Abstract
OBJECTIVE
To test the hypothesis that single nucleotide polymorphisms (SNPs) in Toll-like receptor (TLR) genes alter susceptibility to bacterial infections and modulate WBC counts during infections in very low birth-weight infants (birth weight <1500g, VLBW).
STUDY DESIGN
VLBW infants recruited in a multi-center study were genotyped for 9 functional TLR SNPs and associations between SNPs and infection rates examined. WBC counts obtained during infections were compared among infants with and without SNPs.
RESULTS
In our cohort (n=408), 90 infants developed bacterial infections. Presence of TLR4 (rs4986790 & 4986791) variants were associated with Gram-negative infections. Female infants heterozygous for the X-linked IRAK1 (rs1059703) SNP had less Gram-negative infections. In regression models controlling for confounders, the TLR4 (rs4986790) SNP was associated with increased Gram-negative infections. The TLR5 (rs5744105) variant was associated with elevated WBC counts during infections.
CONCLUSION
TLR genetic variants can contribute to increased risk of bacterial infections and altered immune responses in VLBW infants.
Keywords: Toll-like receptors, infections, SNP, premature infants, genetic susceptibility
INTRODUCTION
Bacterial infections remain a major cause of morbidity and mortality in very low birth weight [VLBW] infants. 1, 2 Perinatal factors such as clinical chorioamnionitis, maternal GBS status, duration of rupture of membranes as well as infant characteristics such as gestational age, birth weight, use of indwelling vascular catheters and ventilation are known risk-factors for bacterial infections in this population. 1, 2, 3 While recent evidence suggests that genetic factors contribute significantly to diseases in VLBW infants, genetic loci that increase susceptibility to infections have not yet been characterized. 4, 5, 6 The pathogenesis of infections in humans is complex, but can arise from a failure of host genome-regulated mucosal immune defenses to overcome invasive pathogens. 7 The VLBW infant’s ability to fight infections is compromised as neutrophil function is inadequate, humoral and T-cell responses are ontogenically restricted and transplacental transfer of immunoglobulin is incomplete. 8, 9 In this setting, intact functioning of the innate immune system is critical to prevent bacterial diseases. We hypothesized that aberrant functioning of the innate immune system arising from functional genetic variation in pathogen recognition receptors will increase the risk of infections in VLBW infants.
The Toll-like receptor (TLR) signaling pathway proteins are pattern recognition receptors that recognize signature microbial motifs and stress ligands. 10 TLR-mediated innate immune responses play an important role in preventing bacterial invasion, maintaining mucosal homeostasis and regulating inflammation. 10, 11 Studies in adults and children demonstrate that TLR genetic variants modulate susceptibility to sepsis, pneumococcal infections and other infections with increased penetrance in childhood. 7, 12, 13 In this study, we investigated the impact of nine functional TLR single nucleotide polymorphisms [SNPs] on bacterial infections in a VLBW infant cohort. Recognition of pathogens by TLRs results in expression of chemokines and cytokines such as C5a, IL-8 and GM-CSF which regulate the release and activation of immune cells (neutrophils and monocytes) from the bone marrow. 14, 15 Infections in VLBW infants are often characterized by changes in the white blood cell [WBC] and absolute neutrophil count [ANC], and WBC indices are used to screen for sepsis in this population. 16, 17 Because the TLRs regulate the immune response to microbes we examined whether WBC/ANC counts obtained during infection episodes are regulated by TLR genotype.
MATERIALS AND METHODS
Patient recruitment
VLBW infants were recruited prospectively from tertiary neonatal intensive care units [NICUs] at Children’s Hospital of Wisconsin (Milwaukee, WI), St. Joseph’s Hospital (Milwaukee, WI), Kosair’s Children’s Hospital (Louisville, KY), Rush University Medical Center (Chicago, IL) and Children’s Hospitals and Clinics of Minnesota (Minneapolis, MN) between October 2006 and January 2012 after institutional review board approval at each center. After consent was obtained 0.5mL of blood was collected in de-identified sample containers, labeled with the study ID number and shipped on ice to Children’s Hospital of Wisconsin where DNA extraction and genotyping was done. Epidemiological data and disease outcomes recorded in a standardized data sheet were extracted and entered into a password-protected database.
Eligibility
Preterm infants with a birth weight ≤ 1500 g [VLBW] admitted to the participating NICUs were eligible if they did not have chromosomal disorders or major congenital anomalies.
Definition of cases
Infants with clinical signs of sepsis were considered to have a culture- positive (C+ve) infection if bacteria or candida spp. were isolated from blood, trachea, urine or cerebrospinal fluid and they were treated with antibiotics for ≥ 5d. For tracheal infections, only episodes associated with radiographic evidence of pulmonary infiltrates or a change in the clinical status were deemed as infections by the attending neonatologist and treated for ≥ 5d.
