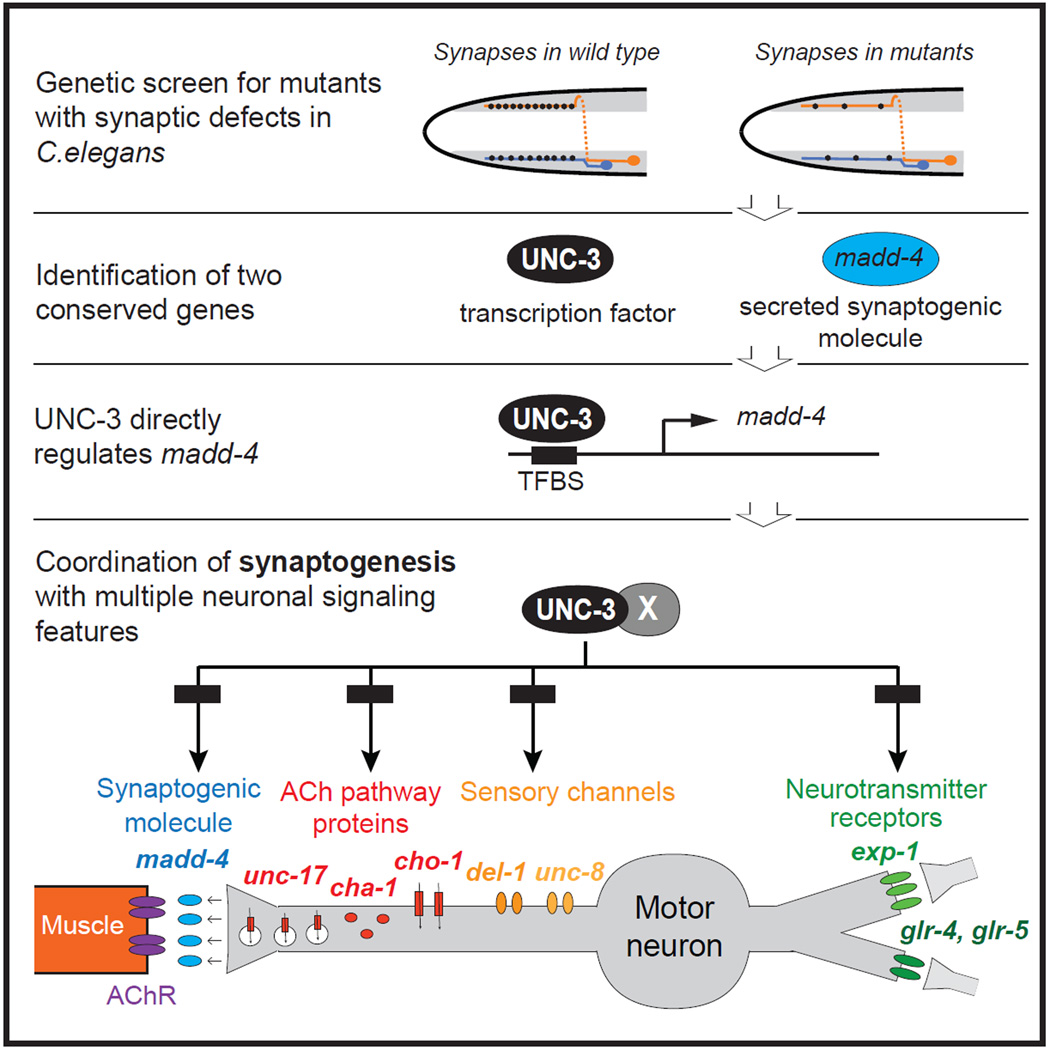
SUMMARY
During nervous system development, postmitotic neurons face the challenge of generating and structurally organizing specific synapses with appropriate synaptic partners. An important unexplored question is whether the process of synaptogenesis is coordinated with the adoption of specific signaling properties of a neuron. Such signaling properties are defined by the neurotransmitter system that a neuron uses to communicate with postsynaptic partners, by the neurotransmitter receptor type used to receive input from presynaptic neurons and, potentially, by other sensory receptors that activate a neuron. Elucidating the mechanisms that coordinate synaptogenesis, neuronal activation and neurotransmitter signaling in a postmitotic neuron represents one key approach to understand how neurons develop as functional units. Using the SAB class of Caenorhabditis elegans motor neurons as a model system, we show here that the phylogenetically conserved COE-type transcription factor UNC-3 is required for synaptogenesis. UNC-3 directly controls the expression of the ADAMTS-like protein MADD-4/Punctin, a presynaptically secreted synapse-organizing molecule that clusters postsynaptic receptors. UNC-3 also controls the assembly of presynaptic specializations and ensures the coordinated expression of enzymes and transporters that define the cholinergic neurotransmitter identity of the SAB neurons. Furthermore, synaptic output properties of the SAB neurons are coordinated with neuronal activation and synaptic input, as evidenced by UNC-3 also regulating the expression of ionotropic neurotransmitter receptors and putative stretch receptors. Our study shows how synaptogenesis and distinct, function-defining signaling features of a postmitotic neuron are hardwired together through coordinated transcriptional control.
INTRODUCTION
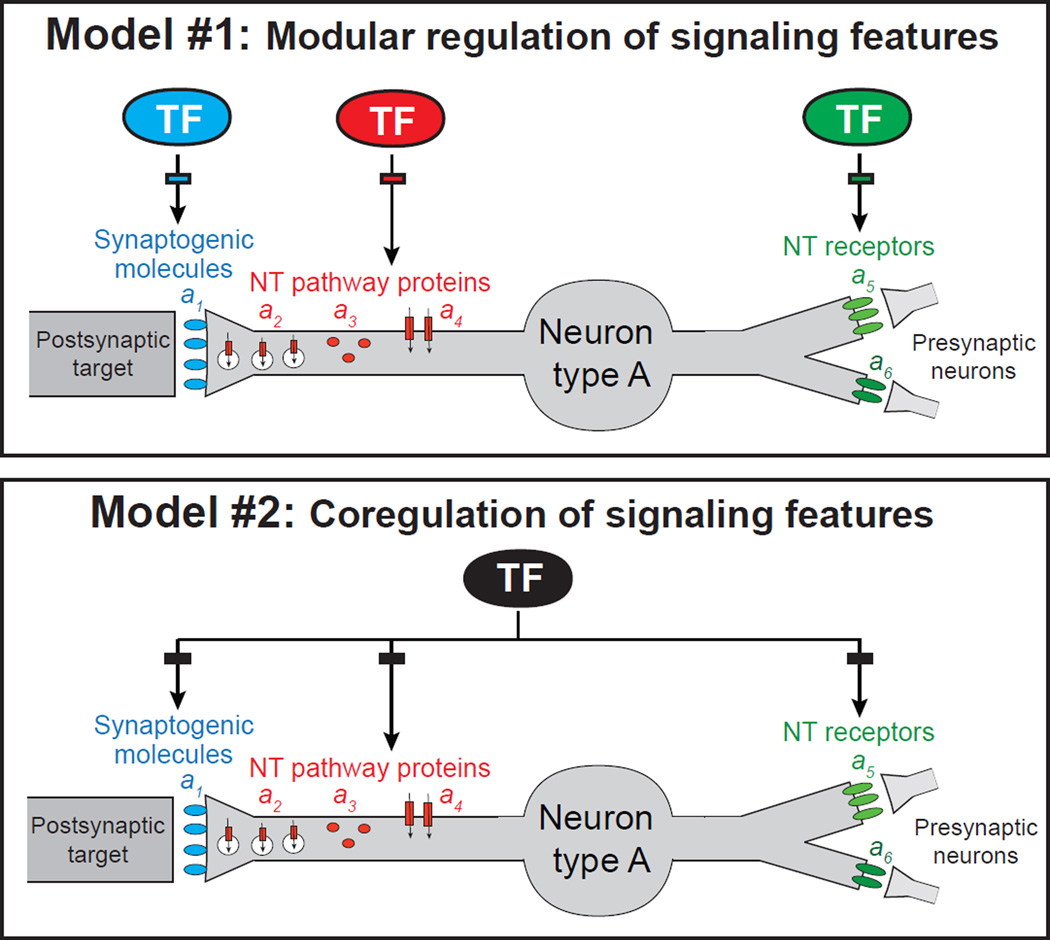
To establish a functional synapse, neurons must choose which neurotransmitter system to use to signal via that synapse to postsynaptic targets. In general, it is not understood whether and how the process of organizing structural features of a specific synapse is linked with the functional properties of that synapse, such as neurotransmitter choice. It is also not known whether or how a neuron coordinates the choice of which neurotransmitter to use to signal to its postsynaptic targets (i.e. synaptic output) with the choice of which neurotransmitter receptor system to implement to receive signals from presynaptic neurons (i.e. synaptic input). Whether the expression of synaptic input features is coordinated with the expression of other activating inputs that a neuron may process (e.g. through sensory receptors) is also not explored. All of these features are of critical importance for the function of a neuron and its assembly into a specific circuit. As illustrated in Figure 1, in principle, the expression of genes that define these activity- and circuit-defining features of a neuron could be independently regulated by distinct transcription factors. Alternatively, these activity- and circuit-defining features could be coordinated through a coregulatory strategy (Figure 1). We investigate here these distinct regulatory strategies in the context of a motor circuit in the nematode Caenorhabditis elegans.
Figure 1. Possible models for the regulation of distinct, function-defining features of a specific neuron type.
Synaptogenic molecules (a1), neurotransmitter (NT) pathway genes (a2, a3, a4), and NT receptors (a5, a6). TF, transcription factor. Rectangles represent TF binding sites and indicate direct regulation. For simplicity, only synaptogenic molecules of the presynaptic side are depicted.
