ely DL. Regulation of tyrosine hydroxylase gene transcription by Sry. Neuroscience Letters. 2004;369:203–207. doi: 10.1016/j.neulet.2004.07.052. [DOI] [PubMed] [Google Scholar]
- Mishima T, Takizawa T, Luo SS, Ishibashi O, Kawahigashi Y, Mizuguchi Y, et al. Microrna (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction. 2008;136:811–822. doi: 10.1530/REP-08-0349. [DOI] [PubMed] [Google Scholar]
- Mosconi L, Berti V, Swerdlow RH, Pupi A, Duara R, de Leon M. Maternal transmission of Alzheimer's disease: Prodromal metabolic phenotype and the search for genes. Human Genomics. 2010;4:170–193. doi: 10.1186/1479-7364-4-3-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EK, Hien A, de Vries GJ, Forger NG. Epigenetic control of sexual differentiation of the bed nucleus of the stria terminalis. Endocrinology. 2009;150:4241–4247. doi: 10.1210/en.2009-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musicco M. Gender differences in the occurrence of Alzheimer's disease. Functional Neurology. 2009;24:89–92. [PubMed] [Google Scholar]
- Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB Journal. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niino M, Hirotani M, Fukazawa T, Kikuchi S, Sasaki H. Estrogens as potential therapeutic agents in multiple sclerosis. Central Nervous System Agents in Medicinal Chemistry. 2009;9:87–94. doi: 10.2174/187152409788452054. [DOI] [PubMed] [Google Scholar]
- Nishino K, Hattori N, Tanaka S, Shiota K. DNA methylation-mediated control of Sry gene expression in mouse gonadal development. Journal of Biological Chemistry. 2004;279:22306–22313. doi: 10.1074/jbc.M309513200. [DOI] [PubMed] [Google Scholar]
- Oh HJ, Li Y, Lau YF. Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biology of Reproduction. 2005;72:407–415. doi: 10.1095/biolreprod.104.034447. [DOI] [PubMed] [Google Scholar]
- Park C, Carrel L, Makova K. Strong purifying selection at genes escaping X chromosome inactivation. Molecular Biology and Evolution. 2010a Jan 9; doi: 10.1093/molbev/msq143. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Bonthius PJ, Tsai HW, Bekiranov S, Rissman EF. Amyloid {beta} precursor protein regulates male sexual behavior. Journal of Neuroscience. 2010b;30:9967–9972. doi: 10.1523/JNEUROSCI.1988-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parma P, Radi O, Vidal V, Chaboissier MC, Dellambra E, Valentini S, et al. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nature Genetics. 2006;38:1304–1309. doi: 10.1038/ng1907. [DOI] [PubMed] [Google Scholar]
- Peng H, Ivanov AV, Oh HJ, Lau YF, Rauscher FJ., III Epigenetic gene silencing by the SRY protein is mediated by a KRAB-O protein that recruits the KAP1 corepressor machinery. Journal of Biological Chemistry. 2009;284:35670–35680. doi: 10.1074/jbc.M109.032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron H, Bernard C, Bertrand JB, Lang AB, Popa I, Sanhadji K, et al. Endogenous retroviral genes, Herpesviruses and gender in Multiple Sclerosis. Journal of the Neurological Sciences. 2009;286:65–72. doi: 10.1016/j.jns.2009.04.034. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Research. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius B, Jazin E. Prenatal sex differences in the human brain. Molecular Psychiatry. 2009;14(987):988–989. doi: 10.1038/mp.2009.79. [DOI] [PubMed] [Google Scholar]
- Reinius B, Saetre P, Leonard JA, Blekhman R, Merino-Martinez R, Gilad Y, et al. An evolutionarily conserved sexual signature in the primate brain. PLoS Genetics. 2008;4:e1000100. doi: 10.1371/journal.pgen.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nature Neuroscience. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos C, Rodriguez-Revenga L, Madrigal I, Badenas C, Pineda M, Mila M. A novel mutation in JARID1C gene associated with mental retardation. European Journal of Human Genetics. 2006;14:583–586. doi: 10.1038/sj.ejhg.5201608. [DOI] [PubMed] [Google Scholar]
- Seo S, Herr A, Lim JW, Richardson GA, Richardson H, Kroll KL. Geminin regulates neuronal differentiation by antagonizing brg1 activity. Genes and Development. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RP, Tun N, Grayson DR. Depolarization induces downregulation of DNMT1 and DNMT3a in primary cortical cultures. Epigenetics. 2008;3:74–80. doi: 10.4161/epi.3.2.6103. [DOI] [PubMed] [Google Scholar]
