Abstract
Direct reprogramming of adult fibroblasts to a pluripotent state has opened new possibilities for the generation of patient- and disease-specific stem cells. However the ability of induced pluripotent stem (iPS) cells to generate tissue that mediates functional repair has been demonstrated in very few animal models of disease to date. Here we present the proof of principle that iPS cells may be used effectively for the treatment of muscle disorders. We combine the generation of iPS cells with conditional expression of Pax7, a robust approach to derive myogenic progenitors. Transplantation of Pax7-induced iPS-derived myogenic progenitors into dystrophic mice results in extensive engraftment, which is accompanied by improved contractility of treated muscles. These findings demonstrate the myogenic regenerative potential of iPS cells and provide rationale for their future therapeutic application for muscular dystrophies.
Keywords: iPS cells, Pax7, Skeletal muscle progenitors, Muscular dystrophy
Introduction
The ability to self-renew and differentiate into all somatic cell types make pluripotent embryonic stem (ES) cells an attractive cell source for therapeutic applications. The recent derivation of induced pluripotent stem (iPS) cells from skin fibroblasts through reprogramming technology [1–3] brings this type of therapy much closer to reality since it allows for the derivation of patient-specific iPS cells, eliminating the ethical and immunological issues associated with ES cells. iPS cells have been shown by several investigators to behave very similarly to ES cells in terms of pluripotency [1–7], best evidenced by their ability to form teratomas. Although many issues need to be addressed before iPS cells can safely be applied therapeutically, including bypassing the use of oncogenes as reprogramming factors, and eliminating the use of retroviral vectors that carry the risk of mutagenesis, the rapid pace of research advance in this field provides rationale for the immediate assessment of the regenerative potential of these cells. For instance, several approaches to generate iPS cells without viral integration into the genome have been reported within the last few years, including the use of safer transient vectors [8–13], or alternatively the transduction of recombinant proteins [14, 15]. One caveat for these methods has been the low efficiency of reprogramming. In this regard, a recent study has demonstrated safe and efficient generation of human iPS cells by the use of synthetic modified mRNA [16], suggesting that this approach might be valuable for future cell-based therapies.
Another fundamental requirement for therapeutic application is the generation of abundant tissue-specific, tumor-free, cell populations that are able to integrate and properly function in the host following transplantation. This has been accomplished for iPS-derived hematopoietic, endothelial, neural, pancreatic, and liver precursor cells, which have had their therapeutic potential assessed in the respective models of disease, sickle cell anemia [17], hemophilia A [18], Parkinson’s disease and spinal cord injury [19, 20], diabetes [21], and deficiency in fumarylacetoacetate hydrolase [22]. However to date, there has been no report of functional skeletal muscle engraftment following the transplantation of iPS-derived progenitors using a mouse model of muscular dystrophy. Although one group has indicated the derivation of myogenic progenitors from iPS cells [23], the nature and function of engrafted cells have not been rigorously studied.
In reality, the derivation of therapeutic skeletal myogenic progenitors from ES cells is challenging since this lineage is inefficiently produced during in vitro differentiation of ES cells into embryoid bodies (EBs). This deficit is due to improper somatogenesis and patterning of paraxial mesoderm within the EB system. In the embryo these processes culminate with activation of the myogenic program, evidenced by up-regulation of the Myogenic Regulatory Factors (MRFs) Myf5, Mrf4, and MyoD, [24, 25], a process led by paired-box transcription factors Pax3 and Pax7 [26–28], key players of embryonic muscle specification [29, 30] and postnatal muscle regeneration [31–33] respectively. By conditional expression of Pax3 [34] or Pax7 [35] in the appropriate developmental window in vitro, we are able to promote efficient myogenesis in vitro in spite of the absence of somitogenesis or paraxial mesoderm patterning cues, eg. notochord and neural tube, during EB differentiation. This results in highly efficient generation of therapeutic myogenic progenitors, which when transplanted into dystrophic mice produce large quantities of functional skeletal muscle tissue that incorporates normally into the host muscle [34].
In the present study, we assessed the myogenic potential of iPS cells obtained from mice bearing inducible expression of Pax7. Our results demonstrate that Pax7-induced iPS cells give rise to a proliferating population of myogenic progenitors that when injected into dystrophic mice promote not only substantial muscle regeneration, but also contribute to improvement of the contractile properties of engrafted muscles. These findings confirm the regenerative potential of iPS cells and give rationale for their future therapeutic application in muscular dystrophies.
Material and Methods
Generation of Inducible Pax7 iPS Cell Lines
An inducible Pax7 ES cell line was generated by targeting Pax7 into the inducible locus of A2Lox.cre ES cells, an improved version of A2Lox [36], in which cre is present at the doxycycline-inducible locus before recombination and catalyzes its replacement by the incoming gene (inducible cassette exchange, ICE, (Iacovino et al., submitted and [37])). Following recombination, the locus bearing the inducible transgene is identical regardless of which cell line (A2Lox or ICE) was used to generate it. Mice were derived from the iPax7 ES cells through blastocyst injection, crossed to C57/BL6 to establish a founder bearing the iPax7 gene, and then bred to Rosa-rtTA2SM2 mice [38] to establish an inducible Pax7 mouse.
ICE iPS cells were derived from ICE mice (Iacovino et al., submitted) bearing the same inducible cassette exchange locus, and additionally carrying a ubiquitously expressed GFP transgene [39]. Tail tip fibroblasts (TTF) isolated from 2 to 3 week old iPax7 or ICE mice were used to generate iPS cells through retroviral transduction with Oct3/4, Sox2 and Klf-4 [1]. One week after spin infection, 10% FBS IMDM medium was switched to mouse ES medium, and 1 to 2 weeks later, iPS colonies were picked individually and clonally expanded. ICE iPS cells were then targeted using a p2Lox-Pax7 and selected in G418. Tet-inducible expression of Pax7 for both cell lines was assessed by western blot using a monoclonal anti-Pax7 antibody (R&D Systems).
