Abstract
Evolutionary systems biology (ESB) is an emerging hybrid approach that integrates methods, models, and data from evolutionary and systems biology. Drawing on themes that arose at a cross-disciplinary meeting on ESB in 2013, we discuss in detail some of the explanatory friction that arises in the interaction between evolutionary and systems biology. These tensions appear because of different modeling approaches, diverse explanatory aims and strategies, and divergent views about the scope of the evolutionary synthesis. We locate these discussions in the context of long-running philosophical deliberations on explanation, modeling, and theoretical synthesis. We show how many of the issues central to ESB’s progress can be understood as general philosophical problems. The benefits of addressing these philosophical issues feed back into philosophy too, because ESB provides excellent examples of scientific practice for the development of philosophy of science and philosophy of biology.
Keywords: Evolutionary biology, Evolutionary systems biology, Explanation, Philosophy of biology, Philosophy of science, Systems biology
The interaction between evolutionary and molecular biology has taken various forms in the last several decades. Many of these interactions have been concerned with individual genes and particular protein products, or with comparative insights that treat whole genomes as static entities. Evolutionary systems biology (ESB) takes both evolutionary and molecular approaches further, by combining dynamic mechanistic models within multilevel evolutionary frameworks. There are several strands of ESB, with some focusing on genome architecture, others concentrating on population-level phenomena, and yet others concerned with the interactions of molecular networks. Both classic and novel evolutionary questions can be addressed by the wide range of tools and approaches deployed under the ESB umbrella. For example, new insights into epistasis may be gained (Segre et al. 2005), older views about network structure and causal properties rectified (Siegal et al. 2007), or the fitness consequences of stochastic gene expression addressed (Wang and Zhang 2011). In this article, which serves as a substantive introduction to a thematic section on ESB, we will not try to provide a general overview of the practices and history of the approach—we have done that in detail elsewhere (O’Malley 2012; Soyer 2012; Soyer and O’Malley 2013). Instead, we will show how valuable ESB is for philosophical discussion of biology and of science in general, and suggest that there may be a feedback loop between scientific and philosophical interpretations of ESB.
We will focus on how ESB illuminates several key topics to do with explanation, modeling, and the application of theory to a range of data. These themes arose out of a recent (2013) workshop on ESB, sponsored by the Konrad Lorenz Institute for Evolution and Cognition Research (The KLI Institute) near Vienna. In that meeting, participants from philosophical and scientific backgrounds engaged in discussions of key modeling strategies, conceptual machinery, and the relationship between evolutionary and systems biology. Three other articles in this thematic section will take particular topics from that workshop and discuss them in depth. In this introductory article, we will build on the rich exchanges witnessed at the workshop to address reasons for and against the existence of a special label of ESB. We will outline some of the “paradigm problems” ESB aims to solve. Once that groundwork is laid, we can address generally and specifically philosophical issues in ESB, and how discussing them might indeed have value for both the relevant science and the philosophy of science.
ESB as a Label
Integrative practice is something that marks ESB as a scientific activity. Its practitioners combine large-scale data analysis with experimentation, and quantitative modeling with qualitative mechanistic explanations. These combinations come in such a variety of forms that some commentators have expressed uncertainty about whether labeling these diverse research activities as a singular approach called ESB is justified or useful. Instead, they hold that existing fields—such as systems biology or population genetics—can simply add new tools and datasets to their repertoires, and thus produce useful knowledge without presuming that a new field or approach is developing. For example,
It seems that ESB covers a lot of ground. First systems biology—whatever it means—itself is pretty broad. And then on top of that, ESB apparently covers even more than SB. Still not sure what ESB is—I am torn about whether it could be interesting or completely flaky. I am a big fan of adding evolutionary approaches to just about any area of biology. So that alone makes me think about reading [these papers] to see whether there is anything] there. (Eisen 2012)
Other commentators have echoed these thoughts, as in some anonymous referee comments received by the authors on manuscripts in 2013. For example, one said that, “ESB has no real agenda of its own and is not exactly a distinct scientific discipline,” while other comments stated that they “just don’t see what type of research actually belongs to the realm of ESB.”
Some of the resistance to the label may arise from skepticism about what ESB could actually accomplish. From this point of view, “how” questions about the function of biological systems are enough; “why” questions about their evolution are just too speculative. This is a very practical concern because understanding the current functions of intracellular networks takes enormous effort. Trying to provide an evolutionary account of why that function and structure evolved may be far too premature. Other reasons against the need for a label are given by people working within the very research area that might be labeled as ESB. These researchers note that they have been practicing variants of ESB for many years, and from this perspective, the term is redundant because the practice preexisted it.
However, there are also several strong reasons on offer for using the label of ESB. Some of these reasons are epistemic, to do with the nature of knowledge production. Others are more sociological, to do with the creation of scientific communities that can advance these epistemic aims. Positive epistemic reasons for ESB revolve around the advantages and even the necessity of an approach that seeks to generalize findings across specific projects. For example, ESB proponents have pointed to common interests across the field, which include “a mechanistic understanding of the genotype-phenotype map, or insights into the evolutionary origin of network- or genome-level properties,” as one anonymous reviewer wrote recently. An associated motivation is that without evolutionary analyses, systems biology will fall short of what it could achieve. In a complementary way, evolutionary biology without systems biology might not make the progress it needs to in a time of rapid scientific change. Sociological reasons for ESB highlight the formation of a like-minded community as the basis for these epistemic rationales.
Since we cannot deny the empirical fact that ESB appears to exist, in that scientists self-describe their research as ESB or are willing to contribute to efforts with that label (e.g., Soyer 2012), we think the main issue is not one of whether there is a “field” but of how its research activities are configured. This configuration can be described as a flexible combination of approaches rather than a settled field of research. There are several different “fundamental questions” driving ESB, and they have not yet been reconciled in its current “turbulent phase of development” (Koonin and Wolf 2008, p. 24).