Motor circuits of various organisms have long served as models to study core principles of synaptic assembly and neurotransmitter signaling. Motor circuits in the nematode C.elegans include ventral nerve cord (VNC) motor neurons (MNs) that control the body musculature along the length of the animal and MNs located in head ganglia that control neck and head muscles [1, 2]. The SAB head MN class is composed of three members, a single MN with two distinct, bilaterally symmetric projections to dorsal head muscles (SABD for SAB dorsal) and a bilateral pair of neurons that project single axons to ventral head muscles (SABVL and SABVR for left and right SAB ventral) [3] (Figure 2 A–C). The SAB MNs use the neurotransmitter acetylcholine (ACh) [3] and, as inferred from the expression of GABA- and glutamate-gated ion channels and their anatomical synaptic connectivity, receive GABAergic and glutamatergic synaptic inputs [2, 4, 5]. Moreover, the SAB neurons are thought to be activated by putative stretch receptors of the DEG/ENaC ion channel family [6, 7]. The SAB neurons therefore constitute an excellent model to address whether and how the expression of genes that define synaptic input, neuronal activation, neurotransmitter choice and synaptic output features of a neuron are coordinated (Figure 1).
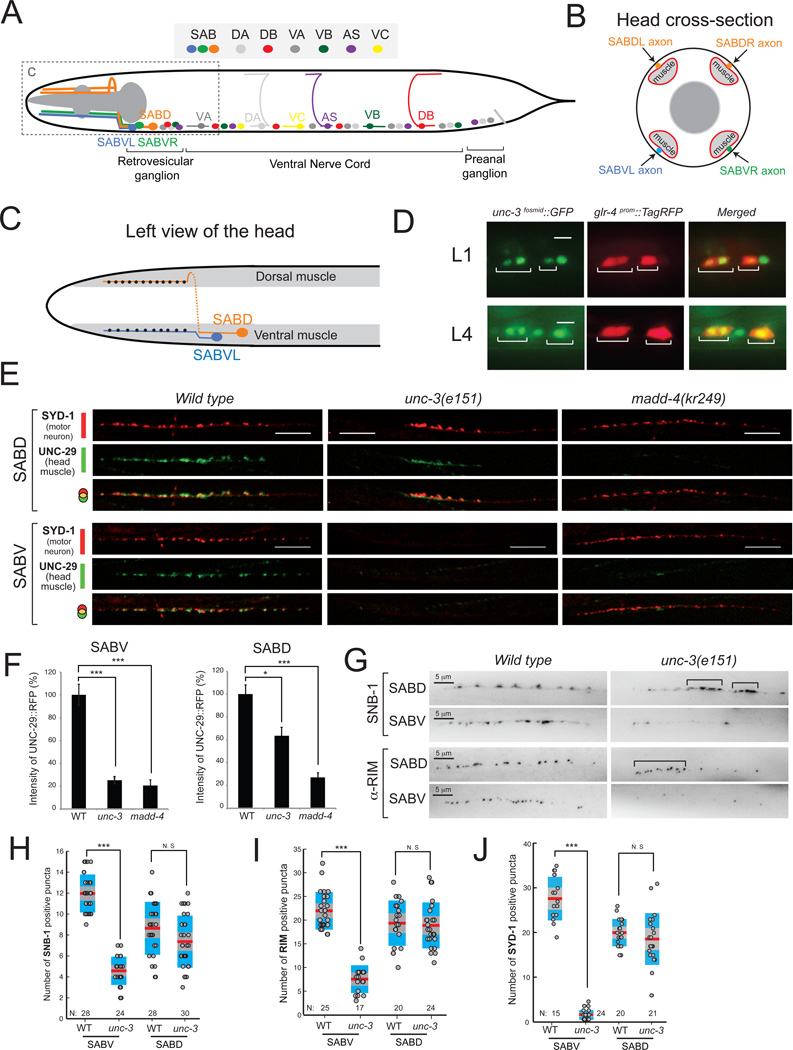
Figure 2. unc-3 controls SAB motor neuron synapse formation.
A: Schematic of SAB and other classes of cholinergic MNs (DA, DB, VA, VB, AS, VC) in C.elegans.
B: Diagram of a cross section of the head showing SAB axons (orange, blue, green), head muscle (light grey), and pharyngeal muscle (dark grey).
C: Left view of the head showing the synapses (black dots) of SABVL onto ventral muscle and the synapses (black dots) of left axon of SABD onto dorsal muscle. Anterior is left.
D: unc-3 is expressed in SAB neurons during larval (L1 and L4) stages. The transgene otIs476[glr-4prom::TagRFP] was used as a marker of SAB neurons. unc-3 expression was monitored using a fosmid-based gfp reporter. Expression of unc-3 was also detected in SABV and SABD neurons during adult stages (data not shown). Scale bar 5 µm. N > 10.
E: Synaptic defects at the SAB innervation zones of unc-3(e151) and madd-4(kr249) mutants. In wild-type (WT) SABD synapses, the presynaptic zone marker SYD-1 is juxtaposed to the AChR marker (UNC-29) expressed in head muscle (20/20 animals examined). In unc-3(e151) mutants, the SABD presynaptic zones are formed but are abnormally clustered as evident by SYD-1 fluorescence. Using SYD-1::GFP as a marker, we observe enlarged, abnormally clustered presynaptic zones in 11/20 (55%) SABD axons in unc-3 mutants compared to 1/20 (5%) SABD axons in WT animals. In the SABV of unc-3(e151) mutants, we barely observe any SYD-1 puncta (20/20 animals). Detailed quantification of SYD-1 positive puncta is provided in J. Post-synaptic clustering of AChR (UNC-29::RFP) is significantly reduced in the SABD and never observed in the SABV neurons of unc-3(e151) mutants (20/20 animals examined). The clustering of AChR in madd-4(kr249) mutants is dramatically reduced in SAB synapses, while the presynaptic zones (labeled by SYD-1::GFP) are normally generated (20/20 animals examined). The line of faint small AChR clusters in SABV of unc-3 and madd-4 mutants corresponds to the boundary of muscle cells (not the SABV innervation zone), which often contains a small amount of extra-synaptic AChRs. As expected, this boundary of muscle cells is also evident in the WT SABV image. For optimal contrast SYD-1::GFP is depicted in red and UNC-29::RFP is in green. Scale bar 10 µm.
F: The degree of L-AChR clustering at the SABV and SABD innervation zones was evaluated by measuring UNC-29::RFP fluorescent intensity. The percentage (%) of fluorescent intensity of UNC-29::RFP intensity was calculated along 55 µm of the SAB innervation zone as previously described in [13]. Error bars show standard error of the mean (sem). We ran a Kruskall-Wallis nonparametric test followed by a Dunn's post test. * = P < 0.05, *** = P < 0.001. SABV: WT (N=16), unc-3 (N=32), madd-4 (N=21); SABD, WT (N=9), unc-3 (N=22), madd-4 (N=15).
G: Visualization of presynaptic boutons using a translational yfp reporter for synaptobrevin-1 (snb-1). A representative image of the boutons along the SABDL and SABVL axons are shown (left view) in WT and unc-3(e151) mutants. Presynaptic boutons often cluster abnormally in the SABD of unc-3 mutants (indicated by brackets), while their number appears normal when compared to WT SABD. Using SNB-1::YFP as a marker, we observe enlarged, abnormally clustered presynaptic zones in 15/24 (62%) SABD axons in unc-3 mutants compared to 3/20 (15%) SABD axons in WT animals. In SABV of unc-3 (e151) mutants, the number of boutons is significantly reduced. Animals at the fourth larval (L4) stage were analyzed. Detailed quantification of SNB-1 positive boutons is provided in H. Visualization of SAB active zones using an antibody for RIM protein. In SABD of unc-3(e151) mutants, the distribution of active zones is abnormal as they often miscluster (indicated by brackets). However, the total number of active zones is not significantly reduced in the SABD of unc-3(e151) mutants. There is a significant reduction of active zones in the SABV of unc-3(e151) mutants. Quantification of RIM staining is provided in I. Scale bar 5 µm. Animals at the first day of adulthood were analyzed. Each dot represents the number of SNB-1 or RIM positive puncta along one SABD or SABV axon. *** : p value < 0.001. N.S, not statistically significant. N = number of axons analyzed. Mean values are represented with a horizontal red line.
RESULTS
unc-3 affects synaptogenesis of the SAB motor neurons
The neuromuscular junctions (NMJs) of the SAB MNs can be visualized in transgenic animals with fluorescently tagged presynaptic (e.g. synaptobrevin) or postsynaptic (acetylcholine receptor/AChR) proteins, which are expressed at the SAB MNs and head muscle, respectively. In wild-type animals approximately a dozen synaptic boutons can be observed along each SAB axonal segment in close apposition with head muscle [3] (“innervation zones”; Figure 2C,G,H). To examine how the SAB neuromuscular junctions are generated, we screened for mutants in which synapses in the innervation zone fail to be appropriately assembled using a reporter strain in which the postsynaptic nicotinic AChR subunit UNC-29 (expressed in head muscle) is fluorescently tagged [8]. We identified two mutant alleles, kr268 and kr249 in which we detected a dramatic decrease in AChR clustering at the innervation zone of the SAB neurons (Figure 2E,F). We mapped the kr268 lesion to the X chromosome and - through whole genome sequencing and complementation testing - found that the kr268 allele affects the unc-3 locus, which codes for the sole homolog of the Collier/Olf/Ebf (COE) transcription factor family in C.elegans [9]. The role of the COE-type transcription factors in synapse formation has not been previously studied.
We find that homozygous unc-3(e151) null mutants display strong SAB synapse defects as visualized with a number of different markers (Figure 2E,F). On the postsynaptic side, there is a strong reduction in the intensity and localization of AChR clusters. In the SABD innervation zone, AChR clusters (as visualized with fluorescently tagged UNC-29) are reduced, but when visible, the clustering is abnormal in terms of size and localization along the SAB innervation zone (Figure 2E,F). In the innervation zones of SABV neurons of unc-3(e151) animals, AChR clustering is even more strongly reduced (Figure 2E,F).