In vitro Characterization of iPS Cells
Following initial expansion, several iPS clones were then characterized for SSEA-1 expression by flow cytometry, as well as Nanog and Alkaline Phosphatase expression by immunostaining. Selected iPS clones were subjected to karyotype analysis by G-banding.
Growth and Differentiation of iPS Cells
iPS cells were maintained and differentiated similarly to ES cells as described [34]. To induce Pax7 expression during EB differentiation, doxycycline (Sigma) was added to the cultures at 0.75 μg/ml beginning at day 2 of EB differentiation. At day 5 of differentiation, myogenic precursors were purified by sorting for the PDGFαR+Flk-1− fraction and expanded for 7–10 days before in vitro analysis or in vivo transplantation experiments [34]. To assess the ability of these cells to undergo final maturation, we discontinued doxycycline and exposed cells to differentiation medium, which consisted of low glucose DMEM supplemented with 2% horse serum. After 1 week in culture, cells were evaluated by immunofluorescence.
Antibody Staining and FACS Analysis
EBs or monolayers were collected after a short incubation with Trypsin or PBS without calcium or magnesium supplemented with 1 mM EDTA and 0.5% BSA, washed twice with staining buffer (PBS 2% FBS), and then suspended in the staining buffer containing Fc block (Pharmingen, 0.25 μg per 106 cells). Antibodies were added at 1 μg per 106 cells and incubated at 4°C for 30 min before washing with staining buffer. For PDGFαR and Flk-1, PE- and APC-conjugated antibodies were used, respectively (eBioscience). We used PE-conjugated anti-Syndecan4 (clone KY/8.2; Pharmingen), anti-Sca-1 (clone D7, eBioscience), and anti-CD44 (eBioscience) antibodies. PE-Cy7-conjugated anti-CD29 (clone KMI6; Pharmingen), APC-conjugated anti-CXCR4 (Pharmingen), and mouse anti-M-cadherin (clone 5; Pharmingen) antibodies. For secondary staining in the case of M-cadherin, we used APC-conjugated goat anti-mouse Ig (Pharmingen). Stained cells were analyzed and sorted on a FACS Aria (Becton–Dickinson) after addition of propidium iodide (Pharmingen) to exclude dead cells.
Real Time PCR Analysis
Real time PCR for muscle specific genes was performed using probe sets from Applied Biosystems.
Mice
All animal experiments were carried out according to protocols approved by the University of Minnesota Institutional Animal Care and Use Committee. Six- to eight-week-old Rag2−/−/IL2Rg−/− immunodeficient and C57 BL/10ScSN-Dmdmdx/j (X-linked muscular dystrophy) mice (Taconic and Jackson Laboratories) were used for the teratomas and transplantation studies, respectively.
Teratoma Assay
In vivo teratoma assay was performed by injecting one million iPS cells subcutaneously into Rag2−/−/IL2Rg−/− immunodeficient mice. Tumor masses formed within 4 to 6 weeks and paraffin sections were analyzed by H&E staining.
Transplantation Studies
Before intramuscular cell transplantation, mice were injured with cardiotoxin, and received immunosuppression (FK-506) daily, as previously described [34]. Briefly, 24 h after cardiotoxin damage into both tibialis anterior muscles, myogenic progenitors from each iPS cell line (1×106 cells/10 μl PBS) were injected into the left TA muscle, while the right leg received the same volume of PBS as negative control (n=7 in each group).
Immunofluorescence Staining of Cultured Cells and Tissue Sections
Engrafted muscles were frozen in isopentane cooled in liquid nitrogen. Serial 8 to 12 μm cryosections were collected. For immunofluorescence staining, cells cultured on slides and tissue cryosections were fixed using acetone, permeabilized with 0.3% Triton X-100 (Sigma), and blocked with 3% BSA, and then incubated with primary antibodies including GFP (Abcam), Pax7 (Hybridoma bank), Myf5, Myogenin (clone F5D), MyoD (clone MoAb 5.8A) (all 3 from BD Biosciences), MHC (MF-20 from Developmental Studies Hybridoma Bank), M-cadherin (Pharmingen), and dystrophin (Abcam). Alexa fluor 555 goat-anti-rabbit (Molecular probes) was used for secondary staining of dystrophin (rabbit anti-dystrophin from Abcam) and Alexa fluor 488 goat-anti-chicken (Invitrogen) was used for secondary staining of GFP (chicken anti-GFP from Abcam). DAPI (4,6-diamidino-2-phenylindole; Sigma) was used to counter-stain nuclei.
Immunostaining Quantification and Calculation of Fusion Index
For immunostaining quantification, total of 4 images (200×) from 2 independent experiments were counted and data are presented as Mean ± S.E. To estimate the fusion index and the mean number of nuclei per myotube, cells were allowed to differentiate in differentiation medium, as described above, and stained for MHC. The efficiency of the fusion was assessed by counting the number of nuclei in differentiated myotubes (>2 myonuclei) as a percentage of the total number of nuclei (mononucleated and plurinucleated). This percentage was determined by counting 4 images (200×) from 2 independent experiments and data are presented as Mean ± S.E.
