Abstract
Bacteria and archaea rely on CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) RNA-guided adaptive immune systems for targeted elimination of foreign nucleic acids. These immune systems have been divided into three main Types, and the first atomic-resolution structure of a Type III RNA-guided immune complex provides new insights into the mechanisms of nucleic acid degradation. Here we compare the crystal structure of a Type III complex to recently determined structures of DNA-targeting Type I CRISPR complexes. Structural comparisons support previous assertions that Type I and Type III systems share a common ancestor and reveal how a conserved structural chassis is used to support RNA, DNA, or both RNA and DNA-targeting mechanisms.
RNA and proteins assemble into sophisticated ribonucleoprotein (RNP) machines that perform cellular functions essential for life. In bacteria and archaea, large RNA-guided protein complexes are essential for mounting an adaptive immune response (Bailey, 2013; Barrangou and Marraffini, 2014; Bondy-Denomy and Davidson, 2014; Gasiunas et al., 2014; Sorek et al., 2013; van der Oost et al., 2014). To acquire immunity, bacteria and archaea integrate short fragments of invading phage and plasmid DNA into Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs). CRISPR loci are transcribed and processed into a library of short CRISPR-derived RNA guides (crRNAs) that assemble with CRISPR-associated (Cas) proteins into large RNP machines. These surveillance machines patrol the intracellular environment and bind foreign nucleic acid targets that are complementary to the crRNA-guide.
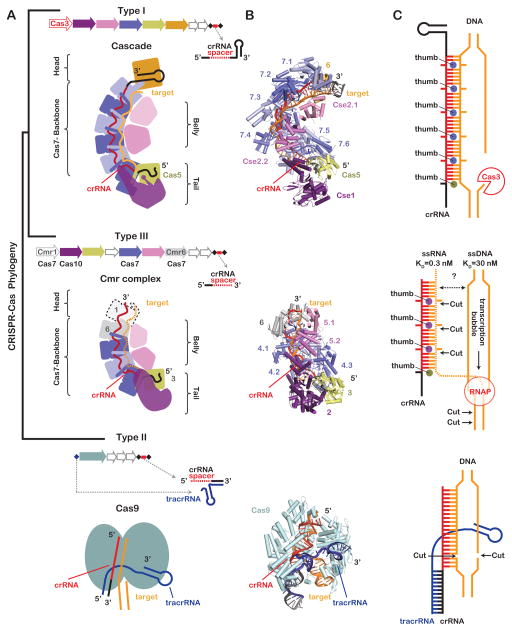
Phylogenetic studies have identified three distinct Types (Type I, II and III) of CRISPR-mediated immune systems that are further divided into at least 11 subtypes (Type IA-F, Type IIA-C and Type IIIA-B) (Makarova et al., 2011). Type I and Type III systems rely on large multi-subunit complexes that assemble around a single crRNA, while the Type II systems rely on a single Cas9 protein and two RNAs (crRNA and tracrRNA) (Figure 1). Distant phylogenetic relationships have been reported for several proteins shared by the Type I and III systems (Koonin and Makarova, 2013; Makarova et al., 2011), and accumulating structural studies now suggest that the Type I and Type III surveillance systems may have evolved from a common ancestor distinct from the Cas9-based Type II surveillance systems.
Figure 1. Re-rooting the CRISPR-Cas phylogenetic tree.
(A) Phylogenetic studies have identified three main CRISPR system types (Type I, II, and III) that are defined by the content and organization of cas genes and CRISPR loci. Type I and Type III systems rely on large multi-subunit complexes composed of a CRISPR-derived RNA (crRNA) and several CRISPR-associated (cas) genes, whereas Type II systems rely on two RNAs (i.e. tracrRNA and crRNA) and a single Cas9 protein. (B) Structures reveal a striking architectural similarity between the Type I and Type III surveillance complexes (PDB 4QYZ and 3X1L); suggesting that these systems evolved from a common ancestor, while the Cas9 protein is structurally and evolutionarily distinct (PDB 4UN3). (C) Type I complexes bind dsDNA and recruit transacting nuclease Cas3, while Type lll complexes bind single-stranded RNA with a 100 fold greater affinity than single-stranded DNA. Type III complexes cleave the RNA target using nuclease active sites that are positioned at discrete intervals along the backbone. These systems also cleave the non-template stand of transcriptionally active DNA. DNA cleavage relies on amino acids in the Cmr2 tail (purple) that are conserved in Cas10 family proteins. Type II systems rely on a single Cas9 protein that binds and cleaves dsDNA targets.