Synaptogenic defects are also evident on the presynaptic side in unc-3 mutants. The synaptic boutons of SABD, visualized with GFP-tagged synaptobrevin, often appear enlarged and mislocalized, forming abnormal clusters along the SABD innervation zone (Figure 2E, G). Apart from SNB-1 clustering defects, the SABD innervation zones of unc-3 mutants also display clustering defects of two other proteins that mark presynaptic zones, the rhoGAP protein SYD-1 [10, 11] (Figure 2E) and the RIM protein UNC-10 (Figure 2G).
Presynaptic zones not only appear to be disorganized in both the SABD and SABV neurons, but there is a striking reduction of the overall number of presynaptic clusters in the SABV neurons of unc-3 mutants. The number of SNB-1, UNC-10 and SYD-1 positive puncta is significantly reduced in unc-3 mutants (Figure 2H–J). This defect is not simply explicable by unc-3 regulating snb-1 or unc-10 gene expression, since transcriptional snb-1 and unc-10 reporters are normally expressed in unc-3 mutants (Figure 4D). Moreover, single molecule fluorescence in situ hybridization (smFISH) [12] shows no effect of unc-3 on snb-1 and unc-10 mRNA abundance (Figure S1).
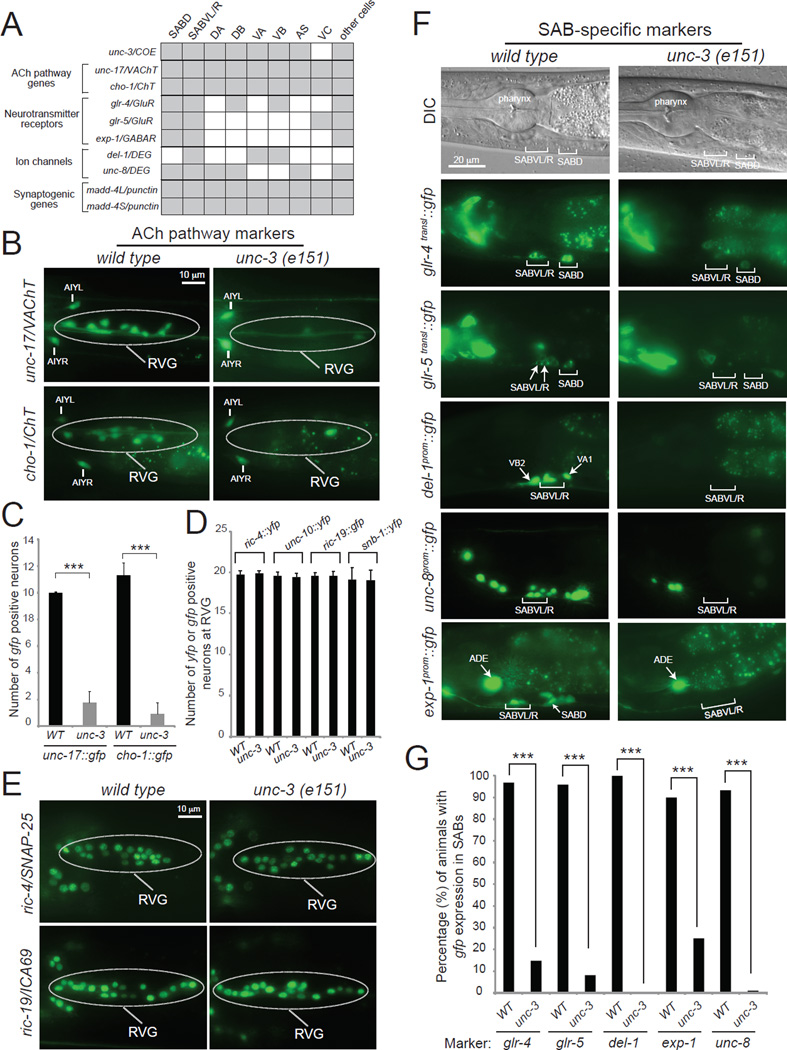
Figure 4. unc-3 is required for multiple aspects of neuronal signaling in the SAB motor neurons.
A: A combinatorial code of gene expression for SAB head MNs based on our findings and WormBase (www.wormbase.org). Grey shaded boxes indicate expression in the respective neuron type; white boxes indicate absence of expression.
B, C: ACh pathway genes (unc-17/VAChT, cho-1/ChT) fail to be expressed at the retrovesicular ganglion (RVG) of unc-3(e151) mutants. The cell bodies of SABs are located at the RVG together with other (around 7–9) cholinergic neurons. Scale bar 10 µm. Error bars represent standard deviation (s.d). *** : p value < 0.001. N > 10 for unc-17prom::gfp in WT and unc-3(e151) animals. N = 22 for cho-1fosmid::gfp in WT and unc-3(e151) animals. Animals at the first day of adulthood were analyzed.
D, E: Transcriptional reporters for panneuronally-expressed genes (ric-4, ric-19, unc-10, and snb-1) are unaffected in unc-3(e151) mutants. The cell bodies of SABs are located at the RVG together with other 17 neurons. Scale bar 10 µm. Detailed Quantification provided in D. Error bars represent standard deviation (s.d). N > 10 for ric-4fosmid::yfp, ric-19prom::gfp, unc-10fosmid::YFP, and snb-1fosmid::YFP in WT and unc-3(e151) animals. Animals at the first day of adulthood were analyzed. See also Figure S1.
F, G: The expression of neurotransmitter receptors (glr-4, glr-5, exp-1) and ion channel-encoding genes (del-1, unc-8) is significantly affected in the SAB neurons of unc-3(e151) mutants. See also Figures S1 and S4. Scale bar 20 µm. Quantification provided in G. N = 25 for glr-4, glr-5, del-1, and unc-8 reporters in WT and unc-3(e151) animals. N = 20 for exp-1 reporter in WT and unc-3(e151) animals. Animals at the fourth larval (L4) stage were analyzed. In the quantification graph (G), the percentage of animals expressing the reporter in SABVL/R and SABD is shown for glr-4 and glr-5, while for del-1 and unc-8 the percentage of animals expressing the reporter in SABVL/R is shown. Similar results were obtained when a fosmid-based reporter was used to monitor UNC-8 protein expression in the SAB neurons (100 % of wdEx948 [unc-8fosmid::gfp] animals (N=35) showed gfp expression in the SABV neurons, while 42.85 % of unc-3(e151); wdEx948 [unc-8fosmid::gfp] showed gfp expression in SABV neurons (N=35). For exp-1, the percentage of animals with expression in SABD is shown. *** : p value < 0.0001.
SAB neurons also show some axon extension defects in unc-3 mutants, but the synaptic defect at the innervation zones is more severe (based on phenotype penetrance). For example, 100% of animals show AChR clustering defects in the SABV neurons, but less than 20% show axon outgrowth defects (Figure S2). To avoid any contribution of the SAB axonal defect phenotype on our synaptic defect analysis, we evaluated synaptic defects only in those unc-3 animals in which the SAB axons clearly follow the correct path of innervation onto head muscle.
Through the examination of a fosmid-based reporter construct containing the entire unc-3 locus, as well as ~ 10kb of upstream and downstream cis-regulatory regions, we find that unc-3 is expressed in all three SAB neurons, but not in postsynaptic muscle cells (Figure 2D and data not shown). Moreover, the pre- and post-synaptic SAB phenotypes of unc-3 mutants can be rescued by the MN expressed fosmid-based unc-3 reporter (Table S1), indicating that unc-3 operates autonomously to control presynaptic assembly and non-autonomously to affect postsynaptic receptor assembly in head muscle. Non-autonomous regulation of postsynaptic receptor clustering suggests that unc-3 may control the expression of one or more postsynaptic organizer molecules that originate from the SAB neurons. We find that the second allele identified in our SAB synapse screen, kr249, defines such an organizing factor through which unc-3 acts to affect postsynaptic receptor clustering in head muscle.