Muscle Preparation for Mechanical Studies
For the measurement of contractile properties, mice were anaesthetized with avertin (250 mg/kg I.P.) and intact tibialis anterior (TA) muscles were dissected and placed in an experimental organ bath filled with mammalian Ringer solution as previously described [34]. The muscles were stimulated by an electric field generated between two platinum electrodes placed longitudinally on either side of the muscle (Square wave pulses 25 V, 0.2 ms in duration, 150 Hz). Muscles were adjusted to the optimum length (Lo) for the development of isometric twitch force and a 5 min recovery period was allowed between stimulations. Optimal muscle length (Lo) and stimulation voltage (25 V) were determined from micromanipulation of muscle length and a series of twitch contractions that produced maximum isometric twitch force. In brief, after determination of optimal muscle length (Lo) and measurement of maximum isometric tetanic force, total muscle cross-sectional area (CSA) was calculated by dividing muscle mass (mg) by the product of muscle length (mm) and 1.06 mg/mm3, the density of mammalian skeletal muscle. Specific force (sFo) was determined by normalizing maximum isometric tetanic force to CSA.
Statistical Analysis
Differences between samples were assessed by using the Student’s t test.
Results
Generation of Pax7-Induced iPS Cells
To derive iPS cells, we took advantage of an inducible Pax7 mouse, which was obtained from iPax7 ES cells (Fig. 1). After crossing these mice with a strain bearing rtTA at the Rosa26 locus on chromosome 6 [38], we obtained derivative mice carrying both transgenes, in which Pax7 expression is dox-dependent (iPax7 mice). A ubiquitously expressed GFP transgene [39] was then crossed in to serve as a marker. Tail tip fibroblasts (TTF) from iPax7 mice were transduced with retroviral vectors expressing Oct4, Sox2 and Klf4 [1] (Fig. 1). Several iPS cells clones emerged after 2–3 weeks. From a total of 40 picked colonies, 10 were passaged, cryopreserved, characterized for pluripotency and ability to differentiate into embryoid bodies (EBs) (Fig. 2, Supplemental Table 1). Based on this screening, 2 iPS clones were selected for further studies (Supplemental Table 1). Alternatively, we also used iPS cells derived from a mouse carrying an Inducible Cassette Exchange (ICE) locus (Iacovino et al., submitted). ICE iPS cells were then directly targeted with p2lox-Pax7 to generate iPax7-ICE-iPS cells (Fig. 1, annotated in red). Nine iPax7 iPS clones were obtained following targeting, and 3 were selected for further differentiation analyses following screening for pluripotency and EB differentiation (Supplemental Table 1). Selected iPS clones presented typical ES morphology (Fig. 2a and Supplemental Fig. 1A), and expressed high levels of SSEA-1, Nanog, and alkaline phosphatase (Fig. 2c–e, Supplemental Fig. 1B–D, and Supplemental Table 1). Inducible expression of Pax7 was detected in both iPax7 and iPax7-ICE iPS cells (Fig. 2b). We confirmed the pluripotency of these 5 iPS cells clones by injecting these clones subcutaneously into immunodeficient mice. Like ES cells, all iPS cell clones gave rise to teratomas (Supplemental Table 1), as evidenced by the presence of ectoderm, endoderm, and mesoderm within the tumors (Fig. 2g and Supplemental Fig. 1F). Karyotype analyses revealed that 2 of the 5 screened iPS clones were entirely without any karyotypically abnormal cells (TTF2 and ICE7; Supplemental Table 1).
Fig. 1.
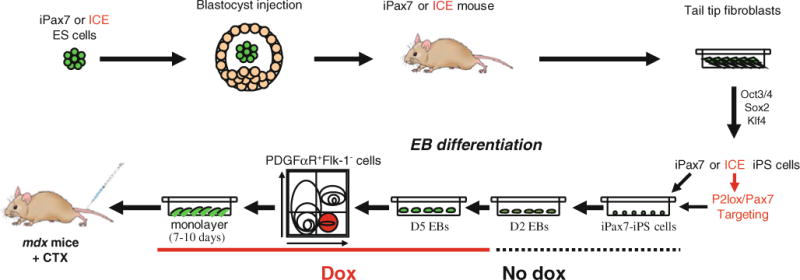
Scheme of the methods utilized to generate inducible Pax7 iPS cells. In one approach, iPS cells were derived from tail tip fibroblasts (TTF) obtained from an iPax7 mouse, which was generated following the blastocyst injection of iPax7 ES cells. In the second approach, iPS cells were derived from TTFs obtained from an ICE mouse, which resulted from the blastocyst injection of ES cells carrying the inducible cassette exchange (ICE) system (A172lox Cre-containing rTTA at Rosa-26 locus and Cre exchange cassette at HPRT locus). ICE iPS clones were then targeted with Pax7 targeting construct (P2Lox-Pax7), and clones were obtained following neo selection (iPax7-ICE-iPS). After screening for pluripotency, selected iPax7 iPS or iPax7 ICE iPS clones were differentiated into EBs and induced with doxycycline to promote skeletal muscle differentiation (Pax7 expression) from day 2 to day 5, at which time point cultures were purified for PDGFαR+Flk-1− cells, expanded, and used for in vitro and in vivo experiments
Fig. 2.