Despite structural similarities between Type I and Type III surveillance complexes, these two systems are mechanistically distinct. Unlike the Type I systems, which target double-stranded DNA (Hochstrasser et al., 2014; Mulepati and Bailey, 2013; Rollins et al., 2015; Sinkunas et al., 2013; Szczelkun et al., 2014; Westra et al., 2012), the Type III systems are multi-functional machines that target single-stranded RNA and transcriptionally active DNA (Benda et al., 2014; Deng et al., 2013; Goldberg et al., 2014; Hale et al., 2009; Osawa et al., 2015; Peng et al., 2015; Ramia et al., 2014a; Samai et al., 2015; Staals et al., 2013; Zhang et al., 2012). However, the structural basis that explains the mechanistic versatility of Type III systems has been unclear. In a recent issue of Molecular Cell, Osawa et al. present a 2.1Å-resolution structure of a chimeric Type III Cmr complex (Osawa et al., 2015). This structure supports the phylogenetic connection between Type I and Type III systems, while providing atomic-resolution details that explain mechanistic distinctions between DNA targeting Type I systems and the Type III systems, which are capable of cleaving both RNA and DNA substrates. Here we present a short overview of the similarities and differences between the Type I and Type III systems, and highlight important new insights from genetic, biochemical, and structural studies that help explain the mechanistic versatility of Type III CRISPR-systems.
Structural Similarities
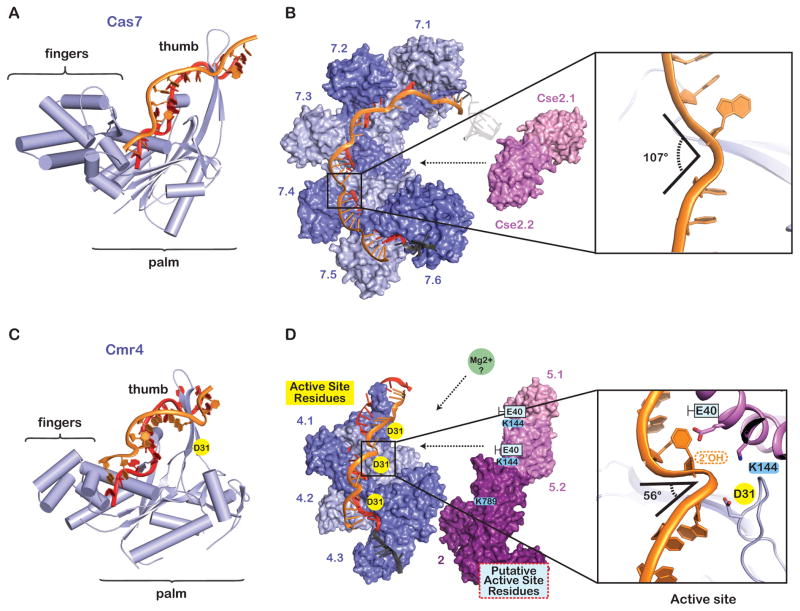
The morphology of the Type I-E crRNA-guided surveillance complex from Escherichia coli K12 (i.e. Cascade) has been likened to a seahorse; with subunits that represent the head, backbone, belly, and tail (Figure 1) (Jore et al., 2011; Wiedenheft et al., 2011; Zhang and Sontheimer, 2014). Although the head and tail features of the seahorse are slightly less pronounced in the Type III complexes, the analogy provides a familiar anatomic reference for comparing the structures of Type I and Type III complexes. In both systems, proteins from the Cas7 superfamily (e.g. Cas7, Cmr4, and Csm3) assemble into a helical backbone capped at either end by head and tail subunits (Benda et al., 2014; Hale et al., 2009; Hrle et al., 2013; Jackson et al., 2014a; Jore et al., 2011; Lintner et al., 2011; Mulepati et al., 2014; Osawa et al., 2015; Ramia et al., 2014a; Rouillon et al., 2013; Spilman et al., 2013; Staals et al., 2013; Staals et al., 2014; Taylor et al., 2015; Wiedenheft et al., 2011; Zhang et al., 2012; Zhao et al., 2014). Amino acid sequences are diverse, but all Cas7 family proteins share a similar “right hand” morphology consisting of fingers, palm, and thumb-shaped domains (Figure 2). In Type I and Type III complexes, the Cas7 palm domain binds to the sugar-phosphate backbone of the crRNA though electrostatic, non-sequence specific interactions, and the thumb folds over the top of the crRNA and across the palm of an adjacent molecule. This pattern of Cas7 oligomerization results in a crRNA that is pinched between the palm of one subunit and the thumb of another, creating a kink in the crRNA at regular 6-nucleotide intervals (Jackson et al., 2014a; Mulepati et al., 2014; Osawa et al., 2015; Zhao et al., 2014). Upon hybridization to a complementary target, the Cas7 thumbs sterically block base pairing at 6-nucleotide intervals, kinking the backbone and splaying nucleobases on both strands in opposite directions.
