Abstract
West Nile virus (WNV) and Dengue virus (DENV) are important human pathogens for which there are presently no vaccine or specific antivirals. We report herein a 5′-silylated nucleoside scaffold derived from 3′-azidothymidine (AZT) consistently and selectively inhibiting WNV and DENV at low micromolar concentrations. Further synthesis of various triazole bioisosteres demonstrated clear structure–activity relationships (SARs) in which the antiviral activity against WNV and DENV hinges largely on both the 5′-silyl group and the substituent of 3′-triazole or its bioisosteres. Particularly interesting is the 5′ silyl group which turns on the antiviral activity against WNV and DENV while abrogating the previously reported antiviral potency against human immunodeficiency virus (HIV-1). The antiviral activity was confirmed through a plaque assay where viral titer reduction was observed in the presence of selected compounds. Molecular modeling and competitive S-adenosyl-L-methionine (SAM) binding assay suggest that these compounds likely confer antiviral activity via binding to methyltransferase (MTase).
Graphical abstract
INTRODUCTION
West Nile virus (WNV) and Dengue virus (DENV) are two important members of the genus Flavivirus in the family Flaviviridae. Endemic in many tropical and subtropical regions of the world and transmitted by infected mosquitos, these viruses infect a large human population and cause significant human morbidity and mortality. WNV is a neurotropic virus with outbreaks on multiple continents. Particularly the epidemics of 1999 and 2012 in the U.S.A. have resulted in thousands of reported human cases, with clinical manifestations ranging from asymptomatic to severe neuroinvasive diseases such as meningitis, flaccid paralysis, and encephalitis. On the other hand, DENV endangers 2.5 billion people worldwide with 50–100 million annual infections and can cause dengue fever, dengue hemorrhagic fever, and dengue shock syndrome. Despite these grave public health threats, currently there are no effective antiviral therapies against either virus. Developing antivirals for the treatment of WNV and DENV infections addresses a critical medical need. Current efforts toward this end target either the nucleoside triphosphate biosynthesis as exemplified by mycophenolic acid (MPA),1,2 ribavirin,1,3 and 6-azauridine;4 or viral proteins including both the helicase5,6 and the protease7–15 activities of NS3, the RNA dependent RNA polymerase16–19 and the MTase20–22 functions of NS5. We have previously reported the first AZT-derived 1,2,3-triazole scaffold (1, Figure 1) potently inhibiting HIV-1.23 Key to the unprecedented antiviral activity with these 1,2,3-triazoles is the incorporation of a bulky group at the C5 position of the triazole ring. A key SAR trend was that the C5 bulk substituent conferred significantly better antiviral potency than the C4 one, reflecting a critical requirement of bulkiness in the region between 3′ and 5′ positions (highlighted). Interestingly, the bulkiness in scaffold 1 is highly reminiscent of the unique TSAO-T chemotype (2, Figure 1), a well-known HIV non-nucleoside reverse transcriptase inhibitor (NNRTI). This observation led us to explore the impact of the 5′ silyl protecting group on the antiviral activity of our scaffold 1. With this aim we synthesized a new series of 5′ silyl protected AZT 3′-1,2,3-triazoles (scaffold 3). Unfortunately, none of the silylated analogues inhibited HIV-1. However, when the antiviral evaluation was extended to a panel of other viruses, these silylated analogues were found to selectively inhibit WNV and DENV without inhibiting influenza virus, human cytomegalovirus, or hepatitis C virus. Such a flavivirus-specific inhibition profile prompted us to expand the SAR by synthesizing a few types of triazole bioisosteres. We report herein the synthesis, antiviral and biochemical studies of these new scaffolds as inhibitors of WNV and DENV.
Figure 1.