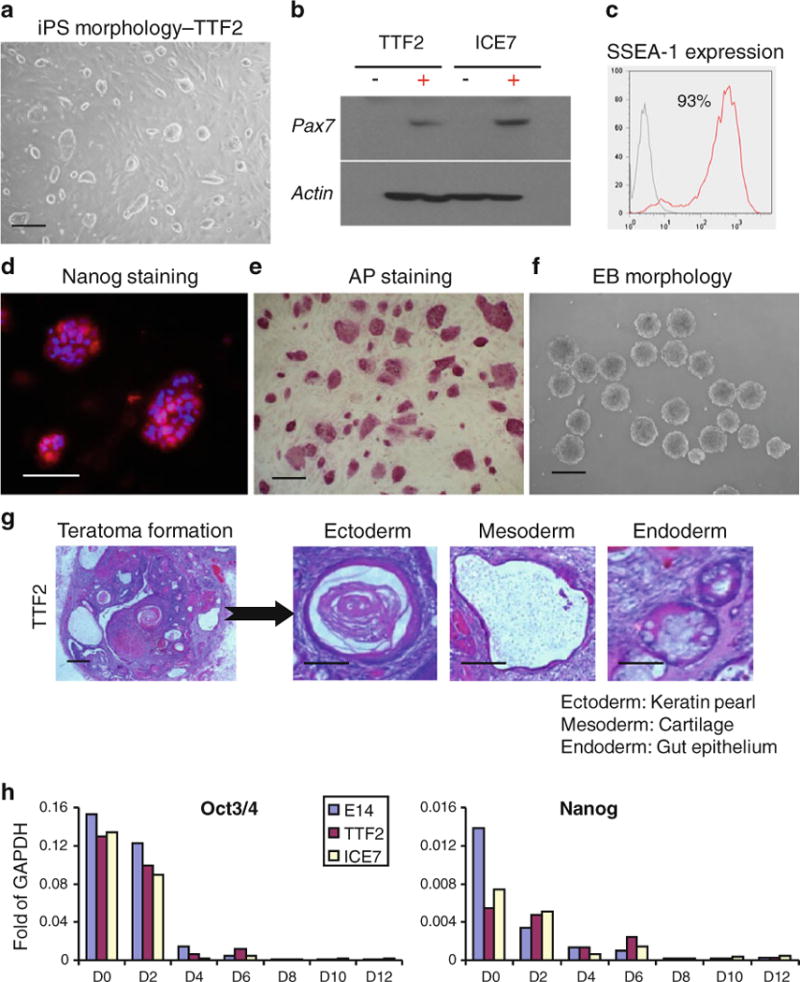
Characterization of iPS clone—TTF2. (a) Morphology of iPax7-iPS clone. (b) Western blot analyses for Pax7 in iPax7-iPS clones TTF2 and ICE7. (c) FACS analysis for SSEA-1 expression. (d) Immunofluorescent staining for Nanog. (e) Staining for alkaline phosphatase (AP). (f) Morphology of iPS-derived embryoid bodies (EBs) (g) H&E staining of teratomas derived from Rag2−/−/IL2Rg−/− immunodeficient mice injected with iPax7 TTF2 iPS cells. (h) Relative levels of gene expression for Oct3/4 and Nanog in iPax7 iPS cells (TTF2 and ICE7, non-induced) and respective EBs (days 2, 4, 6, 8, 10 and 12). Transcripts are normalized to GAPDH. E14 ES cells are used as reference
iPax7 and iPax7-ICE iPS cells were then differentiated into EBs using the hanging drop method previously described [34] (Fig. 1). The majority of these iPS clones differentiated efficiently into EBs (Fig. 2f and Supplemental Fig. 1E), and as expected, this process was accompanied by down-regulation of pluripotency-associated genes (Fig. 2h and Supplemental Table 1, no dox added). When Pax7 induction was applied from day 2 to day 5 of EB differentiation, myogenic progenitors could be greatly enriched by sorting for presumptive paraxial mesoderm PDGFαR+Flk-1− cells on day 5 (Fig. 3a, C, and Supplemental Table 1), as observed with Pax3- and Pax7-induced ES cells (Darabi et al., 2008 and [35]). Under proliferation conditions, PDGFαR+Flk-1− cells sorted from both iPax7 and iPax7-ICE iPS cells gave rise to an expanding myogenic population characterized by the expression of Pax7 (93.7%±0.6% and 94.6%±1.1%, respectively), and scant Myosin Heavy Chain (MHC) (9.5%±1.0% and 13.1%±2.1%, respectively), a marker of terminal muscle differentiation (Fig. 3b, d, upper panel). This profile switched when cells were exposed to differentiation medium (2% horse serum and dox withdrawal). Under these conditions, Pax7 signal faded (3.0%±0.7% and 3.3%±1.2%, respectively) and most cells expressed MHC (87.8%±2.3% and 81.6%±5.0%, respectively). Accordingly, cells exhibited a typical and uniform morphology of multinucleated myotubes (Fig. 3b, d, lower panel), and presented a high fusion index (Supplemental Fig. 2A). Gene expression analyses confirmed these results (Fig. 3e), as iPS-derived monolayers exposed to differentiation conditions expressed higher levels of MyoD, Myogenin, Dystrophin, and MHC but much lower levels of Myf5 than their proliferating Pax7-induced counterparts maintained in the presence of dox (Fig. 3e and Supplemental Fig. 2B). This pattern overall applied to all 5 clones, except for TTF1 (one of the abnormal clones) that in average presented lower gene expression levels for markers of differentiation (Supplemental Fig. 2B). The majority of these Pax7-induced iPS-derived muscle progenitors expressed homogenously the surface markers CD44, M-cadherin, CD29, and Sca-1 (Supplemental Fig. 3).
Fig. 3.
