Abstract
Fluorescent proteins are increasingly being used to analyze cellular gene expression and to facilitate tracking of cell lineages in vivo. One of these, enhanced yellow fluorescent protein (EYFP) has several properties such as intense fluorescence and little to no toxicity in cells, which makes it an excellent molecule to label proteins and cells of interest. In live cells, visualization of EYFP has been highly successful; however, detection of EYFP in lymphoid tissue sections, particularly in combination with other markers of interest has been difficult. This is because of the enhanced solubility of EYFP in the absence of fixation. When extended fixation protocols are employed, EYFP is preserved but detection of other cellular antigens becomes problematic due to over fixation. Here we demonstrate that EYFP-expressing T and B cells can be efficiently visualized in lymphoid tissue sections without compromising the ability to detect other cellular markers.
Keywords: EYFP, Immunofluorescence microscopy, Lymphoid organs, Tissue sections
1. Introduction
The detection of gene products in relation to the cells that produce them and their subcellular localization are often the focus of research projects. In the past, proteins have been tagged with FLAG or hemagglutinin (HA) epitopes. The tagged protein of interest can be studied by staining for the FLAG or HA tag by using commercially available antibodies. However, in recent years fluorescent proteins have become a popular method for tagging proteins. The advantage of these molecules is that they fluoresce when exposed to certain wavelengths of light. Therefore instead of indirectly visualizing a protein through antibody staining, researchers can use the endogenous fluorescence of these molecules to monitor expression. Fluorescent proteins have been applied to a wide range of studies including analyzing gene expression within tissues, the cellular location of a protein, and tracking a cell population of interest.
Green fluorescent protein (GFP, Shimomura et al., 1962) was one of the first fluorescent proteins to be extensively used in biological research. One downfall of GFP was its low intensity of fluorescence (Patterson et al., 1997). A significant amount of research was done to analyze GFP's structure and to generate mutants that would be more advantageous to use. One GFP variant, enhanced yellow fluorescent protein (originally called GFP-10), has been an extremely promising replacement (Ormo et al., 1996). Enhanced yellow fluorescent protein (EYFP) was generated by introducing the following four mutations in to GFP: T203Y, S65G, V68L, and S72A. Together these mutations alter the spectral properties and fluorescence intensity of EYFP as compared to GFP.
EYFP's spectral properties and intense fluorescence make it a desirable marker to use in gene expression and cell-tracking studies. It has been expressed under the control of specific promoters or induced via cre-mediated recombination as a nontoxic, soluble molecule in neurons (Sawamoto et al., 2001; Metzger et al., 2002; Schmid et al., 2006) or hematopoietic cells (de Boer et al., 2003) and even in all cell types in mice (Srinivas et al., 2001). EYFP has also been used to study subcellular localization of proteins by fusing it with a variety of proteins including histones (Fraser et al., 2005), Phospholipase C-ζ (Coward et al., 2006), β-actin (Gertzberg et al., 2007), and nyctalopin (Gregg et al., 2007). Once expressed, various assays have been adopted to analyze the EYFP expression profile. Flow cytometric analysis has been highly successful in analyzing EYFP expression within bulk tissue samples. Fluorescent microscopy has been successfully used to detect EYFP expression in live tissue samples and live cells cultured in vitro. However, attempts to analyze the expression of EYFP in fixed tissue sections have met with limited success. This is because EYFP is highly soluble, and the extensive fixation protocols used to preserve EYFP lead to over fixation of other cellular antigens thereby precluding their detection. Here we have expanded our initial description of EYFP preservation in situ (Dauner et al., 2008), and further evaluated a variety of fixation processes and their ability to subsequently preserve EYFP. We show that cardiac perfusion of mice with a paraformaldehyde solution allows for the visualization of EYFP in situ in lymphoid tissues while maintaining staining of a variety of phenotypic markers and structural features.
2. Materials and methods
2.1. Mice
ROSA26R-EYFP (Srinivas et al., 2001) mice were crossed with either the Granzyme B-Cre mouse (Jacob and Baltimore, 1999) or germinal center-cre mouse (Chappell and Jacob, 2006). In double transgenic offspring of these matings, activated T cells and B cells, respectively, express EYFP after cre-mediated recombination. Mice were maintained at Emory University following the guidelines of the Institutional Animal Care and Use Committee and used at an age of 4–8 weeks.
