Abstract
DNA repair systems are essential for cellular functions. Defects due to sequence variations in DNA repair genes can lead severe failure of cell functions and causing many cancer types including leukemia. The aim of this study was to investigate the relationship between XRCC1 Arg399Gln and XRCC3 Thr241Met polymorphisms and susceptibility to chronic lymphocytic leukemia (CLL) in Turkish patients. In addition, genotype distribution of these polymorphisms was compared with other populations. The frequencies of Arg399Gln and Thr241Met single nucleotide polymorphisms were studied in 25 CLL patients and 30 healthy individuals. Single nucleotide polymorphisms were genotyped by PCR–RFLP method. The genotype and allele frequencies of Arg399Gln and Thr241Met polymorphisms were not statistically different between the CLL patients and control group. The allelic frequency similarities were found between Turkish and Brazilian populations for Arg399Gln polymorphism. On the other hand, similarities were found between Turkish and other Caucasian populations for Thr241Met polymorphism. Marked differences were observed between American African versus Turkish and Chinese versus Turkish populations for Arg399Gln and Thr241Met polymorphisms respectively. These results indicate that Arg399Gln and Thr241Met polymorphisms were not associated with the development of CLL in Turkish population and ethnic differences is one of the most important factor for allele frequency differences.
Keywords: Chronic lymphocytic leukemia, SNP, XRCC1, XRCC3
Introduction
DNA repair systems maintain genome integrity and protect DNA from damage, due to both endogenous and exogenous sources [1, 2]. Defects in repair pathways lead severe failure of cell functions resulting in many different types of diseases, including leukemia [3]. Polymorphisms in DNA repair genes may result in deficient DNA repair, which in turn leads to increased cancer susceptibility [4].
There are various mechanisms of DNA repair. XRCC (X-ray cross-complementing) genes have played an important role in mammalian DNA repair processes. They protect mammalian cells from damage caused by ionizing radiations [5]. DNA repair protein XRCC1 functions in the repair of single-strand DNA breaks in mammalian cells and forms a repair complex with DNA ligase III, polymerase beta and poly (ADP-ribose) polymerase (PARP) to participate in the base excision repair pathway [6]. XRCC3 encodes a member of the RecA/Rad51-related protein family that participates to maintain chromosome stability and repair DNA damage.
Recently, association studies between XRCC polymorphisms and different types of cancers including hematological malignancies have been performed [2, 7–14]. However, depending on cancer type and ethnic groups, the risk genotype of these polymorphisms are controversial [14].
In human; within 80 genes, which are related to DNA repair pathways, over 400 single nucleotide polymorphisms (SNPs) were characterized [15]. The Arg399Gln (rs25487) polymorphism (G → A at nucleotide 28152 in exon 10, Arg → Gln at codon 399) in the XRCC1 gene occurs in the PARP binding domain. This amino acid substitution occur in evolutionary conserved regions suggesting potential functional significance [16, 17]. Most important polymorphism identified for XRCC3 gene is Thr241Met (rs861539) polymorphism (C → T at nucleotide 18607 in exon 7, Thr → Met at codon 241) which may affect the coding enzyme’s function and/or its interaction with other proteins involved in the DNA repair. Some studies demostrated that Thr241Met variant allele is associated with relatively high DNA adduct levels in lymphocyte DNA, indicating relatively low DNA repair capacity [16, 18].
In this study, we investigated the relationship between Arg399Gln and Thr241Met polymorphisms in CLL patients and in controls to determine the association between these polymorphisms and disease outcome in Turkish patients. The reason of chosing XRCC1 Arg399Gln and XRCC3 Thr241Met SNPs is due to their position in the evolutionary conserved regions and having a potential functional significance in the protein structure. On the other hand, both of them are mostly studied polymorphisms in cancer susceptibility including hematological malignancies but having contraversial results. In addition, the genotype distribution of these polymorphisms was compared with other populations that were reported previously.