Figure 2. The helical backbone is a versatile architectural platform with mechanistic plasticity.
(A) The Cas7 protein from E. coli (Type I-E) is shaped like a right hand. The palm of Cas7 binds the crRNA through non-sequence specific interactions and the thumb pierces the crRNA/DNA duplex (PDB 4QYZ). (B) The Cascade backbone is composed of six interwoven Cas7 proteins (Cas7.1–7.6). The Cas7 thumbs distort the backbone geometry of the single-stranded DNA target, introducing ~107-degree kinks at 6-nucleotide intervals. (C) The Cmr4 protein from A. fulgidus is a Cas7 family protein with a “right hand” morphology. The crRNA is non-specifically bound by the palm and the thumbs pierce the crRNA/target duplex at 6-nucleotide intervals (PDB 3X1L). (D) The catalytic residue (D31) on Cmr4 is adjacent to ~56-degree kinks in the phosphate backbone. Conserved residues on Cmr5 (K144) and Cmr2 (K789) interact with the kinked out phosphate, and the side chain of a non-conserved residues (e.g. Cmr5E40) nudges the kinked bases towards the major groove of the adjacent duplex.
The belly of Type I and Type III complexes is composed of a strictly alpha helical protein (e.g. Cse2, Cmr5, or Csm2), sometimes referred to as the “small subunit” (Makarova et al., 2011). Structures of Cascade (Type I-E) before and after target binding reveal major conformational changes in the belly (Cse2) and the tail (Cse1) subunits, while the Cas7 backbone is a rigid structure (Hochstrasser et al., 2014; Jackson et al., 2014a; Mulepati et al., 2014; Wiedenheft et al., 2011; Zhao et al., 2014). Similarly, near-atomic resolution cryo-electron microscopy (cryo-EM) reconstructions of the Type III Cmr complex from Thermus thermophilus reveal that the Cas7-like backbone (Cmr4 subunits) forms a rigid structure while the belly subunits (Cmr5) and tail (Cmr2) are repositioned upon target binding (Taylor et al., 2015). The conformational change of the belly subunits in Cascade has been implicated in a “locking” mechanism that increases the grip of Cascade on a DNA target (Szczelkun et al., 2014). However, motion of the analogous belly subunits in the Cmr complex appears to be slightly different, and the implications of this conformational change await further experimental investigation.
In both Type I and Type III systems, Cas6 processes the primary transcript of the CRISPR locus into a library of crRNAs that have an 8-nucleotide repeat-derived “handle” on the 5′ end (Brouns et al., 2008; Carte et al., 2008). Notably, the distinctive S-shape of the 5′-handle, first reported in the Cascade structures, is conserved in the Cmr complex (Jackson et al., 2014a; Mulepati et al., 2014; Osawa et al., 2015; Wiedenheft et al., 2011). In both systems, the three 5′-terminal nucleotides fit into binding pockets on either Cas5 (Type I) or Cmr3 (Type III). In both systems, the next three nucleotides stack into a well ordered triplet and the thumb of either Cas5 (Type I) or Cmr3 (Type III) folds over the last nucleotide of the 5′-handle creating a structural partition that separates the 5′-handle from nucleotides in the crRNA-guide sequence.