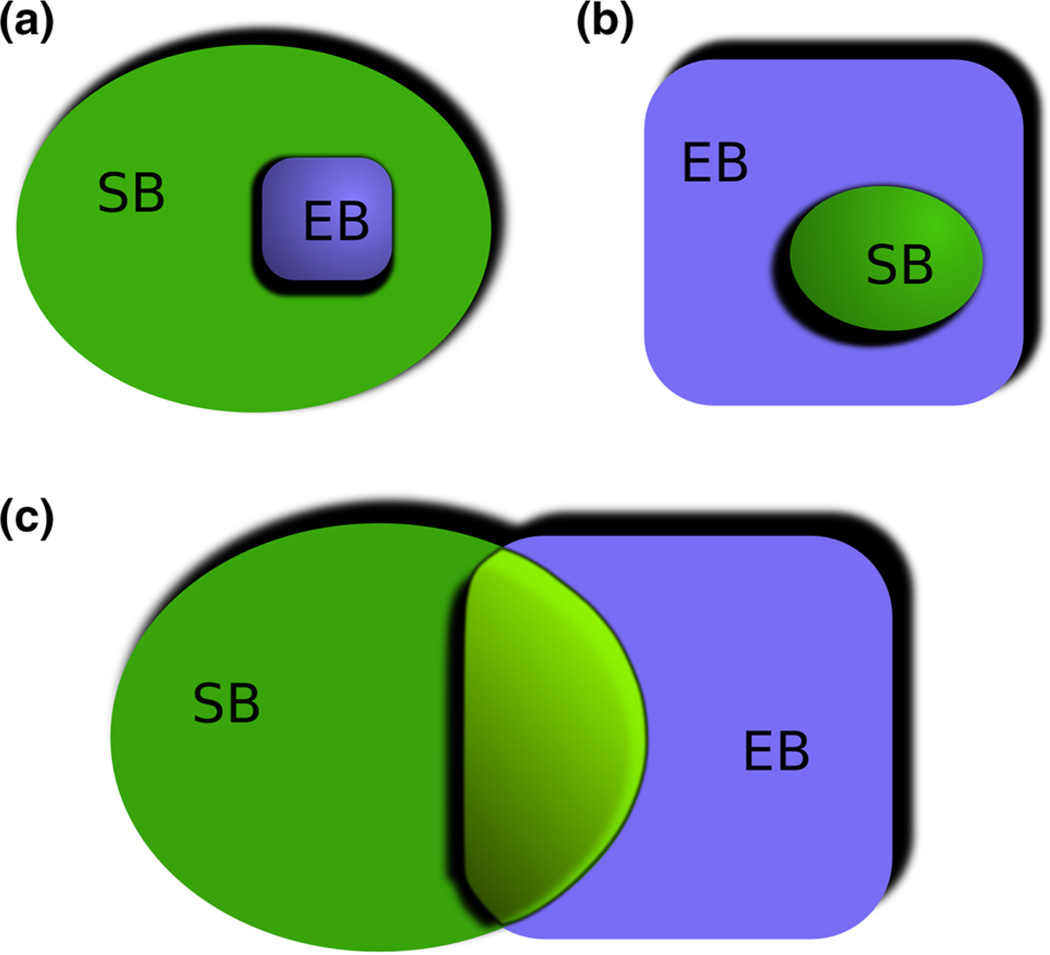
Advocates for ESB cite the possibility of unifying and synthesizing biological approaches as important motivations behind the field. In the workshop at the KLI, participants tended to focus on how systems biology could contribute to evolutionary biology (Fig. 1a). Many of the themes below continue in that vein. However, an alternative focus could be how an evolutionary perspective contributes to systems biology (Fig. 1b), with the emphasis on how that would change systems biology. But as some participants in the meeting concluded, in reality, the situation is more like the representation in Fig. 1c, in which there is overlap between evolutionary and systems biology, and this overlap is inhabited by different combinations of methods and data. As one participant noted, even if this overlap is only 1 % of the total activity going on in either systems biology or evolutionary biology, it is likely to be very valuable. Total fusion of evolutionary and systems biology is not only highly unlikely, but also undesirable: not all research goals will be shared. There are many different practical implications that arise from how the integration of systems and evolutionary biology happens, but our focus in the workshop and this article is on issues common to both areas of research. As another participant at the meeting observed, understanding what ESB actually is might best be achieved by understanding the core problems that attract researchers to the general area.
Fig. 1.
Interactions between systems biology (SB) and evolutionary biology (EB). Image created by Arno Steinacher. a Systems biology as a subset of practices contributing to evolutionary biology. b Evolutionary biology as a subset of practices contributing to systems biology. c Evolutionary biology and systems biology as largely distinct but fruitfully overlapping approaches, with overlap likely to increase with time
Paradigm Problems of ESB
There are several central problems that ESB sets itself the task of resolving or at least illuminating. These are issues on which systems biology or evolutionary biology on its own is not able to make sufficient headway. One of these core issues is the development of more detailed genotype-phenotype maps. While such mappings have been produced for decades now, they have so far lacked mechanistic elaboration and predictive capacity (Dean and Thornton 2007; Loewe 2009), with the exception of very specific cases such as RNA secondary structure (e.g., Wagner 2011). General statements about population size, mutation frequency, and fitness are immensely valuable for guiding experimental work and understanding evolution in the abstract. But for explaining and predicting evolutionary outcomes, more detail is needed about mutational effects on phenotypes, and how such effects are produced through mechanistic effects on underlying systems. Not only will this require far more detailed understandings of epistasis and pleiotropy, but also more extensive insight into phenotypic variability. Mechanistic models of cellular networks are key to filling in these gaps.
Whichever way genotype-phenotype maps are elaborated will require a combination of modeling strategies with experimental work. As some of the discussion below will demonstrate, the integration of “multilevel” models is a major theme in ESB. While at the meeting we discussed mostly mathematical models of different sorts, other modeling strategies in ESB also involve the material construction of intracellular networks—an activity known as synthetic biology—which are important modeling tools in their own right (Soyer and O’Malley 2013). These synthetic biology constructions add to the capacity of experimental evolutionary approaches by demonstrating alternative or potential evolutionary pathways. Enhancing the experimental contribution to genotype-phenotype mapping is of major importance, but the question we will focus on is when and how to integrate modeling strategies, and whether certain types of modeling should dominate ESB. While some of these questions can be answered contextually in relation to specific research questions, they may also be guided by general reflections on how evolutionary explanations are structured.
The distinction between “how” and “why” explanations has often been thought of as central to evolutionary biology, with “how” explanations relegated to physiological and developmental research (e.g., Mayr 1961). ESB represents the antithesis of this separation, because one of its characteristic activities is embedding proximate causal explanations within ultimate evolutionary explanations, and showing that there is in fact a two-way interaction between such causal bases. This topic has been the focus of recent philosophical and scientific attention (e.g., Laland et al. 2013; plus associated commentaries). One conclusion of this discussion is that,
Mayr’s dichotomous formulation [of proximate and ultimate causes] has now run its useful course and … evolutionary biology would be better served by a concept of reciprocal causation, in which causation is perceived to cycle through biological systems recursively. (Laland et al. 2013, p. 719)
Such claims would seem to lie at the heart of ESB and are of considerable importance for the Modern Synthesis of evolutionary biology. Applying these emerging philosophical arguments to ESB will be helpful for evaluating positions that urge new interpretations of the how-why or proximate-ultimate distinction.
Philosophical Themes in ESB
ESB offers a wealth of opportunity for both philosophy of science and philosophy of biology to advance some of their long-running deliberations on scientific practice. The issues we focus on are explanation, modeling, and theoretical synthesis. In addition to enriching philosophy, philosophical analyses of ESB probe and potentially make contributions to several of the issues that are central to ESB’s own development.
Explanation and Prediction
Philosophy of science has a particularly strong historical investment in the nature of explanation. Despite earlier efforts to give a mutually entailed account of explanation and prediction within a “covering law account” (Hempel and Oppenheim 1948), philosophers began to think of the two activities quite separately in the 1950s. Evolutionary biology was in fact one of the examples used by philosophers to show that explanation is not necessarily predictive (Scriven 1959). Explanation took over as the philosophical focus, with prediction becoming its lesser, dubiously related cousin (e.g., Rescher 1958). Today, explanation—and especially mechanistic explanation—rule the roost. However, the standard philosophical formulation of mechanistic explanation asserts that mechanistic explanations generate predictions, and that failed predictions can help refine the mechanistic explanatory model (Machamer et al. 2000; Douglas 2009).