Materials and Methods
Study Subjects
Twenty five unrelated CLL patients diagnosed clinically and according to peripheral blood and/or bone marrow evaluation and immunophenotyping at Gülhane Military Medical Academy. The immunophenotype of typical B cell CLL includes the coexpression of weak monotypic surface immunoglobulin, CD5, CD19, CD23 and weak or absent CD79B, CD22 and FMC7 markers [19]. Control group of 30 unrelated healthy volunteers frequency-matched to the patients by age and sex. The controls were also patients at Gülhane Military Medical Academy but none of them has no history of cancer. Both patients and control cases were military personel and selected from different geographic regions of Turkey in order to represent a homogenous Turkish population. Peripheral blood samples collected from both patients and control cases for genomic DNA isolation. Consent of local ethics committee obtained from Gülhane Military Medical Academy. The study was conducted in accordance with the guidelines of the Declaration of Helsinki.
DNA Isolation and Polymerase Chain Reaction (PCR)–Restriction Fragment Length Polymorphism (RFLP)
Genomic DNA was isolated from peripheral blood of patients and of controls by phenol–chloroform procedure. The Arg399Gln and Thr241Met SNPs were genotyped by using PCR–RFLP method. Primer sequences, restriction enzymes and fragment lenghts are given in Table 1. DNA amplification was carried out under similar conditions for both of the polymorphisms on a Techne TC-512 PCR system in a 50 µl reaction mixture containing 10× PCR Buffer (MBI Fermentas), 25 mM magnesium chloride (MBI Fermentas), 200 µM of dNTP (Sigma, St. Louis, MO), 10 pmol of forward (F) and reverse (R) primers (Iontek, İstanbul, Turkey), 0,5 µU Taq DNA Polymerase (MBI Fermentas) and 50 ng genomic DNA. The PCR cycling conditions consisted of an initial denaturation step at 95 °C for 5 min followed by 40 cycles of 95 °C for 1 min, 60 °C for 1 min, 72 °C for 1 min and final extension step at 72 °C for 5 min. Then 10 µl PCR products of XRCC1 (403 bp) and XRCC3 (456 bp) was digested with 10 U Msp I and Nla III restriction endonuclease enzyme (New England Biolabs, Hertfordshire, UK) respectively with 4 h incubation at 37 °C. The digested and undigested PCR products were seperated on a 2,5 % agarose gel electrophoresis at 120 V, stained with ethidium bromide (0,5 ug/ml) and visualized under a UV transilluminator (Vilber Lourmat, Marne-la-Vallée, France).
Table 1.
Primer sequences, restriction enzymes and RFLP fragment sizes of variant alleles of XRCC1 and XRCC3 polymorphisms
| Gene name | SNP | Primer sequence | PCR product (bp) | Restriction enzyme | PCR fragment lenghts (bp) after digestion |
|---|---|---|---|---|---|
| XRCC1 | Arg399Gln | F: 5′-TCTCCCTTGGTCTCCAACCT-3′ | 403 | Msp I | Arg/Arg (wild type): 133 + 270 |
| R: 5′-AGTAGTCTGCTGGCTCTGG-3′ | Gln/Gln (mutant): 403 | ||||
| XRCC3 | Thr241Met | F: 5′-GGTCGAGTGACAGTCCAAAC-3′ | 456 | Nla III | Thr/Thr (wild type): 315 + 141 |
| R: 5′-TGCAACGGCTGAGGGTCTT-3′ | Met/Met (mutant): 210 + 141 + 105 |
Statistical Analysis
The Statistical Package for Social Sciences (SPSS) version 16.0 software was used for the statistical analysis. The frequencies of XRCC1 and XRCC3 alleles and genotypes were obtained by direct count, and the departure from the Hardy–Weinberg equilibrium was evaluated by the χ2 analysis. Statistical significance was defined as P < 0.05. Odd ratios (OR) with a confidence of interval (CI) of 95 % were calculated for the comparison of the allele frequencies observed in patients and controls.
Results
Genotypes of the CLL patients and controls were determined by using PCR–RFLP genotyping methodology for Arg399Gln and Thr241Met polymorphisms. Figures 1 and 2 show the gel photographs which contain DNA fragments representing homozygote wild type, heterozygote and homozygote mutant genotype for both of the polymorphisms.
Fig. 1.