Selection of SNPs
TLR pathway genes essential for mediating immune responses against known bacterial and fungal pathogens in this population were targeted. SNPs in candidate genes were identified by searching databases such as PubMed and dBSNP and selected based on; a) whether they were reported to modulate infectious disease susceptibility, b) demonstration of altered functionality with the variant, and c) mean allele frequency of > 2% among Caucasians.
Genotyping
DNA was extracted from blood samples using the FlexiGene DNA kit (Qiagen, CA). The nine TLR pathway SNPs [TLR2 (rs5743708), TLR4 (rs4986790 & rs4986791), TLR5 (rs5744168), TLR9 (rs352140), IRAK1 (rs1059703), TIRAP (rs8177374), NFKB1 (28362491) and NFKBIA (rs3138053)] were genotyped by developing a multiplexed single-base extension (SBE) assay using the ABI PRISM SNaPshot Multiplex kit (Applied Biosystems, Foster City, CA) and performed essentially as described before. 6
Quality control
7% of samples were re-genotyped by a technician blinded to prior results. There was 100% concordance for all samples.
Estimation of WBC/ANC counts
WBC counts acquired when blood, urine, cerebrospinal fluid or tracheal cultures were obtained were used. WBC counts were quantified using automated cell analyzers such as Cell-Dyn Sapphire (Abbott Diagnostics, CA) and Beckman-Coulter (Brea, CA). ANC was calculated based on the proportion of neutrophils in the differential WBC count.
Statistical analysis
Categorical demographic variables were analyzed using Chi-square tests. Wilcoxon-Mann-Whitney rank sum test was used for continuous variables like birth weight and central venous line [CVL] days.
Power and Sample size
A genetic dominance model was used for power calculations. Assuming a rate of 24% for C+ve infections, a sample size of 400 would give us 80% power to detect a 12–15 % difference in the prevalence of SNPs between infants with and without infections assuming a P <0.05. Rates of C+ve, Gram-positive (G+ve), and Gram-negative (G-ve) infections were compared among infants with and without TLR SNPs using chi-square tests or Fisher’s exact test. Bonferroni correction was applied to adjust for multiple comparisons [9 SNPs]. To control for potential confounders we developed temporal logistic regression models in which risk factors present at birth [birth-weight, race, antenatal steroids, gender, chorioamnionitis, gestational age, sex, Apgar-5min etc.,] along with TLR SNPs were examined for association with infections. Non-significant variables [p<0.10] were removed in a stepwise fashion until only those associated (p<0.05) with a phenotype remained. CVL days was then added to the model and stepwise elimination done to identify variables significantly associated with each infection phenotype. WBC and ANC counts obtained during the first episode of C+ve infection were compared between VLBW infants with TLR variants and those without using Wilcoxon-Mann-Whitney rank sum test. Linear regression was used to explore interrelationships between genetic variants, epidemiological variables and WBC counts. SPSS 19.0 [SPSS Inc., Chicago, Illinois and SAS 9.3 [SAS Inc., Cary, NC] were used for data analysis.
RESULTS
Infections in VLBW cohort
In our cohort (n=408), 90 infants developed C+ve infections. Forty infants had blood stream infections, 13 had blood and tracheal infections, 4 had blood and urinary tract infections, 26 had tracheal infections, 3 developed urinary tract infections and 4 had tracheal and urinary tract infections. Early-onset infection (≤ 3d) was detected in 3.4% (14/408) of infants while late-onset infections (>3d) occurred in 19.8% (81/408). There were 129 C+ve infection episodes with 25 infants (5.6%) having two or more episodes. Bacteria isolated during C+ve infections are shown in the supplement section (S.1). Infants with infections had a lower birth weight, were more premature, and required more central venous line (CVL) days when compared to infants without infections (Table 1). Comparisons between infants with G-ve or G+ve infections and infants without infections showed similar results as above (data not shown).
Table 1. Comparison of epidemiological risk-factors among infants with and without C+ve infections in the VLBW cohort.
Data is represented as mean ± SD or as percentages. Comparisons between gestational age and birth-weight were done using non-parametric tests. Other comparisons were done using Chi-Square or Fisher’s exact tests. Chorioamnionitis was diagnosed by the presence of maternal fever >38°C plus one additional criteria (uterine tenderness, malodorous vaginal discharge, maternal leukocytes >15,000 cells/mm3 or fetal heart-rate of >160/min).