Structural Differences
The conserved helical core of Type I and Type III complexes are capped by structurally distinct head and tail subunits. Unlike the Type I-E complex, which is capped at the head (3′-end) and the tail (5′-end) by proteins that specifically interact with repeat-derived portions of the crRNA, the Type III complexes do not maintain a conserved recognition signal on the 3-end of the crRNAs (Figure 3). In Type III systems, the 3′-end of the crRNA is removed by a nuclease, and the complex is capped by subunits that are structurally similar to the Cas7 backbone proteins (i.e. Cmr1 and Cmr6 or Csm5). The number of backbone and belly subunits in Type III systems varies according to the length of the crRNA, suggesting that the crRNA participates in templated assembly of the helical core.
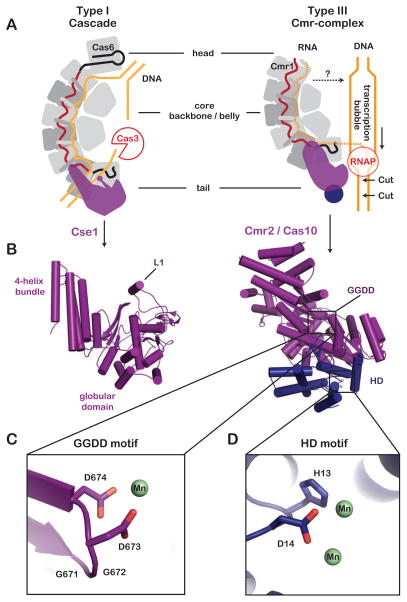
Figure 3. Tail subunits participate in the destruction of DNA targets.
(A) Type I and Type III surveillance complexes share a multi-subunit sea-horse-shaped architecture. The tail subunits of both Types are involved in DNA cleavage. The Cse1 protein of Type I complexes recruit the trans-acting nuclease Cas3 for DNA cleavage, and the Csm2 protein of Type III complexes is critical for DNA target degradation. (B) Structural comparison of the distinct tail subunits (PDB 4TVX and 4W8Y). (C) Close-up of the conserved the GGDD motif and a coordinated manganese ion (green). (D) Closeup of the histidine-aspartate (HD)-motif and two manganese ions (green) in the N-terminal domain.
Another major structural difference between Type I and Type III complexes is in the tail. While both complexes have large tail subunits, these proteins are structurally distinct. The Cse1 tail in Type I-E complexes is composed of an N-terminal globular domain juxtaposed to a C-terminal 4-helix bundle (Figure 3). A short alpha-helix (L1) in the globular domain participates in foreign DNA recognition by binding to an antigenic sequence motif in foreign DNA targets called the PAM (protospacer adjacent motif) (Sashital et al., 2012). In contrast, the Type III Cmr2 tail protein, which belongs to the Cas10 family, is composed of a central RRM (RNA Recognition Motif) flanked by three alpha-helical domains and a conserved N-terminal histidine-aspartate (HD)-domain (Makarova et al., 2011). Although, these tail subunits are structurally different, they both appear to play a critical role in DNA target interference.
Functional versatility
The major functional difference between the Type I and Type III systems is that Type I systems bind double-stranded DNA and recruit a trans-acting nuclease-helicase (Cas3) for DNA degradation (Jackson et al., 2014b), whereas Type III systems target single-stranded RNA and transcriptionally active DNA (Figure 1C). Type III systems are divided into two subtypes, III-A (Csm) and III-B (Cmr). Genetic experiments in Staphylococcus epidermidis provided indirect evidence that III-A systems target DNA (Marraffini and Sontheimer, 2008). Target recognition relied on base paring between the crRNA-guide and the target, but base pairing that extended beyond the guide and into the repeat derived 5′-handle of the crRNA blocked target elimination (Marraffini and Sontheimer, 2010). Sequence in the 5′-handle is derived from the CRISPR repeat sequence, so base pairing between the 5′-handle and the target offered a rational mechanism that explained how this DNA targeting system might distinguish complementary DNA sequences in the host CRISPR locus (self) from non-self sequences in foreign DNA, which are not flanked by CRISPR repeat sequence. However, the mechanism of DNA cleavage remained undetermined and in vitro cleavage assays preformed using the purified Type III-A and III-B complex demonstrated that complementary single-stranded RNA, but not single-stranded DNA targets were cleaved in a metal dependent reaction resulting in cleavage products separated by 6-nucleotides (Benda et al., 2014; Hale et al., 2014; Hale et al., 2009; Osawa et al., 2015; Ramia et al., 2014a; Staals et al., 2013; Tamulaitis et al., 2014; Zhu and Ye, 2015). Staals et al speculated that this activity might be indicative of an active site present on each of the backbone or belly subunits and subsequent comparative structural analyses revealed a conserved acidic residue on the backbone subunits in both the Cmr (PfCmr4D26) and Csm (StCsm3D33) complexes (Benda et al., 2014; Staals et al., 2013; Staals et al., 2014; Tamulaitis et al., 2014). Alanine substitutions at this position in either the Cmr or Csm complexes abrogated RNA cleavage (Benda et al., 2014; Ramia et al., 2014a; Tamulaitis et al., 2014; Zhu and Ye, 2015).