While it might be reasonably straightforward to posit mechanistic explanations as the background to the foregrounded predictive aspects of systems biology, will this be the case for ESB? A perennial question of evolutionary biology has been its capacity for predictiveness, which— prior to the advent of large molecular datasets—was often decreed to be impossible (e.g., Mayr 1961; Gould 1990). ESB gives evolutionary biology new means by which to generate evolutionary predictions. The mechanistic models that underpin network biology are set out as mathematical equations. These models can generate predictions about future system states given additional knowledge about ecological conditions over time (Soyer 2010; Papp et al. 2011). However, philosophy of mechanism has so far tended to focus on concrete and easily visualized mechanisms, meaning philosophical accounts of explanation relate best to graphical representation and qualitative causal reasoning. This might work well for some molecular-physiological models and proximate explanation, but becomes problematic for probabilistic population-level phenomena and ultimate explanation (Matthewson and Calcott 2011; Levy 2013). Evolutionary models are very commonly concerned with these aggregated phenomena and their explanations. How then, do mechanistic models fit what is going on in evolutionary biology? Exploring this question is one of the primary challenges for philosophical analyses of ESB, and ESB research itself.
While systems biology’s very rationale is to combine explanation and prediction, evolutionary biology is currently framed by ultimate explanatory aims (Mayr 1961). By integrating systems biology, which requires precise models of mechanisms, ESB is attempting to model those mechanisms as they operate across stretches of evolutionary time. In its efforts to develop mechanistic explanations of evolutionary processes and outcomes, ESB brings in causal details to generate a fine-grained understanding of evolution. But it also aims to predict phenotypic consequences of genetic and environmental changes on evolving lineages (Papp et al. 2011). What ESB can do, therefore, is give philosophers a variety of ways in which to think about the explanation-prediction connection, including that of explanation on different timescales, with special reference to modeling strategies.
Modeling and Explanation
Because ESB arises at the confluence of different approaches and research questions, modeling tensions can emerge between different levels of explanatory model (Calcott et al. 2015, this issue). These tensions are especially strong between detailed mechanistic and law-based explanations, as one of the articles in this collection discusses (Green et al. 2014, this issue; see below). The philosophy of modeling has undergone an enormous upsurge in philosophy of science as it has replaced adherence to law-based theory by a focus on mechanistic models. Although there are diverse discussions of modeling in philosophy of science, one area that has developed very rapidly in the last decade is that of how models explain. This is of particular importance when different modeling strategies are used for the same research questions— something highly characteristic of ESB. In systems biology, the aim is to go beyond phenomenological descriptions and qualitative causal models, and towards prediction and control via mathematical modeling. But in addition to prediction, explanation is sought: “the structure of the model should somehow reflect the underlying mechanisms in the biological system” (Cedersund and Roll 2009, p. 904).
In philosophical accounts of mechanism, explanatory models describe in detail mechanisms and their behaviors. More abstract dynamic mathematical modeling is deemed by some philosophers to be explanatory only when it describes a mechanism in a very concrete way (Kaplan and Bechtel 2011; Kaplan and Craver 2011). While many biologists are entirely focused on such mechanisms—for good reason, because such models enable intervention—a more abstracted level of modeling (e.g., dynamical systems theory, which allows generalizable inferences to be made about biological systems over time) is also at work in ESB (for other fields, see Fagan 2012). Some workshop participants saw ESB’s goal as one of being able to combine these modeling approaches in order to give general high-level accounts that are of relevance to experimentalists. A key challenge for any integration of such models is to account for the influence of evolutionary forces on cellular and subcellular processes. Most approaches and tools from dynamical systems theory assume time invariance of the environment and hence constant parameter values. There are few practical tools for the analysis of nonlinear time-invariant dynamical models. From a philosophical perspective, particular formulations of dynamical systems theory, such as mathematical general systems theory (Wolkenhauer et al. 2012), may offer opportunities for philosophers of science to develop accounts of non-mechanistic explanation.
Philosophy of Evolution: Explanatory Synthesis
As well as philosophy of science issues, ESB addresses some of the core concerns of philosophy of biology. Historically, philosophy of biology has focused on evolutionary biology, and especially evolutionary theory in the form of the conceptual machinery of Darwinian evolutionary theory (rather than population genetics). Particular attention has been paid to conceptualizing units of selection and species. ESB is not of particular relevance to those efforts, but it does tread a path that is just as well worn in philosophy of biology. This shared interest has to do with evolutionary explanation. ESB highlights several important explanatory conflicts in evolutionary biology. They include those between non-adaptional and adaptive evolutionary explanation (see de Visser et al. 2003; Lynch 2012; Landry et al. 2013; Siegal 2013), proximate and ultimate explanation (see above), optimality and non-optimality explanation (Flamholz et al. 2013), and engineering versus evolutionary explanation (Calcott 2014).
A general theme that is of great relevance to the philosophy of evolution is whether and how ESB contributes to the quest for a revised Modern Synthesis of evolutionary biology. This contribution does not need to be in the sense of overthrowing established knowledge, but may simply have the aim of enhancement. A conservative way of thinking about ESB is that it merely adds new molecular tools and analyses to existing evolutionary approaches. A more radical view would hold that in this very integration of systems and evolutionary biology, the old framework changes into something different. But does this simply involve throwing a bit of evolutionary theory into systems biology, and a bit of systems biology into evolutionary biology? A point raised very strongly at the meeting was that evolutionary analyses in ESB need to be, as one participant said, “integral, not just tacked on.” What might this mean in terms of integrating a population genetics approach with a systems one?
Lately, additions to the Modern Synthesis have been described by philosophers and philosophically oriented scientists as an “Extended Synthesis,” in which there is an “ongoing shift from a population-dynamic account to a causal-mechanistic theory of phenotype evolution” (Pigliucci and Müller 2010, p. 12). This rebalancing is in part what is driving ESB, but not quite in the way these commentators suggest. Our view of ESB research so far is that it is not a matter of going from population-dynamic accounts to causal-mechanistic ones, but of integrating them or working out their relationships to a particular research goal. This is what is of central philosophical importance in ESB, and requires an elaboration of how to interpret Fig. 1.
Population and quantitative genetics offer formal models that describe average fitness in populations and statistical associations between genotypes and phenotypes, whereas systems biology provides quantitative models of the function of intracellular networks in individual organisms. There is no hard line between the two approaches, however, and crossover work already exists between systems biology and population genetics. Ultimately, the aim of integrating the two approaches is to have complete understanding of the fitness effects of variation in molecular networks at a populational level (Loewe 2009). For example, combining large-scale mutational analyses with mechanistic models would allow evolutionary predictions to be made at different levels of biological organization (Papp et al. 2011; Landry and Rifkin 2012). Very often, these sorts of proposals are made on the basis of metabolic networks, for which there is a great deal of data and characterization, as well as assumptions about steady states.