| Clinical Variable | No infection (n=318) Median (25%, 75%) or % |
Infection present (n=90) Median (25%, 75%) or % |
|---|---|---|
|
| ||
| Gestational age (weeks) | 29 (27, 30) | 26 (24, 27) * |
|
| ||
| Birth-weight (grams) | 1130 (869, 1311) | 755 (630, 982)* |
|
| ||
| Race - Caucasians | 68.9 | 73 |
| African American | 28.3 | 25.5 |
| Others | 2.8 | 2.2 |
|
| ||
| Antenatal steroids use | 84 | 91 |
|
| ||
| Male sex | 50.3 | 57.8 |
|
| ||
| Clinical chorioamnionitis | 10.4 | 17.8 ¶ |
|
| ||
| Prenatal Care | 96.3 | 95.6 |
|
| ||
| Inborn | 90 | 86.7 |
|
| ||
| 5-min Apgar score | 8 (6, 9) | 7 (6, 8) # |
|
| ||
| CVL days (Mean ± SD) | 14.6 ± 16 | 35.4 ± 26 * |
P<0.001,
P=0.06,
P=0.004
TLR pathway variants and infection outcomes in VLBW cohort
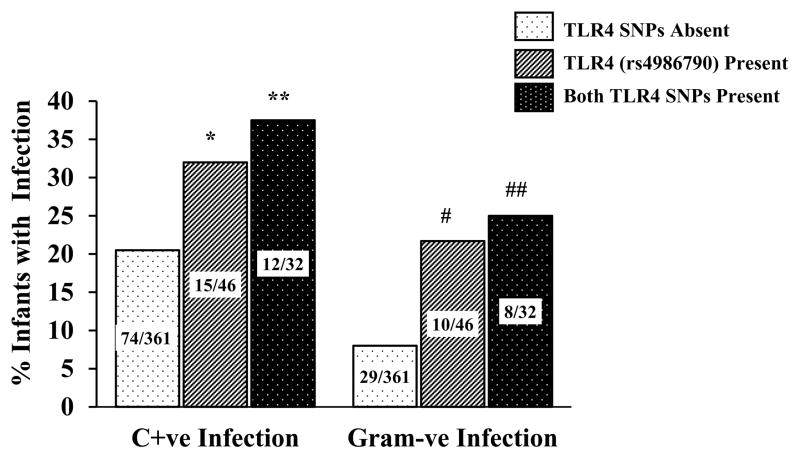
Hardy-Weinberg equilibrium was confirmed for all SNPs except for the IRAK-1 variant which is X-linked. The NFKB1, NFKBIA, TIRAP, TLR2, TLR5, and TLR9 SNPs were not associated with infection phenotypes (Table 2). Functional studies have shown that presence of both TLR4 variants disrupts signaling more profoundly than either variant. 18 Presence of both TLR4 SNPs was associated with C+ve infections (OR = 2.32, 95% CI; 1.02 – 5.25, p=0.026) while presence of the TLR4 (rs4986790) SNP alone showed a marginal (p=0.06) association with increased infections (Figure 1). Because TLR4 recognizes lipopolysaccharide from G-ve bacteria we examined associations between TLR4 SNPs and G-ve infections (Figure 1). Infants who had both TLR4 variants had increased G-ve infections (OR = 3.7, 95% CI; 1.4 – 9.6, p=0.002). Presence of the TLR4 (rs4986790) variant alone was associated with increased G-ve infections (OR = 3.2, 95% CI; 1.3 – 7.5, p=0.003). The above associations met the Bonferroni statistic of p<0.0055.
Table 2. Distribution of TLR SNPs.
Variant allele frequency stratified by infection phenotypes in our cohort. rs number; reference SNP accession ID number. Genotyping data was not obtained in one infant for 5 variants and another infant for 2 variants.
| SNP | Alleles | Genotype prevalence N (%)
|
|||
|---|---|---|---|---|---|
| No Infection (n=318) | C+ve Infection (n=90) | Gram +ve Infection (n=61) | Gram −ve Infection (n=39) | ||
|
| |||||
| TLR4 rs4986790 | AA | 287 (90.3) | 74 (83.1)* | 52 (86.7) | 29 (74.3)# |
| AG | 31 (9.7) | 15 (16.9) | 8 (13.3) | 10 (25.7) | |
|
| |||||
| TLR4 rs4986791 | CC | 298 (93.7) | 77 (86.5)** | 53 (88.3) | 31 (79.5)## |
| CT | 20 (6.3) | 12 (13.5) | 7 (11.7) | 8 (20.5) | |
|
| |||||
| TLR2 rs5743708 | GG | 306 (96.1) | 89 (100) | 60 (100) | 39 (100) |
| GA | 12 (3.9) | 0 | 0 | 0 (0) | |
|
| |||||
| TLR5 rs5744168 | CC | 296 (93.1) | 80 (89.9) | 54 (90) | 35 (89.7) |
| CT | 21 (6.6) | 9 (10.1) | 6 (10) | 4 (10.3) | |
| TT | 1 (0.3) | 0 | 0 | 0 | |
|
| |||||
| TLR9 rs352140 | CC | 85 (26.7) | 26 (29.2) | 17 (28.3) | 13 (33.3) |
| CT | 155 (48.7) | 43 (48.3) | 30 (50) | 16 (41.1) | |
| TT | 78 (24.6) | 20 (22.5) | 13 (21.7) | 10 (25.6) | |
|
| |||||
| IRAK1 rs1059703 | TT | 212 (66.7) | 59 (66.3) | 41 (68.3) | 28 (71.8) |
