Abstract
Cytogenetic abnormalities in chromosomal number and structure are common in pediatric ALL and some have prognostic significance. One interesting association between cytogenetic status and treatment response involves the metabolism of methotrexate. Hyperdiploid lymphoblasts accumulate increased amounts of MTX and MTX polyglutamates, and they have higher basal apoptotic rates compared with leukemic cells with lower ploidy and normal cells. These characteristics may contribute to the better outcomes observed for patients with hyperdiploid lymphoblasts. A number of recurrent chromosomal abnormalities have been shown to have prognostic significance, especially in B-precursor ALL. Some chromosomal abnormalities are associated with more favorable outcomes, such as high hyperdiploidy (51–65 chromosomes) and the ETV6–RUNX1 fusion. Others are associated with a poorer prognosis, including the Philadelphia chromosome [t(9;22)], rearrangements of the MLL gene (chromosome 11q23), and intrachromosomal amplification of the AML1 gene (iAMP21).
Keywords: ALL, Prognosis, Cytogenetics
Techniques for the Detection of Significant Genetic Abnormalities in ALL
Molecular Technologies
Molecular technologies make use of DNA or RNA extracted directly from bone marrow. With the increasing numbers of significant chromosomal translocations identified, techniques of reverse transcriptase polymerase chain reaction (PCR) have provided a rapid, accurate and sensitive method for the detection of their fusion transcripts. One major disadvantage is that this approach is highly specific and cooperating abnormalities cannot be simultaneously identified. Quantitative PCR measures the relative expression levels, usually of upregulated genes.
Cytogenetics
Cytogenetic analysis of bone marrow blasts has provided the gold standard technique for the identification of significant numerical and structural chromosomal abnormalities in leukaemia since the 1970s. Some 40 years later it remains integral to the diagnosis of ALL in many laboratories. The main advantage of cytogenetics is that it is well established into clinical practice, it provides a global screen of the entire genome, while the disadvantages include its low resolution and restriction to the analysis of abnormalities visible in meta-phase.
Flow Cytometry for Determination of DNA Index
In a small number of centres, determination of DNA index by flow cytometry is used to indicate changes in chromosome number. Although it will not identify the specific chromosomes gained, the presence of high hyperdiploid clones can be detected [1]. Similarly, this approach has identified near-haploid and low-hypodiploid clones [2]. However, there are examples where measurement of DNA index has failed to detect high hyperdiploidy, possibly due to the variable stages of the cell cycle among leukaemic blasts [3].
Fluorescence In Situ Hybridization
It is based on the visualization of specific DNA probes annealed to their complementary sequences in a target genome. Many probes are now commercially available, providing rapid and accurate screening for most of the significant chromosomal abnormalities in leukaemia. FISH offers a valuable substitute for cytogenetics when used to indicate the presence of specific chromosomal rearrangements, as well as deletion and gain, usually in a dual colour arrangement.
High Resolution Array Based Technologies
These technologies include aCGH, single nucleotide polymorphism (SNP) arrays and gene expression profiling. Arrays for genome-wide microRNA expression, methylation status and mutation screening using deep sequencing approaches are now available. Currently these techniques provide valuable research tools in the study of the cancer genome [4].
Multiplex Ligation-Dependent Probe Amplification (MLPA)
Multiplex ligation-dependent probe amplification (MLPA) is a rapid multiplex PCR-based technique that allows the comparative quantification of multiple sites in a single test [5]. It targets very small sequences and can distinguish those differing in a single nucleotide. Thus MLPA is able to identify those focal genetic aberrations that are too small to be detected by FISH. Although MLPA is not suitable for genome-wide research screening, it provides a low cost, simple alternative to array-based techniques for many routine applications. It is now used routinely to identify copy number alterations in haematological malignancies for which specific copy number changes are clinically significant [6].
Cytogenetic Abnormalities in ALL
Cytogenetic abnormalities in chromosomal number and structure are common in pediatric ALL and some have prognostic significance. One interesting association between cytogenetic status and treatment response involves the metabolism of methotrexate. Hyperdiploid lymphoblasts accumulate increased amounts of MTX and MTX polyglutamates, and they have higher basal apoptotic rates compared with leukemic cells with lower ploidy and normal cells. These characteristics may contribute to the better outcomes observed for patients with hyperdiploid lymphoblasts.
A number of recurrent chromosomal abnormalities have been shown to have prognostic significance, especially in B-precursor ALL. Some chromosomal abnormalities are associated with more favorable outcomes, such as high hyperdiploidy (51–65 chromosomes) and the ETV6–RUNX1 fusion. Others are associated with a poorer prognosis, including the Philadelphia chromosome [t(9;22)], rearrangements of the MLL gene (chromosome 11q23), and intrachromosomal amplification of the AML1 gene (iAMP21).
Prognostically significant chromosomal abnormalities in childhood ALL include the following (Table 1).
Table 1.
Prognostic relevance and frequency of cytogenetic abnormalities in paediatric precursor B-cell ALL
| Gene abberation | Frequency (%) | Prognosis |
|---|---|---|
| High hyperdiploidy (51–65 chromosomes) | 20–25 | Good prognosis (7, 8) |
| Hypodiploidy | 1–2 | Poor prognosis (17) |
| t(12:21) | 20–25 | Good prognosis (15, 21–24) |
| t(9:22) | 3 | Poor prognosis (13, 30–32) |
| MLL translocation | 5 | Poor prognosis (35–37) |
| t(1:19) | 5 | Poor prognosis (41, 42, 43) |
| 1AM P21 | 1–2 | Poor prognosis (51, 52) |
| IKZF1 deletion | 15 | Poor prognosis (54, 55) |
Chromosome Number
High Hyperdiploidy
High hyperdiploidy, defined as 51–65 chromosomes per cell or a DNA index greater than 1.16, occurs in 20 % to 25 % of cases of precursor B cell ALL, but very rarely in cases of T-cell ALL [7]. Hyperdiploidy can be evaluated by measuring the DNA content of cells (DNA index) or by karyotyping. In cases with a normal karyotype or in which standard cytogenetic analysis was unsuccessful, interphase FISH may detect hidden hyperdiploidy. High hyperdiploidy generally occurs in cases with clinically favorable prognostic factors (patients aged 1–<10 years with a low WBC count) and is itself an independent favorable prognostic factor [7, 8]. Within the hyperdiploid range of 51–66 chromosomes, patients with higher modal numbers (58–66) appeared to have a better prognosis in one study. Hyperdiploid leukemia cells are particularly susceptible to undergoing apoptosis and accumulate higher levels of methotrexate and its active polyglutamate metabolites [9], which may explain the favorable outcome commonly observed in these cases.