These analyses can be extended to molecular phenomena with unknown fitness effects, such as “noise” in gene expression. Noise refers generally to molecular fluctuations in biological processes; with regard to gene expression it is the stochastic variability of messenger RNA and protein levels in cells with identical genomes and environments. Very little is known about how such noise percolates through regulatory networks, although it is likely to be attenuated or amplified by various nonlin-earities in those networks (see, e.g., Raj et al. 2010). Noisy processes can be captured by mechanistic models of molecular activity on cellular biomass production (e.g., Wang and Zhang 2011), whereas the classic abstract models of population genetics do not incorporate effects that arise from intracellular noise. These classic models also shy away from nonlinear processes—even deterministic ones. In addition, mechanistic models can make predictions about those effects and what their alterations mean for evolving systems. The inevitability of noise and its effects on fitness make it clear that “molecular stochas-ticity must be included in a comprehensive evolutionary theory” (Wang and Zhang 2011, p. E74). Importantly, however, integrating mechanistic models into population genetic theory might not lead to complete incorporation of one sort of model by the other, but to the mutual revision of both population-genetic and mechanistic models.
The key place where traditional population/quantitative genetic theory and ESB intersect is the genotype-pheno-type map. Theorists have always made simplifying assumptions about the nature of this map. This is understandable because without simplifying assumptions, the models would be mathematically intractable and generalizations would be hard to come by. The danger, of course, is that if the assumptions are unfounded then so are the generalizations. Yet it must be said that in general, population genetics is not in crisis; indeed, it is experiencing a massive resurgence as genotype data on populations of humans and other species are being generated at a rapid and accelerating pace. Population genetics has been crucial to extracting meaning from these data by revealing, for example, recent adaptive events or demographic histories.
The situation for quantitative genetics is a bit different. The outpouring of human data from genome-wide association studies (GWAS), although leading to exciting discoveries of genes associated with important human traits, has revealed a consistent problem of “missing herit-ability.” This problem refers to the realization that the genetic loci found to contribute to variation in a particular trait typically account for a minor fraction of the variability expected to be explained by genetic causes. This gap raises the major problem of just how explanatory quantitative genetic explanations of traits are (Turkheimer 2011). The reason or reasons for the missing heritability remain to be determined, but several hypotheses concern aspects of the genotype-phenotype map that are central to ESB, such as robustness and epistasis (Zuk et al. 2012). Other proposed explanations for the missing heritability focus on the low power of (even extremely large) GWAS to detect all relevant loci: the missing heritability could be explained by genetic architectures in which very many loci of very small effect contribute to phenotypic variation (Rockman 2011). These explanations are not mutually exclusive. For example, the small effects could be context dependent and small only on average. This is a place where ESB has the potential to contribute substantially to evolutionary theory—by forcing the adoption of more complicated models of the genotype-phenotype map but at the same time constraining the potential forms those models could take.
The abstract connection in quantitative genetics between genotype and phenotype might not, however, be neatly supplemented by systems biological approaches. Much successful systems biology takes a modular approach, in which genes with a common function are conceptualized as having some independence from genes that do not contribute to that function (Wagner et al. 2007). The way in which modules of genes are experimentally mapped to functional phenotypes greatly reduces the complexity of the mapping process (Landry and Rifkin 2012). But unfortunately, module-based mapping is not the same sort of mapping that quantitative genetics aims at, due to the mutational complexities it addresses via statistical associations. Systems biology and quantitative genetics thus deploy different idealizations of gene interactions such as pleiotropy and epistasis. The modular approach of systems biology is unlikely to capture the subtleties of quantitative trait maps, even if the latter maps do not causally describe multiple levels of biological interaction (Landry and Rifkin 2012). However, despite the imperfect correspondence between such mapping approaches, there is considerable optimism in ESB that integrated mapping is feasible and valuable, even if it is more difficult than inserting one mapping methodology or result inside the other.
But ongoing attempts at this integration still leave untouched the epistemic challenges posed by the missing heritability problem: if robustness, epistasis, and/or weak causation are widespread, is there a limit to what we can know about the causes of phenotypic variation even with extremely large studies? Although this is a fascinating conundrum for philosophers to reflect on further, we suspect the biologists confronting this challenge will continue to attack it regardless of any theorized limit to knowledge. In fact, that limit is not clear. Missing heritability is comprised at least in part by hidden heritability (Gibson 2010) and what may still be missing are the appropriate methodologies to reveal that hidden fraction. And indeed, if ESB has one message for observers of the life sciences, it would be that problems that are understood as methodological limits to evolutionary biology are merely the best starting places for novel approaches in ESB.
A similarly challenging situation arises when evolutionary theory and ESB intersect in attempts to explain features of system organization. These are both global features, such as scale-free architectures, and local features, such as over-represented regulatory motifs. So far, many of these explanatory efforts have been adaptationist, with network structures and properties (e.g., robustness) being explained by selective advantage (see Siegal 2013). What is still lacking is a fully elaborated “non-adaptional theory”—incorporating but not restricted to neutral theory— of the evolution of biological systems. Some important work is already pointing toward ways in which to do this (e.g., de Visser et al. 2003; Solé and Valverde 2006; Soyer and Bonhoeffer 2006; Lynch 2007 a, b; Crombach and Hogeweg 2008; Wagner 2008b), but further development of appropriate null models for the evolution of complex networks is likely to become a major goal not only for ESB but also evolutionary biology as a whole.
However, the residual general issue embedded in questions about modifications to the Modern Synthesis is the fact that for some evolutionary biologists, the very idea that anything “revolutionary” could be underway with regard to the Modern Synthesis is suspect.
I think the theoretical tools—the idea of natural selection and mathematical population genetics—are already at hand, and although we may get new analytical tools (such as genomics!), the rest is just hard grinding work trying to understand speciation and natural selection in the wild. I may be wrong, but I don’t scent revolution in the wind yet. (Coyne 2009)
EvoDevo is sometimes cited as a truly novel contributor to the Modern Synthesis (e.g., Miiller 2007; Carroll 2008), but according to many other commentators, its inclusion does not greatly perturb the core conceptual apparatus of evolutionary biology (e.g., Sterelny 2000; Minelli 2010). Here, we are not particularly interested in that debate as it stands; far more compelling is the question of how EvoDevo itself can be understood in light of ESB. Some very promising results have been achieved along those lines already, with—for example—the development of a conceptual framework that has the scope for understanding phenotypic variability both quantitatively and dynamically (Jaeger et al. 2012). One of the articles in this collection will delve more deeply into epistemic questions about EvoDevo and dynamic mechanistic models (Jaeger et al. 2015, this issue—see below).