madd-4, a synaptic organizer, is a direct target of unc-3
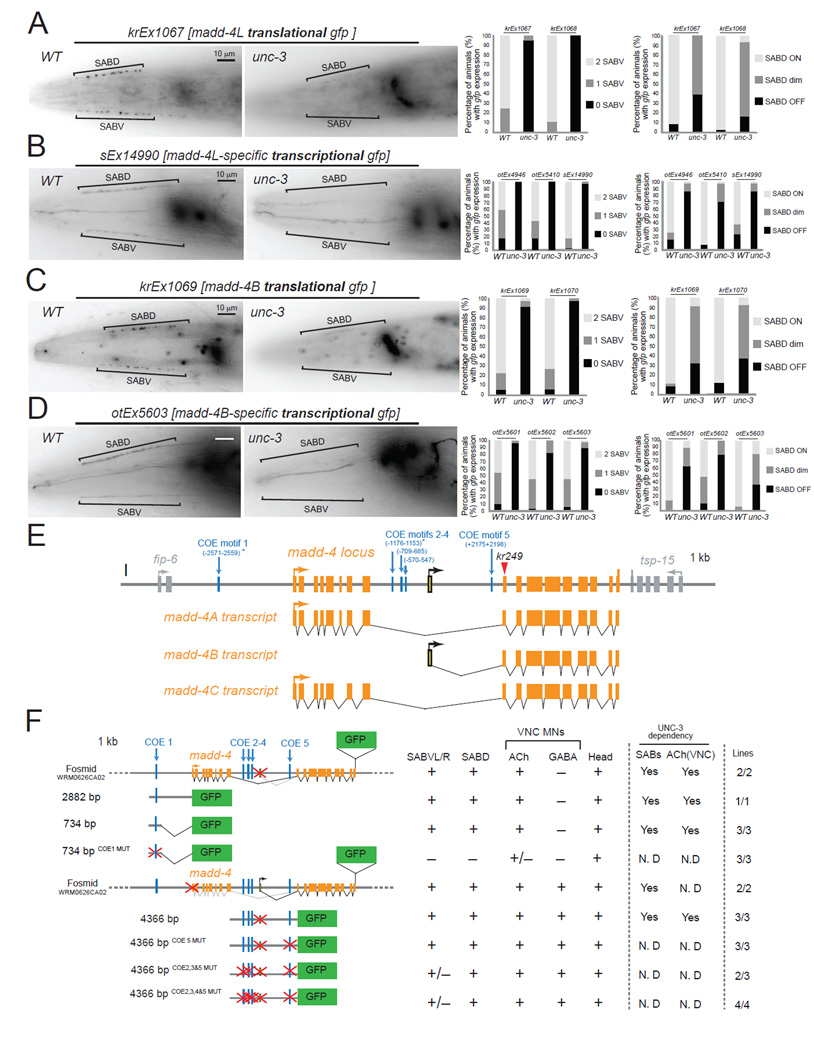
Through a combination of SNP mapping, whole genome sequencing and transformation rescue, we found that the second mutant allele retrieved from our screen, kr249, carries an opal mutation (W374stop) in the madd-4 locus. madd-4, the C.elegans ortholog of mammalian Punctin-1 and Punctin-2, encodes a member of the ADAMTS-Like family, which are extracellular matrix proteins related to ADAM proteases but devoid of proteolytic activity [13, 14]. In a screen for molecules that affect synapse formation in the C.elegans ventral nerve cord, madd-4 was recently identified as a gene required for synaptogenesis of various MN classes [13]. MADD-4 protein is secreted from cholinergic MNs and acts as an anterograde synapse-organizing signal that controls AChR clustering on the muscle without affecting presynaptic assembly on the MN side [13]. The madd-4 locus encodes three isoforms, which are all affected by the kr249 (W374stop) allele retrieved from our screen (Figure 3E). MADD-4A and MADD-4C are the longest isoforms, which differ by alternative splicing of a two-amino acid encoding exon, and will be referred to as MADD-4L (Long) hereafter (Figure 3E). MADD-4B is the shortest isoform and is likely generated by the use of an alternative promoter ([14] and this study). Animals lacking all madd-4 isoforms (kr249 W374stop allele, kr270 deletion allele of entire locus) display severe AChR clustering defects at the SAB neuromuscular synapse that are reminiscent of the postsynaptic defects observed in unc-3 mutants (Figure 2 E). While MADD-4B-specific mutants display no apparent SAB phenotype, MADD-4L-specific mutants display milder SAB AChR clustering defects when compared to madd-4 null mutants (data not shown and [13]), suggesting that MADD-4B might partially compensate for the absence of MADD-4L activity in the SAB neurons.
Figure 3. unc-3 controls madd-4 expression.
A: The protein expression of madd-4L is significantly affected in the SAB neurons of unc-3(e151) mutants. Results for two independent extra-chromosomal arrays (kr1067 and kr1068 are madd-4L translational fusions) are quantified on the right. Scale bar 10 µm. N = 20 for krEx1067 in WT and unc-3(e151) animals. N >30 for krEx1068 in WT and unc-3(e151) animals. Left side view of the head is shown. Anterior is to the left. madd-4L is also expressed in head ganglia, which appear out of focus to the right of each image.
B: The expression of madd-4L transcriptional reporters is significantly affected in the SAB neurons of unc-3(e151) mutants. Results for three independent extrachromosomal arrays (otEx4946, otEx5410, sEx14990) are quantified on the right. Scale bar 10 µm. N >30 for otEx4946, otEx5410, and sEx14990 in WT and unc-3(e151) animals.
C: The protein expression of MADD-4B is significantly affected in the SAB neurons of unc-3(e151) mutants. Results for two independent extra-chromosomal arrays (kr1069 and kr1070 are MADD-4B translational fusions) are quantified on the right. Scale bar 10 µm. N > 33 for krEx1069 and krEx1070 in WT and unc-3(e151) animals. MADD-4B is also expressed in head ganglia, which appear out of focus to the right of the image.
D: The expression of MADD-4B transcriptional reporters is significantly affected in unc-3(e151) mutants. Results for three independent extra-chromosomal arrays (otEx5601, otEx5602, otEx5603) are quantified on the right. Scale bar 10 µm. N > 30 for otEx5601, otEx5602 and otEx5603 in WT and unc-3(e151) animals.
E: Schematic representation of the madd-4 locus. The location of the kr249 molecular lesion (W374stop) is indicated with a red triangle.
F: Mutational analysis of the cis-regulatory region of madd-4. Multiple transgenic lines were analyzed for each construct. See Table S2 for detailed quantification (number of animals and number of transgenic lines) of the promoter analysis data. Lines indicate genomic region fused to gfp (green). (+) indicates robust expression in the SAB neurons, or ventral nerve cord MNs, or head (at least 80% of the animals) in at least 2 independent transgenic lines. (+/−) indicates significant reduction in the number of neurons expressing the reporter gene or in the fluorescent intensity of the reporter gene in at least 50% of the animals (in at least 2 independent transgenic lines) when compared to transgenic animals carrying longer genomic fragments of the cis-regulatory region. (−) indicates complete loss of reporter gene expression in the SAB neurons, or ventral nerve cord MNs, or head in at least 90% of the animals (in at least 2 independent transgenic lines) when compared to transgenic animals carrying longer genomic fragments of the cis-regulatory region. (*) indicates that the COE motif (vertical light blue line) is conserved in at least 3 other nematode species. MUT indicates that COE motif has been mutated by always substituting the same 2 nucleotides in the core sequence (for example, wild-type COE site: TCCCNNGGGA >> COE MUT site: TGGCNNGGGA). Animals at the fourth larval (L4) stage were analyzed. The madd-4B-transcriptional reporter of 4366 bp cis-regulatory region does not contain the first exon of madd-4B.
As unc-3 and madd-4 null animals both display AChR clustering defects, we asked whether the synaptogenic defects of unc-3 mutants could be indicative of unc-3 regulating madd-4 expression. To this end, we first monitored the expression of MADD-4L and MADD-4B proteins in the SAB MNs of unc-3 mutants using fosmid-based translational gfp reporter strains that specifically indicate expression of each of these madd-4 isoforms [13] (Figure 3A,C,E). Unlike wild-type controls, animals lacking unc-3 gene activity fail to properly express MADD-4L and MADD-4B proteins in any of the SAB innervation zones (Figure 3A,C,F). The residual MADD-4L and MADD-4B expression in the SAB innervation zones of unc-3 mutants (Figure 3A,C) suggests that as yet unknown factors cooperate with unc-3 to induce madd-4 expression in the SAB neurons.
Next, we examined whether madd-4 may be a direct target of the UNC-3 protein by scanning the madd-4 locus for the occurrence of phylogenetically conserved UNC-3 binding sites, so-called COE motifs, using several bioinformatic tools (see Supplementary Methods for more details). This UNC-3 binding motif was defined through previous analysis of both C. elegans UNC-3 and its vertebrate orthologs [15, 16]. We found one COE motif (COE1) less than 3kb upstream of the start codon of the madd-4A isoform (which encodes the MADD-4L protein), three COE motifs (COE2–4) in the 5’ region of madd-4B and one COE motif (COE5) in the first intron of the madd-4B isoform (which encodes the MADD-4B protein) (Figure 3E). In the context of transgenic animals, small regulatory regions upstream of madd-4A that contain the COE1 motif (2882bp or 734 bp) are sufficient to drive expression of a reporter gene in SAB MNs (Figure 3F, Table S2) and, as expected, expression of these smaller fragments is unc-3 dependent (Figure 3 B, F). Mutation of this phylogenetically conserved COE1 motif in the context of a 734bp fragment (734bp COE1MUT) abrogates expression in the SAB neurons, while expression in other head neurons remains unaffected (Figure 3F, Table S2). We followed a similar strategy to analyze the cis-regulatory information of the madd-4B isoform. A 4.4 kb regulatory region that contains both the 1.9kb sequence upstream of the first madd-4B exon and the 2.5kb first intron is able to drive expression of a reporter gene in SAB MNs and other neuronal types (Figure 3D,F and Table S2) and this expression is unc-3-dependent (Figure 3D, F and Table S2). Mutating three or four COE motifs in this 4.4kb fragment diminishes expression in the SABV MNs (Figure 3F, Table S2). All together, these findings provide the first insights into the gene regulatory mechanisms that control expression of an anterograde synapse-organizing signal (madd-4).
Temporal and regulatory coupling of synaptogenesis and neurotransmitter signaling
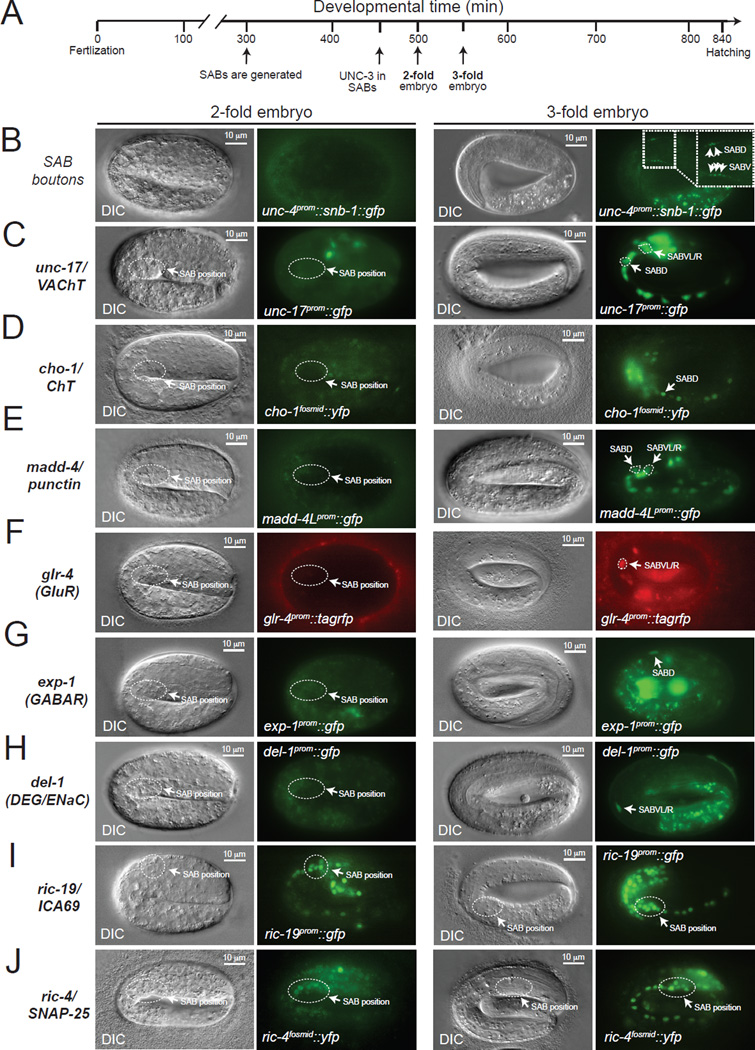
It is unknown in any system to date whether the process of structural organization of a specific synapse is directly coupled to the choice of the neurotransmitter that signals through this synapse. The SAB MNs are cholinergic, as determined by the expression of the evolutionarily conserved ACh pathway genes, unc-17/VAChT [17, 18], cha-1/ChAT [17] and cho-1/ChT [19] (Figure 4A–B). We first examined whether the adoption of cholinergic neurotransmitter identity is temporally linked to synaptogenesis. We found that the onset of expression of unc-17/cha-1 and cho-1 in the SAB neurons occurs at 3-fold stage embryos and coincides with the onset of madd-4 expression in these neurons (Figure 5C–E). Moreover, SAB presynaptic SNB-1 clusters can also be first observed in 3-fold stage embryos (Figure 5B). These findings indicate that cholinergic neurotransmitter identity and synaptogenesis are indeed temporally coupled.