While the overall outcome of patients with high hyperdiploidy is considered to be favorable, factors such as age, WBC count, specific trisomies, and early response to treatment have been shown to modify its prognostic significance [10].
Patients with trisomies of chromosomes 4, 10, and 17 (triple trisomies) have been shown to have a particularly favorable outcome as demonstrated by both Pediatric Oncology Group (POG) and Children’s Cancer Group analyses of NCI standard-risk ALL [11]. POG data suggest that NCI standard-risk patients with trisomies of 4 and 10, without regard to chromosome 17 status, have an excellent prognosis [12].
Chromosomal translocations may be seen with high hyperdiploidy, and in those cases, patients are more appropriately risk-classified based on the prognostic significance of the translocation. For instance, in one study, 8 % of patients with the Philadelphia chromosome [t(9;22)] also had high hyperdiploidy [13], and the outcome of these patients (treated without tyrosine kinase inhibitors) was inferior to that observed in non-Philadelphia chromosome-positive (Ph+) high hyperdiploid patients.
Near triploidy (68–80 chromosomes) and near tetraploidy (>80 chromosomes) are much less common and appear to be biologically distinct from high hyperdiploidy [14]. Unlike high hyperdiploidy, a high proportion of near tetraploid cases harbor a cryptic ETV6–RUNX1 fusion [14–16]. Near triploidy and tetraploidy were previously thought to be associated with an unfavorable prognosis, but later studies suggest that this may not be the case [14, 16].
Hypodiploidy (<44 Chromosomes)
Precursor B-cell ALL cases with fewer than the normal number of chromosomes have been subdivided in various ways, with one report stratifying based on modal chromosome number into the following four groups:
Near-haploid: 24–29 chromosomes (n = 46).
Low-hypodiploid: 33–39 chromosomes (n = 26).
High-hypodiploid: 40–43 chromosomes (n = 13).
Near-diploid: 44 chromosomes (n = 54).
Most patients with hypodiploidy are in the near-haploid and low-hypodiploid groups, and both of these groups have an elevated risk of treatment failure compared with nonhypodiploid cases [17]. Patients with fewer than 44 chromosomes have a worse outcome than do patients with 44 or 45 chromosomes in their leukemic cells.
Chromosomal Translocations
ETV6–RUNX1 [t(12;21) Cryptic Translocation, Formerly Known as TEL-AML1]
Fusion of the ETV6 gene on chromosome 12 to the RUNX1 gene on chromosome 21 can be detected in 20–25 % of cases of B-precursor ALL but is rarely observed in T-cell ALL [15]. The t(12;21) occurs most commonly in children aged 2–9 years [18, 19]. Hispanic children with ALL have a lower incidence of t(12;21) than do white children [20].
Reports generally indicate favorable EFS and OS in children with the ETV6–RUNX1 fusion; however, the prognostic impact of this genetic feature is modified by the following factors [21–24]:
Early response to treatment.
NCI risk category (age and WBC count at diagnosis).
Treatment regimen.
In one study of the treatment of newly diagnosed children with ALL, multivariate analysis of prognostic factors found age and leukocyte count, but not ETV6–RUNX1, to be independent prognostic factors [21]. There is a higher frequency of late relapses in patients with ETV6–RUNX1 fusion compared with other B-precursor ALL [21, 25]. Patients with the ETV6–RUNX1 fusion who relapse seem to have a better outcome than other relapse patients [26], with an especially favorable prognosis for patients who relapse more than 36 months from diagnosis [27]. Some relapses in patients with t(12;21) may represent a new independent second hit in a persistent preleukemic clone (with the first hit being the ETV6–RUNX1 translocation) [28, 29].
Philadelphia Chromosome [t(9;22) Translocation]
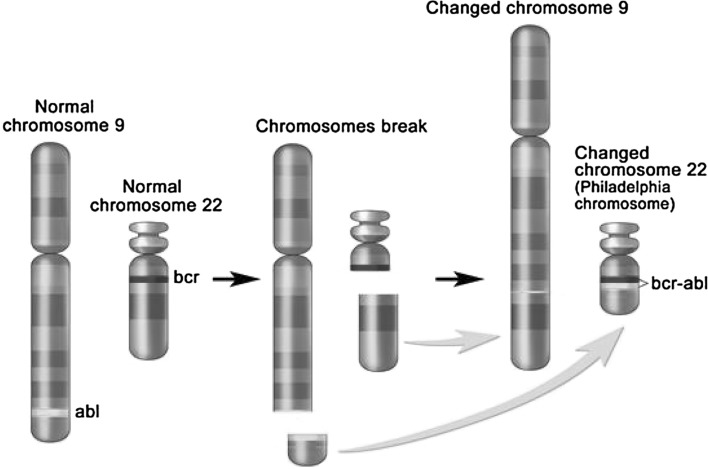
The Philadelphia chromosome t(9;22) is present in approximately 3 % of children with ALL and leads to production of a BCR–ABL1 fusion protein with tyrosine kinase activity (see Fig. 1).
Fig. 1.
The Philadelphia chromosome is a translocation between the ABL1 oncogene (on the long arm of chromosome 9) and the breakpoint cluster region (BCR; on the long arm of chromosome 22), resulting in the fusion gene BCR–ABL. BCR–ABL encodes an oncogenic protein with tyrosine kinase activity
This subtype of ALL is more common in older children with precursor B-cell ALL and high WBC count.
Historically, the Philadelphia chromosome t(9;22) was associated with an extremely poor prognosis, and its presence had been considered an indication for allogeneic hematopoietic stem cell transplantation in patients in first remission [13, 30–32]. Inhibitors of the BCR–ABL tyrosine kinase, such as imatinib mesylate, are effective in patients with Ph+ ALL [33]. A study by the COG, which used intensive chemotherapy and concurrent imatinib mesylate given daily, demonstrated a 3-year EFS rate of 80.5 %, which was superior to the EFS rate of historical controls in the pre-tyrosine kinase inhibitor (imatinib mesylate) era [33, 34]. Longer follow-up is necessary to determine whether this treatment improves the cure rate or merely prolongs DFS.