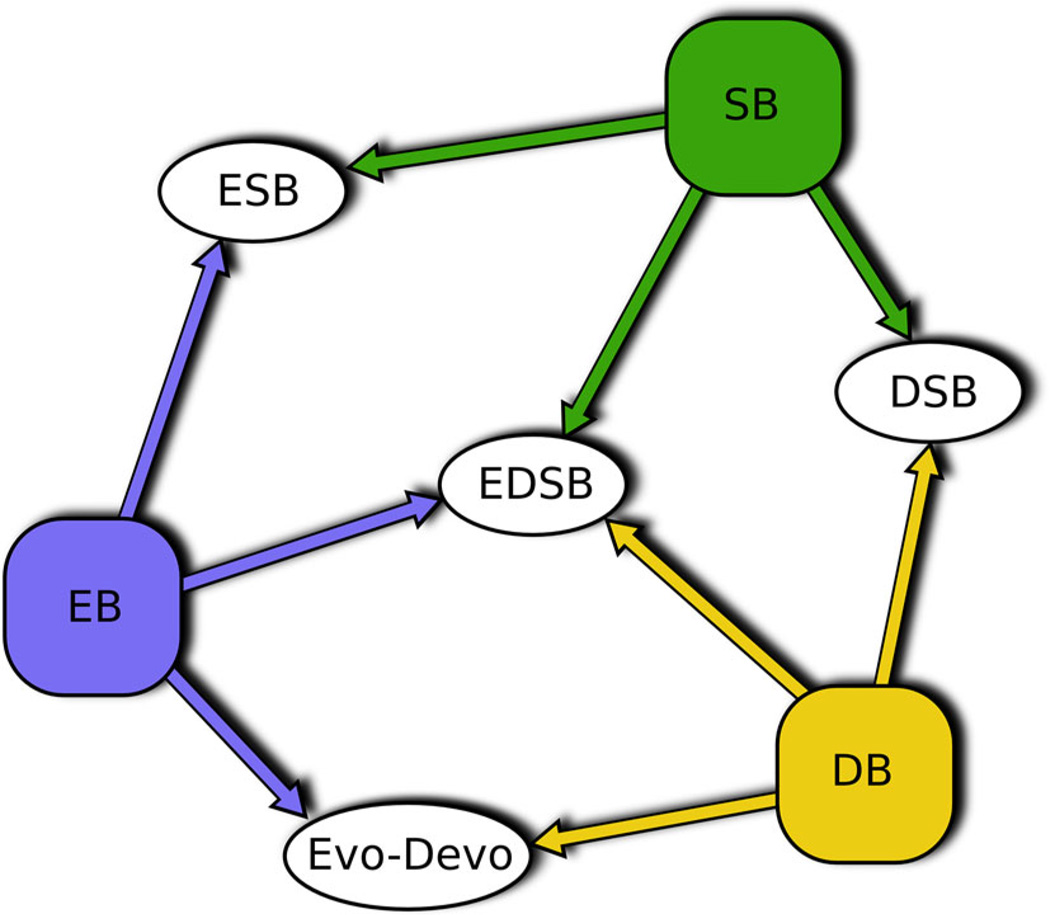
In addition to these advances, it is clear that developmental biology on its own is increasingly incorporating systems biology approaches, as the former increases its attention to modeling and genome-scale quantitative data (see Jaeger et al. 2015, this issue). Figure 2 expands Fig. 1 to represent the interactions between these approaches. This trend in itself is worth watching, to see the transformations brought about in developmental systems biology, and how it becomes “evolutionized” (Busser et al. 2008; Wunderlich and DePace 2011). This outcome, of developmental and systems approaches becoming integrated with evolutionary analyses, is also reached by developmental approaches that are already evolutionary (EvoDevo), and which integrate systems biological approaches to achieve multilevel explanations of particular developmental phenomena (Fig. 2). But as in the population-genetic and network biology intersection discussed above, the adaptiveness of organizational features in development cannot be taken for granted. Developmental systems drift and null models of genome evolution will have to be ruled out by rigorous analysis (True and Haag 2001; Siegal 2013), and consideration of constraints at different levels taken into account (de Visser et al. 2003).
Fig. 2.
Interactions between systems biology (SB), evolutionary biology (EB), and developmental biology (DB)
Figure 2 depicts general research approaches, in which methods, aims, and interests are shared in various configurations. Practitioners located in any general research area (developmental biology (DB), evolutionary biology (EB), systems biology (SB)) can combine elements of approaches from their neighbors. We see ESB, evolutionary developmental systems biology (EDSB), and developmental systems biology (DSB) as comprised of these overlaps rather than being some sort of fixed area into which practitioners move. EvoDevo, however, has a somewhat more defined identity. Moreover, we do not mean to indicate any hard boundaries between ESB and EDSB. We are merely suggesting that not all ESB is going to be concerned with metazoan (and sometimes plant) development, and that issues arising in EDSB—or for that matter, EvoDevo and DSB—may not always be shared with EB or SB. In this article, however, we have focused on issues we think are common to both ESB and EDSB.
Philosophy of Evolution: Scope
The final broad question raised by ESB is whether all biology must necessarily be evolutionary. Theodosius Dobzhansky’s famous quote about all biology being understood in the light of evolution can be interpreted as biological understanding being achieved “only in light of evolution.” However, this interpretation is a rather unnu-anced claim that that can be easily resisted. The other view is more difficult to reject: that non-evolutionary research approaches have made progress and will continue to do so without any consideration of evolutionary processes, thus refuting Dobzhanksy’s claim. For example, research into physiological function is often thought to be exempt from evolutionary questions, in that the focus is what a system does now rather than how it came to be that way (e.g., Boogerd et al. 2007). Once a good mechanistic account of such a system has been developed, this view agrees that there may be some epistemic and therapeutic benefits from applying evolutionary analyses to that system, in order to understand malfunction. However, according to this interpretation, evolutionary analysis is not necessary to achieve explanations of current function.
We think this position is balanced by numerous biological phenomena in which both mechanistic and evolutionary accounts are valuably combined in order to grasp current function as well as dysfunction. A classic example is that of the heart as a pump. There is a fairly complete understanding of the function of the heart from a “plumbing” perspective, but high levels of heart disease require an integrated evolutionary perspective that addresses dysfunction within the context of explaining the evolution of healthy function. The increasing rates of heart disease in industrialized societies is one of the prime examples used in discussions of evolutionary medicine (e.g., Nesse and Williams 1998). A more molecular but still familiar illustration can be drawn from adaptive immunity and the production of antibody diversity, which is understood in deep molecular detail. Despite this detail, the existence of a diversity-generating mechanism for antibodies requires an evolutionary explanation, and understanding the evolutionary forces shaping that diversity has important clinical implications (James and Tawfik 2003; Burton et al. 2005).
Heart function and immunity are both helpful examples because they are familiar to most people. In ESB, examples are more complicated but they illustrate this explanatory interdependence even more convincingly. For instance, mechanistic explanation of a process known as stochastic switching in gene regulation—whereby intracellular networks flip between two phenotypic states—turns out to require an evolutionary explanation and vice versa. Fluctuating selection, which arises in relation to variable environments, is not enough on its own to drive the emergence of stochastic switching. Noise or stochastic variability in gene expression is what tips intracellular systems into one state or the other, and this capacity to switch has consequences for fitness and evolvability (Kuwahara and Soyer 2012). Or, viewed from the perspective of robustness, mechanistic explanations alone cannot adequately characterize how systems respond robustly in different environments, and what the role of noise is in those responses (Felix and Wagner 2008). Only by combining both these explanations can we understand the switch mechanism, the contexts in which it will operate, and the fitness consequences for the entities involved.
In Summary
Overall, ESB can be understood as a meeting ground from which the mutual enrichment of systems and evolutionary approaches can result. Some of these benefits are to do with breadth. ESB enables attention to a greater diversity of evolved systems, and this comparative potential leads inevitably to evolutionary analyses across different levels of variation. But there is also the advantage of depth. By providing detailed mechanistic explanations of system properties that are embedded in a dynamic landscape, greater explanatory depth is achieved—not to mention predictive potential. But in addition, the discussions above demonstrate very clearly that some of the central issues in ESB are also core issues in philosophy of science and philosophy of biology. This alignment of interests indicates a potentially productive interplay between ESB and philosophical analysis.