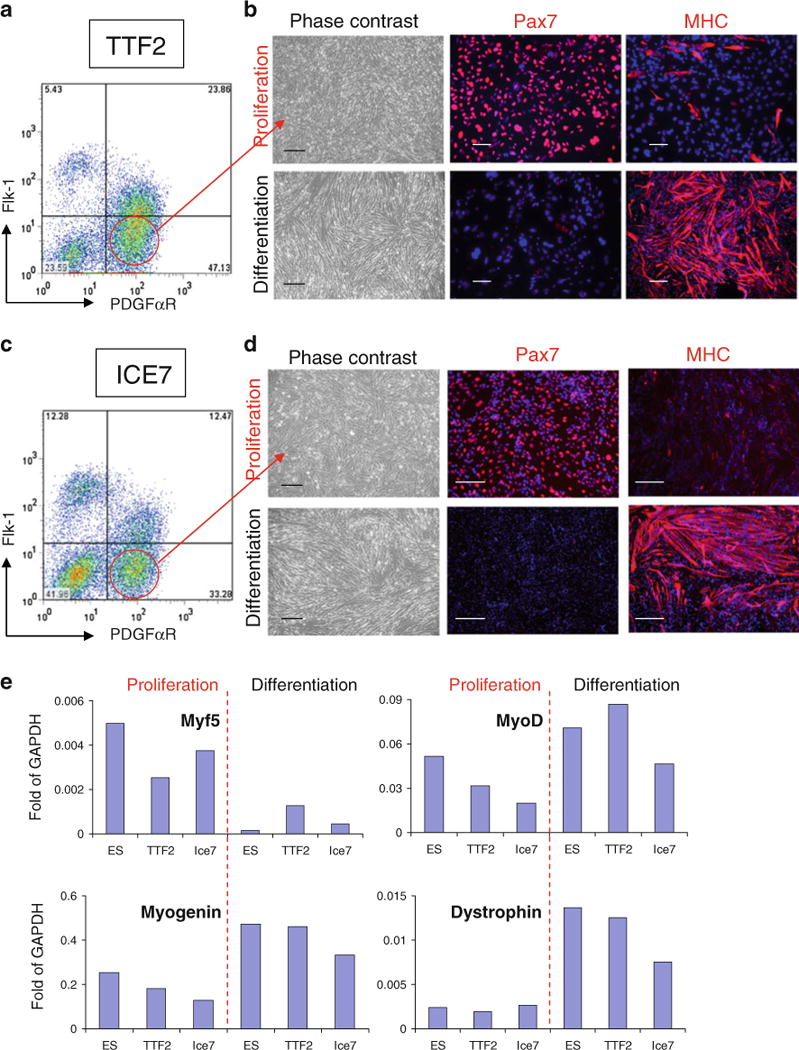
In vitro myogenic differentiation of iPax7 iPS cells. (a, c) Representative FACS profile of day 5 EBs from TTF2 (a) and ICE7 (c) iPax7 iPS cells for PDGFαR and Flk-1 expression. Dox was added to the EB medium beginning at day 2. Fluorescence intensity for Flk-1 (APC-conjugated) is indicated on the y axis and PDGFαR (PE-conjugated) on the x axis. (b, d) Representative images of TTF2 (b) and ICE7 (d) iPS cell-derived monolayers resulting from the sorting for PDGFαR+Flk-1− (a, c) under proliferation (upper panel) and differentiation (lower panel) conditions, and respective immunofluorescent staining for Pax7 and MHC. Cells are co-stained with DAPI (blue). Scale bar is 100 μm. (e) Real time RT-PCR expression analysis for myogenic markers in iPax7 iPS- (TTF2 and ICE7) PDGFαR+Flk-1−-derived cells under proliferation and differentiation conditions indicates comparable levels of myogenesis between iPax7 ES and iPax7 iPS cells. Transcripts are normalized to GAPDH
The regenerative potential of Pax7-induced iPS-derived myogenic progenitors was assessed by transplanting these cells into tibialis anterior (TA) muscles from mdx mice, a mouse model for Duchenne muscular dystrophy characterized by the deficiency of dystrophin (7 mice per iPS cell clone). Control TA muscles were injected with PBS (contralateral leg). In these experiments, we have pre-injured the TA muscle with cardiotoxin one day prior to transplantation, as it has been previously observed that pre-injury facilitates regeneration and the engraftment of donor cells [34, 40–42] by killing recipient’s mature myofibers while maintaining the basal lamina intact for regeneration [43, 44]. Mice were treated with daily IP injection of an immunosuppressive agent (tacrolimus) to prevent the rejection of iPS-derived cells, which are non-isogenic. Four weeks following transplantation, TAs were harvested and analyzed for evaluation of engraftment and contractile properties. While PBS-injected mdx mice presented only sporadic revertant Dystrophin+ myofibers (Fig. 4a), considerable engraftment was observed in mice that had been transplanted with iPS-derived myogenic progenitors (Fig. 4b–c), as evidenced by the presence of Dystrophin+ fibers that co-expressed GFP (Fig. 4b–c, right panel). It is important to note that although the majority of donor-derived cells are GFP+Dystrophin+, there are few myofibers that are single positive for one or another. In the case of GFP+Dystrophin− fibers, this is probably due to the differences in nuclear domains of membrane (dystrophin) versus cytoplasmic (GFP) proteins in hybrid fibers, as previously reported [45, 46]. The detection of very few dystrophin+ fibers with weak or no GFP signal is possibly due to: i) loss of GFP signal in unfixed frozen sections which is necessary for dystrophin antigen preservation and immunostaining and/or ii) silencing of GFP virus following differentiation of injected cells into myofibers. Importantly, quantification of Dystrophin+ fibers in TA muscles from mdx mice that had been transplanted with Pax7-induced iPS-derived (TTF2 and ICE7 clones) myogenic progenitors show similar levels of engraftment (Fig. 4d). However the other analyzed iPS clones, TTF1, ICE3, and ICE15, showed limited engraftment with only sporadic foci of Dystrophin+ myofibers (Supplemental Fig. 4A–B and Supplemental Table 1). Interestingly, the two iPS clones endowed with superior in vivo regenerative potential (TTF2 and ICE7) were the ones with normal karyotype (Supplemental Table 1). We also observed the presence of donor-derived satellite cells, as evidenced by co-expression of GFP with Pax7 or M-cadherin (Supplemental Fig. 4D). It is critical to emphasize that no tumors were detected in mdx mice transplanted with ES- or iPS-derived myogenic progenitors.
Fig. 4.