- Simunovic F, Yi M, Wang Y, Stephens R, Sonntag KC. Evidence for gender-specific transcriptional profiles of nigral dopamine neurons in Parkinson disease. PLoS One. 2010;5:e8856. doi: 10.1371/journal.pone.0008856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nature Genetics. 2009;41:488–493. doi: 10.1038/ng.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svotelis A, Gevry N, Grondin G, Gaudreau L. H2A.Z overexpression promotes cellular proliferation of breast cancer cells. Cell Cycle. 2010;9:364–370. doi: 10.4161/cc.9.2.10465. [DOI] [PubMed] [Google Scholar]
- Tang P, Park DJ, Marshall Graves JA, Harley VR. ATRX and sex differentiation. Trends in Endocrinology and Metabolism. 2004;15:339–344. doi: 10.1016/j.tem.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Thomas NS, Sharp AJ, Browne CE, Skuse D, Hardie C, Dennis NR. Xp deletions associated with autism in three females. Human Genetics. 1999;104:43–48. doi: 10.1007/s004390050908. [DOI] [PubMed] [Google Scholar]
- Tsai HW, Grant PA, Rissman EF. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;4:47–53. doi: 10.4161/epi.4.1.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature Genetics. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter MP, Evans S, Choudary P, Tomita H, Meador-Woodruff J, Molnar M, et al. Gender-specific gene expression in post-mortem human brain: Localization to sex chromosomes. Neuropsychopharmacology. 2004;29:373–384. doi: 10.1038/sj.npp.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigod SN, Stewart DE. Emergent research in the cause of mental illness in women across the lifespan. Current Opinion in Psychiatry. 2009;22:396–400. doi: 10.1097/YCO.0b013e3283297127. [DOI] [PubMed] [Google Scholar]
- Wang JK, Tsai MC, Poulin G, Adler AS, Chen S, Liu H, et al. The histone demethylase UTX enables RB-dependent cell fate control. Genes and Development. 2010;24:327–332. doi: 10.1101/gad.1882610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, Elashoff M, Richards AB, Sinclair D, Bahn S, Paabo S, et al. Transcriptome analysis of male-female differences in prefrontal cortical development. Molecular Psychiatry. 2009;14:558–561. doi: 10.1038/mp.2009.5. [DOI] [PubMed] [Google Scholar]
- Westberry JM, Trout AL, Wilson ME. Epigenetic regulation of estrogen receptor alpha gene expression in the mouse cortex during early postnatal development. Endocrinology. 2010;151:731–740. doi: 10.1210/en.2009-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JB, Chen K, Li Y, Lau YF, Shih JC. Regulation of monoamine oxidase A by the SRY gene on the Y chromosome. FASEB Journal. 2009;23:4029–4038. doi: 10.1096/fj.09-139097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Age-related changes in Usp9x protein expression and DNA methylation in mouse brain. Brain Research. Molecular Brain Research. 2005;140:17–24. doi: 10.1016/j.molbrainres.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Xu J, Burgoyne PS, Arnold AP. Sex differences in sex chromosome gene expression in mouse brain. Human Molecular Genetics. 2002;11:1409–1419. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- Xu J, Deng X, Watkins R, Disteche CM. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. Journal of Neuroscience. 2008;28:4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Taya S, Kaibuchi K, Arnold AP. Sexually dimorphic expression of Usp9x is related to sex chromosome complement in adult mouse brain. European Journal of Neuroscience. 2005a;21:3017–3022. doi: 10.1111/j.1460-9568.2005.04134.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Taya S, Kaibuchi K, Arnold AP. Spatially and temporally specific expression in mouse hippocampus of Usp9x, a ubiquitin-specific protease involved in synaptic development. Journal of Neuroscience Research. 2005b;80:47–55. doi: 10.1002/jnr.20429. [DOI] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from x inactivation by RNA-sequencing in mouse. Genome Research. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Research. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Fotaki V, Mason JO, Price DJ. Analysis of early ventral telencephalic defects in mice lacking functional Gli3 protein. Journal of Comparative Neurology. 2009;512:613–627. doi: 10.1002/cne.21918. [DOI] [PubMed] [Google Scholar]
- Zhang X, Jeyakumar M, Petukhov S, Bagchi MK. A nuclear receptor corepressor modulates transcriptional activity of antagonist-occupied steroid hormone receptor. Molecular Endocrinology. 1998;12:513–524. doi: 10.1210/mend.12.4.0089. [DOI] [PubMed] [Google Scholar]