| TC | 49 (15.4) | 11 (12.4) | 10 (16.7) | 1 (2.6)¶ | |
| CC | 57 (17.9) | 19 (21.3) | 9 (15) | 10 (25.6) | |
|
| |||||
| TIRAP rs8177374 | CC | 260 (81.8) | 73 (81.1) | 49 (80.2) | 31 (79.2) |
| CT | 55 (17.3) | 16 (17.8) | 11 (18.1) | 7 (18.2) | |
| TT | 3 (0.9) | 1 (1.1) | 1 (1.7) | 1 (2.6) | |
|
| |||||
| NFKB1 rs28362491 | ins/ins | 109 (34.3) | 28 (31.1) | 19 (31.1) | 11 (28.2) |
| ins/del | 150 (47.2) | 38 (42.2) | 27 (44.3) | 17 (43.6) | |
| del/del | 59 (18.5) | 24 (26.7) | 15 (24.6) | 11 (28.2) | |
|
| |||||
| NFKBIA rs3138053 | AA | 173 (54.4) | 49 (55.1) | 33 (55) | 20 (51.3) |
| AG | 126 (39.6) | 31 (34.8) | 21 (35) | 15 (38.5) | |
| GG | 19 (6) | 9 (10.1) | 6 (10) | 4 (10.2) | |
P=0.06 (C+ve infection vs. no infection),
P=0.026 (C+ve infection vs. no infection),
P=0.003 (G-ve infection vs. infants without G-ve infection),
P=0.002 (G-ve infection vs. infants without G-ve infection),
P=0.02 (G-ve infection in infants with T/C at IRAK1 (rs1059703) vs. infants with C/C or T/T).
Figure 1. C+ve and G-ve infections stratified by the presence or absence of TLR4 variants in VLBW infants.
Data labels represent number of infants in each category. * - P=0.06, ** - P=0.026, # - P=0.003, ## - P=0.002.
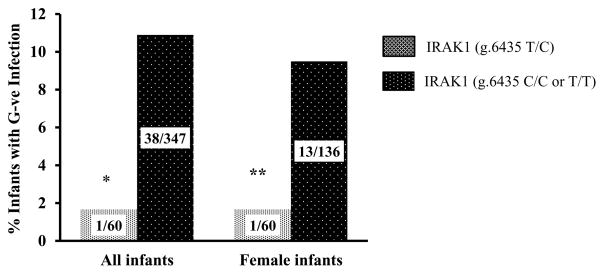
Because studies in adults have reported gender-dependent effects of the X-linked IRAK1 SNP (rs1059703) on inflammation, 19 we examined the gender-dependent effects of this variant on infection outcomes. In our cohort, there was a trend towards decreased G-ve infections in females (male vs. female; 25/212 vs. 14/196, p=0.10). Female infants with the C/T genotype had decreased rates of G-ve infections (Figure 2) when compared to other infants (male or female) who were T/T or C/C at the variant IRAK1 locus (Odds ratio [OR] = 0.14, 95% CI; 0.007 – 0.96, p=0.02). Female infants with the C/T genotype showed a trend towards less G-ve infections (Figure 2) when compared to female infants with T/T or C/C genotypes (p=0.067).
Figure 2. G-ve infection outcomes stratified by IRAK1 (g.6435 T>C) genotype in VLBW cohort.
Data labels represent number of infants in each category. * - P=0.02, ** - P=0.067
TLR4 SNPs are associated with G-ve infections
In regression models that adjusted for potential confounders, only birth-weight (p<0.001) and CVL days (p=0.001) were associated with C+ve or G+ve infection. In models that examined risk-factors for G-ve infections we noted that the TLR4 (rs4986790) variant, birth-weight and CVL days were associated with increased G-ve infections while presence of the T/C genotype at the variant IRAK1 locus had a protective effect (Table 3). Among Caucasian infants (n=285), the largest racial group, 65 (23%) developed C+ve infections (44 G+ve and 28 G-ve). Both TLR4 variants were inherited together and in complete linkage disequilibrium. Infants with TLR4 SNPs had increased G-ve infections (8/30 vs. 20/254; OR = 4.3, 95% CI; 1.5 – 11.7, p=0.004) but not G+ve infections.
Table 3. Logistic regression model for G-ve infections in our cohort.