2.2. Fixation methods and tissue section preparation
Spleens were harvested from transgenic mice and embedded in OCT (Sakura) by flash freezing them in liquid nitrogen-chilled isopentane (Fisher Scientific). The frozen tissue blocks were stored at −80 °C until use. When needed, frozen sections were cut at a thickness of 6 µm and mounted onto to glass slides (Fisher Scientific). The tissue sections were fixed either with cold acetone or 1:1 acetone:methanol for 10 min, 4% paraformaldehyde (PFA) for 5 min, or by formaldehyde vapors for 2 h at −20 °C (Jockusch et al., 2003). In separate experiments, whole organs were fixed by overnight immersion in either PBS containing 4% PFA (Fisher Scientific) or IHC Zinc Fixative (BD Pharmingen). The following day, tissues were embedded in OCT and processed for frozen sectioning as described above. Cohorts of mice were also fixed by cardiac perfusion (Dauner et al., 2008). Briefly, mice were deeply anesthetized with a single, lethal, intraperitoneal injection of 2.5 mg ketamine (Hospira, Inc.) and 0.25 mg X-ject SA (Phoenix Scientific, Inc.). The hearts of anesthetized mice were exposed and the right atrium was clipped with surgical scissors. A 26-gauge needle was inserted into the left ventricle of the heart and 8 ml of PBS (pH. 7.4) was administered with a 10-milliliter syringe. This was followed by the administration of 8 ml of 4% PFA dissolved in PBS (pH7.4). On average, 8ml of solution were administered over a 3-minute interval. Tissues were removed from the deceased mice and embedded in OCT and the blocks were stored at −80 °C. When needed, 6 µm thick frozen tissue sections were cut with a cryostat and allowed to dry overnight at room temperature. Dried tissues were stored at −80 °C for future use.
2.3. Immunofluorescent staining and antibodies
On the day of staining, slides were removed from the −80 °C freezer and thawed at room temperature. Sections were equilibrated in PBS and blocked with Blocking Buffer consisting of PBS containing 2% normal goat serum (Vector Labs) and 0.05% Tween-20 (Fisher Scientific) for 30 min. Antibodies were diluted in Blocking Buffer and all staining was done at room temperature using Shandon Coverplate disposable immunostaining chambers (Thermo Scientific). Primary and secondary antibodies were incubated with sections for 2 and 1 h, respectively. To detect EYFP rabbit polyclonal anti-GFP serum (Molecular Probes) was used as a primary stain and visualized with fluorochrome conjugated goat anti-rabbit IgG. Sections were co-stained with 1–5 µg/ml of fluorochrome- or biotin-labeled antibodies against B220, CD4, CD8, CD11b, CD11c, F4/80 (eBioscience), ER-TR9, MOMA-1 (BMA Biomedical), PNA (Vector Labs), Syndecan-1, Thy1.2 (BD Pharmingen), SLC (R&D Systems), IgM, IgD (Molecular Probes), MOMA-2, MARCO (Serotech) or the NP hapten coupled to chicken γ globulin (NP-CG). Fluorochrome conjugated anti-rat IgG was used to detect purified anti-CD169 (Serotech) and ER-TR7 (BMA Biomedical). In experiments aimed at localizing LCMV antigens we used anti-LCMV sera from infected guinea pigs and visualized the bound antibodies with fluorochrome-conjugated anti-guinea pig antibodies (Molecular Probes). Following staining, we mounted the sections using ProLong Gold antifade with or without DAPI (Molecular Probes). Images were collected using a Nikon E600 fluorescent microscope and Spot Advanced software. The number of pixels composing a DAPI-based nuclear stain was used to compare the relative size of nuclei from different tissue preparation.