Overview of Articles in the Thematic Section
As we noted above, this special collection of articles grew out of a conference on ESB, when philosophers and evolutionary systems biologists met to discuss this research area in depth. The aim was more to engage in dialogue than it was to select particular issues or reach a general conclusion. Nevertheless, we found our discussions repeatedly returned to the same themes and the articles we have now produced collaboratively echo those shared concerns. Presentations at the workshop initially looked as if they would cover many more diverse topics (see http://www.kli.ac.at/events/event-detail/1378332000/evolutionary-systemsbiology), but these naturally converged on the core topics below. One agreement reached by the whole group was that this particular collection of papers on ESB should in fact be driven by the philosophical issues and input from philosophers, rather than specific scientific themes being chosen as the foci. But as is obvious, philosophical interests overlap considerably with some of the more abstract scientific interests.
The first article following this introduction is a good demonstration of this overlap. In an explanation of the tensions between developmental biology and neo- Darwinian evolutionary biology, Sara Green, Melinda Fagan, and Johannes Jaeger suggest that these two fields have not only different causal attributions but also quite different explanatory aims. The authors elucidate these distinctions by depicting both a failed and a successful case of explanatory integration between the two fields. The article focuses on the contrast between the mechanistic models emerging from experimental work, and the dynamical systems models that feature in research seeking general principles. The reconciliation of these different aims and strategies offers a valuable example of how ESB can work when different explanatory projects are recognized, and of how both approaches combined can take the research further than they could when isolated from each other. The article also provides additional grist for the philosophical mill of explanation, and shows how ESB can advance current discussions in philosophy of modeling.
This theme, of explanatory integration, is pursued further by Jaeger, this time with coauthors Manfred Laubichler and Werner Callebaut. Jaeger et al. focus on evolving developmental systems and how these can be understood in light of the perspective afforded by dynamical systems theory. They argue that EvoDevo investigation need not be explanatorily constrained by traditional comparative methodology or more recent molecular experimental investigation. Both of these are reaching the limits of what they can achieve. Extended insight into the evolution of developmental systems can be gained by incorporating the quantitative mechanistic modeling central to ESB, so that development, ecology, and evolution can be addressed cohesively and predictively. Using the metaphor of exploration, Jaeger et al. suggest a local, targeted approach to the configuration space of a diversity of real-world developmental systems in their environments. While such an approach will not produce a single universal theory—something the paper justifies on the grounds of a perspectivist epistemology—it will enable a more integrated and generalizable understanding of evolving developmental systems. The implementation of this methodology and associated conceptual machinery could be understood as the culmination of the EvoDevo synthesis, in that it helps unify and extend existing more constrained findings. The authors urge optimism and a considerably shorter time frame for this synthesis than other commentators outside ESB have.
A nice variation on the explanatory theme is addressed by the final article in the collection, by Brett Calcott, Arnon Levy, Mark Siegal, Orkun Soyer, and Andreas Wagner. They examine the relationship in ESB between engineering and evolutionary methods and explanatory structures. Philosophers and scientists alike have often criticized analogies between engineering and evolution, with the former given connotations of perfected design and rational planning, and the latter those of tinkering (Jacob 1977). Many disanalogies between the two have been mooted (e.g., Lewontin 1996). A perspective advanced at the ESB meeting was that rather than thinking of a direct relationship between engineering and evolution, with a tendency to think of one approach as somehow more “fundamental,” it would be better to understand the relationship as a mediated one (Calcott 2014). Both engineering and evolution can be viewed as drawing on a pool of shared principles that belong to neither approach. Various principles can be taken back to the home discipline, modified appropriately, and formalized by different modeling languages. In their argument for the compatibilities between engineering (especially software engineering) and evolutionary approaches in an ESB context, Calcott et al. erode the traditional distinction between “genuine” models and “mere” metaphors in scientific practice. In the course of this analysis, they refine and elaborate the very concept of model itself, and thus show how investigations of ESB can contribute to philosophy of modeling.
In addition to the concepts and issues discussed in these articles, ESB is also a source for the debate and refinement of other philosophically important concepts: robustness, evolvability, and modularity. While these topics were discussed intensively at the meeting (particularly robustness and evolvability), they are only incidental to the papers in this collection simply because of the choice of focus. However, it is worth noting that ongoing analyses of robustness are of huge importance to ESB, which is developing not only mechanistic accounts of how robust systems evolve but also richer conceptualizations of robustness itself (e.g., Soyer and Pfeiffer 2010; Levy and Siegal 2012; Wagner 2012). Likewise, evolvability is a crucial ESB topic (e.g., Crombach and Hogeweg 2008; Wagner 2008a; Kuwahara and Soyer 2012) and philosophers will find ESB a treasure trove for future refinements of the current philosophical literature. Modularity, as noted above in the discussion of systems biology and quantitative genetics, is central to an understanding of the integration of these approaches, and the obstacles such integration faces (Landry and Rifkin 2012). The success or failure of ongoing ESB efforts to bring together different modeling strategies will be very useful to philosophical discussions of modularity (e.g., Callebaut 2005).
Benefits of this Workshop Encounter Between ESB and Philosophy of Science
All the topics discussed above demonstrate clearly that ESB exemplifies in nuanced and often new ways many of the issues philosophers of science and biology are eager to understand more fully. We think it should also be obvious that attention to ESB will allow philosophical discussions of biological explanation in particular to go further, and for philosophers to develop more sophisticated analyses of explanatory integration. Are there benefits to these philosophical advances for the scientists engaged in ESB? We think so, at several levels.
The first basic one chimes with what we called the “sociological” reason for using the label of ESB. Talking about what is distinctive, strong, and problematic in ESB with philosophers (and historians) of science is likely to function as an outreach exercise in addition to helping with community building. Furthermore, one of the main complaints made about ESB is that some of its key research topics—such as robustness and evolvability—are ill-defined. Whether or not this is really the case we will leave aside; we accept the practical challenge of posing research questions in language that is broadly accepted as precisely defined and logically sound. In their traditional role of clarifying imprecise concepts, philosophers could collaborate with ESB practitioners to devise conceptual tools that communicate more effectively. But there is more to philosophy than clarification, and philosophical model-building and evaluation of scientific models is something philosophers of science see as a disciplinary strength. An important area of cross-disciplinary interaction, therefore, concerns modeling, what it is meant to achieve, and how it achieves those goals. This article has outlined more specifically some of the key areas in ESB modeling where this interaction could be productive on both sides.
This philosophical-ESB interaction would not be confined to research that self-identifies as ESB. As we noted above, there are large areas of evolutionary biology and systems biology that do not necessarily see a need for integration (see Fig. 1). We suggest that a philosophy-ESB interaction helps provide insight into both the distinctions between those approaches and the reasons why in some important research contexts integration is not only desirable but necessary. As meeting participants noted, this integration cannot be superficial and may result in adjustments to the contributing approaches. Philosophers of biology, in virtue of their practical distance from the everyday science and yet a long preoccupation with the general issues of bringing different levels of explanation together, are in a position to at least comment in informed and potentially constructive ways about ongoing integration between evolutionary and systems biology. The converse is true too: philosophy of ESB should be able to propose and assess situations in which the integration of systems and evolutionary biology is neither useful nor necessary.