Clinical variables available at birth along with TLR variants were investigated by logistic regression for association with G-ve infections. A step-wise elimination was used to discard non-significant variables. Then CVL days (postnatal variable) was entered into the model and stepwise elimination performed till only significant (P < 0.05) risk factors associated with G-ve infections remained. The final model representing variables significantly associated with the outcome is depicted.
| Variable | Odds Ratio | 95% CI | Alpha |
|---|---|---|---|
| Birth-weight (gm) | 0.998 | 0.996 – 0.999 | 0.002 |
| TLR4 variant presence | 3.1 | 1.2 – 7.8 | 0.018 |
| IRAK1 T/C genotype | 0.09 | 0.01 – 0.77 | 0.028 |
| CVL days (day) | 1.03 | 1.02 – 1.05 | <0.001 |
TLR variants and risk of blood stream or urinary tract infection
We examined the relationship between TLR SNPs and infections excluding infants with tracheal infection. In this group (n=64), 39 infants had G+ve infection and 27 developed G-ve infection. TLR SNPs were not associated with increased C+ve or G+ve infection except for a trend towards more C+ve infection with presence of both TLR4 SNPs (p=0.07). Presence of the TLR4 (rs4986790) SNP (OR = 3.3, 95% CI; 1.3 – 8.4, p=0.008) or presence of both TLR4 SNPs (OR = 4.6, 95% CI; 1.6 – 12.6, p=0.002) was associated with increased G-ve but not G+ve infections (Figure 3). In regression models for G-ve infection, only birth-weight (p<0.001), CVL days (p<0.001) and the TLR4 (rs4986790) variant (p=0.04) were associated with increased blood stream or urinary tract infection.
Figure 3. G+ve and G-ve infections stratified by the presence or absence of TLR4 variants in infants with only blood stream or urinary tract infections.
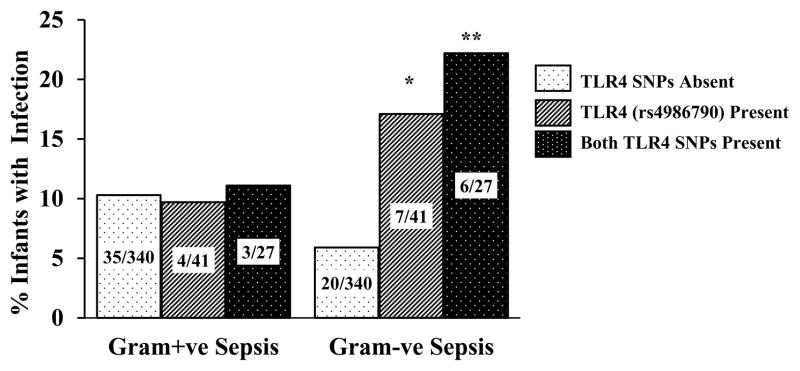
Data labels represent number of infants in each category * - P=0.02, ** - P=0.004.
TLR SNPs and WBC counts
WBC and ANC counts obtained during the first episode of C+ve infection in 90 infants who had infections were compared among infants with and without TLR SNPs (Supplement S.2). Infants with the TLR5 SNP (rs5744168) had higher WBC (p=0.02) and ANC counts (p=0.08) during C+ve infection. Linear regression models exploring associations between clinical variables (such as sex, race, etc.) and TLR variants showed that the TLR5 SNP modified the relationship between birth-weight and WBC count (Figure 4). In infants without the TLR5 variant, there was no correlation between birth-weight and WBC counts while in infants with the TLR5 SNP, an increasing birth-weight was associated with higher WBC count (p=0.01).
Figure 4. Interaction between TLR5 variant (rs5744168), birth-weight and WBC counts (103/μL).
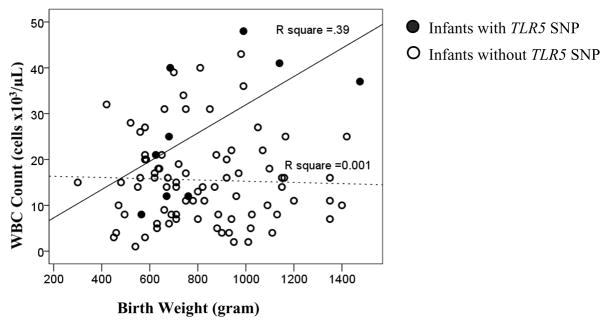
The relationship between WBC counts obtained during C+ve sepsis, genetic variants and epidemiological variables were examined using linear regression. Only birth-weight and the TLR5 variant were associated with WBC counts. The dotted line represents the relationship between WBC counts and birth-weight in infants who did not have the TLR5 variant. The bold line represents the relationship between WBC counts and birth-weight in infants who had the TLR5 variant. The R square value represents the amount of variability of WBC count explained by birth weight in the presence (solid line) or absence (hatched line) of the TLR5 SNP.