MLL Translocations
Translocations involving the MLL (11q23) gene occur in up to 5 % of childhood ALL cases and are generally associated with an increased risk of treatment failure [35–37]. The t(4;11) translocation is the most common translocation involving the MLL gene in children with ALL and occurs in approximately 2 % of cases [35].
Patients with the t(4;11) translocation are usually infants with high WBC counts; they are more likely than other children with ALL to have CNS disease and to have a poor response to initial therapy. While both infants and adults with the t(4;11) translocation are at high risk of treatment failure, children with the t(4;11) translocation appear to have a better outcome than either infants or adults [35]. Irrespective of the type of MLL gene rearrangement, infants with leukemia cells that have MLL gene rearrangements have a worse treatment outcome than older patients whose leukemia cells have an MLL gene rearrangement [35]. Deletion of the MLL gene has not been associated with an adverse prognosis [38].
Of interest, the t(11;19) translocation involving MLL and MLLT1/ENL occurs in approximately 1 % of ALL cases and occurs in both early B-lineage and T-cell ALL [39]. Outcome for infants with the t(11;19) translocation is poor, but outcome appears relatively favorable in older children with T-cell ALL and the t(11;19) translocation [39].
TCF3-PBX1 [E2A–PBX1; t(1;19) Translocation]
The t(1;19) translocation occurs in approximately 5 % of childhood ALL cases and involves fusion of the E2A gene on chromosome 19 to the PBX1 gene on chromosome 1. The t(1;19) translocation may occur as either a balanced translocation or as an unbalanced translocation and is primarily associated with pre-B ALL immunophenotype (cytoplasmic Ig positive). Black children are more likely than white children to have pre-B ALL with the t(1;19) [40].
The t(1;19) translocation had been associated with inferior outcome in the context of antimetabolite-based therapy [41], but the adverse prognostic significance was largely negated by more aggressive multiagent therapies [42]. However, in a trial conducted by SJCRH on which all patients were treated without cranial radiation, patients with the t(1;19) translocation had an overall outcome comparable to children lacking this translocation, with a higher risk of CNS relapse and a lower rate of bone marrow relapse, suggesting that more intensive CNS therapy may be needed for these patients [43].
Other Genomic Alterations
Numerous new genetic lesions have been discovered by various array comparative hybridization and next-generation sequencing methods. Appreciation of these submicroscopic genomic abnormalities and mutations is redefining the subclassification of ALL [44–50].
Intrachromosomal Amplification of Chromosome 21 (iAMP21)
iAMP21 with multiple extra copies of the RUNX1 (AML1) gene occurs in 1–2 % of precursor B-cell ALL cases and may be associated with an inferior outcome [51, 52].
IKZF1 Deletions
IKZF1 deletions, including deletions of the entire gene and deletions of specific exons, are present in approximately 15 % of precursor B-cell ALL cases. Cases with IKZF1 deletions tend to occur in older children, have a higher WBC count at diagnosis, and are therefore, more common among NCI high-risk patients compared with NCI standard-risk patients [53, 54]. A high proportion of BCR–ABL1 cases have a deletion of IKZF1 [54, 55], and ALL arising in children with Down syndrome appears to have elevated rates of IKZF1 deletions. IKZF1 deletions are also common in cases with CRLF2 genomic alterations and in Philadelphia chromosome-like (Ph-like) ALL (see below) [44, 54, 56].
Multiple reports have documented the adverse prognostic significance of a IKZF1 deletion; there are differences between studies in the magnitude of effect and in whether the IKZF1 deletion maintains significance when other prognostic factors are considered using multivariate analysis [44, 54, 56–59].
CRLF2 and JAK Mutations
Genomic alterations in CRLF2, a cytokine receptor gene located on the pseudoautosomal regions of the sex chromosomes, have abnormalities may have adverse prognostic significance, although studies differ on whether CRLF2 maintains significance when other prognostic factors are considered using multivariate analysis [47, 60–62]. However, point mutations within kinase genes are uncommon among Ph-like cases, except for JAK1 and JAK2 [63]. Additionally, there is controversy about whether prognosis should be analyzed based on CRLF2 overexpression or on the presence of CRLF2 genomic alterations [47].
Ph-like ALL
BCR–ABL1-negative patients with a gene expression profile similar to BCR–ABL1-positive patients have been referred to as Ph-like ALL [56, 57]. This occurs in 10–15 % of pediatric ALL patients, increasing in frequency with age, and is associated with a poor prognosis and with IKZF1 deletion/mutation [49, 56, 57, 63]. The hallmark of this entity is activated kinase signaling, with 50 % containing CRLF2 genomic alterations [62] and 25 % concomitant JAK mutations. Many of the remaining cases have been noted to have a series of translocations with a common theme of involvement of either ABL1, JAK2, PDGFRB, or EPOR [49]. Fusion proteins from these gene combinations have been noted in some cases to be transformative and have responded to tyrosine kinase inhibitors both in vitro and in vivo [49], suggesting potential therapeutic strategies for these patients. Point mutations in kinase genes, aside from those in JAK1 and JAK2, are uncommon in Ph-like ALL cases [63].
Cytogenetic Abnormalities in T-Progenitor Acute Lymphoblastic Leukemia
Prognostic Factors Based on Cytogenetics
T-ALL is a malignant disorder of T-cell lymphoid progenitor cells that affects 15 % of children and 25 % of adults with ALL. Structural chromosomal aberrations are identified in approximately 50 % of cases and frequently involve the juxtaposition of strong promoter and enhancer elements from T-cell receptor (TCR) genes with transcription factor genes as consequence of an illegitimate event during V(D)J recombination in normal T-cell development. This leads to the aberrant expression of the fusion partners resulting in thymocytes differentiation block at various stages of maturation.