Figure 5. Formation of SAB synapses coincides with the onset of acetylcholine pathway-, synaptogenic-, neurotransmitter receptor- and ion channel-encoding gene expression.
A: Schematic showing key events (SAB generation, onset of UNC-3 expression in SABs, 2-fold embryo, 3-fold embryo) relevant to this paper during C.elegans embryonic development. Developmental time is shown in minutes (min).
B: First synaptic boutons between the SAB MNs and head muscle (as visualized with SNB-1::GFP driven by the unc-4 gene promoter) are observed at 3-fold embryos. SABD and SABV synaptic boutons are shown with arrows (inset).
C–H: The onset of expression of unc-17/VAChT, cho-1/ChT, madd-4/Punctin, glr-4 (GluR), exp-1 (GABAR), and del-1 (DEG/ENaC) occurs at late 3-fold stage embryos. Transgenic animals carrying transcriptional reporters for unc-17 (vsIs48), cho-1 (otIs321), madd-4 (sEx14990), del-1 (wdIs3), exp-1 (wyIs75), and glr-4 (otIs476) were analyzed. These transcriptional reporters are likely to faithfully monitor the endogenous expression pattern of these genes as they contain large cis-regulatory fragments (See Table S6 for details on these reporter strains). To unambiguously distinguish the SAB MNs from other neurons in panels C, D and E, an unc-4prom::mCherry transgene was used as an SAB marker. Arrows indicate cell body position for SAB neurons.
I–J: Expression of synaptic genes shared by all neurons (ric-4 and ric-19) is detected at 2-fold stage embryos using ric-4/SNAP-25 (otIs350) and ric-19/ICA69 (otIs381) reporter strains. See Table S6 for details on these reporter strains.
On the left of each fluorescence image, a differential interference contrast (DIC) image of the worm embryo is shown. The relative position of the SAB cell bodies is shown (dotted circle) in 2-fold and 3-fold embryo panels. Scale bar, 10 µm.
Since synaptogenesis is unc-3-dependent, we asked whether adoption of the cholinergic phenotype of the SAB neurons is also unc-3 dependent. To this end, we crossed reporter transgenes that monitor expression of these cholinergic features (unc-17/cha-1 and cho-1) into an unc-3(e151) null mutant background. We found that the cholinergic phenotype of these neurons is lost in the absence of unc-3 (Figure 4B, C). The loss of acetylcholine signaling is not the cause of the structural synaptic defects of unc-3 mutants, since disruption of cholinergic neurotransmission does not affect assembly of SAB presynaptic structures [3]. Supporting this conclusion, we found that general synaptic transmission mutants (animals lacking the DAG/phorbol ester-binding protein unc-13, or the syntaxin-binding protein unc-18, or the synaptic vesicle-associated protein snb-1) do not display any major pre- or post-synaptic defects in the SAB neurons (Figure S3). Moreover, silencing of SAB neuronal activity through ectopic expression and activation of a histamine-gated chloride channel [20] did not lead to SAB synaptic defects but resulted, as expected, in head movement defects (Figure S3). Furthermore, unc-3 expression was unaffected in SAB neurons when general neurotransmission was abrogated (Figure S3). Taken together, these results suggest that unc-3-dependent synapse formation is an activity-independent process.
Other neuronal signaling features are also unc-3-dependent
The SAB neurons receive synaptic inputs from the GABAergic AVL inter/motorneuron [2] and express the GABA-gated ion channel EXP-1 (Figure 4A, F and Figure S4) [4]. SAB neurons also receive glutamatergic synaptic inputs as asssessed by expression of ionotropic glutamate receptor subunits encoded by the glr-4 and glr-5 loci (Figure 4 A, F and Figure S4) [5]. We asked whether SAB neurons coordinate the formation of their NMJs (via MADD-4) and their cholinergic phenotype with synaptic input-defining neurotransmitter receptor choice. We found this to indeed be the case. First, we found that the onset of expression of these neurotransmitter receptors (glr-4 and exp-1) at the embryonic 3-fold stage is similar to the onset of synaptogenic and neurotransmitter choice genes (Figure 5C–G). Second, the basis of this temporal coordination again seems to be coregulation by unc-3, since the expression of all three ionotropic neurotransmitter receptors, exp-1, glr-4 and glr-5 is lost in unc-3 mutant animals (Figure 4F, G and Figure S4), and unc-3 expression precedes the expression of these genes (Figure 5A and data not shown). The SAB neurons are also known to express two ion channels of the DEG/ENaC family, del-1 and unc-8, which have been speculated to form stretch-sensitive heteromeric ion channels [7, 21], a notion that would be consistent with a putative proprioceptive function of the SAB neurons. We found that unc-8 and del-1 reporter gene expression was significantly affected in the SAB neurons of unc-3 mutants (Figure 4F,G). Consistent with unc-8 and del-1 being transcriptional targets of unc-3, we found that the number of del-1 and unc-8 mRNA molecules is significantly reduced in the SAB neurons of unc-3 mutants (Figure S1). The onset of del-1 and unc-8 expression is co-incident with the onset of expression of neurotransmitter and synaptic genes at 3-fold stage embryos (Figure 5H and [21]), demonstrating that del-1- and unc-8-mediated signaling is temporally coordinated with neurotransmitter signaling and synaptogenesis. Consistent with a putative role of del-1 and unc-8 in proprioception and unc-3 controlling the expression of these molecules, unc-3, del-1 and unc-8 mutant animals show similar defects in the manner they swing their head, as assessed by automated worm tracking (Figure S5) [22].
The synaptogenic (madd-4/punctin) and neuronal signaling proteins (unc-17/VAChT, cho-1/ChT, glr-4/GluR, glr-5/GluR, exp-1/GABAR, del-1/DEG, unc-8/DEG) described above are expressed in a neuron type-specific manner. unc-3 regulates these neuron-type specific synaptogenic and signaling features, as described above, but unc-3 does not regulate the expression of synaptic signaling molecules that are shared by all neuron types. Specifically, we examined the expression of four panneuronally expressed genes: i) ric-4 which encodes the ortholog of the synaptosomal protein SNAP-25 [23], ii) unc-10 which encodes the ortholog of the presynaptic protein RIM1 [24], iii) snb-1 which encodes synaptobrevin, a synaptic-vesicle associated protein orthologous to human vesicle-associated membrane protein 1 and 2 (VAMP1–2) [25], and iv) ric-19 which encodes the ortholog of the human diabetes autoantigen ICA69, involved in dense core vesicle maturation [26]. We find that expression of these four genes is unaffected in the SAB MNs of unc-3 mutants (Figure 4D–E and Figure S1). Moreover, we observe an earlier onset of expression of these synaptic signaling genes that are shared by all neurons, such as ric-4 and ric-19 (Figure 5I, J), when compared with signaling molecules expressed in a neuron type-specific manner (unc-17, cho-1, glr-4, del-1, exp-1) (Figure 5C,D,F–H). These results demonstrate that the adoption of neuron type-specific synaptic features can be temporally separated from the adoption of generic synaptic features. These findings also show that the absence of expression of neuron type-specific features in unc-3 mutants (Figure 4C,F,G) is not merely an indication of a failure to generate the SAB neurons; rather, the SAB neurons are generated in unc-3 mutants, adopt neuron-like features but fail to adopt neuron type-specific (SAB-specific) signaling properties.