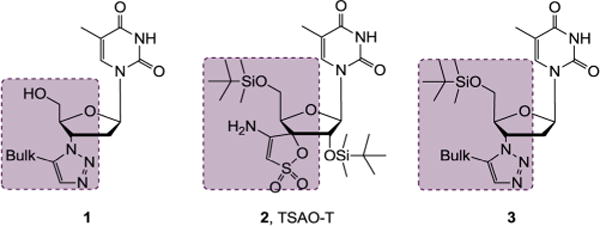
Generation of scaffold 3: AZT-derived 1,2,3-triazole 1 showed potent antiviral activity against HIV-1. Structural comparison between 1 and NNRTI TSAO-T (2) led to the introduction of a 5′ silyl group to generate scaffold 3 which was identified to selectively inhibit WNV and DENV.
RESULTS AND DISCUSSION
Chemistry
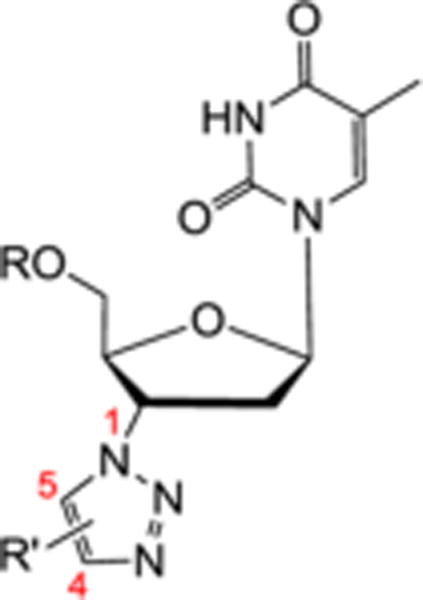
The 3′ azido group of AZT (4) provides an excellent synthetic handle for chemical modifications. All inhibitor scaffolds studied herein were synthetically accessed from AZT as outlined in Scheme 1. The first set of inhibitors were generated via click chemistry between the azido functional group of AZT 4 and alkynes to yield 1,4 (8) and 1,5 (10) disubstituted triazoles according to our reported procedure.23 As previously noted, the copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC)24 worked with substantially higher efficiency than the ruthenium(II)-catalyzed variant (RuAAC).25 All triazole analogues (8a–e and 10a–i) were further derivatized by protecting the 5′-OH group with tert-butyldimethylsilyl chloride (TBSCl) in the presence of imidazole to produce the corresponding 5′-silylated 1,4 and 1,5 disubstituted 1,2,3-triazoles (9a–b and 11a–i) in good yields. Additional sets of inhibitors all feature a bioisostere of the 3′ triazole functionalities. The common 3′-amino intermediate 6 required for the synthesis of these bioisoteres was easily prepared by 5′-TBS protection of AZT followed by reducing the 3′ azido group via catalytic hydrogenation. The thiazolidinone derivatives 13 were synthesized via a one-pot two-step reaction sequence with an aromatic aldehyde and 2-mercaptoacetic acid following a reported procedure.26 The amide analogues 15 were prepared through coupling with a carboxylic acid mediated by 1-ethyl-3-(3-(dimethylamino)-propyl)carbodiimide (EDCI) and hydroxybenzotriazole (HOBt). All other scaffolds were readily accessed by reacting the amino intermediate with various electrophiles, including sulfonyl chloride for sulfonamides 17, isocyanate/isothiocyanate for ureas or thioureas 19, and sulfonyl isocyanate for sulfonylureas 21. Finally, deprotection of the 5′ TBS group using TBAF efficiently generated corresponding 5′-OH analogues (12a–c), (14a–i), (16a–c), (18a–d), and (20a).
Scheme 1. Synthesis of AZT-Derived Scaffolds 8–21a,b.