TCR Gene Rearrangements
The most common rearrangements include TCR alpha/delta chain at 14q11.2, TCR beta chain at 7q34, and TCR gamma chain at 7p14. With few exceptions, the involved gene on the partner chromosome encodes a cell cycle inhibitor or a transcription factor whose expression is deregulated or activated as a result of the rearrangement. TAL1 (1p32) is ectopically expressed in T-ALL as consequence of t(1;14) (p32;q11) [64] (3 % in childhood T-ALL) and more frequently as a consequence of the intrachromosomal deletion resulting in SIL–TAL1 fusion gene, while LYL1 (19p13), TAL2 (9q32), and BHLH1 (21q22) are up-regulated in the rare translocations t(7;19)(q34;p13), t(7;9)(q34;q32), and t(14;21)(q11;q22), respectively. Their aberrant expression may contribute to leukemia through the formation of heterodimers with class I basic helix-loop-helix members that regulate T-cell specific genes, with consequent differentiation and proliferation impairment. LIM domain only genes LMO1 (11p15) and LMO2 (11p13) are involved in t(11;14)(p15;q11) and t(11;14)(p13;q11) with TCR alpha/delta loci and in translocations with TCR beta. LMO abnormal expression associates with deregulation of LYL1 and TAL1, even in absence of specific translocations [65]. The homeo-box (HOX) genes are a highly conserved family of transcription factors that play an important role in morphogenesis during embryonic development and in normal hemopoiesis [66]. The inv(7)(p15q34) and t(7;7)(p15;q34) bring the TCR beta regulatory elements in the vicinity of the HOXA genes cluster disrupting the normal regulatory elements of the cluster with subsequent up-regulation, especially of HOXA9, HOXA10, and HOXA11 [67, 68]. TLX1 (HOX11) is expressed at high level in more than 30 % of adult T-ALL as consequence of t(10;14)(q24;q11) and t(7;10)(q34;q24), while TLX3 (HOX11L2) is involved in t(5;14)(q35;q32) with the fusion partner BCL11B in about 20 % of children and 4 % rarely involved in TCR loci rearrangements are LCK [69, 70], CCND2 [71], and IRS4 [72].
Non-TCR Loci
A variety of cytogenetic abnormalities can occur in T-cell ALL that do not involve the TCR loci. These include del(6q), t(11q23) (MLL gene), t(14q32), trisomy 8, and t(10;11). A favorable prognostic correlation has been assessed for t(10;14), which is extremely rare in adult patients, and more frequent in the pediatric subset. Trisomy of chromosome 8 and monosomy of chromosome 7 usually carry a bad prognosis [73]. The cryptic deletion del(9) (q34.11q34.13) results in the SET-NUP214 fusion product, which transcriptionally activates specific members of the HOXA cluster maybe contributing to T-ALL pathogenesis by the inhibition of T-cell maturation [74]. The ABL1 cytoplasmic tyrosine kinase plays a role in T-cell signaling, leading to the induction of IL-2 production and proliferation following TCR activation [75].
Prognostic Factors Based on Genomic Profiling
Using SNP array platforms, many novel genomic alterations have recently been identified, including focal deletions of RB1, duplications of the proto-oncogene MYB, deletion of 9p21.2 in more than 70 % of patients [76], deletion and mutation of PTEN [77], and deletion or mutation of the U3 ubiquitin ligase FBXW7 [78, 79]. Thus far, mutations of NOTCH1 and FBXW7 have generally been associated with a favorable prognosis [80]. NOTCH1 role in leukemogenesis was of adults [69]. The high frequency of NOTCH1 mutations in T-ALL has sparked an interest in the development of anti-NOTCH1 targeted therapies for the treatment of T-ALL. Other mutated genes in T-ALL are WT1, NRAS, the negative regulator of the RAS pathway, NF1 that is inactivated because of deletions or mutations [81], and rarely FLT3 [82, 83] and PTPN2 [84], that are affected by activating mutations and focal deletions, respectively. All these genes play crucial roles as regulators and alterations in their function may affect critically different signal transduction pathways. Of note, identification of 6q15-16.1 deletion containing CAS-P8AP2 gene represents a novel prognostic factor that defines a high-risk group (Table 2) [85].
Table 2.
Genetic alterations occurring in T-progenitor ALL and their correlation with outcome
| Gene names | Alteration | Frequency | Prognosis | References |
|---|---|---|---|---|
| NOTCH1 | Sequence mutation | ~50 % of T-ALL | Associated with favourable outcome | [89–92] |
| FBXW7 | Sequence mutation | ~20 % of T-ALL | Associated with favourable outcome | [85, 86] |
| PTEN | Deletion or sequence mutation | 6–8 % of T-ALL | Associated with poor response to chemotherapy | [86] |
| CDKN2A/B | Deletion | 30–70 % of T-ALL | Associated with poor outcome | [93, 94] |
| CDKN1A | Deletion or sequence mutation | 12 % of T-ALL | Not known | [85, 95] |
| 6q15-16.1 | Deletion | 12 % of T-ALL | Associated with poor outcome | [85] |
| PHF6 | Deletion or sequence mutation | 16 % of pediatric T-ALL cases 38 % of adult T-ALL cases |
Associated with poor outcome | [86] |
| WT1 | Frameshift mutation | 13 % of pediatric T-ALL cases 12 % of adult T-ALL cases |
No association with outcome | [96, 97] |
| LEF1 | Focal deletion or sequence mutation | 15 % of pediatric T-ALL cases | Associated with better overall survival | [98] |
| JAK1 | Sequence mutation | 18 % of adult T-ALL cases | Associated with poor outcome | [94] |
| FLT3 | Internal tandem duplication | 4 % of adult T-ALL cases: 3 % of pediatric T-ALL case | No association with outcome | [88, 89] |
| PTPN2 | Deletion | 6 % of T-ALL | Down-regulation of PTPN2 expression results in prolonged survival of ALL-SIL cells after imatinib treatment | [99] |
Using exon capture of chromosome X a recent study by Van Vlierberghe et al. [86] identified inactivating mutations of the X-linked plant homeodomain finger 6 (PHF6) in 16 % of pediatric and 38 % of adult T-ALL cases. PHF6 mutations were almost exclusively found in male and were associated with leukemias driven by aberrant expression of TLX1 and TLX3. Although the prognostic significance remains to be determined, PHF6 has emerged as a new X-linked tumor suppressor in T-ALL. Recently, the application of whole-genome sequencing (WGS) to the characterization of “early T-cell precursor” (ETP) ALL, that comprises up to 15 % of T-ALL and is associated with a high risk of treatment failure, has provided potential new molecular targets for therapy [87]. Zhang et al. [87] performed WGS of 12 ETPALL cases identifying activating mutations in genes regulating cytokine receptor and RAS signaling, such as NRAS, KRAS, FLT3, IL7R, JAK3, JAK1, SH2B3, and BRAF (67 %), inactivating lesions disrupting hematopoietic development, such as GATA3, ETV6, RUNX1, IKZF1, and EP300 (58 %), and histone-modifying genes, such as EZH2, EED, SUZ12, SETD2, and EP300 (48 %). Moreover, they identified new targets of recurrent mutation in DNM2, ECT2L, and RELN genes. Importantly, the gene expression profile of ETPALL resulted similar to that of normal and myeloid leukemia hematopoietic stem cells, suggesting the possibility that myeloid-directed therapies might improve the poor outcome of ETPALL.