3. Results
3.1. EYFP is lost from tissue sections fixed with fixatives that do not crosslink proteins
Tissues for these studies were from the offspring of ROSA26R-EYFP mice bred with one of two transgenic cre mice our lab has generated. The ROSA26R-EYFP mouse is a cre-reporter strain designed to permanently express EYFP after a cre-mediated recombination event (Srinivas et al., 2001). Crossing the ROSA26R-EYFP mouse with the Granzyme B Cre (GBC) mouse (Jacob and Baltimore, 1999) produces mice in which activated T cells express the transgenic marker EYFP. Similarly, we visualized germinal centers and the memory B cells they generate within the offspring of germinal center-cre (GCC) mice (Chappell and Jacob, 2006) mated with the ROSA26R-EYFP strain. These two mouse model systems will be referred to as GBC×RYFP and GCC×RYFP, respectively.
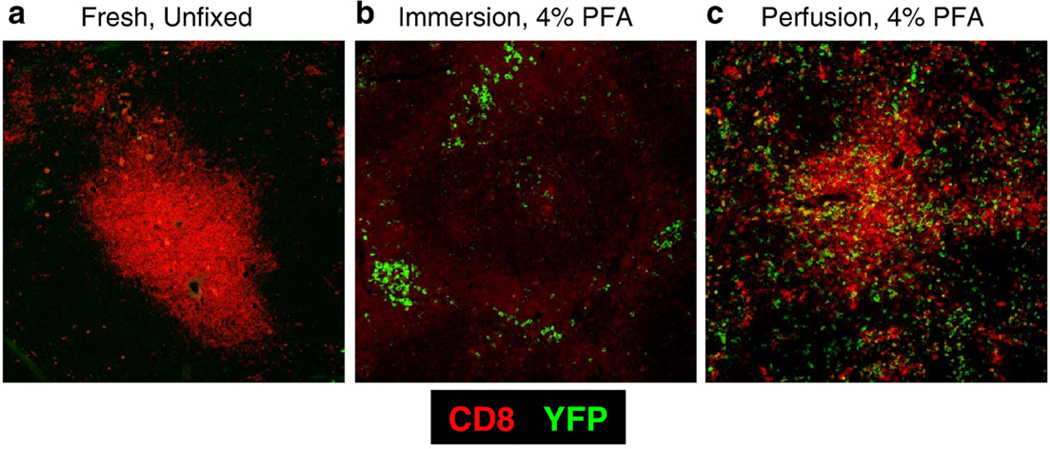
The objective of this study was to optimize the fixation protocol such that soluble EYFP as well as other cellular antigens would be preserved for in situ detection. First, we assessed standard preparations of fresh frozen sections for their ability to preserve EYFP within tissue sections. Sections exposed to cold acetone or a 1:1 mixture of acetone:methanol resulted in an inability to visualize EYFP, but as expected anti-CD8 staining was quite robust (Fig. 1a). One possible explanation for these results was that soluble EYFP was not being maintained in fresh tissues sections. Therefore, we reasoned that crosslinking EYFP to cellular proteins might be necessary to subsequently visualize EYFP in situ. Therefore, we attempted to crosslink EYFP to cellular proteins by post-fixing frozen sections with 4% PFA at room temperature, but this failed to fix EYFP as well (data not shown). It has been reported that exposing tissue sections to formaldehyde vapors could lead to preservation of cellular GFP (Jockusch et al., 2003). However, this method, in our hands, did not succeed in preserving EYFP (data not shown). In conclusion, none of the fixation protocols we tried on fresh cryostat-cut frozen sections led to preservation and subsequent visualization of EYFP in situ.
Fig. 1.
Cardiac perfusion with PFA retains the ability to visualize both EYFP and CD8+ cells in situ. Representative images depicting EYFP and CD8 staining within fresh, unfixed (a), PFA immersion-fixed (b), and PFA perfusion-fixed tissues (c) are presented. Images shown are at a magnification of 20×.
3.2. Whole tissue fixation with PFA efficiently preserves cellular EYFP levels but prevents detection of other cellular markers
Earlier fixation attempts on frozen tissue sections led to the loss of EYFP. So we fixed the entire organ before embedding in OCT and cutting frozen sections. Briefly, we immersed whole spleens in one of two fixative solutions overnight — formalin free, IHC Zinc Fixative from BD Pharmingen or 4% PFA. The Zinc based fixative was unable to preserve EYFP in tissue sections (data not shown). Interestingly, whole organ fixation in 4% PFA resulted in excellent preservation of cellular EYFP (Fig. 1b). To our disappointment, the anti-CD8 antibody tested did not stain cellular markers in these sections (Fig. 1b).