Transcriptional coregulation of synaptic organization and neuron type-specific signaling features
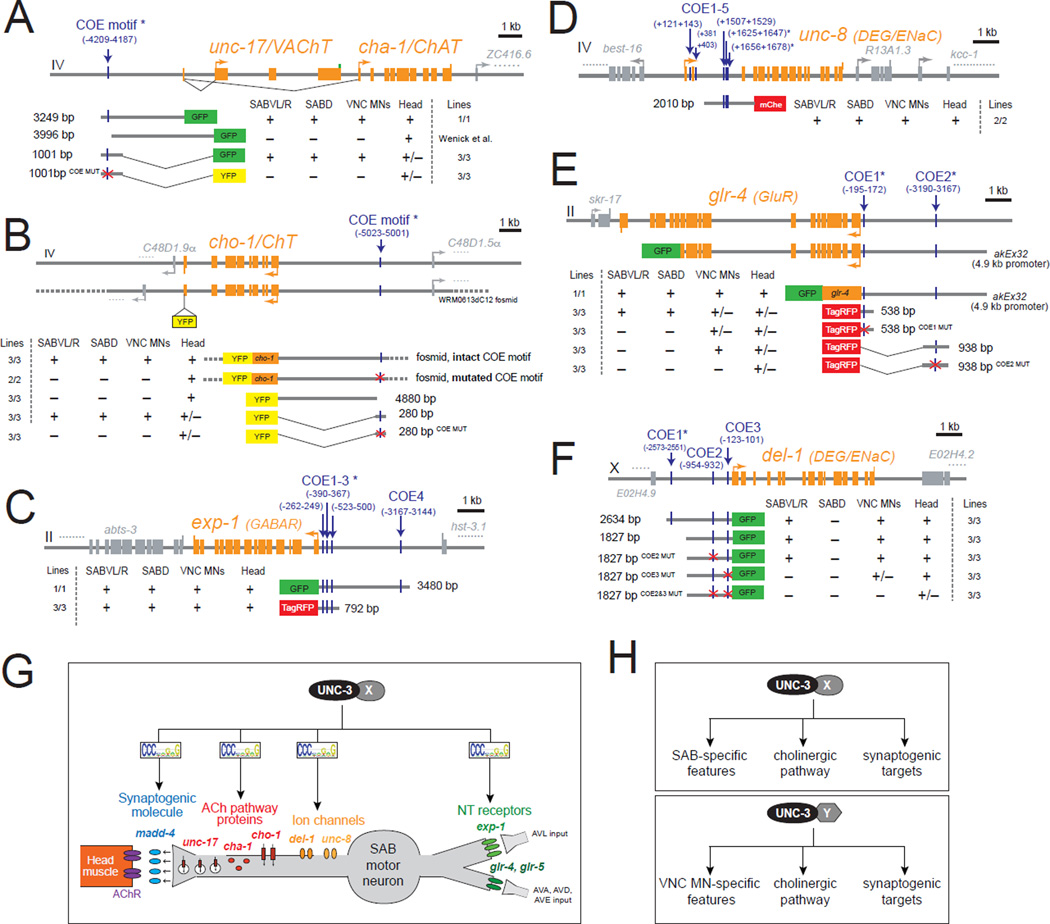
To understand the mechanistic basis of the coordination of SAB synaptogenesis and SAB-specific signaling features by UNC-3, we inspected the loci encoding the neuronal signaling genes (cholinergic pathway genes, neurotransmitter receptors, putative stretch receptors) for the presence of COE motifs, the UNC-3 binding site. We found at least one copy of the COE motif in the cis-regulatory region of all seven unc-3-dependent, SAB-expressed neuronal signaling genes. Most of these COE motifs are phylogenetically conserved in other nematode species (Table S3). We focused on 6 of these 7 genes (the cholinergic identity determinants unc-17/cha-1 and cho-1, the DEG/ENaC family of ion channel-encoding genes del-1 and unc-8, and the ionotropic neurotransmitter receptors glr-4 and exp-1) and performed mutational analysis in the context of transgenic animals. In the case of the cholinergic genes unc-17/cha-1, and cho-1, we found that small regulatory regions (unc-171001bp prom::gfp and cho-1280bp prom::yfp) that contain the COE motif are sufficient to drive expression of a reporter gene in the SAB MNs (Figure 6A–B and Table S4) and mutation of the COE motif in these regulatory regions resulted in a loss of expression in the SAB MNs while expression in other head neurons remained unaltered (Figure 6A–B and Table S4). Mutation of the COE motif in the context of a large (~30kb) fosmid-based reporter of cho-1 results in loss of reporter gene expression in the SAB neurons (Figure 6B and Figure S6), suggesting that a single COE motif is essential for cho-1 expression in these neurons.
Figure 6. unc-3 directly controls the expression of genes required for SAB neuronal signaling.
A–F: Cis-regulatory mutational analysis is shown for genes expressed in SAB neurons [unc-17/VAChT, cho-1/ChT, exp-1(GABAR), unc-8 (DEG/ENaC), glr-4 (GluR), del-1(DEG/ENaC)]. Multiple transgenic lines were analyzed for each construct. See Table S4 for detailed quantification of the promoter analysis data. Lines indicate genomic region fused to gfp (green), tagrfp (red), or yfp (yellow). (+) indicates robust expression in the SAB neurons, or ventral nerve cord MNs, or head (at least 70% of the animals) in at least 2 independent transgenic lines. (+/−) indicates significant reduction in the number of neurons expressing the reporter gene in at least 50% of the animals (in at least 2 independent transgenic lines) when compared to transgenic animals carrying longer genomic fragments of the cis-regulatory region. (−) indicates complete loss or faint expression in the SAB neurons, or ventral nerve cord MNs, or head in at least 50% of the animals (in at least 2 independent transgenic lines) when compared to transgenic animals carrying longer genomic fragments of the cis-regulatory region. (*) indicates that the COE motif (vertical light blue line) is conserved in at least 3 other nematode species. MUT indicates that COE motif has been mutated by always substituting the same 2 nucleotides in the core sequence (for example, COE wild-type site: TCCCNNGGGA >> COE MUT site: TGGCNNGGGA). See also Figure S6 and Table S3.
G: Schematic that summarizes the key findings of this paper. The transcription factor unc-3 (likely together with unknown factors “X”) regulates synaptogenesis by controlling madd-4/Punctin expression, which is a pre-synaptically secreted molecule required for AChR clustering in head muscle. UNC-3 also coregulates the expression of ACh pathway genes (unc-17/cha-1, cho-1), ionotropic neurotransmitter receptor (NT) genes (glr-4, glr-5, exp-1), and ion channel genes (del-1, unc-8). The core sequence (8 nucleotide positions) of the sequence logo of the UNC-3 binding site (COE motif) is shown to indicate direct regulation by UNC-3. The entire sequence logo of the COE motif is presented in Table S3 and was generated by combining 22 COE motifs found in all SAB-expressed genes shown here. The post-synaptic target of SAB MNs is head muscle (depicted on the left). SAB MNs receive input form the AVL interneuron, as well as the command interneurons (AVA, AVD, AVE) depicted on the right.
H: unc-3 likely cooperates with distinct regulatory factors (factor X in SAB neurons, factor Y in VNC motor neurons) to activate the expression of MN type-specific target genes, as well as the expression of shared genes such as cholinergic pathway genes and synaptogenic genes.
The cis-regulatory region of del-1 contains multiple COE motifs. A 1.8 kb cis-regulatory fragment that contains two COE motifs (COE2&COE3) is able to drive reporter gene expression in the ventral SAB neurons and other ventral nerve cord MNs (VA, VB) (Figure 6F and Table S4). While mutation of one COE motif (1827 bp COE2MUT) had no effect in reporter gene expression, mutation of the other COE motif (1827 bp COE3MUT) resulted in complete loss of expression in the SABV MNs and partial loss in ventral nerve cord MNs, which also express unc-3. Only the combined mutation of both COE motifs in the context of this 1.8 kb fragment (1827 bp COE2&3MUT) results in loss of expression in ventral nerve cord MNs (Figure 6F and Table S4). Moreover, the cis-regulatory region of unc-8 contains several COE motifs and a 2kb intronic fragment that has 3 COE motifs is able to drive reporter gene expression in the SAB as well as the ventral nerve cord MNs (Figure 6D and Table S4).
Similar to del-1 and unc-8, the cis-regulatory region of the ionotropic glutamate receptor-encoding gene glr-4 contains multiple COE motifs (Figure 6E and Table S4). A translational glr-4 gfp fusion that includes 4.9kb of cis-regulatory elements immediately upstream of ATG as well as most of the glr-4 genomic locus (lacking only the last 6 exons) contains two COE motifs and is expressed in SAB and ventral nerve cord MNs (DB and VB classes) (Figure 6 E and Table S4). However, a smaller fragment (538 bp) that contains only one COE motif drives expression in SAB neurons and other head neurons, but not ventral nerve cord neurons. Mutation of the COE motif in this 538 bp regulatory fragment (538 bp COE1MUT) specifically affects reporter gene expression in the SAB neurons (Figure 6 E and Table S4). Notably, another glr-4 regulatory fragment (938 bp) that contains a COE motif is sufficient to drive expression in the ventral nerve cord neurons but not in SAB MNs, and mutation of that COE motif (938 bp COE2MUT) abolishes ventral nerve cord neuronal expression (Figure 6E and Table S4). Since these glr-4 regulatory fragments (538 bp and 938 bp) both contain a COE motif and yet show distinct expression pattern in different classes of MNs (SAB versus ventral nerve cord MNs), we hypothesize that UNC-3 may operate with distinct, yet unknown factors to generate MN diversity. Similar to glr-4, the cis-regulatory region of the ionotropic GABA receptor-encoding gene exp-1 contains four COE motifs. A small regulatory fragment (792 bp) that contains three of these evolutionarily conserved motifs (COE 1–3) is sufficient to drive expression in SAB MNs (Figure 6C and Table S4). Taken together, our results suggest that in the SAB class of MNs synaptogenesis, cholinergic neurotransmitter identity and other signaling features (e.g. expression of ionotropic glutamate and GABA receptors) are co-regulated through the transcription factor UNC-3 and its cognate binding site, the COE motif.
Identification of more unc-3 target genes
To assess how broadly unc-3 affects SAB differentiation, we embarked on a small-scale screen to identify novel terminal identity genes expressed in SAB MNs. Since SAB-expressed genes, such as glr-4, unc-8, and del-1, are also expressed in other MN subtypes in the C.elegans ventral nerve cord (VNC) (see Figure 4A), we obtained from the Caenorhabditis Genetics Center (CGC) 30 transgenic reporter worm strains that have been reported to be expressed in VNC MNs and found two of them to be expressed in SAB neurons, a receptor tyrosine kinase (encoded by ddr-2) and a TGFβ-like molecule (encoded by unc-129). Both genes are expressed in a restricted number of neuron types, including the SAB neurons and their expression in the SAB neurons is maintained throughout the life of the animal. In unc-3 mutants, the expression of ddr-2 and unc-129 is lost in the SAB MNs (Figure S7). ddr-2 and unc-129 contain well-conserved COE motifs in their cis-regulatory control regions. When we mutated these motifs in the context of a 395bp fragment derived from the unc-129 locus, we found that expression in SAB MNs was lost, providing further evidence for direct regulation of unc-129 by unc-3 (Figure S7). Taken together, unc-3 affects all known terminal identity features of mature SAB neurons.