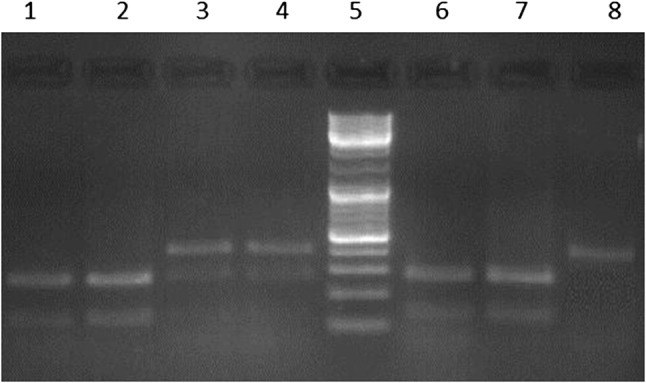
A representative agarose gel image of digested and undigested PCR products with Msp I (Lane 5: 100 bp ladder, lanes 1, 2, 6 and 7 homozygote wild genotype (GG), lanes 3 and 4 heterozygote genotype (GA), lane 8: homozygote mutant type genotype (AA)
Fig. 2.
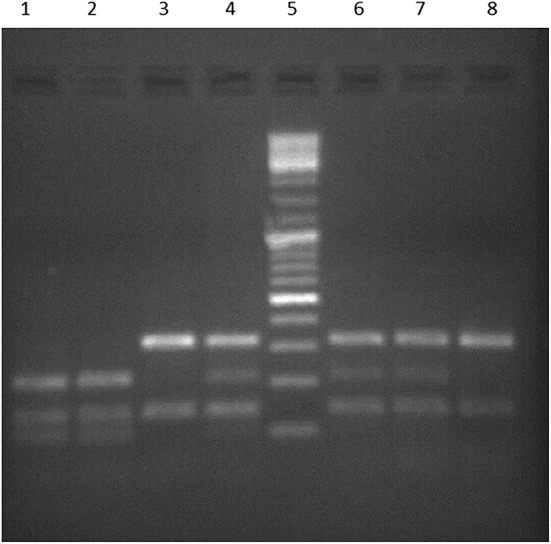
A representative agarose gel image of digested and undigested PCR products with Nla III (Lane 5: 100 bp ladder, lanes 1 and 2 homozygote mutant genotype (TT), lanes 4, 6 and 7 heterozygote genotype (CT), lanes 3 and 8: homozygote wild type genotype (CC)
XRCC1 gene polymorphism (Arg399Gln) in CLL patients and control group subjects were listed in Table 2. Among the CLL patients 32 % were found to be homozygote for wild (GG) type, 44 % were heterozygote (GA) and 24 % were homozygote mutant (AA) type. The G allele frequency was 54 % and A allele was 46 %. Among the control group subjects, 40 % were found to be homozygote for wild type, 27 % were heterozygote and 33 % were homozygote mutant type. The G allele frequency was found as 53 % whereas A allele frequency was 47 %.
Table 2.
Genotype frequencies of XRCC1 gene polymorphism in CLL patients and controls
| Arg399Gln | Controls (30 subjects/60 alleles) (% genotypes and alleles) | CLL patients (25 subjects/50 alleles) (% genotypes and alleles) | P value | OR (CI 95 %) |
|---|---|---|---|---|
| Genotypes | ||||
| Arg/Arg | 10/20 (% 40) | 6/12 (% 32) | 0.400 | – |
| Arg/Gln | 8/16 (% 27) | 11/22 (% 44) | ||
| Gln/Gln | 12/24 (% 33) | 8/16 (% 24) | ||
| Alleles | ||||
| Arg | 18/28 (% 53) | 17/23 (% 54) | 0.539 | 0.800 (0.389–1.644) |
| Gln | 20/32 (% 47) | 18/27 (% 46) | 1.133 (0.762–1.686) | |
Table 3 shows XRCC3 gene polymorphism (Thr241Met) in CLL patients and control group subjects. The Thr241Met genotype frequencies in CLL patients and controls were found higly similar. In both of the groups, approximately 37 % were found to be homozygote wild type, 47 % heterozygote and 16 % were homozygote mutant. The C allele frequency of the CLL and control group was 60 % and the T allele frequency was 40 %.
Table 3.