3.3. Cardiac perfusion of mice with PFA efficiently preserves both EYFP as well as other cellular markers
In neuroscience, it is routine to employ cardiac perfusion with PFA as a fixation step to preserve tissues for subsequent EYFP fluorescent imaging (Ressler et al., 2002; Hasegawa et al., 2004; Rossner et al., 2006). We adopted this approach to determine the extent to which this could preserve EYFP in lymphoid tissues. For these studies we used spleen tissue from GBC×RYFP mice previously infected with LCMV to activate and induce EYFP in effector and memory CD8 T cells. Briefly, we infected GBC×RYFP with LCMV and in the memory phase (>day 30 post-infection)we analyzed their spleens for the expression of EYFP in memory CD8 T cells (Fig. 1c). Interestingly, cardiac perfusion led to superb preservation of EYFP. In contrast to previous fixation strategies we were able to detect the cellular marker CD8 in the same tissue sections.
We then analyzed the integrity of the tissues following various fixation protocols by comparing the relative size of nuclei visualized through DAPI staining. Nuclei within tissues immersed or perfused with PFA were compared to those observed within fresh, unfixed tissue sections. When we analyzed the relative size of individual nuclei from the three samples a reduction of 50% was observed in tissues immersed in PFA as compared to fresh, unfixed tissues. In contrast only a 14% reduction relative to fresh tissues was observed following cardiac perfusion with PFA (Fig. 2).
Fig. 2.
Cardiac perfusion maintains tissue integrity. We measured tissue integrity by comparing the size of nuclei following fixation to that from fresh, unfixed tissue. Nuclei were visualized using DAPI staining and then measured. The relative size of individual nuclei from fresh, unfixed, PFA immersion-fixed and PFA perfusion-fixed tissues are plotted. Immersion fixation led to a decrease in the nuclei sizes by 50% while cardiac perfusion fixation decreased nuclei size only modestly (14%). A total of 2 slides and 5 microscopic fields per slide were analyzed from two mice per fixation method. Error bars denote the mean±SD.
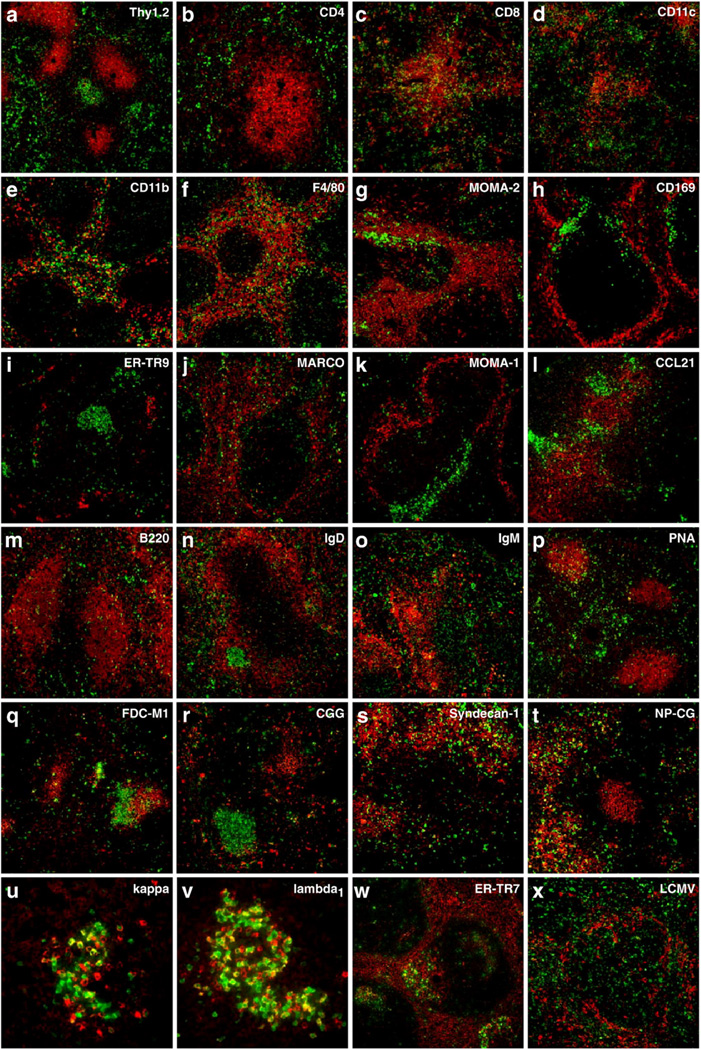
Following the promising results in Figs. 1 and 2, we then assessed the extent in which other cellular markers were preserved via cardiac perfusion with PFA by testing a large array of cellular markers. The data shown in Fig. 3 are two-color images of EYFP in combination with the indicated markers. Within perfused spleens from GBC×RYFP mice, bulk T cells could easily be detected with an anti-Thy1.2 antibody (Fig. 3a) and these could be further segregated into CD4 (Fig. 3b) and CD8 T cell populations (Fig. 3c). Antigen presenting cells, such as dendritic cells and macrophages, which are important in the initiation of T cell responses could be detected (Fig. 3d–g). Dendritic cells were visualized by virtue of their expression