Mechanistic insights from the 2.1Å-resolution Cmr structure
RNA cleavage by Type III-A and Type III-B systems is well established, but the mechanism of cleavage has remained elusive. Now the 2.1Å-resolution crystal structure of a chimeric Cmr complex, bound to a non-cleavable DNA target, provides an atomic-resolution view of the active sites in the Cmr complex (Osawa et al., 2015). The structure shows how residues on the Cmr4 thumbs team-up with residues on Cmr5 to kink the sugar-phosphate backbone of the target at 6-nucleotide intervals (Figure 2). Nucleobases on either side of each kink are constrained by hydrogen bonding to complementary bases on the crRNA-guide, while the scissile phosphate is pulled to one side by a positively charged residue on either Cmr5 (K144) or Cmr2 (K789) (Figure 2).
The degree of the kink is exaggerated by the side chain of a non-conserved amino acid on Cmr5 (E40), which nudges the displaced nucleobase in the opposite direction of the scissile phosphate. Together the push (Cmr5E40) and pull (Cmr5K144) of the Cmr active site create kinks in the backbone that are significantly sharper than the kinks induced by the Cas7 thumbs in Cascade (Type I-E) (Figure 2). Distortion of the backbone may help position the scissile phosphates directly between the 2′ hydroxyl of the adjacent ribose and the strictly conserved aspartate (Cmr4D31), which is critical for RNA cleavage. The structure suggests a mechanism in which the 2′ oxygen acts as a nucleophile on the scissile phosphate and the conserved aspartate acts as a general acid to stabilize the leaving group. Osawa et. al. support this hypothesis by showing that removal of the 2′ hydroxyl at every sixth nucleotide abolishes RNA cleavage (Osawa et al., 2015).
The crystal structure provides important new mechanistic insights, but there are several unanswered questions. Divalent metal ions are necessary for RNA cleavage, but no metals are observed at the Cmr4 active sites. Thus the function of metal ions in RNA cleavage remains unclear. The 2′ hydroxyl on the sugar of the kinked nucleobase may be involved in metal ion coordination and the single-stranded DNA in this structure may preclude metal ion association. The Cmr4D31A mutation kills RNase activity, and this mutant may be an attractive target for structural studies of this complex bound to RNA. An RNA bound structure will include new information about the location of the 2′ hydroxyls and may offer new insight about the role of metal ions in RNA cleavage. Additionally, the importance of the residues involved in the push (Cmr5E40) and pull (Cmr4K144 and Cmr2K789) that contribute to target kinking remain untested.
Not one, but two targets for a single Type III complex
Accumulating structural (Cmr complex), biochemical (cleavage assays), and genetic (MS2 phage protection assay) data indicate that most, if not all Type III systems degrade RNA targets, but the mechanism of DNA targeting has been less clear. In a recent issue of Cell, Samia et al. provide direct evidence for both RNA and DNA cleavage by the Type III-A complex from S. epidermidis (Samai et al., 2015). Using in vitro and in vivo systems, they show that the Csm complex cleaves the complementary strand of nascent RNA and the non-template strand of a transcriptionally active DNA. RNA and DNA targets are cleaved by independent active sites located on either the backbone (RNA targeting) or the tail (DNA targeting) of the Csm complex, and immunity against plasmids and DNA viruses requires DNA, but not RNA cleavage activity. The transcriptional dependence of DNA targeting may explain why previous in vitro studies observed RNA, but not DNA cleavage.