Genotype frequencies of XRCC3 polymorphism in CLL patients and controls
| Thr241Met | Controls (30 subjects/60 alleles) (% genotypes and alleles) | CLL patients (25 subjects/50 alleles) (% genotypes and alleles) | P value | OR (CI 95 %) |
|---|---|---|---|---|
| Genotypes | ||||
| Thr/Thr | 13/26 (% 38) | 9/18 (% 36) | 0.697 | – |
| Thr/Met | 11/22 (% 47) | 12/24 (% 48) | ||
| Met/Met | 6/12 (% 15) | 4/8 (% 16) | ||
| Alleles | ||||
| Thr | 24/37 (% 62) | 21/30 (% 60) | 0.580 | 0.831 (0.428–1.613) |
| Met | 17/23 (% 38) | 16/20 (% 40) | 1.129 (0.735–1.735) | |
The allele frequencies of both of the polymorphisms were consistent with Hardy–Weinberg equilibrium (P = 0.69 for Arg399Gln, P = 1.0 for Thr241Met). The genotype and allele frequencies of Arg399Gln and Thr241Met polymorphisms were not statistically different between the CLL patients and control group (P > 0.05). Carrying mutant allele, 399Gln or 241Met, was not associated with the risk of CLL when compared patients with controls (OR 1.133; 95 % CI 0.762–1.686 for 399Gln and OR 1.129; 95 % CI 0.735–1.735 for 241Met). These results indicate that Arg399Gln and Thr241Met polymorphisms were not associated with the development of chronic lymphocytic leukemia in Turkish population.
In the second part of the study, 25 CLL patients and 30 control group subjects were estimated together forming a Turkish population of 55 individuals. The genotype and allele frequencies of Arg399Gln and Thr241Met polymorphisms were determined in Turkish population and compared with other populations reported in the literature previously (Tables 4 and 5).
Table 4.
Population differences in genotype and allele frequencies for Arg399Gln polymorphism
| References | Population | n | G allele frequency | A allele frequency | Genotype frequency | ||
|---|---|---|---|---|---|---|---|
| Arg/Arg | Arg/Gln | Gln/Gln | |||||
| This study (2013) | Turkish | 55 | 0.54 | 0.46 | 0.36 | 0.35 | 0.29 |
| Lee et al. [22] | South Korea | 190 | 0.77 | 0.33 | 0.58 | 0.37 | 0.05 |
| Bachmann Hagen et al. [23] | German | 142 | 0.65 | 0.35 | 0.46 | 0.38 | 0.16 |
| Meza-Espinoza et al. [2] | Mexican | 120 | 0.74 | 0.26 | 0.54 | 0.39 | 0.07 |
| El-Din et al. [10] | Egyptian | 60 | 0.75 | 0.25 | 0.60 | 0.30 | 0.10 |
| Chiyomaru et al. [34] | Japanese | 93 | 0.66 | 0.34 | 0.55 | 0.23 | 0.22 |
| dos Reis et al. [27] | Brazilian | 150 | 0.59 | 0.41 | 0.41 | 0.36 | 0.23 |
| Duel et al. [21] | African American | 36 | 0.85 | 0.15 | 0.69 | 0.31 | 0 |
Table 5.
Population differences in genotype and allele frequencies for Thr241Met polymorphism
| References | Population | n | C allele frequency | T allele frequency | Genotype frequency | ||
|---|---|---|---|---|---|---|---|
| Thr/Thr | Thr/Met | Met/Met | |||||
| This study (2013) | Turkish | 55 | 0.61 | 0.39 | 0.40 | 0.42 | 0.18 |
| dos Reis et al. [27] | Brazilian | 150 | 0.63 | 0.37 | 0.36 | 0.54 | 0.10 |
| Banescu et al. [26] | Romanian | 78 | 0.66 | 0.34 | 0.46 | 0.39 | 0.15 |
| Wang et al. [35] | USA | 342 | 0.65 | 0.35 | 0.43 | 0.43 | 0.14 |
| Luo et al. [36] | Chinese | 415 | 0.75 | 0.25 | 0.55 | 0.40 | 0.05 |
| Voso et al. [1] | Italian | 161 | 0.59 | 0.41 | 0.30 | 0.58 | 0.12 |
| Mucha et al. [28] | Polish | 209 | 0.63 | 0.37 | 0.38 | 0.50 | 0.12 |
Discussion
There are a lot of studies in the literature about DNA repair gene polymorphisms and their association with different types of cancers. However, the results are contradictory [2, 7, 9–12, 20–31]. Moreover, the effect of genetic polymorphisms on the risk of cancer development may vary from one population to the other due to exposed carcinogenes, and differences in genotype and allele frequencies [14].