of the CD11c marker (Fig. 3d). Macrophage populations in the red pulp could be identified using CD11b, F4/80, and MOMA-2 markers (Fig. 3e–g). Specialized macrophages that reside in the marginal zones of the spleen could be easily detected using CD169, ER-TR9, MARCO and MOMA-1 markers (Fig. 3h–k). We also tested whether chemokines were preserved by this fixation. Interestingly CCL21, a chemokine that is important in maintaining T cell zones within the spleen (Gunn et al.,1999), could also be visualized (Fig. 3l).
Fig. 3.
Cardiac perfusion preserves EYFP and cellular markers within spleen tissue. GCC×RYFP or GBC×RYFP mouse spleens were used as a source of EYFP-positive tissues. Images shown are the results of two-color immunofluorescent microscopy. EYFP appears as green staining and the indicated molecules are visualized in red. Images were collected at a magnification of 20×, except in panels (u) and (v) which were at a magnification of 60×.
We used the GCC×RYFP transgenic model to extend the analysis of EYFP preservation and various B cell markers. Briefly, GCC×RYFP mice were given a primary or secondary immunization with NP-CG. Immunizations with NP-CG/alum led to the development of EYFP-expressing plasma cells, germinal center B cells and memory B cells in GCC×RYFP mice. B cells could be detected using antibodies against B220 (Fig. 3m), and these cells could further be discriminated by expression of either IgD (Fig. 3n) or IgM (Fig. 3o). We were also able to visualize germinal centers by virtue of their binding to the lectin, peanut agglutinin (PNA, Fig. 3p). Germinal centers are the site where antigen-specific antibodies undergo affinity maturation. Follicular dendritic cells (FDCs) and antigen in the form of immune complexes are two components important in this process (Szakal et al., 1988; Barrington et al., 2002). Both follicular dendritic cell networks (FDC-M1, Fig. 3q) and immune complexes of CG (anti-CG, Fig. 3r) could readily be identified within the borders of germinal centers. Following secondary immunization, we could visualize plasma cells using the syndecan-1 marker (Fig. 3s) and we could also determine the specificity of these plasma cells through their ability to bind NP-CG (Fig. 3t). Immunoglobulin light chain usage by plasma cells could be further discriminated between kappa (Fig. 3u) and lambda1 (Fig. 3v).
In addition to the markers discussed above we have successfully used a variety of other markers including the ERTR7marker (Fig. 3w), which differentially stains stromal cells in the red and white pulp (Van Vliet et al., 1986). We were also successful in detecting viral antigens in the spleens of GBC × RYFP mice 5 days following infection with LCMV (Fig. 3x). Taken together, cardiac perfusion with 4% PFA preserved EYFP as well as all cellular markers that we tested. We are yet to come across a cellular antigen that is not preserved by this method.