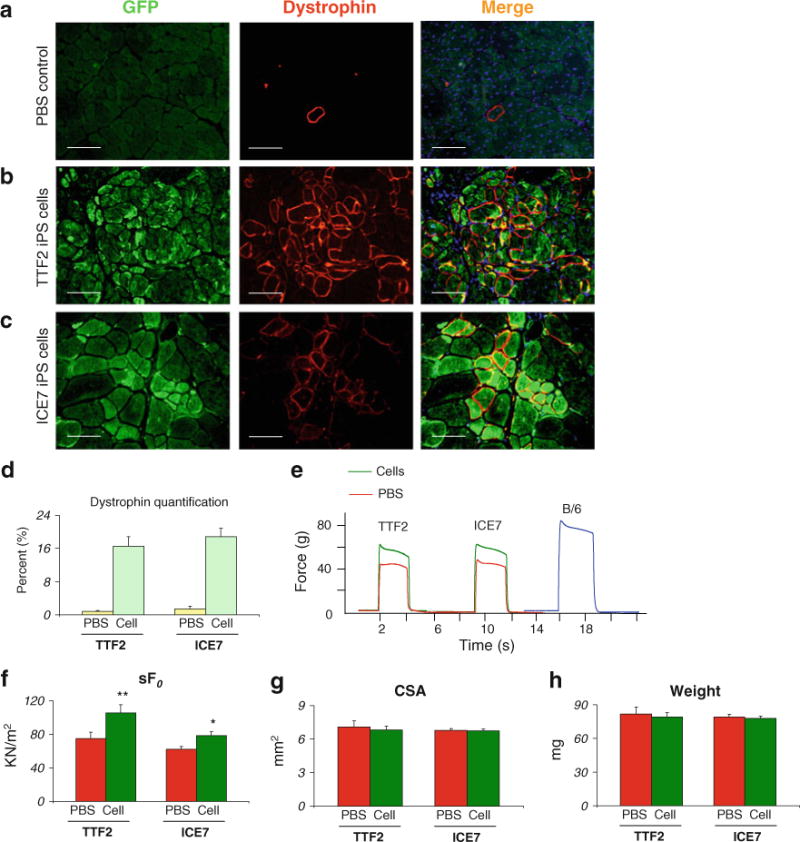
Engraftment ability of iPax7 iPS-derived myogenic progenitors into mdx mice. Pax7-induced (dox) cell monolayers resulting from PDGFαR+Flk-1− sorted cells from day 5 iPax7 and iPax7-ICE iPS embryoid bodies were transplanted via intramuscular into the left TA muscle of CTX-injured mdx mice. (a) Immunofluorescent staining of muscle sections from PBS-injected mdx mice for dystrophin (red) reveals only sporadic Dystrophin+ revertant myofibers. GFP signal was not detected. (b–c) Immunofluorescent staining of muscle sections that had been transplanted with iPax7-TTF2 (b) and iPax7-ICE (c) iPS-derived myogenic progenitors show significant dystrophin restoration that is accompanied by cytoplasmic GFP co-expression, which indicates iPS donor origin of dystrophin+ myofibers in both cases. Dystrophin in red (middle panel), GFP in green (left panel), and merge (right panel). Scale bar is 100 μm. (d) Quantification of Dystrophin+ myofibers in these engrafted muscles. For each muscle, 5 representative cross-sections at 2 mm intervals were counted. For PBS control groups, we examined 20 random sections to enumerate the sporadic revertant Dystrophin+ myofibers. (e) Representative example of force tracing in TA muscles from CTX-injured mdx mice. Green and red lines show force tracing from muscles that had received cell transplantation or PBS (control, contra-lateral leg), respectively. As reference, force tracing of an age-matched B/6 mouse is shown (blue). Muscles at L0 were stimulated for 2 s with a 150 Hz, 25 V, 0.2 ms square pulse and isometric tetanic force was recorded. (f) Effect of cell transplantation on specific force (sF0: F0 normalized to CSA). (g–h) Average CSA and weight of analyzed muscles, respectively. Values shown are the results of experiments on seven animals per group ±SEM. *P<0.05, **P<0.01
Consistently with engraftment data, only muscles treated with myogenic progenitors derived from clones TTF2 and ICE7 showed significant superior isometric tetanic force, both absolute and specific when compared to their respective contralateral PBS-injected leg (Fig. 4e–f and Supplemental Fig. 4). CSA and weight were not affected (Fig. 4g–h), as previously observed following intramuscular transplantation of Pax3-induced ES-derived myogenic progenitors [34].
Discussion
Stem cell therapies have been postulated as an attractive approach for the treatment of neurodegenerative disorders, including muscular dystrophies. Several adult cell populations with ascribed myogenic regenerative potential have been investigated over the last two decades. Although most of these cell populations are able to produce significant engraftment when transplanted into dystrophic mice [41, 47–54], only mesoangioblasts and satellite cells have been documented to produce functional recovery of treated muscles [41, 52, 53]. Despite these encouraging results, a number of caveats limit their therapeutic application, in particular the difficulty in expanding ex vivo large numbers of transplantable myogenic progenitors, which is required since both cell types are present at very low frequency in adult muscle. The major hurdles associated with in vitro cell expansion are senescence and the loss of self-renewal observed for mesoangioblasts and satellite cells, respectively [54, 55].
Because pluripotent stem cells are unique in terms of proliferation and differentiation potential, they represent an advantageous option for therapeutic application. The ethical concern associated with the use of embryonic stem cells has been eased with the revolutionary discovery of reprogramming somatic cells to a pluripotent state by Yamanaka and colleagues [1, 2]. This method has been reproduced successfully by many investigators [3–5], and progress has been made in terms of improving reprogramming efficiency [56, 57] as well as the safety [58, 59] of this approach. Importantly, patient-specific iPS cells have been derived for several genetic diseases [6, 60–63], including muscular dystrophy [6], providing an excellent tool with which to model these genetic diseases as well as testing the possibility of generating autologous cells that could be corrected ex vivo and used for patient-specific treatment. However, before one foresees the use of iPS cells for future therapeutic applications in muscular dystrophies, it will be necessary to assess both their safety as well as their ability to generate functional therapeutic myogenic progenitors in vivo using animal models.