What does the requirement for transcription tell us about the mechanism of DNA cleavage? Unlike Type I systems, neither Type III-A nor III-B complexes will bind to a complementary target sequence in a double-stranded DNA duplex (Hale et al., 2014; Staals et al., 2014; Tamulaitis et al., 2014). This suggests that accessibility may be important and that DNA unwinding by the RNA polymerase may be necessary for DNA targeting by the Type III-A systems. However, access may not be a limiting factor for targeting DNA substrates. Previous biochemical studies have shown that the Type III-A complex from T. thermophilus binds single-stranded RNA (KD =0.3 nM) 100 times tighter than single-stranded DNA (KD=30nM) (Tamulaitis et al., 2014). While this does not exclude crRNA-guided hybridization to a complementary single-stranded DNA, it does indicate that the transcribed RNA may be preferentially bound over DNA. Additional support for the crRNA-guide binding to the nascent RNA, rather than DNA, is that DNA targeting is strand specific (Samai et al., 2015). Targeting the non-template strand of a transcriptionally active DNA results in target DNA elimination, whereas targeting the template stand does not. Together these data suggest that newly transcribed RNA may be the recognition substrate that recruits Type III complexes to DNA targets.
DNA targeting by Type III systems appears to be critical for protecting the host from plasmids and DNA viruses, but the mechanism of DNA cleavage remains speculative. The N-terminal histidine-aspartate (HD)-domain of the Cas10 tail protein was initially predicted to be the nuclease active site, but in vivo and in vitro studies have found this domain is not the primary nuclease in target degradation (Cocozaki et al., 2012; Hatoum-Aslan et al., 2014; Ramia et al., 2014b; Samai et al., 2015). Instead, several studies have shown that the GGDD motif, which is conserved in the tail subunit of Type III Cas10 proteins (i.e., Cmr2 and Csm1), is required for DNA cleavage (Figure 1 and 3) (Hatoum-Aslan et al., 2014; Ramia et al., 2014b; Samai et al., 2015). The GGDD motif is surface exposed in the structure of the Cmr complex, but the N-terminal HD-domain is removed from the Cas10 protein in this structure (Osawa et al., 2015). By modeling a previously determined structure of the complete Cas10 protein into the Cmr complex we noticed that the GGDD motif is buried by a series of short alpha-helices and the N-terminal HD-domain (Figure 3) (Benda et al., 2014). These results suggest that binding to a physiologically relevant target may induce a conformational change that exposes or activates the nuclease active site(s) in Cas10. Alternatively, it is possible that Cas10 is involved in recruitment of a trans-acting nuclease. Several studies have shown that trans-acting Cas proteins (e.g. csx1 and csm6) are required for immunity against DNA targets (Deng et al., 2013; Hatoum-Aslan et al., 2014). The requirement of these proteins for DNA degradation in type III systems is reminiscent of Cas3 proteins in type I systems, though the function of these trans-acting proteins remains undetermined in type III systems.
DNA targeting systems must be able to distinguish complementary DNA targets in the host CRISPR loci (self) from complementary sequences in foreign DNA (non-self). Type I and Type II systems rely on protein mediated recognition of a short nucleotide sequence, known as the protospacer adjacent motif (PAM), which is found adjacent to targets in foreign DNA, but not in the host genome (Anders et al., 2014; Rollins et al., 2015; Sashital et al., 2012). In contrast, Type III systems distinguish self from non-self by differential base pairing between nucleotides in the 5′-handle (Marraffini and Sontheimer, 2010). Base pairing in this region signals self and prevents target elimination. In the Cmr structure, nucleotides in the 5′ handle are inaccessible (Osawa et al., 2015). Hybridization between the target and nucleotides in the 5′-handle would require a conformational change that moves a loop from Cmr3 away from the nucleotides (−2 to −4) involved in self-recognition. This conformational change may inactivate the DNA cleavage mechanisms. As the story continues to unfold, we are certain that structural, computational, biochemical, and genetic techniques will collectively contribute to a complete functional understanding of these remarkable CRISPR RNA-guided machines.