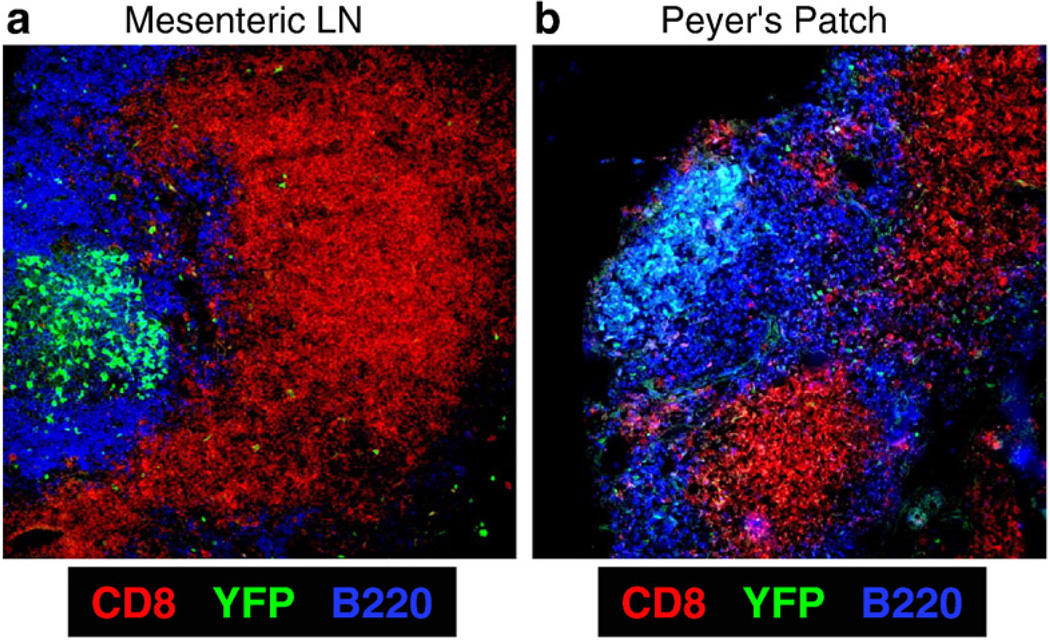
3.4. Cardiac perfusion of mice with PFA efficiently preserves EYFP in Peyer's patch and mesenteric lymph nodes
The spleen is a highly vascular organ, and this may explain the efficient preservation of EYFP-expressing cells within it following cardiac perfusion. Next we evaluated the maintenance of EYFP within other secondary lymphoid organs such as Peyer's patches and mesenteric lymph nodes. We perfused cohorts of immunized GCC×RYFP mice and prepared their Peyer's patches and mesenteric lymph nodes for immunofluorescent microscopy. EYFP, CD8 and B220 were successfully visualized in both tissue types (Fig. 4). Taken together these data suggest that PFA delivered via cardiac perfusion is able to enter a variety of secondary lymphoid tissues and preserves EYFP and other cellular proteins for subsequent visualization.
Fig. 4.
Cardiac perfusion with PFA preserves EYFP and other cellular markers not just in spleen but also in other secondary lymphoid organs such as mesenteric lymph nodes and Peyer's patches. Representative images depicting EYFP, CD8 and B220 staining within a mesenteric lymph node (a) and a Peyer's patch (b) harvested from mice perfused with PFA are shown (magnification is 20×).
4. Discussion
In this report, we compared the ability of a variety of fixation protocols to preserve EYFP as well as other cellular antigens in multiple lymphoid tissues. Attempts to post-fix frozen sections failed in their ability to preserve EYFP for in situ staining. Immersion of whole spleen tissue in PFA resulted in excellent preservation of cellular EYFP, but destroyed other cellular markers of interest. Cardiac perfusion of mice with a 4% PFA solution performed the best as both EYFP as well a variety of markers tested could be visualized in situ.