Using two distinct approaches to generate iPS cells with inducible expression of Pax7 (Fig. 1), we demonstrate that induction of Pax7 from day 2 of EB differentiation enables the generation of proliferating early myogenic progenitors. These are greatly enriched by sorting for paraxial mesoderm PDGFα+Flk-1− in day 5 EBs (Fig. 3a, c), in the same manner we have observed for ES cells [34, 35]. Pax7-programmed cells are able to undergo terminal myogenic differentiation in vitro upon dox and FBS withdrawal and addition of horse serum to the culture medium (Fig. 3b, d). Our proof of principle experiment used an integrated, doxycycline-inducible Pax7 transgene. As this work is translated clinically, it will be important to explore non-genetic methods of inducing Pax7 expression, perhaps similar to methods now in use to generate iPS cells [14–16].
Importantly, when transplanted into dystrophic mice, proliferating skeletal myogenic progenitors are able to promote significant regeneration as well as to improve the contractile parameters of engrafted muscles. Through systematic analyses and extensive screening, our results demonstrate for the first time proof-of-principle that iPS-derived myogenic progenitors can improve contractility in transplanted muscle. Combined with gene correction, this approach has great potential for the future therapeutic application of iPS cells to muscular dystrophy.
Supplementary Material
Acknowledgments
The project was supported by NIH grants RC1AR058118 to RCRP and R01 HL081186 to MK and by the Dr. Bob and Jean Smith Foundation. The monoclonal antibody to MHC was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa.
Footnotes
Authorship Statement Contribution R.D. designed and conducted experiments, performed final analysis of the data and contributed to writing the paper. W.P. generated iPS cells and assisted R.D. with experiments. D.B. and M.K. generated the A2Lox.cre ES cells, the ICE mice and derivative ICE iPS cells. J. B. conducted the breeding to generated appropriate strains. R.C.R. P. supervised the overall project, designed experiments, analyzed the data and wrote the paper.
Conflict of Interest The authors declare no potential conflicts of interest.
Electronic supplementary material The online version of this article (doi:10.1007/s12015-011-9258-2) contains supplementary material, which is available to authorized users.
Contributor Information
Radbod Darabi, Lillehei Heart Institute, Department of Medicine, University of Minnesota, 4-124 Nils Hasselmo Hall, 312 Church St. S.E., Minneapolis 55455 MN, USA.
Weihong Pan, Lillehei Heart Institute, Department of Medicine, University of Minnesota, 4-124 Nils Hasselmo Hall, 312 Church St. S.E., Minneapolis 55455 MN, USA.
Darko Bosnakovski, Lillehei Heart Institute, Department of Pediatrics, University of Minnesota, Minneapolis, MN, USA.
June Baik, Lillehei Heart Institute, Department of Medicine, University of Minnesota, 4-124 Nils Hasselmo Hall, 312 Church St. S.E., Minneapolis 55455 MN, USA.
Michael Kyba, Lillehei Heart Institute, Department of Pediatrics, University of Minnesota, Minneapolis, MN, USA.
Rita C. R. Perlingeiro, Email: perli032@umn.edu, Lillehei Heart Institute, Department of Medicine, University of Minnesota, 4-124 Nils Hasselmo Hall, 312 Church St. S.E., Minneapolis 55455 MN, USA.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. (Translated from Eng) Cell. 2007 doi: 10.1016/j.cell.2007.11.019. (in Eng) [DOI] [PubMed] [Google Scholar]
- 3.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. (Translated from Eng) Science. 2007 doi: 10.1126/science.1151526. (in Eng) [DOI] [PubMed] [Google Scholar]
- 4.Blelloch R, Venere M, Yen J, Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell. 2007;1:254–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 6.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 7.Byrne JA, et al. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- 8.Chang CW, et al. Polycistronic lentiviral vector for ’hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. (Translated from eng) Stem Cells. 2009;27(5):1042–1049. doi: 10.1002/stem.39. (in eng) [DOI] [PubMed] [Google Scholar]
- 9.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. (Translated from eng) Nature. 2009;458(7239):771–775. doi: 10.1038/nature07864. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. (Translated from eng) Science. 2008;322(5903):949–953. doi: 10.1126/science.1164270. (in eng) [DOI] [PubMed] [Google Scholar]
- 11.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. (Translated from eng) Science. 2008;322(5903):945–949. doi: 10.1126/science.1162494. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. (Translated from eng) Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. (Translated from eng) Nature. 2009;458(7239):766–770. doi: 10.1038/nature07863. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. (Translated from eng) Cell Stem Cell. 2009;4(6):472–476. doi: 10.1016/j.stem.2009.05.005. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H, et al. Generation of induced pluripotent stem cells using recombinant proteins. (Translated from eng) Cell Stem Cell. 2009;4(5):381–384. doi: 10.1016/j.stem.2009.04.005. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. (Translated from eng) Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanna JWM, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 18.Xu D, et al. Phenotypic correction of murine hemophilia A using an iPS cell-based therapy. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:808–813. doi: 10.1073/pnas.0812090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wernig M, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuji O, et al. Therapeutic potential of appropriately evaluated safe-induced pluripotent stem cells for spinal cord injury. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12704–12709. doi: 10.1073/pnas.0910106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alipio Z, et al. Reversal of hyperglycemia in diabetic mouse models using induced-pluripotent stem (iPS)-derived pancreatic beta-like cells. (Translated from eng) Proceedings of the National Academy of Sciences of the United States of America. 2010;107(30):13426–13431. doi: 10.1073/pnas.1007884107. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espejel S, et al. Induced pluripotent stem cell-derived hepatocytes have the functional and proliferative capabilities needed for liver regeneration in mice. (Translated from eng) Journal of Clinical Investigation. 2010;120(9):3120–3126. doi: 10.1172/JCI43267. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuno Y, et al. Generation of skeletal muscle stem/progenitor cells from murine induced pluripotent stem cells. The FASEB Journal. 2010;24:2245–2253. doi: 10.1096/fj.09-137174. [DOI] [PubMed] [Google Scholar]