In the present study, we investigated the association between Arg399Gln and Thr241Met polymorphisms and chronic lymphocytic leukemia in Turkish population. Both Arg399Gln and Thr241Met polymorphisms occur in evolutionary conserved regions, so they are suggesting to have a potential functional significance in the protein structure. However, our results suggest that independently Arg399Gln and Thr241Met polymorphisms do not associate with the development of CLL in Turkish population. In addition carriers of both of the polymorphic alleles had not an elevated risk of CLL.
According to the literature, an increased risk related to the Arg399Gln polymorphism was found for acute myeloid leukemia in Egyptian patients [10], chronic myeloid leukemia in India population [7], acute lymphoblastic leukemia in Asian populations [30], pancreatic adenocarcinoma in American population [21], ovarian cancer in Chinese population [12] and gastric cancer in Turkish population [9]. However, some of these reports are contraversial in the literature since some authors have reported negative results for ALL in Mexican patients [2] and gastric cancer in Korean population [22]. On the other hand, there are also same reports in which no association was found between Arg399Gln polymorphism and multiple myeloma [25], B cell lymphoma [24], lung cancer [11], colorectal cancer in Turkish patients [9] and renal cell carcinoma in Caucasian patients [23]. In addition, there are studies indicating that the presence of Met/Met genotype for Thr241Met polymorphism is a risk factor for AML in Romanian population [26] and glioma in Chinese population [31] with contraversial reports for AML in Egyptian population [29]. However, some of the studies failed to find a significant Thr241Met polymorphism association with lung cancer in Caucasians and African-Americans [20], colorectal cancer in Polish population [28] and oral cavity cancers in Brazilian patients [27].
The contraversial results are due to many factors such as cancer type, selection criteria, ethnic differences and size of the population. In general, the risk of cancer depends on the involvement of several factors rather than the presence of a certain polymorphism [32]. Ethnic differences is one of the most important factor for allele frequency differences and knowledge of SNP frequencies in a population is a critical parameter for the estimation of interindividual differences and for the development of new individualize treatment and diagnostic approaches.
In the current study, allele frequencies which were observed for Arg399Gln and Thr241Met polymorphisms in Turkish population were compared with other ethnic groups. Compared to other populations (South Korea, German, Mexican, Egyptian, Japanese and African American), there are marked differences in genotype and allele frequencies for XRCC1 Arg399Gln polymorphism. The incidence of mutant A allele (15 %) was too low in African–Americans when compared to other populations. Turkish population, like other Caucasian and Asian populations, is significantly different than African population for Arg399Gln polymorphism. The Brazilian population, like in the case of Turkish population, is very heterogeneous in the world with a mixture of immigrants from all ethnic groups and native indigenous people [33]. The allelic frequency similarities between Turkish and Brazilian populations can be due to this ethnic heterogeneity. Turkish and Brazilian populations that carry the A allele more frequently compared to African population and other Caucasian/Asian populations may be expected to be more sensitive to exposure to environmental carcinogens.
On the other hand, T allele frequency for Thr241Met polymorphism in Turkish population (39 %) is similar with the other populations (Brazilian, Romania, USA, Italian and Polish). However, in Chinese population T allele frequency is low (25 %). From this data it can be speculated that Chinese population which carry the C allele more frequently than the other populations may be expected to be more resistant to environmental carcinogens.