DISCUSSION
Although complete loss of function mutations in TLR pathway genes such as IRAK4 are a cause of primary immunodeficiency, the role of TLR signaling pathway SNPs in altering susceptibility to infections in the VLBW infant population remains understudied. 13 In this study, we report a novel association between presence of non-synonymous TLR4 variants and increased G-ve infection in VLBW infants. Our data also suggest an association between the presence of the C/T genotype at the IRAK1 (g.6435C>T) locus and decreased G-ve infections. Further, the TLR5 (Arg392X) variant was associated with a higher WBC count among infants who developed C+ve infections. The use of a genetically heterogeneous population, examination of a limited number of TLR pathway variants, and lack of a replication cohort are limitations of this study.
Studies by Agnese et al. and Lorenz et al. 20, 21 showed that TLR4 SNPs are associated with increased G-ve sepsis and shock in adult intensive-care patients. We noted similar conclusions in VLBW infants. However, our results differ from Abu-Maziad et al. 22 who did not find an association between the TLR4 variant (rs4986791) and infections in a retrospective case-control study done in premature infants. In their study, the control population did not constitute entirely of VLBW infants and there were three times as many males among cases while the gender composition was more balanced among controls. Ahrens et al. 23 did not demonstrate an association between the TLR4 (rs4986790) variant and sepsis in VLBW infants. In our study TLR4 variants were more robustly associated with G-ve infections rather than C+ve infections; an association not investigated by this group. Szebeni et al. 24 noted no association between TLR4 variants and NEC in VLBW infants but did not examine relationships between TLR4 SNPs and sepsis. The study population was predominantly Caucasian in the above studies while about 30% of the infants in our cohort were African-American. However, the association between TLR4 variants and G-ve infections was robust among Caucasian infants in our cohort, and remained significant after correcting for race and other confounders. Because TLR function is critical for immune responses in the lung, urinary tract and blood our initial analysis included tracheal infections along with blood stream and urinary tract infections. In our study, tracheal infections needing prolonged antibiotic treatment were usually associated with radiographic evidence of pneumonia or a change in the clinical status. However, this is a potential limitation of our study. In analysis that excluded infants with tracheal infections, TLR4 SNPs were still associated with G-ve blood stream or urinary tract infection.
TLR4 recognizes lipopolysaccharide (LPS), a cell-membrane component of G-ve bacteria such as Klebsiella pneumoniae and Escherichia coli. 10 The variants examined in this study alter the LPS recognition domain of TLR4 which would result in aberrant pathogen recognition and sepsis susceptibility. 10, 25 Rallabhandi et al. 18 have demonstrated that both TLR4 variants examined in this study decrease LPS-mediated cytokine responses in monocytes and interestingly, presence of both variants compromises TLR4 function more than either variant. In our cohort, presence of both variants was more robustly associated with C+ve and G-ve infections compared to presence of the TLR4 SNP (rs4986790) alone. Further, Arbour et al. and Tulic et al. 26, 27 have demonstrated that adults and children who are heterozygous for these TLR4 SNPs have impaired responses to endotoxin and LPS respectively, supporting impaired function with haploinsufficiency. Identification of genetic markers associated with higher risk of G-ve infections in VLBW infants is important as mortality associated with G-ve sepsis is >25%. 1, 3
All-cause neonatal mortality and sepsis-related mortality are higher in male VLBW infants. 3, 28 The IRAK1 variant (rs1059703) was shown to be associated with increased C-reactive protein in females from the Diabetes Heart Study suggesting a gender-dependent effect. 19 In our cohort, there were less G-ve infections among females of Caucasian descent and female infants with the T/C genotype at the IRAK1 locus (g.6435T>C) appeared to be protected against G-ve infections. In a study examining associations between this variant and sepsis in adults, Arcaroli et al. 29 reported that patients who were homozygous for the IRAK1 variant had increased sepsis-related mortality. We did not find increased infections in VLBW infants homozygous for this variant. Premature infants differ from adults in having dysregulated TLR-mediated responses to bacteria and this potentially can explain differences in sepsis outcomes. 8, 30 The effect of the heterozygous state on immune responses against G-ve bacteria as well as replication of our findings remains topics for future research. In contrast to adult studies and a study in preterm infants <37 weeks we did not find an association between the TLR2 (rs5743708) variant and G+ve infections. 22, 31 A potential explanation could be that TLR2 SNPs are a marker of premature birth rather than infection. In analysis that corrected for gestational age, the association between TLR2 SNPs and G+ve infections were no longer evident. 22
Neutrophils are a critical component of the immune system and play a key role in the acute-phase response to infections. 15, 16, 32 We therefore took an exploratory approach to examine relationships between TLR variants and WBC counts obtained during infections. Infants with the TLR5 variant had increased WBC/ANC counts during episodes of bacterial infections and birth weight was associated with higher WBC count only in infants with the TLR5 variant. As far as we are aware, this is the first study to examine associations between functional TLR variants and WBC counts during infections. Previously, Arbour et al. 26 have reported that adults with TLR4 SNPs showed decreased bronchial responsiveness to inhaled endotoxin. Michel et al. 33 demonstrated that adults with the TLR4 (rs4986790 or rs4986791) variants had decreased CRP levels after systemic challenge with LPS. We speculate that loss of TLR5 function arising from the TLR5 variant may exaggerate immune responses through other TLRs and alter WBC counts. It is possible that our results could have been different if we had a larger cohort and as such our results should be considered pilot in nature. Whether TLR SNPs alter in-vivo cellular and cytokine responses during sepsis is a key area for future research.