- 24.Rudnicki MA, et al. Myod or myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 25.Tajbakhsh S, Rocancourt D, Buckingham M. Muscle progenitors cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature. 1996;384:266–270. doi: 10.1038/384266a0. [DOI] [PubMed] [Google Scholar]
- 26.Kassar-Duchossoy L, et al. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes & Development. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 28.Bajard L, et al. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes & Development. 2006;20:2450–2464. doi: 10.1101/gad.382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goulding M, Lumsden A, Paquette AJ. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development. 1994;120:957–971. doi: 10.1242/dev.120.4.957. [DOI] [PubMed] [Google Scholar]
- 30.Tremblay JP, et al. A crucial role for Pax3 in the development of the hypaxial musculature and the long-range migration of muscle precursors. Developmental Biology. 1998;203:49–61. doi: 10.1006/dbio.1998.9041. [DOI] [PubMed] [Google Scholar]
- 31.Ordahl CP, Le Douarin NM. Two myogenic lineages within the developing somite. Development. 1992;114:339–353. doi: 10.1242/dev.114.2.339. [DOI] [PubMed] [Google Scholar]
- 32.Tajbakhsh S. Stem cells to tissue: molecular, cellular and anatomical heterogeneity in skeletal muscle. Current Opinion in Genetics & Development. 2003;13:412–422. doi: 10.1016/s0959-437x(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 33.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in posnatal myogenesis. Developmental Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 34.Darabi R, et al. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Natural Medicines. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- 35.Darabi R, et al. Assessment of the myogenic stem cell compartment following transplantation of Pax3/Pax7-induced embryonic stem cell-derived progenitors. Stem Cells. 2011 doi: 10.1002/stem.625. “in press”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iacovino M, et al. A conserved role for Hox paralog group 4 in regulation of hematopoietic progenitors. Stem Cells and Development. 2009;18:783–792. doi: 10.1089/scd.2008.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosnakovski D, et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. (Translated from eng) EMBO Journal. 2008;27(20):2766–2779. doi: 10.1038/emboj.2008.201. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121(3):465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Letters. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 40.Yan Z, et al. Highly coordinated gene regulation in mouse skeletal muscle regeneration. (Translated from eng) Journal of Biological Chemistry. 2003;278(10):8826–8836. doi: 10.1074/jbc.M209879200. (in eng) [DOI] [PubMed] [Google Scholar]
- 41.Cerletti M, et al. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles.+ Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couteaux R, Mira JC, d’Albis Regeneration of muscles after cardiotoxin injury I. Cytological aspects. Biology of the Cell. 1988;62:171–182. [PubMed] [Google Scholar]
- 43.Harris JB, Johnson MA. Further observations on the responses of rat skeletal muscle to single subcutaneous injection of a toxin isolated from the venom of the Astralian tiger snake Notechis scutatos scutatus. Clinical and Experimental Pharmacology and Physiology. 1978;5:587–600. doi: 10.1111/j.1440-1681.1978.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 44.Harris JB. Myotoxic phospholipases A2 and the regeneration of skeletal muscles. Toxicon. 2003;42:933–945. doi: 10.1016/j.toxicon.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 45.Chretien F, et al. In vivo fusion of circulating fluorescent cells with dystrophin-deficient myofibers results in extensive sarcoplasmic fluorescence expression but limited dystrophin sarcolemmal expression. (Translated from eng) American Journal of Pathology. 2005;166(6):1741–1748. doi: 10.1016/S0002-9440(10)62484-4. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auda-Boucher G, et al. Fetal muscle-derived cells can repair dystrophic muscles in mdx mice. (Translated from eng) Experimental Cell Research. 2007;313(5):997–1007. doi: 10.1016/j.yexcr.2006.12.021. (in eng) [DOI] [PubMed] [Google Scholar]
- 47.Asakura A, Seale P, Girgis-Gabardo A, Rudnicki MA. Myogenic specification of side population cells in skeletal muscle. The Journal of Cell Biology. 2002;159:123–134. doi: 10.1083/jcb.200202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachrach E, et al. Muscle engraftment of myogenic progenitor cells following intraarterial transplantation. Muscle & Nerve. 2006;34:44–52. doi: 10.1002/mus.20560. [DOI] [PubMed] [Google Scholar]
- 49.Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. The Journal of Cell Biology. 1994;125(6):1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JY, et al. Clonal isolation of muscle-derived cells capable of enhancing muscle regeneration and bone healing. The Journal of Cell Biology. 2000;150:1085–1000. doi: 10.1083/jcb.150.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sampaolesi M, et al. Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301:487–492. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- 53.Sampaolesi M, et al. Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 2006;444:574–579. doi: 10.1038/nature05282. [DOI] [PubMed] [Google Scholar]
- 54.Montarras D, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309(5743):2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 55.Gálvez BG, et al. Human cardiac mesoangioblasts isolated from hypertrophic cardiomyopathies are greatly reduced in proliferation and differentiation potency. Cardiovascular Research. 2009;83:707–716. doi: 10.1093/cvr/cvp159. [DOI] [PubMed] [Google Scholar]
- 56.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maherali N, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moretti A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. New England Journal of Medicine. 2010 Jul 21; doi: 10.1056/NEJMoa0908679. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 61.Carvajal-Vergara X, et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465 doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agarwal S, et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maehr R, et al. Generation of pluripotent stem cells from patients with type 1 diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15768–15773. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