While EYFP is an excellent marker of proteins and cells, in the field of immunological research, its use has been limited to flow cytometry and live cell microscopy analyses (de Boer et al., 2003; Lindquist et al., 2004; Schmid et al., 2006). In situ analysis of EYFP+ cells in tissue sections has lagged behind because of the described difficulties in preserving both cellular markers and EYFP. In one report, researchers were able to visualize EYFP expressing cells, but visualization of other cellular markers required a harsh antigen retrieval step to “unmask” the over crosslinked epitopes (Casola et al., 2006). Randall and colleagues analyzed GFP but not EYFP expression within tissue sections by fixing the tissues using PFA perfusion (Kusser and Randall, 2003). This report described a three-stage perfusion in which 20 ml each of PBS containing heparin, 4% PFA and 10% sucrose were administered for a total of 60 ml of fluid. This fixation preserved GFP expression but it disrupted the splenic architecture. Since the spleen is directly connected to the bloodstream, this disruption was attributed to the pressure of the perfusing fluids as it flowed through blood vessels. In contrast, we observed excellent preservation of the splenic architecture and nuclear size as compared to other PFA based fixations. This could be because we employed a two-stage perfusion where we delivered just 16 ml of solution (8 ml each of PBS and 4% PFA) to preserve EYFP within lymphoid tissues. Taken together, our approach simplifies and reduces the time of tissue preparation. The resulting sections can be stained using standard immunofluorescent techniques, without a need for antigen retrieval methods. One explanation for the success of this perfusion technique is that PFA is delivered via the vasculature, effectively reaching most tissues and preserving some level of EYFP within cells. This fixation process was so rapid that the entire body of the mouse was rigid in a matter of minutes as opposed to the more lengthy PFA exposures employed during tissue immersion. These longer exposure times may result in the destruction of PFA-sensitive epitopes.
Several novel genes have been identified using genomic and proteomic analyses. It would be of great interest to determine the expression profile of these genes and in situ localization of cells that express them. Introducing EYFP either under the control of a specific promoter or as a fusion protein will facilitate this endeavor and our cardiac perfusion protocol provides a simple method to secure EYFP within tissues for valuable and informative in situ analysis.
Acknowledgements
We thank Ms. Leela Thomas for maintaining the mouse colony and the American Cancer Society for research funds.
Abbreviations
- GFP
green fluorescent protein
- EYFP
enhanced yellow fluorescent protein
- PNA
peanut agglutinin
- NP
(4-hydroxy-3-nitrophenyl) acetyl
- NP-CG
(4-hydroxy-3-nitrophenyl)acetyl chicken γ globulin
- GBC
Granzyme B-cre
- GCC
Germinal Center-Cre
- RYFP
ROSA26R-EYFP.
References
- Barrington RA, Pozdnyakova O, Zafari MR, Benjamin CD, Carroll MC. B lymphocyte memory: role of stromal cell complement and FcgammaRIIB receptors. J. Exp. Med. 2002;196:1189. doi: 10.1084/jem.20021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, Waisman A, Egert A, Ghitza D, Rajewsky K. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7396. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell CP, Jacob J. Identification of memory B cells using a novel transgenic mouse model. J. Immunol. 2006;176:4706. doi: 10.4049/jimmunol.176.8.4706. [DOI] [PubMed] [Google Scholar]
- Coward K, Kubota H, Hibbitt O, McIlhinney J, Kohri K, Parrington J. Expression of a fluorescent recombinant form of sperm protein phospholipase C zeta in mouse epididymal sperm by in vivo gene transfer into the testis. Fertil. Steril. 2006;85(Suppl 1):1281. doi: 10.1016/j.fertnstert.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Dauner JG, Williams IR, Jacob J. Differential microenvironment localization of effector and memory CD8 T cells. J. Immunol. 2008;180:291. doi: 10.4049/jimmunol.180.1.291. [DOI] [PubMed] [Google Scholar]