References
- 1.Forestier E, Holmgren G, Roos G. Flow cytometric DNA index and karyotype in childhood lymphoblastic leukemia. Anal Cell Pathol. 1998;17:145–156. doi: 10.1155/1998/712042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark B, Jeison M, Gobuzov R, Krug H, Glaser-Gabay L, Luria D, El Hasid R, Harush MB, Avrahami G, Fisher S, Stein J, Zaizov R, Yaniv I. Near haploid childhood acute lymphoblastic leukemia masked by hyperdiploid line: detection by fluorescence in situ hybridization. Cancer Genet Cytogenet. 2001;128:108–113. doi: 10.1016/S0165-4608(01)00411-3. [DOI] [PubMed] [Google Scholar]
- 3.Bungaro S, Dell’Orto MC, Zangrando A, Basso D, Gorletta T, Lo Nigro L, Leszl A, Young BD, Basso G, Bicciato S, Biondi A, te Kronnie G, Cazzaniga G. Integration of genomic and gene expression data of childhood ALL without known aberrations identifies subgroups with specific genetic hallmarks. Genes Chromosomes Cancer. 2009;48:22–38. doi: 10.1002/gcc.20616. [DOI] [PubMed] [Google Scholar]
- 4.Izraeli S. Application of genomics for risk stratification of childhood acute lymphoblastic leukemia: from bench to bed-side? Br J Haematol. 2010 doi: 10.1111/j.1365-2141.2010.08312.x. [DOI] [PubMed] [Google Scholar]
- 5.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coll-Mulet L, Santidrian AF, Cosialls AM, Iglesias-Serret D, de Frias M, Grau J, Menoyo A, Gonzalez-Barca E, Pons G, Domingo A, Gil J. Multiplex ligation-dependent probe amplification for detection of genomic alterations in chronic lymphocytic leukaemia. Br J Haematol. 2008;142:793–801. doi: 10.1111/j.1365-2141.2008.07268.x. [DOI] [PubMed] [Google Scholar]
- 7.Paulsson K, Johansson B. High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2009;48(8):637–660. doi: 10.1002/gcc.20671. [DOI] [PubMed] [Google Scholar]
- 8.Aricò M, Valsecchi MG, Rizzari C, et al. Long-term results of the AIEOP-ALL-95 Trial for Childhood Acute Lymphoblastic Leukemia: insight on the prognostic value of DNA index in the framework of Berlin–Frankfurt–Muenster based chemotherapy. J Clin Oncol. 2008;26(2):283–289. doi: 10.1200/JCO.2007.12.3927. [DOI] [PubMed] [Google Scholar]
- 9.Synold TW, Relling MV, Boyett JM, et al. Blast cell methotrexate–polyglutamate accumulation in vivo differs by lineage, ploidy, and methotrexate dose in acute lymphoblastic leukemia. J Clin Investig. 1994;94(5):1996–2001. doi: 10.1172/JCI117552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moorman AV, Richards SM, Martineau M, et al. Outcome heterogeneity in childhood high-hyperdiploid acute lymphoblastic leukemia. Blood. 2003;102(8):2756–2762. doi: 10.1182/blood-2003-04-1128. [DOI] [PubMed] [Google Scholar]
- 11.Sutcliffe MJ, Shuster JJ, Sather HN, et al. High concordance from independent studies by the Children’s Cancer Group (CCG) and Pediatric Oncology Group (POG) associating favorable prognosis with combined trisomies 4, 10, and 17 in children with NCI Standard-Risk B-precursor Acute Lymphoblastic Leukemia: a Children’s Oncology Group (COG) initiative. Leukemia. 2005;19(5):734–740. doi: 10.1038/sj.leu.2403673. [DOI] [PubMed] [Google Scholar]
- 12.Harris MB, Shuster JJ, Carroll A, et al. Trisomy of leukemic cell chromosomes 4 and 10 identifies children with B-progenitor cell acute lymphoblastic leukemia with a very low risk of treatment failure: a Pediatric Oncology Group study. Blood. 1992;79(12):3316–3324. [PubMed] [Google Scholar]
- 13.Heerema NA, Harbott J, Galimberti S, et al. Secondary cytogenetic aberrations in childhood Philadelphia chromosome positive acute lymphoblastic leukemia are nonrandom and may be associated with outcome. Leukemia. 2004;18(4):693–702. doi: 10.1038/sj.leu.2403324. [DOI] [PubMed] [Google Scholar]
- 14.Raimondi SC, Zhou Y, Shurtleff SA, et al. Near-triploidy and near-tetraploidy in childhood acute lymphoblastic leukemia: association with B-lineage blast cells carrying the ETV6–RUNX1 fusion, T-lineage immunophenotype, and favorable outcome. Cancer Genet Cytogenet. 2006;169(1):50–57. doi: 10.1016/j.cancergencyto.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Attarbaschi A, Mann G, König M, et al. Incidence and relevance of secondary chromosome abnormalities in childhood TEL/AML1+ acute lymphoblastic leukemia: an interphase FISH analysis. Leukemia. 2004;18(10):1611–1616. doi: 10.1038/sj.leu.2403471. [DOI] [PubMed] [Google Scholar]
- 16.Lemez P, Attarbaschi A, Béné MC, et al. Childhood near-tetraploid acute lymphoblastic leukemia: an EGIL study on 36 cases. Eur J Haematol. 2010;85(4):300–308. doi: 10.1111/j.1600-0609.2010.01493.x. [DOI] [PubMed] [Google Scholar]
- 17.Harrison CJ, Moorman AV, Broadfield ZJ, et al. Three distinct subgroups of hypodiploidy in acute lymphoblastic leukaemia. Br J Haematol. 2004;125(5):552–559. doi: 10.1111/j.1365-2141.2004.04948.x. [DOI] [PubMed] [Google Scholar]
- 18.Rubnitz JE, Wichlan D, Devidas M, et al. Prospective analysis of TEL gene rearrangements in childhood acute lymphoblastic leukemia: a Children’s Oncology Group study. J Clin Oncol. 2008;26(13):2186–2191. doi: 10.1200/JCO.2007.14.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanerva J, Saarinen-Pihkala UM, Niini T, et al. Favorable outcome in 20-year follow-up of children with very-low-risk ALL and minimal standard therapy, with special reference to TEL-AML1 fusion. Pediatr Blood Cancer. 2004;42(1):30–35. doi: 10.1002/pbc.10417. [DOI] [PubMed] [Google Scholar]
- 20.Aldrich MC, Zhang L, Wiemels JL, et al. Cytogenetics of Hispanic and White children with acute lymphoblastic leukemia in California. Cancer Epidemiol Biomark Prev. 2006;15(3):578–581. doi: 10.1158/1055-9965.EPI-05-0833. [DOI] [PubMed] [Google Scholar]