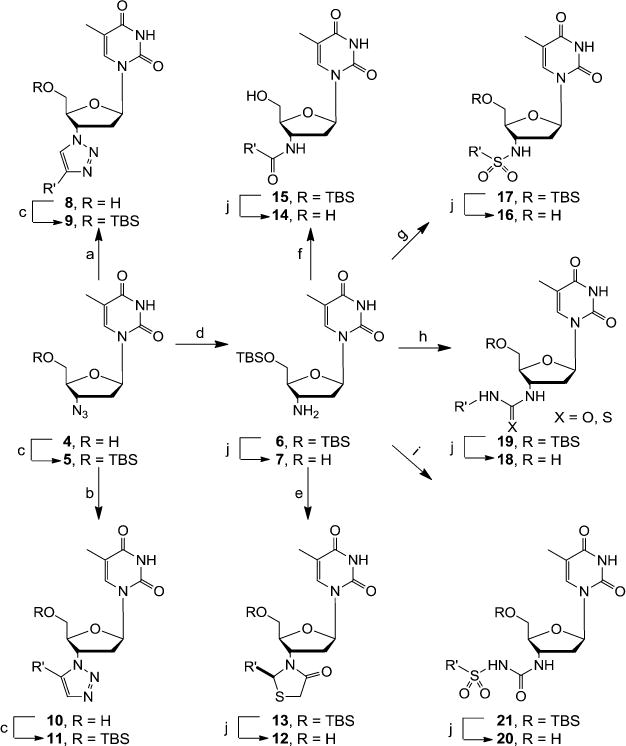
aReagents and conditions: (a) R’ substituted alkyne, sodium ascorbate, CuSO4•5H2O, THF/H2O (3:1), rt, 12 h, 54–81%; (b) R’ substituted alkyne, Cp*RuCl(PPh3)2, THF, 60°C, 1–2 d, 30–52%; (c) TBSCl, imidazole, THF, rt, 12 h, 91–76%; (d) Pd/C, H2, MeOH, rt, overnight, 78%; (e) (1) R’CHO, MeOH, 50°C, 2–3 h (2) SHCH2COOH, tolune, reflux, 12 h, 51–65%; (f) R’COOH, HOBt, EDCI, DMF, rt, 6–10 h, 70–80%; (g) R’SO2Cl, Et3N, CH2Cl2, 0°C–rt, 81–86%; (h) R’NCO/R’NCS, CH2Cl2, 0°C–rt, overnight, 63–72%; (i) R’SO2NCO, CH2Cl2, 40°C, 1 h, then rt, 10 h, 60–70%; (j) 1N TBAF, THF, rt, 5–10 h, 80–92%. bR’ for all analogues is defined in Tables 2 and 3.
To explore the effect of 5′-OH protecting group, eight different analogues of triazole 10a were prepared as depicted in Scheme 2. The protecting group featured in these analogues ranges from small methyl (25) and acyl (24) groups to medium sized acetal functionalities (26 and 27) to bulky dimethoxytrityl (28) and silyl groups (11a, 22, and 23). Synthetically, silylation of the 5′-OH was effected by treating compound 10a with silylating agents TBSCl, TIPSCl, and TBDPSCl to yield corresponding 5′-O-tertbutyldimethylsilyl ether (TBS) 11a, 5′-O-triisopropylsilyl ether (TIPS) 22 and 5′-O-tertbutyldiphenylsilyl ether (TBDPS) 23 respectively in good yields. However, 5′-O-trimethylsilyl ether (TMS) and 5′-O-triethylsilyl ether (TES) were found to be unstable and could not be isolated in the pure form. The protection of the 5′-OH with various alkyl ethers was achieved with ethoxymethyl chloride in the presence of DIPEA, 3,4-dihydropyran in the presence of catalytic amount of pTSA and 4,4′-dimethoxytrityl chloride in pyridine solvent, resulting in corresponding 5′-O-ethoxymethyl ether 26, 5′-O-tetrahydropyranyl ether (THP) 11a and 5′-O-dimethoxytrityl ether (DMT) 28 respectively in moderate yields. It must be pointed out that reacting 6 with methyl iodide in the presence of sodium hydride resulted in the 5′-O and N-3 bismethylated analogue 25. Lastly, 5′-OH of 10a was easily acylated using acetic anhydride in pyridine to furnish 5′-O-acyl compound 24 in 87% yield.
Scheme 2. Preparation of 5′ OH Protected Analogues of 10aa.