Data from this and other studies support our hypothesis that functional TLR signaling pathway SNPs will be penetrant in VLBW infants and contribute to morbidity from pathogen-mediated diseases. 6, 22 Further research is needed to validate the findings of this study, identify other genetic markers in immune genes that alter sepsis susceptibility, and to understand mechanisms by which variants contribute to disease pathogenesis. Characterization of robust genetic biomarkers that can predict susceptibility/severity of infections in this population will represent a major advance in this field and aid the development of immune-based prevention and treatment strategies to decrease the burden of microbial disease in this population.
Supplementary Material
Acknowledgments
Funding support: This study was partly supported by the NIEHS Children’s Environmental Health Sciences Core (ES004184) Center pilot and 8KL2TR000056 grants to V. Sampath
The authors acknowledge the contribution of research nurses Kathleen Meskin and Laura Lane for their help with recruitment of study infants.
Footnotes
Conflict of Interest
All authors confirm that they have not competing financial interests in relation to the work described in this manuscript.
References
- 1.Stoll BJ, Hansen N. Infections in VLBW infants: studies from the NICHD Neonatal Research Network. Semin Perinatol. 2003;27:293–301. doi: 10.1016/s0146-0005(03)00046-6. [DOI] [PubMed] [Google Scholar]
- 2.Johnson TJ, Patel AL, Jegier BJ, Engstrom JL, Meier PP. Cost of Morbidities in Very Low Birth Weight Infants. The Journal of pediatrics. 2013 doi: 10.1016/j.jpeds.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fanaroff AA, Stoll BJ, Wright LL, Carlo WA, Ehrenkranz RA, Stark AR, et al. Trends in neonatal morbidity and mortality for very low birthweight infants. Am J Obstet Gynecol. 2007;196:147, e141–148. doi: 10.1016/j.ajog.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Bhandari V, Bizzarro MJ, Shetty A, Zhong X, Page GP, Zhang H, et al. Familial and genetic susceptibility to major neonatal morbidities in preterm twins. Pediatrics. 2006;117:1901–1906. doi: 10.1542/peds.2005-1414. [DOI] [PubMed] [Google Scholar]
- 5.Lavoie PM, Dube MP. Genetics of bronchopulmonary dysplasia in the age of genomics. Current opinion in pediatrics. 22:134–138. doi: 10.1097/MOP.0b013e328336eb85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampath V, Garland JS, Le M, Patel AL, Konduri GG, Cohen JD, et al. A TLR5 (g. 1174C > T) variant that encodes a stop codon (R392X) is associated with bronchopulmonary dysplasia. Pediatric pulmonology. 2012;47:460–468. doi: 10.1002/ppul.21568. [DOI] [PubMed] [Google Scholar]
- 7.Netea MG, Wijmenga C, O’Neill LA. Genetic variation in Toll-like receptors and disease susceptibility. Nature immunology. 13:535–542. doi: 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
- 8.Levy O. Innate immunity of the human newborn: distinct cytokine responses to LPS and other Toll-like receptor agonists. J Endotoxin Res. 2005;11:113–116. doi: 10.1179/096805105X37376. [DOI] [PubMed] [Google Scholar]
- 9.Wynn JL, Levy O. Role of innate host defenses in susceptibility to early-onset neonatal sepsis. Clinics in perinatology. 37:307–337. doi: 10.1016/j.clp.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Casanova JL, Abel L, Quintana-Murci L. Human TLRs and IL-1Rs in host defense: natural insights from evolutionary, epidemiological, and clinical genetics. Annual review of immunology. 29:447–491. doi: 10.1146/annurev-immunol-030409-101335. [DOI] [PubMed] [Google Scholar]
- 13.Picard C, von Bernuth H, Ghandil P, Chrabieh M, Levy O, Arkwright PD, et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine. 89:403–425. doi: 10.1097/MD.0b013e3181fd8ec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo RF, Ward PA. Role of C5a in inflammatory responses. Annual review of immunology. 2005;23:821–852. doi: 10.1146/annurev.immunol.23.021704.115835. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, Kubes P. Molecular mechanisms of leukocyte recruitment: organ-specific mechanisms of action. Thrombosis and haemostasis. 2003;89:213–220. [PubMed] [Google Scholar]