Acknowledgments
R.N.J is a recipient of the Ruth L. Kirschstein National Research Service Award from the National Institutes of Health (F32 GM108436). Research in the Wiedenheft lab is supported by the National Institutes of Health (P20GM103500 and R01GM108888), the National Science Foundation EPSCoR (EPS-110134), the M.J. Murdock Charitable Trust, and the Montana State University Agricultural Experimental Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S. The Cmr complex: an RNA-guided endoribonuclease. Biochem Soc Trans. 2013;41:1464–1467. doi: 10.1042/BST20130216. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA. CRISPR-Cas Systems: Prokaryotes Upgrade to Adaptive Immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benda C, Ebert J, Scheltema RA, Schiller HB, Baumgartner M, Bonneau F, Mann M, Conti E. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell. 2014;56:43–54. doi: 10.1016/j.molcel.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Davidson AR. To acquire or resist: the complex biological effects of CRISPR-Cas systems. Trends in microbiology. 2014;22:218–225. doi: 10.1016/j.tim.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes and Development. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocozaki AI, Ramia NF, Shao Y, Hale CR, Terns RM, Terns MP, Li H. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure. 2012;20:545–553. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Sinkunas T, Siksnys V. Molecular mechanisms of CRISPR-mediated microbial immunity. Cell Mol Life Sci. 2014;71:449–465. doi: 10.1007/s00018-013-1438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Cocozaki A, Li H, Terns RM, Terns MP. Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev. 2014;28:2432–2443. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Samai P, Marraffini LA. Genetic Characterization of Antiplasmid Immunity through a Type III-A CRISPR-Cas System. Journal of Bacteriology. 2014;196:310–317. doi: 10.1128/JB.01130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci U S A. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrle A, Su AA, Ebert J, Benda C, Randau L, Conti E. Structure and RNA-binding properties of the type III-A CRISPR-associated protein Csm3. RNA Biol. 2013;10:1670–1678. doi: 10.4161/rna.26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, Van Der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014a;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Lavin M, Wiedenheft B. Fitting CRISPR-associated Cas3 into the Helicase Family Tree. Current Opinion in Structural Biology. 2014b;24:106–114. doi: 10.1016/j.sbi.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013;10:679–686. doi: 10.4161/rna.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, et al. Structural and Functional Characterization of an Archaeal Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated Complex for Antiviral Defense (CASCADE) J Biol Chem. 2011;286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR Interference Limits Horizontal Gene Transfer in Staphylococci by Targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. In Vitro Reconstitution of an Escherichia coli RNA-guided Immune System Reveals Unidirectional, ATP-dependent Degradation of DNA Target. J Biol Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Heroux A, Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Inanaga H, Sato C, Numata T. Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.03.018. this issue. [DOI] [PubMed] [Google Scholar]
- Peng W, Feng M, Feng X, Liang YX, She Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015;43:406–417. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramia NF, Spilman M, Tang L, Shao Y, Elmore J, Hale C, Cocozaki A, Bhattacharya N, Terns RM, Terns MP, et al. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR-Cas complex. Cell Rep. 2014a;9:1610–1617. doi: 10.1016/j.celrep.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramia NF, Tang L, Cocozaki AI, Li H. Staphylococcus epidermidis Csm1 is a 3′-5′ exonuclease. Nucleic Acids Research. 2014b;42:1129–1138. doi: 10.1093/nar/gkt914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins MF, Schuman JT, Paulus K, Bukhari HS, Wiedenheft B. Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, Zhou M, Zhang J, Politis A, Beilsten-Edmands V, Cannone G, Graham S, Robinson CV, Spagnolo L, White MF. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell. 2015 doi: 10.1016/j.cell.2015.04.027. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;48:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. Embo Journal. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, Terns R, Terns M, Li H, Stagg S. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell. 2013;52:146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas C, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V. Programmable RNA Shredding by the Type III-A CRISPR-Cas System of Streptococcus thermophilus. Mol Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- Taylor DW, Zhu Y, Staals RH, Kornfeld JE, Shinkai A, van der Oost J, Nogales E, Doudna JA. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science. 2015 doi: 10.1126/science.aaa4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Nilges B, van Erp PB, van der Oost J, Dame RT, Brouns SJ. Cascade-mediated binding and bending of negatively supercoiled DNA. RNA Biol. 2012;9 doi: 10.4161/rna.21410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sontheimer EJ. Structural biology. Cascading into focus. Science. 2014;345:1452–1453. doi: 10.1126/science.1260026. [DOI] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, Wang Y. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–150. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- Zhu X, Ye K. Cmr4 is the slicer in the RNA-targeting Cmr CRISPR complex. Nucleic Acids Res. 2015;43:1257–1267. doi: 10.1093/nar/gku1355. [DOI] [PMC free article] [PubMed] [Google Scholar]