- de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- Fraser ST, Hadjantonakis AK, Sahr KE, Willey S, Kelly OG, Jones EA, Dickinson ME, Baron MH. Using a histone yellow fluorescent protein fusion for tagging and tracking endothelial cells in ES cells and mice. Genesis. 2005;42:162. doi: 10.1002/gene.20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertzberg N, Gurnani T, Neumann P, Forbes AK, Jean-Louis N, Johnson A. Tumor necrosis factor-{alpha}(TNF) causes barrier dysfunction mediated by tyrosine198 & tyrosine218 in {beta}-actin. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007;293:1219. doi: 10.1152/ajplung.00083.2007. [DOI] [PubMed] [Google Scholar]
- Gregg RG, Kamermans M, Klooster J, Lukasiewicz PD, Peachey NS, Vessey KA, McCall MA. Nyctalopin expression in retinal bipolar cells restores visual function in a mouse model of complete X-linked congenital stationary night blindness. J. Neurophysiol. 2007;98:3023. doi: 10.1152/jn.00608.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn MD, Kyuwa S, Tam C, Kakiuchi T, Matsuzawa A, Williams LT, Nakano H. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J. Exp. Med. 1999;189:451. doi: 10.1084/jem.189.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, Ashigaki S, Takamatsu M, Suzuki-Migishima R, Ohbayashi N, Itoh N, Takada S, Tanabe Y. Laminar patterning in the developing neocortex by temporally coordinated fibroblast growth factor signaling. J. Neurosci. 2004;24:8711. doi: 10.1523/JNEUROSCI.3070-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Baltimore D. Modelling T-cell memory by genetic marking of memory T cells in vivo. Nature. 1999;399:593. doi: 10.1038/21208. [DOI] [PubMed] [Google Scholar]
- Jockusch H, Voigt S, Eberhard D. Localization of GFP in frozen sections from unfixed mouse tissues: immobilization of a highly soluble marker protein by formaldehyde vapor. J. Histochem. Cytochem. 2003;51:401. doi: 10.1177/002215540305100315. [DOI] [PubMed] [Google Scholar]
- Kusser KL, Randall TD. Simultaneous detection of EGFP and cell surface markers by fluorescence microscopy in lymphoid tissues. J. Histochem. Cytochem. 2003;51:5. doi: 10.1177/002215540305100102. [DOI] [PubMed] [Google Scholar]
- Lindquist RL, Shakhar G, Dudziak D, Wardemann H, Eisenreich T, Dustin ML, Nussenzweig MC. Visualizing dendritic cell networks in vivo. Nat. Immunol. 2004;5:1243. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- Metzger F, Repunte-Canonigo V, Matsushita S, Akemann W, Diez-Garcia J, Ho CS, Iwasato T, Grandes P, Itohara S, Joho RH, Knopfel T. Transgenic mice expressing a pH and Cl-sensing yellow-fluorescent protein under the control of a potassium channel promoter. Eur. J. Neurosci. 2002;15:40. doi: 10.1046/j.0953-816x.2001.01837.x. [DOI] [PubMed] [Google Scholar]
- Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 1997;73:2782. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Paschall G, Zhou XL, Davis M. Regulation of synaptic plasticity genes during consolidation of fear conditioning. J. Neurosci. 2002;22:7892. doi: 10.1523/JNEUROSCI.22-18-07892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner MJ, Hirrlinger J, Wichert SP, Boehm C, Newrzella D, Hiemisch H, Eisenhardt G, Stuenkel C, von Ahsen O, Nave KA. Global transcriptome analysis of genetically identified neurons in the adult cortex. J. Neurosci. 2006;26:9956. doi: 10.1523/JNEUROSCI.0468-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K, Yamamoto A, Kawaguchi A, Yamaguchi M, Mori K, Goldman SA, Okano H. Direct isolation of committed neuronal progenitor cells from transgenic mice coexpressing spectrally distinct fluorescent proteins regulated by stage-specific neural promoters. J. Neurosci. Res. 2001;65:220. doi: 10.1002/jnr.1145. [DOI] [PubMed] [Google Scholar]
- Schmid RS, Yokota Y, Anton ES. Generation and characterization of brain lipid-binding protein promoter-based transgenic mouse models for the study of radial glia. Glia. 2006;53:345. doi: 10.1002/glia.20274. [DOI] [PubMed] [Google Scholar]
- Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962;59:223. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakal AK, Kosco MH, Tew JG. A novel in vivo follicular dendritic cell-dependent iccosome-mediated mechanism for delivery of antigen to antigen-processing cells. J. Immunol. 1988;140:341. [PubMed] [Google Scholar]
- Van Vliet E, Melis M, Foidart JM, Van Ewijk W. Reticular fibroblasts in peripheral lymphoid organs identified by a monoclonal antibody. J. Histochem. Cytochem. 1986;34:883. doi: 10.1177/34.7.3519751. [DOI] [PubMed] [Google Scholar]