In conclusion, XRCC1 and XRCC3 polymorphisms do not associate with development of CLL in Turkish population. In addition carriers of both of the polymorphic alleles had not an elevated risk of CLL. Ethnic differences is one of the most important factor for allele frequency differences. The allelic frequency similarities were found between Turkish and Brazilian populations for Arg399Gln polymorphism. On the other hand, similarities were found between Turkish and other Caucasian populations for Thr241Met polymorphism. Marked differences were observed between American African versus Turkish and Chinese versus Turkish populations for Arg399Gln and Thr241Met polymorphisms respectively. However, in spite of these conclusions it should be kept in mind that 25 subjects/50 alleles is a very small sample size for a common malignancy like CLL and that 55 subjects/110 alleles may be insufficient to be a representative of Turkish population. These results can be valuable as a pilot study for the comparison of the similar studies in Turkey and also in other populations. On the other hand, although no significant relationship was identified, the number of cases in our study is limited and would need to be supported with additional studies with higher number of Turkish CLL patients.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Voso MT, Fabiani E, D’Alo’ F, Guidi F, Di Ruscio A, Sica S, Pagano L, Greco M, Hohaus S, Leone G. Increased risk of acute myeloid leukemia due to polymorphisms in detoxification and DNA repair enzymes. Ann Oncol. 2007;18:1523–1528. doi: 10.1093/annonc/mdm191. [DOI] [PubMed] [Google Scholar]
- 2.Meza-Espinoza JP, Peralta-Leal V, Gutierrez-Angulo M, Macias-Gomez N, Ayala-Madrigal ML, Barros-Nuñez P, Duran-Gonzalez J, Leal-Ugarte E. XRCC1 polymorphisms and haplotypes in Mexican patients with acute lymphoblastic leukemia. Genet Mol Res. 2009;8:1451–1458. doi: 10.4238/vol8-4gmr687. [DOI] [PubMed] [Google Scholar]
- 3.Seedhouse C, Baintan R, Lewis M, Hording A, Russell N, Das-Gupta E. The genotype distribution of the XRCC1 gene indicates a role for base excision repair in the development of therapy-related acute myeloblastic leukemia. Blood. 2002;100:3761–3766. doi: 10.1182/blood-2002-04-1152. [DOI] [PubMed] [Google Scholar]
- 4.Krajinovic M, Labuda D, Mathonnet G, Labuda M, Moghrabi A, Champagne J, Sinnett D. Polymorphisms in genes encoding drugs and xenobiotic metabolizing enzymes, DNA repair enzymes and response to treatment of childhood acute lymphoblastic leukemia. Clin Cancer Res. 2002;8:802–810. [PubMed] [Google Scholar]
- 5.Thacker J, Zdzienicka MZ. The mammalian XRCC genes: their roles in DNA repair and genetic stability. DNA Repair (Amst) 2003;2:655–672. doi: 10.1016/S1568-7864(03)00062-4. [DOI] [PubMed] [Google Scholar]
- 6.Rice PA. Holding damaged DNA together. Nat Struct Biol. 1999;6:805–806. doi: 10.1038/12257. [DOI] [PubMed] [Google Scholar]
- 7.Annamaneni S, Gorre M, Kagita S, Addepalli K, Digumarti RR, Satti V, Battini MR. Association of XRCC1 gene polymorphisms with chronic myeloid leukemia in the population of Andhra Pradesh, India. Hematology. 2012;18:163–168. doi: 10.1179/1607845412Y.0000000040. [DOI] [PubMed] [Google Scholar]
- 8.Duman N, Aktan M, Ozturk S, Palanduz S, Cakiris A, Ustek D, Ozbek U, Nalcaci M, Cefle K. Investigation of Arg399Gln and Arg194Trp polymorphisms of the XRCC1 (x-ray cross-complementing group 1) gene and its correlation to sister chromatid exchange frequency in patients with chronic lymphocytic leukemia. Genet Test Mol Biomark. 2012;16:287–291. doi: 10.1089/gtmb.2011.0152. [DOI] [PubMed] [Google Scholar]