Coupling of neurotransmitter identity and synaptogenesis in other motor neuron types
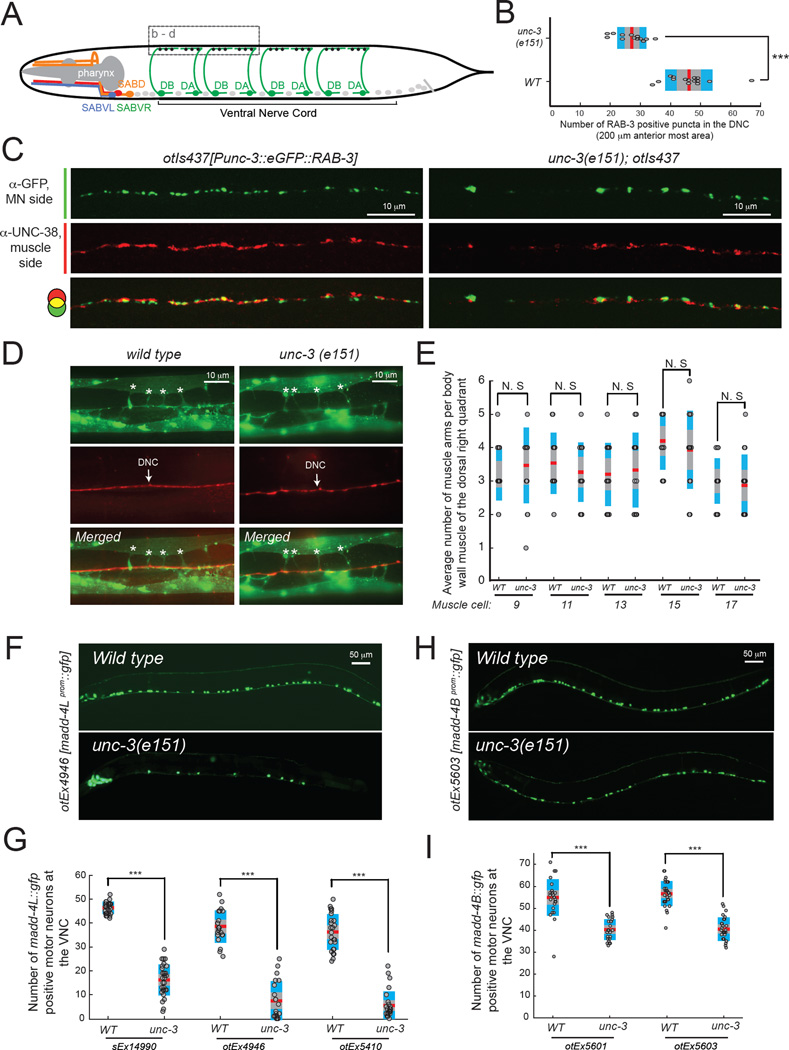
To examine whether the coupling of neurotransmitter choice and synaptogenesis is a common theme in MN differentiation, we turned to cholinergic MNs in the ventral nerve cord (VNC), which form synapses with body wall muscles along the body of the animal. We have previously shown that unc-3 controls cholinergic neurotransmitter identity in multiple distinct types of VNC MNs [15]. By visualizing synaptic specializations in wild type and unc-3 mutant animals, we found a significant reduction in the number of presynaptic specializations in unc-3 mutants in VNC MNs (Figure 7A–C). This reduction in the number of VNC synapses is not a reflection of a general neuromuscular defect as we found that muscle arm development and extension occurs normally in unc-3 mutants (Figure 7D–E). Similarly to the SAB MNs, unc-3 affects the expression of both madd-4 isoforms in VNC MNs (Figure 7F–I). We conclude that unc-3 coordinates synaptogenesis and cholinergic neurotransmitter identity in distinct MN types of the C.elegans motor circuit.
Figure 7. unc-3 coordinates synaptogenesis and neurotransmitter choice in ventral nerve cord (VNC) cholinergic motor neurons.
A: For simplicity, the axons of four DA and four DB cholinergic VNC MNs are shown in green. In total, there are 9 DA and 7 DB MNs along the C.elegans VNC. The cell bodies of the rest of cholinergic VNC MNs (VA, VB, AS, VC) are shown as grey dots. The synapses of DA and DB MNs onto dorsal body wall muscle (BWM) are highlighted with black dots. Dotted box shows the region of the dorsal nerve cord where synapses were analyzed in Figure 7 B–D. Dorsal BWM is not shown.
B: The number of presynaptic specializations of the DA and DB MNs onto dorsal BWM is significantly reduced in unc-3(e151) animals. Presynapse was visualized by driving RAB-3 protein expression (fused to gfp) in DA and DB MNs using the unc-3 promoter (transgene otIs437). Presynaptic boutons were counted in a 200 µm region located at the most anterior part of the dorsal nerve cord (DNC). *** : p value < 0.001. N = 15. Mean values are represented with a vertical red line.
C: Double immunofluorescence staining for gfp and the AChR subunit UNC-38 allows to visualize the presynaptic specializations of DA and DB MNs (RAB-3::GFP, green signal), as well as the post-synaptic specializations of dorsal BWM (UNC-38, red signal) in WT and unc-3(e151) animals.
D–E: The average number of muscle arms (asterisks) per BWM of the dorsal right quadrant was evaluated in WT and unc-3(e151) animals. The transgenic strain trIs30 allows for visualization of the muscle cell in yellow (him-4prom::MB::YFP) and the dorsal nerve cord in red (hmr-1bprom::DsRed2 and unc-129prom::DsRed2). No significant differences in the number of muscle arms per BWM between WT and unc-3(e151) animals were observed. Five different muscle cells (#9, #11, #13, #15, #17) were analyzed. N = 10. N.S, not significant. Mean values are represented with a horizontal red line.
F–I: The expression of madd-4L (F) and madd-4B (H) transcriptional reporters is significantly affected in VNC cholinergic MNs of unc-3(e151) mutants. Scale bar 50 µm. Anterior is to the left. Results for three independent transgenic madd-4L reporter lines (otEx4946, otEx5410, sEx14990) are quantified in G. Red horizontal lines represent the mean value. N > 20 for otEx4946, otEx5410, and sEx14990 in WT and unc-3(e151) animals. Results for two independent transgenic madd-4B reporters lines (otEx5601, otEx5603) are quantified in I. Red horizontal lines represent the mean value. N = 25 – 30 for otEx5601 and otEx5603 in WT and unc-3(e151) animals. *** : p value < 0.001.
DISCUSSION
This study describes a gene regulatory program that operates in a previously little studied C. elegans head MN class to instruct the adoption of a specific set of function-defining neuronal signaling features. While it has become clear that the acquisition of biochemically linked features of a neuron, such as the expression of enzymes and transporters for the synthesis and synaptic vesicle transport of a specific neurotransmitter, is coordinated by transcriptional coregulatory mechanisms [15, 27–29], other questions of coordination of neuronal signaling features have remained poorly explored. Specifically, it was previously not known whether synapse formation is coordinated with neurotransmitter choice and whether the expression of a neurotransmitter system that a neuron uses to signal to postsynaptic targets is coordinated with the expression of neurotransmitter receptors through which the same neuron receives signaling inputs (Figure 1). The results shown here demonstrate that the adoption of these distinct properties of a neuron is temporally coordinated and that this coordination is achieved via a transcriptional coregulatory mechanism (Figure 6G). In this coregulatory routine, unc-3 controls the expression of a synaptogenic molecule, MADD-4, that organizes postsynaptic acetylcholine receptor clustering; the expression of genes required for acetylcholine synthesis and transport (unc-17/cha-1, cho-1); and the expression of neurotransmitter receptors (exp-1, glr-4, glr-5) that receive inputs from neurons that innervate the SAB neurons (Figure 6G).
Animals lacking unc-3 gene activity not only display postsynaptic receptor clustering defects in head muscle, but also show severe defects in presynaptic organization. We therefore surmise that unc-3 also regulates the expression of a protein that assembles presynaptic machinery. In the HSN MNs of C.elegans, an immunoglobulin superfamily member has such presynaptic organizing function [30, 31] and we hypothesize that an analogously acting protein may operate at the SAB NMJs and may be under the control of unc-3. We can exclude the possibility that the presynaptic structural defects in unc-3 mutants are a mere secondary consequence of the loss of cholinergic phenotype observed in unc-3 mutants, because the presynaptic structures of SAB axons appear normal in animals in which synaptic signaling is genetically disrupted [3] (this manuscript).
The results presented here go beyond the demonstration that synaptogenesis and neurotransmitter signaling in a specific neuron type are temporally coordinated through a single transcription factor. We further show that this same transcription factor also regulates the expression of two ion channel-encoding genes (del-1 and unc-8) of the DEG/ENaC family of proteins (Figure 4F–G), which have been proposed to be stretch-sensitive, sensory channels [7, 21]. Together these findings demonstrate that distinct function-defining, yet not biochemically linked hallmarks of a postmitotic neuron – synaptogenesis, neurotransmitter input and output, and sensory channel expression – are temporally and transcriptionally coregulated (Figure 6G) and, hence, organized in a “regulon” akin to classic bacterial biosynthetic pathways [32, 33]. Such organization into a regulon ensures that all the features that are required together will appear together. Expressing neurotransmitter synthesizing enzymes and transporters at a different time than building a synapse would be wasteful. Synthesis of neurotransmitter receptors that receive synaptic input (or synthesis of other receptor systems that activate a neuron, such as the putative stretch receptors UNC-8 or DEL-1) is pointless if the neuron does not have the means to transmit this signal via a neurotransmitter to downstream effector cells (muscle in this case).