- 21.Loh ML, Goldwasser MA, Silverman LB, et al. Prospective analysis of TEL/AML1-positive patients treated on Dana-Farber Cancer Institute Consortium Protocol 95-01. Blood. 2006;107(11):4508–4513. doi: 10.1182/blood-2005-08-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borowitz MJ, Devidas M, Hunger SP, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children’s Oncology Group study. Blood. 2008;111(12):5477–5485. doi: 10.1182/blood-2008-01-132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madzo J, Zuna J, Muzíková K, et al. Slower molecular response to treatment predicts poor outcome in patients with TEL/AML1 positive acute lymphoblastic leukemia: prospective real-time quantitative reverse transcriptase-polymerase chain reaction study. Cancer. 2003;97(1):105–113. doi: 10.1002/cncr.11043. [DOI] [PubMed] [Google Scholar]
- 24.Bhojwani D, Pei D, Sandlund JT, et al. ETV6–RUNX1-positive childhood acute lymphoblastic leukemia: improved outcome with contemporary therapy. Leukemia. 2012;26(2):265–270. doi: 10.1038/leu.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forestier E, Heyman M, Andersen MK, et al. Outcome of ETV6/RUNX1-positive childhood acute lymphoblastic leukaemia in the NOPHO-ALL-1992 protocol: frequent late relapses but good overall survival. Br J Haematol. 2008;140(6):665–672. doi: 10.1111/j.1365-2141.2008.06980.x. [DOI] [PubMed] [Google Scholar]
- 26.Seeger K, Stackelberg AV, Taube T, et al. Relapse of TEL-AML1-positive acute lymphoblastic leukemia in childhood: a matched-pair analysis. J Clin Oncol. 2001;19(13):3188–3193. doi: 10.1200/JCO.2001.19.13.3188. [DOI] [PubMed] [Google Scholar]
- 27.Gandemer V, Chevret S, Petit A, et al. Excellent prognosis of late relapses of ETV6/RUNX1-positive childhood acute lymphoblastic leukemia: lessons from the FRALLE 93 protocol. Haematologica. 2012;97(11):1743–1750. doi: 10.3324/haematol.2011.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuna J, Ford AM, Peham M, et al. TEL deletion analysis supports a novel view of relapse in childhood acute lymphoblastic leukemia. Clin Cancer Res. 2004;10(16):5355–5360. doi: 10.1158/1078-0432.CCR-04-0584. [DOI] [PubMed] [Google Scholar]
- 29.van Delft FW, Horsley S, Colman S, et al. Clonal origins of relapse in ETV6–RUNX1 acute lymphoblastic leukemia. Blood. 2011;117(23):6247–6254. doi: 10.1182/blood-2010-10-314674. [DOI] [PubMed] [Google Scholar]
- 30.Aricò M, Schrappe M, Hunger SP, et al. Clinical outcome of children with newly diagnosed Philadelphia chromosome-positive acute lymphoblastic leukemia treated between 1995 and 2005. J Clin Oncol. 2010;28(31):4755–4761. doi: 10.1200/JCO.2010.30.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schrappe M, Aricò M, Harbott J, et al. Philadelphia chromosome-positive (Ph+) childhood acute lymphoblastic leukemia: good initial steroid response allows early prediction of a favorable treatment outcome. Blood. 1998;92(8):2730–2741. [PubMed] [Google Scholar]
- 32.Ribeiro RC, Broniscer A, Rivera GK, et al. Philadelphia chromosome-positive acute lymphoblastic leukemia in children: durable responses to chemotherapy associated with low initial white blood cell counts. Leukemia. 1997;11(9):1493–1496. doi: 10.1038/sj.leu.2400749. [DOI] [PubMed] [Google Scholar]
- 33.Biondi A, Schrappe M, De Lorenzo P, et al. Imatinib after induction for treatment of children and adolescents with Philadelphia-chromosome-positive acute lymphoblastic leukaemia (EsPhALL): a randomised, open-label, intergroup study. Lancet Oncol. 2012;13(9):936–945. doi: 10.1016/S1470-2045(12)70377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz KR, Bowman WP, Aledo A, et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a Children’s Oncology Group study. J Clin Oncol. 2009;27(31):5175–5181. doi: 10.1200/JCO.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson B, Moorman AV, Haas OA, et al. Hematologic malignancies with t(4;11)(q21;q23)—a cytogenetic, morphologic, immunophenotypic and clinical study of 183 cases. European 11q23 Workshop participants. Leukemia. 1998;12(5):779–787. doi: 10.1038/sj.leu.2401012. [DOI] [PubMed] [Google Scholar]
- 36.Raimondi SC, Peiper SC, Kitchingman GR, et al. Childhood acute lymphoblastic leukemia with chromosomal breakpoints at 11q23. Blood. 1989;73(6):1627–1634. [PubMed] [Google Scholar]
- 37.Harrison CJ, Moorman AV, Barber KE, et al. Interphase molecular cytogenetic screening for chromosomal abnormalities of prognostic significance in childhood acute lymphoblastic leukaemia: a UK Cancer Cytogenetics Group study. Br J Haematol. 2005;129(4):520–530. doi: 10.1111/j.1365-2141.2005.05497.x. [DOI] [PubMed] [Google Scholar]
- 38.Pui CH, Gaynon PS, Boyett JM, et al. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet. 2002;359(9321):1909–1915. doi: 10.1016/S0140-6736(02)08782-2. [DOI] [PubMed] [Google Scholar]
- 39.Rubnitz JE, Camitta BM, Mahmoud H, et al. Childhood acute lymphoblastic leukemia with the MLL-ENL fusion and t(11;19)(q23;p13.3) translocation. J Clin Oncol. 1999;17(1):191–196. doi: 10.1200/JCO.1999.17.1.191. [DOI] [PubMed] [Google Scholar]
- 40.Pui CH, Sandlund JT, Pei D, et al. Results of therapy for acute lymphoblastic leukemia in black and white children. JAMA. 2003;290(15):2001–2007. doi: 10.1001/jama.290.15.2001. [DOI] [PubMed] [Google Scholar]
- 41.Crist WM, Carroll AJ, Shuster JJ, et al. Poor prognosis of children with pre-B acute lymphoblastic leukemia is associated with the t(1;19)(q23;p13): a Pediatric Oncology Group study. Blood. 1990;76(1):117–122. [PubMed] [Google Scholar]
- 42.Andersen MK, Autio K, Barbany G, et al. Paediatric B-cell precursor acute lymphoblastic leukaemia with t(1;19)(q23;p13): clinical and cytogenetic characteristics of 47 cases from the Nordic countries treated according to NOPHO protocols. Br J Haematol. 2011;155(2):235–243. doi: 10.1111/j.1365-2141.2011.08824.x. [DOI] [PubMed] [Google Scholar]