- 9.Engin AB, Karahalil B, Karakaya AE, Engin A. Association between XRCC1 ARG399GLN and P53 ARG72PRO polymorphisms and the risk of gastric and colorectal cancer in Turkish population. Arh Hig Rada Toksikol. 2011;62:207–214. doi: 10.2478/10004-1254-62-2011-2098. [DOI] [PubMed] [Google Scholar]
- 10.El-Din MS, Raslan H, Abdel-Hamid S, Makhlouf M. Detection of XRCC1 gene polymorphisms in Egyptian patients with acute myeloid leukemia. Comp Clin Pathol. 2012;21:505–513. doi: 10.1007/s00580-010-1120-4. [DOI] [Google Scholar]
- 11.Karakucak M, Yakut T, Evrensel T, Deligonul A, Gulten T, Ocakoglu G, Kurt E, Kanat O, Cubukcu E, Sehitoglu İ, et al. XRCC1 gene polymorphisms and risk of lung cancer in Turkish Patients. Int J Hum Genet. 2012;12:113–117. [Google Scholar]
- 12.Miao J, Zhang X, Tang QL, Wang XY, Kai L. Prediction value of XRCC 1 gene polymorphism on the survival of ovarian cancer treated by adjuvant chemotherapy. Asian Pac J Cancer Prev. 2012;13:5007–5010. doi: 10.7314/APJCP.2012.13.10.5007. [DOI] [PubMed] [Google Scholar]
- 13.Liu YT, Shi JP, Fu LY, Zhou B, Wang HL, Wu XM. Gene polymorphism of XRCC1 Arg399Gln and cervical carcinoma susceptibility in Asians: a meta-analysis based on 1,759 cases and 2,497 controls. Asian Pac J Cancer Prev. 2013;14:189–193. doi: 10.7314/APJCP.2013.14.1.189. [DOI] [PubMed] [Google Scholar]
- 14.Celkan T, Guven M, Batar B, Alhaj S. The difference between pre-B cell acute lymphoblastic leukemia and Burkitt lymphoma in relation to DNA damage repair gene polymorphisms in childhood. Leuk Lymphoma. 2008;49:1638–1640. doi: 10.1080/10428190802140063. [DOI] [PubMed] [Google Scholar]
- 15.Mohrenweiser HW, Wilson DM, 3rd, Jones IM. Challenges and complexities in estimating both the functional impact and the disease risk associated with the extensive genetic variation in human DNA repair genes. Mutat Res. 2003;526:93–125. doi: 10.1016/S0027-5107(03)00049-6. [DOI] [PubMed] [Google Scholar]
- 16.Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–608. [PubMed] [Google Scholar]
- 17.Park JY, Lee SY, Jeon HS, Bae NC, Chae SC, Joo S, Kim CH, Park JH, Kam S, Kim IS, et al. Polymorphism of the DNA repair gene XRCC1 and risk of primary lung cancer. Cancer Epidemiol Biomark Prev. 2002;11:23–27. [PubMed] [Google Scholar]
- 18.Matullo G, Palli D, Peluso M, Guarrera S, Carturan S, Celentano E, Krogh V, Munnia A, Tumino R, Polidoro S, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32) p-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 19.Oscier D, Dearden C, Eren E, Fegan C, Follows G, Hillmen P, Illidge T, Matutes E, Milligan DW, Pettitt A, et al. Guidelines on the diagnosis, investigation and management of chronic lymphocytic leukemia. Br J Haematol. 2004;159:541–564. doi: 10.1111/bjh.12067. [DOI] [PubMed] [Google Scholar]
- 20.David-Beabes GL, Lunn RM, London SJ. No association between the XPD (Lys751G1n) polymorphism of the XRCC3 (Thr241Met) polymorphism and lung cancer risk. Cancer Epidemiol Biomark Prev. 2001;10:911–912. [PubMed] [Google Scholar]
- 21.Duel EJ, Holly EA, Bracci PM, Wiencke JK, Kelsey KT. A population-based study of the Arg399Gln polymorphism in X-ray repair cross-complementing group 1 (XRCC1) and risk of pancreatic adenocarcinoma. Cancer Res. 2002;62:4630–4636. [PubMed] [Google Scholar]
- 22.Lee SG, Kim B, Choi J, Kim C, Lee I, Song K. Genetic polymorphisms of XRCC1 and risk of gastric cancer. Cancer Lett. 2002;187:53–60. doi: 10.1016/S0304-3835(02)00381-6. [DOI] [PubMed] [Google Scholar]
- 23.Bachmann HS, Rübben H, Schmid KW, Siffert W, Riemann K. DNA repair gene XRCC1 polymorphisms and outcome of renal cell carcinoma in caucasian patients. Anticancer Res. 2009;29:5131–5135. [PubMed] [Google Scholar]