Previous observations in other systems point to a role of neuronal activity in synaptogenesis and in the regulation of neurotransmitter and neurotransmitter receptor expression (reviewed in [34–37]). Our finding of a “hardwired” coordination of synaptogenesis, neurotransmitter signaling, and ion channel expression through a developmental neuron type-specific transcription factor in a simple invertebrate organism suggests that activity-dependent mechanisms of synaptic and signaling properties may have been superimposed onto a more ancient hard-wired regulatory program that may still operate in many instances in the vertebrate nervous system.
EXPERIMENTAL PROCEDURES
Experimental Procedures can be found in Supplemental Information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nikolaos Stefanakis for kindly providing panneuronal and cho-1 fosmid-based reporter strains, Ines Carrera for providing panneuronal reporter strains, Michael Hart for providing unc-8 reporter strains, Yishi Jin for providing juIs40 strain, Navin Pokala and Cornelia Bargmann for providing the kyEx5161 transgenic animals, Sarah Petersen, Tyne Miller and David Miller for generating and providing the wdEx948 transgenic animals, Qi Chen for expert assistance in generating strains, Alexander Boyanov for whole genome sequencing, Marie Pierron for analysis of whole genome sequencing data, Lori Glenwinkel for bioinformatic analysis, Ev Yemini for expert assistance on automated worm tracking and statistical analysis, and members of the Hobert lab for comments on the manuscript. We are grateful to Caenorhabditis Genetics Center (University of Minnesota) for providing strains. This work was funded by the National Institutes of Health [R01NS039996-05 and R01NS050266-03 to O.H.; 1K99NS084988-01 to P.K.], the Howard Hughes Medical Institute, the Association Française contre les Myopathies (post-doctoral fellowship to B.P.-L.), the Programme Avenir Lyon Saint-Etienne (doctoral fellowship to A.W.), and the Agence Nationale de la Recherche (ANR-11-BSV4-019 to J-L.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication., As a service to our customers we are providing this early version of the manuscript., The manuscript will undergo copyediting, typesetting, and review of the resulting proof, before it is published in its final citable form. Please note that during the production, process errors may be discovered which could affect the content, and all legal disclaimers, that apply to the journal pertain.
Author contributions
P.K and S.Y.K performed experiments under the supervision of O.H. B.P.L and A.W performed experiments under the supervision of J.L.B. P.K and O.H wrote the manuscript. All authors edited the manuscript.
Contributor Information
Paschalis Kratsios, Email: pk2357@columbia.edu.
Oliver Hobert, Email: or38@columbia.edu.
References
- 1.Von Stetina SE, Treinin M, Miller DM., 3rd The motor circuit. Int. Rev. Neurobiol. 2006;69:125–167. doi: 10.1016/S0074-7742(05)69005-8. [DOI] [PubMed] [Google Scholar]
- 2.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 3.Zhao H, Nonet ML. A retrograde signal is involved in activity dependent remodeling at a C. elegans neuromuscular junction. Development. 2000;127:1253–1266. doi: 10.1242/dev.127.6.1253. [DOI] [PubMed] [Google Scholar]
- 4.Pinan-Lucarre B, Tu H, Pierron M, Cruceyra PI, Zhan H, Stigloher C, Richmond JE, Bessereau JL. C. elegans Punctin specifies cholinergic versus GABAergic identity of postsynaptic domains. Nature. 2014;511:466–470. doi: 10.1038/nature13313. [DOI] [PubMed] [Google Scholar]
- 5.Beg AA, Jorgensen EM. EXP-1 is an excitatory GABAgated cation channel. Nat. Neurosci. 2003;6:1145–1152. doi: 10.1038/nn1136. [DOI] [PubMed] [Google Scholar]
- 6.Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J. Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tavernarakis N, Driscoll M. Molecular modeling of mechanotransduction in the nematode Caenorhabditis elegans. Annu. Rev. Physiol. 1997;59:659–689. doi: 10.1146/annurev.physiol.59.1.659. [DOI] [PubMed] [Google Scholar]
- 8.Driscoll M, Tavernarakis N. Molecules that mediate touch transduction in the nematode Caenorhabditis elegans. Gravit. Space Biol. Bull. 1997;10:33–42. [PubMed] [Google Scholar]
- 9.Richard M, Boulin T, Robert VJ, Richmond JE, Bessereau JL. Biosynthesis of ionotropic acetylcholine receptors requires the evolutionarily conserved ER membrane complex. Proc. Natl. Acad. Sci. USA. 2013;110:E1055–E1063. doi: 10.1073/pnas.1216154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prasad BC, Ye B, Zackhary R, Schrader K, Seydoux G, Reed RR. unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development. 1998;125:1561–1568. doi: 10.1242/dev.125.8.1561. [DOI] [PubMed] [Google Scholar]
- 11.Hallam SJ, Goncharov A, McEwen J, Baran R, Jin Y. SYD-1, a presynaptic protein with PDZ, C2 and rhoGAP-like domains, specifies axon identity in C. elegans. Nat. Neurosci. 2002;5:1137–1146. doi: 10.1038/nn959. [DOI] [PubMed] [Google Scholar]
- 12.Patel MR, Lehrman EK, Poon VY, Crump JG, Zhen M, Bargmann CI, Shen K. Hierarchical assembly of presynaptic components in defined C. elegans synapses. Nat. Neurosci. 2006;9:1488–1498. doi: 10.1038/nn1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seetharaman A, Selman G, Puckrin R, Barbier L, Wong E, D’Souza SA, Roy PJ. MADD-4 is a secreted cue required for midline oriented guidance in Caenorhabditis elegans. Dev. Cell. 2011;21:669–680. doi: 10.1016/j.devcel.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 15.Kratsios P, Stolfi A, Levine M, Hobert O. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 2012;15:205–214. doi: 10.1038/nn.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treiber T, Mandel EM, Pott S, Györy I, Firner S, Liu ET, Grosschedl R. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription-independent poising of chromatin. Immunity. 2010;32:714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Duerr JS, Han HP, Fields SD, Rand JB. Identification of major classes of cholinergic neurons in the nematode Caenorhabditis elegans. J. Comp. Neurol. 2008;506:398–408. doi: 10.1002/cne.21551. [DOI] [PubMed] [Google Scholar]
- 18.Lickteig KM, Duerr JS, Frisby DL, Hall DH, Rand JB, Miller DM., 3rd Regulation of neurotransmitter vesicles by the homeodomain protein UNC-4 and its transcriptional corepressor UNC-37/groucho in Caenorhabditis elegans cholinergic motor neurons. J. Neurosci. 2001;21:2001–2014. doi: 10.1523/JNEUROSCI.21-06-02001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullen GP, Mathews EA, Vu MH, Hunter JW, Frisby DL, Duke A, Grundahl K, Osborne JD, Crowell JA, Rand JB. Choline transport and de novo choline synthesis support acetylcholine biosynthesis in Caenorhabditis elegans cholinergic neurons. Genetics. 2007;177:195–204. doi: 10.1534/genetics.107.074120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pokala N, Liu Q, Gordus A, Bargmann CI. Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. Proc. Natl. Acad. Sci. USA. 2014;111:2770–2775. doi: 10.1073/pnas.1400615111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tavernarakis N, Shreffler W, Wang S, Driscoll M. unc-8, a DEG/ENaC family member, encodes a subunit of a candidate mechanically gated channel that modulates C. elegans locomotion. Neuron. 1997;18:107–119. doi: 10.1016/s0896-6273(01)80050-7. [DOI] [PubMed] [Google Scholar]
- 22.Yemini E, Jucikas T, Grundy LJ, Brown AE, Schafer WR. A database of Caenorhabditis elegans behavioral phenotypes. Nat. Methods. 2013;10:877–879. doi: 10.1038/nmeth.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc. Natl. Acad. Sci. USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koushika SP, Richmond JE, Hadwiger G, Weimer RM, Jorgensen EM, Nonet ML. A post-docking role for active zone protein Rim. Nat. Neurosci. 2001;4:997–1005. doi: 10.1038/nn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nonet ML, Saifee O, Zhao H, Rand JB, Wei L. Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J. Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sumakovic M, Hegermann J, Luo L, Husson SJ, Schwarze K, Olendrowitz C, Schoofs L, Richmond J, Eimer S. UNC-108/RAB-2 and its effector RIC-19 are involved in dense core vesicle maturation in Caenorhabditis elegans. J. Cell Biol. 2009;186:897–914. doi: 10.1083/jcb.200902096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Eastman C, Horvitz HR, Jin Y. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J. Neurosci. 1999;19:6225–6234. doi: 10.1523/JNEUROSCI.19-15-06225.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu. Rev. Cell Dev. Biol. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- 31.Shen K, Bargmann CI. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell. 2003;112:619–630. doi: 10.1016/s0092-8674(03)00113-2. [DOI] [PubMed] [Google Scholar]
- 32.Shen K, Fetter RD, Bargmann CI. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell. 2004;116:869–881. doi: 10.1016/s0092-8674(04)00251-x. [DOI] [PubMed] [Google Scholar]
- 33.Maas WK, Clark AJ. Studies on the mechanism of repression of arginine biosynthesis in Escherichia coli: II. Dominance of repressibility in diploids. J. Mol. Biol. 1964;8:365–370. doi: 10.1016/s0022-2836(64)80200-x. [DOI] [PubMed] [Google Scholar]
- 34.Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Polleux F, Ince-Dunn G, Ghosh A. Transcriptional regulation of vertebrate axon guidance and synapse formation. Nat. Rev. Neurosci. 2007;8:331–340. doi: 10.1038/nrn2118. [DOI] [PubMed] [Google Scholar]
- 36.Shen K, Scheiffele P. Genetics and cell biology of building specific synaptic connectivity. Annu. Rev. Neurosci. 2010;33:473–507. doi: 10.1146/annurev.neuro.051508.135302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfram V, Baines RA. Blurring the boundaries: developmental and activity-dependent determinants of neural circuits. Trends Neurosci. 2013;36:610–619. doi: 10.1016/j.tins.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.