- 43.Jeha S, Pei D, Raimondi SC, et al. Increased risk for CNS relapse in pre-B cell leukemia with the t(1;19)/TCF3-PBX1. Leukemia. 2009;23(8):1406–1409. doi: 10.1038/leu.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harvey RC, Mullighan CG, Wang X, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116(23):4874–4884. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Mullighan CG, Harvey RC, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2011;118(11):3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunger SP, Raetz EA, Loh ML, et al. Improving outcomes for high-risk ALL: translating new discoveries into clinical care. Pediatr Blood Cancer. 2011;56(6):984–993. doi: 10.1002/pbc.22996. [DOI] [PubMed] [Google Scholar]
- 47.Chen IM, Harvey RC, Mullighan CG, et al. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2012;119(15):3512–3522. doi: 10.1182/blood-2011-11-394221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pui CH, Mullighan CG, Evans WE, et al. Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood. 2012;120(6):1165–1174. doi: 10.1182/blood-2012-05-378943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Investig. 2012;122(10):3407–3415. doi: 10.1172/JCI61203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moorman AV, Richards SM, Robinson HM, et al. Prognosis of children with acute lymphoblastic leukemia (ALL) and intrachromosomal amplification of chromosome 21 (iAMP21) Blood. 2007;109(6):2327–2330. doi: 10.1182/blood-2006-08-040436. [DOI] [PubMed] [Google Scholar]
- 52.Attarbaschi A, Mann G, Panzer-Grümayer R, et al. Minimal residual disease values discriminate between low and high relapse risk in children with B-cell precursor acute lymphoblastic leukemia and an intrachromosomal amplification of chromosome 21: the Austrian and German acute lymphoblastic leukemia Berlin–Frankfurt–Munster (ALL-BFM) trials. J Clin Oncol. 2008;26(18):3046–3050. doi: 10.1200/JCO.2008.16.1117. [DOI] [PubMed] [Google Scholar]
- 53.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 54.Schwab CJ, Chilton L, Morrison H, et al. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica. 2013;98(7):1081–1088. doi: 10.3324/haematol.2013.085175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mullighan CG, Miller CB, Radtke I, et al. BCR–ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 56.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10(2):125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krentz S, Hof J, Mendioroz A, et al. Prognostic value of genetic alterations in children with first bone marrow relapse of childhood B-cell precursor acute lymphoblastic leukemia. Leukemia. 2013;27(2):295–304. doi: 10.1038/leu.2012.155. [DOI] [PubMed] [Google Scholar]
- 59.Feng J, Tang Y. Prognostic significance of IKZF1 alteration status in pediatric B-lineage acute lymphoblastic leukemia: a meta-analysis. Leuk Lymphoma. 2013;54(4):889–891. doi: 10.3109/10428194.2012.723212. [DOI] [PubMed] [Google Scholar]
- 60.Cario G, Zimmermann M, Romey R, et al. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 2010;115(26):5393–5397. doi: 10.1182/blood-2009-11-256131. [DOI] [PubMed] [Google Scholar]
- 61.Ensor HM, Schwab C, Russell LJ, et al. Demographic, clinical, and outcome features of children with acute lymphoblastic leukemia and CRLF2 deregulation: results from the MRC ALL97 clinical trial. Blood. 2011;117(7):2129–2136. doi: 10.1182/blood-2010-07-297135. [DOI] [PubMed] [Google Scholar]
- 62.Harvey RC, Mullighan CG, Chen IM, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115(26):5312–5321. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loh ML, Zhang J, Harvey RC, et al. Tyrosine kinome sequencing of pediatric acute lymphoblastic leukemia: a report from the Children’s Oncology Group TARGET Project. Blood. 2013;121(3):485–488. doi: 10.1182/blood-2012-04-422691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carroll AJ, Crist WM, Link MP, et al. The t(1;14)(p34;q11) is nonrandom and restricted to T-cell acute lymphoblastic leukemia: a Pediatric Oncology Group study. Blood. 1990;76:1220–1224. [PubMed] [Google Scholar]
- 65.Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/S1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 66.van Oostveen J, Bijl J, Raaphorst F, et al. The role of homeobox genes in normal hematopoiesis and hematological malignancies. Leukemia. 1999;13:1675–1690. doi: 10.1038/sj.leu.2401562. [DOI] [PubMed] [Google Scholar]
- 67.Soulier J, Clappier E, Cayuela JM, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005;106:274–286. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 68.Speleman F, Cauwelier B, Dastugue N, et al. A new recurrent inversion, inv(7)(p15q34), leads to transcriptional activation of HOXA10 and HOXA11 in a subset of T-cell acute lymphoblastic leukemias. Leukemia. 2005;19:358–366. doi: 10.1038/sj.leu.2403657. [DOI] [PubMed] [Google Scholar]
- 69.Bernard OA, Busson-LeConiat M, Ballerini P, et al. A new recurrent and specific cryptic translocation, t(5;14)(q35;q32), is associated with expression of the Hox11L2 gene in T acute lymphoblastic leukemia. Leukemia. 2001;15:1495–1504. doi: 10.1038/sj.leu.2402249. [DOI] [PubMed] [Google Scholar]
- 70.Tycko B, Smith SD, Sklar J. Chromosomal translocations joining LCK and TCRB loci in human T cell leukemia. J Exp Med. 1991;174:867–873. doi: 10.1084/jem.174.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clappier E, Cuccuini W, Cayuela JM, et al. Cyclin D2 dysregulation by chromosomal translocations to TCR loci in T-cell acute lymphoblastic leukemias. Leukemia. 2006;20:82–86. doi: 10.1038/sj.leu.2404008. [DOI] [PubMed] [Google Scholar]