- 24.Baris S, Celkan T, Batar B, Guven M, Ozdil M, Ozkan A, Apak H, Yildiz I. Association between genetic polymorphism in DNA repair genes and risk of B-cell lymphoma. Pediatr Hematol Oncol. 2009;26:467–472. doi: 10.3109/08880010903096201. [DOI] [PubMed] [Google Scholar]
- 25.Cifci S, Yılmaz M, Pehlivan M, Sever T, Okan V, Pehlivan S. DNA repair genes polymorphisms in multiple myeloma: no association with XRCC1 (Arg399Gln) polymorphism, but the XRCC4 (VNTR in intron 3 and G-1394T) and XPD (Lys751Gln) polymorphisms is associated with the disease in Turkish patients. Hematology. 2011;16:361–367. doi: 10.1179/102453311X13127324303399. [DOI] [PubMed] [Google Scholar]
- 26.Bănescu C, Tilinca M, Benedek EL, Demian S, Macarie I, Duicu C, Dobreanu M. XRCC3 THr241Met polymorphism and risk of acute myeloid leukemia in a Romanian population. Gene. 2013;526:478–483. doi: 10.1016/j.gene.2013.05.054. [DOI] [PubMed] [Google Scholar]
- 27.Dos Reis MB, Losi-Guembarovski R, de Souza Fonseca Ribeiro EM, Cavalli IJ, Morita MC, Ramos GH, de Oliveira BV, Mizuno LT, Rogatto SR, de Syllos Cólus IM. Allelic variants of XRCC1 and XRCC3 repair genes and susceptibility of oral cancer in Brazilian patients. J Oral Pathol Med. 2013;42:80–85. doi: 10.1111/j.1600-0714.2012.01192.x. [DOI] [PubMed] [Google Scholar]
- 28.Mucha B, Przybylowska-Sygut K, Dziki AJ, Dziki L, Sygut A, Majsterek I. Association of Thr241Met polymorphism of XRCC3 gene with risk of colorectal cancer in the Polish population. Pol J Pathol. 2013;64:185–190. doi: 10.5114/pjp.2013.38137. [DOI] [PubMed] [Google Scholar]
- 29.Sorour A, Ayad MW, Kassem H. The genotype distribution of the XRCC1, XRCC3 and XPD DNA repair genes and their role for the development of acute myeloblastic leukemia. Genet Test Mol Biomark. 2013;17:195–201. doi: 10.1089/gtmb.2012.0278. [DOI] [PubMed] [Google Scholar]
- 30.Wang R, Hu X, Zhou Y, Feng Q, Su L, Long J, Wei B. XRCC1 Arg399Gln and Arg194Trp polymorphisms in childhood acute lymphoblastic leukemia risk: a meta-analysis. Leuk Lymphoma. 2013;54:153–159. doi: 10.3109/10428194.2012.704031. [DOI] [PubMed] [Google Scholar]
- 31.Zhao B, Ye J, Li B, Ma Q, Su G, Han R. DNA repair gene XRCC3 Thr241Met polymorphism and glioma risk: a meta-analysis. Int J Clin Exp Med. 2013;6:438–443. [PMC free article] [PubMed] [Google Scholar]
- 32.Naccarati A, Pardini B, Hemminki K, Vodicka P. Sporadic colorectal cancer and individual susceptibility: a review of the association studies investigating the role of DNA repair genetic polymorphisms. Mutat Res. 2007;635:118–145. doi: 10.1016/j.mrrev.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Arruda VR, Grignolli CE, Gonçalves MS, Soares MC, Menezes R, Saad ST, Costa FF. Prevalence of homozygosity for the deleted alleles of glutathione S-transferase mu (GSTM1) and theta (GSTT1) among distinct ethnic groups from Brazil: relevance to environmental carcinogenesis? Clin Genet. 1998;54:210–214. doi: 10.1111/j.1399-0004.1998.tb04286.x. [DOI] [PubMed] [Google Scholar]
- 34.Chiyomaru K, Nagano T, Nishigori C (2012) XRCC1 Arg194Trp polymorphism, risk of nonmelanoma skin cancer and extramammary Paget's disease in a Japanese population. Arch Dermatol Res 305:363–370. doi:10.1007/s00403-012-1245-1 [DOI] [PMC free article] [PubMed]
- 35.Wang LE, Bondy ML, Shen H, El-Zein R, Aldape K, Cao Y, Pudavalli V, Levin VA, Yung WK, Wei Q (2004) Polymorphisms of DNA repair genes and risk of glioma. Cancer Res 64:5560–5563. doi:10.1158/0008-5472.CAN-03-2181 [DOI] [PubMed]
- 36. Luo KQ, Mu SQ, Wu ZX, Shi YN, Peng JC (2013) Polymorphisms in DNA repair genes and risk of glioma and meningioma. Asian Pac J Cancer Prev 14:449–452 [DOI] [PubMed]