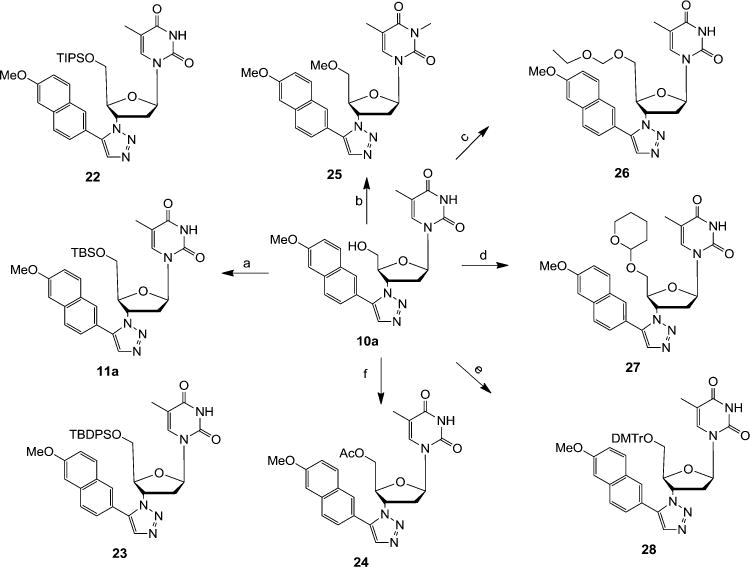
aReagents and conditions: (a) TIPSCl/TBSCl/TBDPSCl, imidazole, THF, rt, 12 h, 76–81%; (b) CH3I, NaH, CH2Cl2, 0°C–rt, 6 h, 78%; (c) C2H5OCH2Cl, DIPEA, CH2Cl2, rt, 2 h, 66%; (d) 3,4-dihydropyran, pTSA, CH2Cl2, 8 h, 72%; (e) DMTrCl, pyridine, 60°C, 6 h, 78%; (f) Ac2O, pyridine, rt, 5 h, 87%.
Antiviral Screening
Since the introduction of a 5′ silyl protecting group was to mimic the bulkiness of NNRTI TSAO-T (2, Figure 1), all analogues of scaffold 3 were first screened in an HIV-1 antiviral assay as well as a biochemical assay against HIV RT. Unfortunately, none of these compounds showed any appreciable activity in either assay (data not shown). Additional antiviral testing against influenza virus, hCMV and HCV did not yield any hit either (data not shown); however, when the antiviral screening was expanded to WNV and DENV, the 5′-silyl-3′-1,2,3-triazole series of scaffold 3 demonstrated consistent inhibitory activities. This observation led to further efforts on antiviral SAR against WNV and DENV in which compounds were evaluated for antiviral properties using a viral subgenomic replicon-containing baby hamster kidney (BHK) replicon cell line. In these assays the level of replicon RNA produced by the respective viral proteins was monitored by measuring the activity of the renilla luciferase that is embedded and expressed within each of the WNV and DENV subgenomic replicons. Lycorine, a natural product and a reported WNV inhibitor27 was used at 1 μM for experiments with the WNV replicon-containing cells. MPA, a published DENV inhibitor,2 was used at 1 μM as a control inhibitory compound for experiments with the DENV replicon-containing cells. Initial screening was done at a single concentration (10 μM) and an inhibition% was calculated after 3 days. In parallel, the cell viability under the same concentration was determined.
It was observed early on during this study that protecting the 5′ OH of the 1, 2, 3-triazole scaffold 1 with TBS completely flipped its antiviral profile. For example compound 10a demonstrated exceptionally potent antiviral activity against HIV-1 (EC50 = 67 nM) as reported previously,23 whereas no appreciable anti-HIV-1 activity was observed with its 5′ TBS analogue 11a; in contrast, 10a did not show any inhibition against WNV or DENV while 11a inhibited both viruses almost completely at 10 μM (Table 1). To confirm this SAR trend and further establish the effect of the 5′ protecting group on antiviral potency against WNV and DENV, we synthesized a panel of seven additional analogues with different 5′ OH protecting groups and tested them against WNV and DENV at 10 μM. The results are summarized in Table 1. The most striking SAR from this series is that analogues with a bulky silyl protecting group including TBS (11a), triisopropylsilyl (TIPS, 22), and tert-butyldiphenylsilyl (TBDPS, 23) all potently inhibited both WNV and DENV with 85–100% inhibition at 10 μM, whereas small nonsilyl protecting groups such as acetyl (24), methyl (25), ethoxymethyl (26), and tetrahydropyran (THP, 27) did not yield appreciable antiviral activity. Clearly, a certain level of bulkiness is required at the 5′ position to achieve antiviral activity. In addition, the nature of the protecting group also appears to substantially impact antiviral potency as a particularly bulky nonsilyl dimethoxytrityl (DMTr, 28) conferred only modest antiviral potencies (23% against WNV and 46% against DENV). The exact reason why Si-based protecting groups offer drastically better antiviral activities than C-based ones is presently unclear. Finally, both TIPS (22) and TBDPS (23) analogues are associated with significant cytotoxicity, rendering TBS as the choice of 5′ protecting group for optimal antiviral activities.