- 72.Karrman K, Kjeldsen E, Lassen C, et al. The t(X;7)(q22;q34) in paediatric T-cell acute lymphoblastic leukaemia results in over-expression of the insulin receptor substrate 4 gene through illegitimate recombination with the T-cell receptor beta locus. Br J Haematol. 2009;144:546–551. doi: 10.1111/j.1365-2141.2008.07453.x. [DOI] [PubMed] [Google Scholar]
- 73.Rowe JM. Prognostic factors in adult acute lymphoblastic leukaemia. Br J Haematol. 2010;150:389–405. doi: 10.1111/j.1365-2141.2010.08246.x. [DOI] [PubMed] [Google Scholar]
- 74.Van Vlierberghe P, van Grotel M, Tchinda J, et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood. 2008;111:4668. doi: 10.1182/blood-2007-09-111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zipfel PA, Zhang W, Quiroz M, et al. Requirement for Abl kinases in T cell receptor signaling. Curr Biol. 2004;14:1222–1231. doi: 10.1016/j.cub.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 76.Hebert J, Cayuela JM, Berkeley J, et al. Candidate tumor-suppressor genes MTS1 (p16INK4A) and MTS2 (p15INK4B) display frequent homozygous deletions in primary cells from T- but not from B-cell lineage acute lymphoblastic leukemias. Blood. 1994;84:4038–4044. [PubMed] [Google Scholar]
- 77.Palomero T, Sulis ML, Cortina M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maser RS, Choudhury B, Campbell PJ, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferrando A. NOTCH mutations as prognostic markers in T-ALL. Leukemia. 2010;24:2003–2004. doi: 10.1038/leu.2010.237. [DOI] [PubMed] [Google Scholar]
- 81.Balgobind BV, Van Vlierberghe P, van den Ouweland AM, et al. Leukemia-associated NF1 inactivation in patients with pediatric T-ALL and AML lacking evidence for neurofibromatosis. Blood. 2008;111:4322–4328. doi: 10.1182/blood-2007-06-095075. [DOI] [PubMed] [Google Scholar]
- 82.Van Vlierberghe P, Meijerink JP, Stam RW, et al. Activating FLT3 mutations in CD4+/CD8− pediatric T-cell acute lymphoblastic leukemias. Blood. 2005;106:4414–4415. doi: 10.1182/blood-2005-06-2267. [DOI] [PubMed] [Google Scholar]
- 83.Paietta E, Ferrando AA, Neuberg D, et al. Activating FLT3 mutations in CD117/KIT(+) T-cell acute lymphoblastic leukemias. Blood. 2004;104:558–560. doi: 10.1182/blood-2004-01-0168. [DOI] [PubMed] [Google Scholar]
- 84.Kleppe M, Lahortiga I, El Chaar T, et al. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:530–535. doi: 10.1038/ng.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Remke M, Pfister S, Kox C, et al. High-resolution genomic profiling of childhood T-ALL reveals frequent copy-number alterations affecting the TGF-beta and PI3K-AKT pathways and deletions at 6q15–16.1 as a genomic marker for unfavorable early treatment response. Blood. 2009;114:1053–1062. doi: 10.1182/blood-2008-10-186536. [DOI] [PubMed] [Google Scholar]
- 86.Van Vlierberghe P, Palomero T, Khiabanian H, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang J, Ding L, Holmfeldt L et al (2012) The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012;481:157–63. Zhang and colleagues performed whole genome sequencing of 12 ETPALL cases identifying activating mutations in genes regulating cytokine receptor and RAS signaling (67%), inactivating lesions disrupting hematopoietic development (58%), and histone-modifying genes (48%). Gene expression profile of ETPALL resulted similar to that of normal and myeloid leukemia hematopoietic stem cells, suggesting the possibility that myeloid-directed therapies might improve the poor outcome of ETPALL [DOI] [PMC free article] [PubMed]
- 88.Flex E, Petrangeli V, Stella L, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205:751–758. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 90.Breit S, Stanulla M, Flohr T, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108:1151–1157. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- 91.Asnafi V, Buzyn A, Le Noir S, et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) study. Blood. 2009;113:3918–3924. doi: 10.1182/blood-2008-10-184069. [DOI] [PubMed] [Google Scholar]
- 92.Baldus CD, Thibaut J, Goekbuget N, et al. Prognostic implications of NOTCH1 and FBXW7 mutations in adult acute T-lymphoblastic leukemia. Haematologica. 2009;94:1383–1390. doi: 10.3324/haematol.2008.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamada Y, Hatta Y, Murata K, et al. Deletions of p15 and/or p16 genes as a poor-prognosis factor in adult T-cell leukemia. J Clin Oncol. 1997;15:1778–1785. doi: 10.1200/JCO.1997.15.5.1778. [DOI] [PubMed] [Google Scholar]
- 94.Fizzotti M, Cimino G, Pisegna S, et al. Detection of homozygous deletions of the cyclin-dependent kinase 4 inhibitor (p16) gene in acute lymphoblastic leukemia and association with adverse prognostic features. Blood. 1995;85:2685–2690. [PubMed] [Google Scholar]
- 95.Markaki EA, Stiakaki E, Zafiropoulos A, et al. Mutational analysis of the cell cycle inhibitor Kip1/p27 in childhood leukemia. Pediatr Blood Cancer. 2006;47:14–21. doi: 10.1002/pbc.20730. [DOI] [PubMed] [Google Scholar]
- 96.Tosello V, Mansour MR, Barnes K, et al. WT1 mutations in T-ALL. Blood. 2009;114:1038–1045. doi: 10.1182/blood-2008-12-192039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Renneville A, Kaltenbach S, Clappier E, et al. Wilms tumor 1 (WT1) gene mutations in pediatric T-cell malignancies. Leukemia. 2010;24:476–480. doi: 10.1038/leu.2009.221. [DOI] [PubMed] [Google Scholar]
- 98.Gutierrez A, Sanda T, Ma W, et al. Inactivation of LEF1 in T-cell acute lymphoblastic leukemia. Blood. 2010;115:2845–2851. doi: 10.1182/blood-2009-07-234377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kleppe M, Soulier J, Asnafi V, et al. PTPN2 negatively regulates oncogenic JAK1 in T-cell acute lymphoblastic leukemia. Blood. 2011;117:7090–7098. doi: 10.1182/blood-2010-10-314286. [DOI] [PubMed] [Google Scholar]