Concluding Thoughts and Future Directions
Drawing attention to the different foci and approaches of ESB helps make sense of the “turbulence” and broad scope of ESB (see above) by depicting it as a complex set of interacting practices and research questions that are productively understood in light of some central philosophical tensions. These tensions are by no means irresolvable, and aiming to address them is likely to lead to advances that are valuable even outside ESB (assuming a restricted definition of its activities). We have shown above some profound issues that are as philosophical as they are scientific. As an emerging hybrid approach, ESB has not been around long enough to paper over its philosophical cracks. We suggest that philosophers take advantage of this transparency, to develop richer philosophical accounts of explanatory integration and multilevel modeling. And we predict that scientists will take advantage of the philosophical depths of ESB (even if they don’t call it philosophy) to continue the ongoing transformation of evolutionary theory in the systems-biology era.
Acknowledgments
Melinda Fagan, Olaf Wolkenhauer, and Michael Lynch made helpful comments on an earlier draft of this paper. Arno Steinacher kindly drew the images. We thank the KLI for funding and hosting this meeting in Konrad Lorenz’s summer mansion in Altenberg, near Vienna. Eva Lackner, Isabella Sarto-Jackson, Werner Callebaut, and Gerd Miiller made every aspect of the meeting run smoothly and effectively. But above all, we thank the participants who invested so much time, thought, and effort in their contributions to this meeting. This was the case not only for the individual presentations, but also—even more so—for the wonderfully enriching discussions that followed each talk and concluded the meeting. MAO thanks the Australian Research Council and University of Sydney for Future Fellowship Funding; OSS acknowledges funding from EPSRC Research Grant EP/1017445/1; MLS acknowledges funding from National Institutes of Health grants R01GM086673 and R01GM097415, and National Science Foundation grant IOS-1258078.
Contributor Information
Maureen A. O’Malley, Email: maureen.omalley@sydney.edu.au, Department of Philosophy, University of Sydney, Sydney, Australia.
Orkun S. Soyer, School of Life Sciences, University of Warwick, Coventry, UK
Mark L. Siegal, Department of Biology, Center for Genomics and Systems, Biology, New York University, New York, NY, USA
References
- Boogerd FC, Bruggeman FJ, Hofmeyr J-HS, Westerhoff HV. Afterthoughts as foundations for systems biology. In: Boogerd FC, Bruggeman FJ, Hofmeyr J-HS, Westerhoff HV, editors. Systems biology: philosophical foundations. Amsterdam: Elsevier; 2007. pp. 321–336. [Google Scholar]
- Burton DR, Stanfield RL, Wilson IA. Antibody vs. HIV in a clash of evolutionary titans. Proc Natl Acad Sci USA. 2005;102:14943–14948. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busser BW, Bulyk ML, Michelson AM. Toward a systems-level understanding of developmental regulatory networks. Curr Opin Genet Dev. 2008;18:521–529. doi: 10.1016/j.gde.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcott B. Engineering and evolvability. Biol Philos. 2014 [Google Scholar]
- Calcott B, Levy A, Siegal ML, et al. Engineering and biology: counsel for a continued relationship. Biol Theory. 2015 doi: 10.1007/s13752-014-0198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut W. The ubiquity of modularity. In: Callebaut W, Rasskin-Gutman D, editors. Modularity: understanding the development and evolution of natural complex systems. Cambridge: MIT Press; 2005. pp. 3–28. [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Cedersund G, Roll J. Systems biology: model based evaluation and comparison of potential explanations for given biological data. FEBS J. 2009;276:903–922. doi: 10.1111/j.1742-4658.2008.06845.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA. Are we ready for an “extended evolutionary synthesis”? [Accessed 11 December 2014];Why evolution is true (blog) 2009 Feb 16; https://whyevolutionistrue.wordpress.com/2009/02/16/are-we-ready-for-an-extended-evolutionary-synthesis/
- Crombach A, Hogeweg P. Evolution of evolvability in gene regulatory networks. PLoS Comput Biol. 2008;4(7):el000112. doi: 10.1371/journal.pcbi.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser JAGM, Hermisson J, Wagner GP, et al. Evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas HE. Reintroducing prediction to explanation. Philos Sci. 2009;76:444–463. [Google Scholar]
- Eisen JA. Not a #badomics word but—”Evolutionary Systems Biology” is—well—pretty complex. [Accessed 11 December 2014];Tree of Life (blog) 2012 Jul 28; http://phylogenomics.blogspot.com.au/2012/07/not-badomics-word-but-evolutionary.html.
- Fagan MB. Waddington redux: models and explanation in stem cell and systems biology. Biol Philos. 2012;27:179–213. [Google Scholar]
- Felix M-A, Wagner A. Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity. 2008;100:132–140. doi: 10.1038/sj.hdy.6800915. [DOI] [PubMed] [Google Scholar]
- Flamholz A, Noor E, Bar-Even A, et al. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc Natl Acad Sci USA. 2013;110:10039–10044. doi: 10.1073/pnas.1215283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Hints of hidden heritability in GWAS. Nat Genet. 2010;42:558–560. doi: 10.1038/ng0710-558. [DOI] [PubMed] [Google Scholar]
- Gould SJ. Wonderful life: The Burgess shale and the nature of history. London: Hutchinson Radius; 1990. [Google Scholar]
- Green S, Fagan M, Jaeger J. Explanatory integration challenges in evolutionary systems biology. Biol Theory. 2014 [Google Scholar]
- Hempel CG, Oppenheim P. Studies in the logic of explanation. Philos Sci. 1948;15:135–175. [Google Scholar]
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- Jaeger J, Irons D, Monk N. The inheritance of process: a dynamical systems approach. J Exp Zool (Mol Dev Evol) 2012;318B:519–612. doi: 10.1002/jez.b.22468. [DOI] [PubMed] [Google Scholar]
- Jaeger J, Laubichler M, Callebaut W. The comet cometh: evolving developmental systems. Biol Theory. 2015 doi: 10.1007/s13752-015-0203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LC, Tawfik DS. Conformational diversity and protein evolution—a 60-year-old hypothesis revisited. Trends Biochem Sci. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- Kaplan DM, Bechtel W. Dynamical models: an alternative or complement to mechanistic explanations? Top Cogn Sci. 2011;3:438–444. doi: 10.1111/j.1756-8765.2011.01147.x. [DOI] [PubMed] [Google Scholar]
- Kaplan DM, Craver CF. The explanatory force of dynamical and mathematical models in neuroscience: a mechanistic perspective. Philos Sci. 2011;78:601–627. [Google Scholar]
- Koonin EV, Wolf YI. Evolutionary systems biology. In: Pagel M, Pomiankowski A, editors. Evolutionary genomics and proteomics. Sunderland, MA: Sinauer; 2008. pp. 11–25. [Google Scholar]