- 16.Newman TB, Puopolo KM, Wi S, Draper D, Escobar GJ. Interpreting complete blood counts soon after birth in newborns at risk for sepsis. Pediatrics. 126:903–909. doi: 10.1542/peds.2010-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy K, Weiner J. Use of leukocyte counts in evaluation of early-onset neonatal sepsis. The Pediatric infectious disease journal. 31:16–19. doi: 10.1097/INF.0b013e31822ffc17. [DOI] [PubMed] [Google Scholar]
- 18.Rallabhandi P, Bell J, Boukhvalova MS, Medvedev A, Lorenz E, Arditi M, et al. Analysis of TLR4 polymorphic variants: new insights into TLR4/MD-2/CD14 stoichiometry, structure, and signaling. J Immunol. 2006;177:322–332. doi: 10.4049/jimmunol.177.1.322. [DOI] [PubMed] [Google Scholar]
- 19.Lakoski SG, Li L, Langefeld CD, Liu Y, Howard TD, Brosnihan KB, et al. The association between innate immunity gene (IRAK1) and C-reactive protein in the Diabetes Heart Study. Experimental and molecular pathology. 2007;82:280–283. doi: 10.1016/j.yexmp.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Archives of internal medicine. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 21.Agnese DM, Calvano JE, Hahm SJ, Coyle SM, Corbett SA, Calvano SE, et al. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. The Journal of infectious diseases. 2002;186:1522–1525. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 22.Abu-Maziad A, Schaa K, Bell EF, Dagle JM, Cooper M, Marazita ML, et al. Role of polymorphic variants as genetic modulators of infection in neonatal sepsis. Pediatric research. 68:323–329. doi: 10.1203/PDR.0b013e3181e6a068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahrens P, Kattner E, Kohler B, Hartel C, Seidenberg J, Segerer H, et al. Mutations of genes involved in the innate immune system as predictors of sepsis in very low birth weight infants. Pediatric research. 2004;55:652–656. doi: 10.1203/01.PDR.0000112100.61253.85. [DOI] [PubMed] [Google Scholar]
- 24.Szebeni B, Szekeres R, Rusai K, Vannay A, Veres G, Treszl A, et al. Genetic polymorphisms of CD14, toll-like receptor 4, and caspase-recruitment domain 15 are not associated with necrotizing enterocolitis in very low birth weight infants. Journal of pediatric gastroenterology and nutrition. 2006;42:27–31. doi: 10.1097/01.mpg.0000192246.47959.b2. [DOI] [PubMed] [Google Scholar]
- 25.Faure K, Sawa T, Ajayi T, Fujimoto J, Moriyama K, Shime N, et al. TLR4 signaling is essential for survival in acute lung injury induced by virulent Pseudomonas aeruginosa secreting type III secretory toxins. Respiratory research. 2004;5:1. doi: 10.1186/1465-9921-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nature genetics. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 27.Tulic MK, Hurrelbrink RJ, Prele CM, Laing IA, Upham JW, Le Souef P, et al. TLR4 polymorphisms mediate impaired responses to respiratory syncytial virus and lipopolysaccharide. J Immunol. 2007;179:132–140. doi: 10.4049/jimmunol.179.1.132. [DOI] [PubMed] [Google Scholar]
- 28.Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002;110:285–291. doi: 10.1542/peds.110.2.285. [DOI] [PubMed] [Google Scholar]
- 29.Arcaroli J, Silva E, Maloney JP, He Q, Svetkauskaite D, Murphy JR, et al. Variant IRAK-1 haplotype is associated with increased nuclear factor-kappaB activation and worse outcomes in sepsis. Am J Respir Crit Care Med. 2006;173:1335–1341. doi: 10.1164/rccm.200603-341OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kollmann TR, Levy O, Montgomery RR, Goriely S. Innate immune function by Toll-like receptors: distinct responses in newborns and the elderly. Immunity. 2012;37:771–783. doi: 10.1016/j.immuni.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arcaroli J, Fessler MB, Abraham E. Genetic polymorphisms and sepsis. Shock. 2005;24:300–312. doi: 10.1097/01.shk.0000180621.52058.e1. [DOI] [PubMed] [Google Scholar]
- 32.Parker LC, Whyte MK, Dower SK, Sabroe I. The expression and roles of Toll-like receptors in the biology of the human neutrophil. Journal of leukocyte biology. 2005;77:886–892. doi: 10.1189/jlb.1104636. [DOI] [PubMed] [Google Scholar]
- 33.Michel O, LeVan TD, Stern D, Dentener M, Thorn J, Gnat D, et al. Systemic responsiveness to lipopolysaccharide and polymorphisms in the toll-like receptor 4 gene in human beings. The Journal of allergy and clinical immunology. 2003;112:923–929. doi: 10.1016/j.jaci.2003.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.