- Kuwahara H, Soyer OSS. Bistability in feedback circuits as a byproduct of evolution of evolvability. Mol Syst Biol. 2012;8:564. doi: 10.1038/msb.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laland KN, Odling-Smee J, Hoppitt W, Uller T. More on how and why: cause and effect in biology. Biol Philos. 2013;28:719–745. doi: 10.1007/s10539-013-9380-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Rifkin SA. The genotype-phenotype maps of systems biology and quantitative genetics: distinct and complementary. In: Soyer OS, editor. Evolutionary systems biology. New York: Springer; 2012. pp. 371–398. [DOI] [PubMed] [Google Scholar]
- Landry CR, Levy ED, Rabbo DA, Tarassov K, Michnick SW. Extracting insight from noisy cellular networks. Cell. 2013;155:983–989. doi: 10.1016/j.cell.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Levy A. Three kinds of new mechanism. Biol Philos. 2013;28:99–114. [Google Scholar]
- Levy SF, Siegal ML. The robustness continuum. In: Soyer OS, editor. Evolutionary systems biology. New York: Springer; 2012. pp. 431–452. [Google Scholar]
- Lewontin RC. Evolution as engineering. In: Collado-Vides J, Magasanik B, Smith TF, editors. Integrative approaches to molecular biology. Cambridge: MIT Press; 1996. pp. 1–10. [Google Scholar]
- Loewe L. A framework for evolutionary systems biology. BMC Syst Biol. 2009;3:27. doi: 10.1186/1752-0509-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci USA. 2007a;104(Suppl 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007b;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- Lynch M. Evolutionary layering and the limits to cellular perfection. Proc Natl Acad Sci USA. 2012;109:18851–18856. doi: 10.1073/pnas.1216130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer P, Darden L, Craver CF. Thinking about mechanisms. Philos Sci. 2000;67:1–25. [Google Scholar]
- Matthewson J, Calcott B. Mechanistic models of population-level phenomena. Biol Philos. 2011;26:737–756. [Google Scholar]
- Mayr E. Cause and effect in biology. Science. 1961;134:1501–1506. doi: 10.1126/science.134.3489.1501. [DOI] [PubMed] [Google Scholar]
- Minelli A. Evolutionary developmental biology does not offer a significant challenge to the neo-Darwinian paradigm. In: Ayala FJ, Arp R, editors. Contemporary debates in philosophy of biology. Maiden: Wiley; 2010. pp. 213–226. [Google Scholar]
- Müller GB. Evo-devo: extending the evolutionary synthesis. Nat Rev Genet. 2007;8:943–949. doi: 10.1038/nrg2219. [DOI] [PubMed] [Google Scholar]
- Nesse RM, Williams GC. Evolution and the origins of disease. Sci Am. 1998;279:86–93. doi: 10.1038/scientificamerican1198-86. [DOI] [PubMed] [Google Scholar]
- O’Malley MA. Evolutionary systems biology: historical and philosophical perspectives on an emerging synthesis. In: Soyer OS, editor. Evolutionary systems biology. New York: Springer; 2012. pp. 1–28. [DOI] [PubMed] [Google Scholar]
- Papp B, Notebaart RA, Pal C. Systems-biology approaches for predicting genomic evolution. Nat Rev Genet. 2011;12:591–602. doi: 10.1038/nrg3033. [DOI] [PubMed] [Google Scholar]
- Pigliucci M, Müller GB. Elements of an extended evolutionary thesis. In: Pigliucci M, Müller GB, editors. Evolution: the Extended Synthesis. Cambridge: MIT Press; 2010. pp. 3–17. [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penentrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescher N. On prediction and explanation. Br J Philos Sci. 1958;8:281–290. [Google Scholar]
- Rockman MV. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution. 2011;66:1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriven M. Explanation and prediction in evolutionary theory. Science. 1959;130:477–482. doi: 10.1126/science.130.3374.477. [DOI] [PubMed] [Google Scholar]
- Segre D, Deluna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- Siegal ML. Evolution of molecular networks. In: Losos JB, Baum DA, Futuyma DJ, et al., editors. The Princeton guide to evolution. Princeton: Princeton University Press; 2013. pp. 428–436. [Google Scholar]
- Siegal ML, Promislow DE, Bergman A. Functional and evolutionary inference in gene networks: does topology matter? Genetica. 2007;129:83–103. doi: 10.1007/s10709-006-0035-0. [DOI] [PubMed] [Google Scholar]
- Solé RV, Valverde S. Are network motifs the spandrels of cellular complexity? Trends Ecol Evol. 2006;8:419–422. doi: 10.1016/j.tree.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Soyer OS. The promise of evolutionary systems biology: lessons from bacterial chemotaxis. Sci Signal. 2010;3:128, e23. doi: 10.1126/scisignal.3128pe23. [DOI] [PubMed] [Google Scholar]
- Soyer OS, editor. Evolutionary systems biology. New York: Springer; 2012. [Google Scholar]
- Soyer OS, Bonhoeffer S. Evolution of complexity in signaling pathways. Proc Natl Acad Sci USA. 2006;103:16337–16342. doi: 10.1073/pnas.0604449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyer OS, O’Malley MA. Evolutionary systems biology: what it is and why it matters. BioEssays. 2013;35:696–705. doi: 10.1002/bies.201300029. [DOI] [PubMed] [Google Scholar]
- Soyer OS, Pfeiffer S. Evolution under fluctuating environments explains observed robustness in metabolic networks. PLoS Comput Biol. 2010;6(8):el000907. doi: 10.1371/journal.pcbi.1000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterelny K. Development, evolution, and adaptation. Philos Sci. 2000;67:S369–S387. (Proc) [Google Scholar]
- True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- Turkheimer E. Still missing. Res Hum Dev. 2011;8:227–241. [Google Scholar]
- Wagner A. Robustness and evolvability: a paradox resolved. Proc R Soc Lond B. 2008a;275:91–100. doi: 10.1098/rspb.2007.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Neutralism and selectionism: a network-based reconciliation. Nat Rev Genet. 2008b;9:965–974. doi: 10.1038/nrg2473. [DOI] [PubMed] [Google Scholar]
- Wagner A. Genotype networks shed light on evolutionary constraints. Trends Ecol Evol. 2011;26:577–584. doi: 10.1016/j.tree.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Wagner A. The role of robustness in phenotypic adaptation and innovation. Proc R Soc Lond B. 2012;279:1249–1258. doi: 10.1098/rspb.2011.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Pavlicev M, Cheverud JM. The road to modularity. Nat Rev Genet. 2007;8:921–931. doi: 10.1038/nrg2267. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang J. Impact of gene expression noise on organismal fitness and the efficacy of natural selection. Proc Natl Acad Sci USA. 2011;108:E67–E76. doi: 10.1073/pnas.1100059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkenhauer O, Shibata D, Mesarović MD. The role of theorem proving in systems biology. J Theor Biol. 2012;300:57–61. doi: 10.1016/j.jtbi.2011.12.023. [DOI] [PubMed] [Google Scholar]
- Wunderlich Z, DePace AH. Modeling transcriptional networks in Drosophila development at multiple scales. Curr Opin Genet Dev. 2011;21:711–718. doi: 10.1016/j.gde.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]