Table 1.
Effect of the 5′ Protecting Group (R) on Antiviral Activities against WNV and DENV
| |||||
|---|---|---|---|---|---|
|
| |||||
| Compound | R | WNV | DENV | ||
|
| |||||
| Inhibition %a | Viability %a | Inhibition %a | Viability %a | ||
| 10a | H | 1.4 | 100 | 0 | 100 |
| 11a |
|
97 | 100 | 85 | 86 |
| 22 |
|
98 | 67 | 98 | 53 |
| 23 |
|
99 | 38 | 100 | 11 |
| 24 |
|
5.3 | 100 | 0 | 97 |
| 25 | Me, N3-Me | 0 | 100 | 0 | 100 |
| 26 |
|
0 | 89 | 13 | 95 |
| 27 |
|
1.0 | 75 | 21 | 92 |
| 28 |
|
23 | 93 | 46 | 93 |
| 4, AZT | – | 0 | 83 | 0 | 89 |
| 5 | – | 17 | 96 | 3.6 | 87 |
Single concentration assay at 10 μM.
The SAR around the substituent on the 3′ triazole ring (R′ group) was explored by testing another series of synthetic triazole compounds (Table 2). The 5′ unprotected analogue (R = H) of each compound was also included in the assay for comparison purpose. Significantly these 5′-TBS triazole analogues typically do not show cytotoxicity at 10 μM (Table 2) except for 11c and 11e, further substantiating TBS as the optimal 5′ protecting group. As for antiviral activity, again none of the 5′ unprotected compounds showed any activity against WNV or DENV while large inhibition was observed with all 5′ protected analogues, with the lone exception of 11h which has a small cyclopropyl substituent on the triazole ring. Another interesting observation was that when the bulky group is connected to the triazole ring through a linker, the resulting compound (e.g., 11g) showed considerably lower potency against both WNV and DENV (11g vs 11b). Furthermore, while the substituent is an aromatic ring in most cases, a bulky alkyl group appears to also confer antiviral activity effectively (compound 11i). Finally, both C5 and C4 substituents seem to confer nearly equal antiviral activities (9a vs 11a and 9b vs 11d), an SAR trend in stark contrast with the previously observed anti-HIV SAR where the antiviral potency was significantly reduced when the bulky substituent is relocated from C5 to C4 triazoles. The overall SAR from this series appears to indicate that the substitution on the 3′ triazole ring may require a bulkiness threshold, and that once the bulkiness is satisfied, antiviral activity may not be sensitive to size changes.
Table 2.
Effect of the Bulky Substituent (R′ Group) on Antiviral Activities against WNV and DENV
| |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Compound | R | R′ | Substitution | WNV | DENV | ||
|
| |||||||
| Position (C4/C5) | Inhibition %a | Viability %a | Inhibition %a | Viability %a | |||
| 11a | TBS |
|
5 | 97 | 100 | 85 | 86 |
| 10a | H |
|
5 | 7.4 | 100 | 0 | 100 |
| 11b | TBS |
|
5 | 93 | 99 | 51 | 93 |
| 10b | H |
|
5 | 4.0 | 99 | 0 | 100 |
| 11c | TBS |
|
5 | 100 | 8.0 | 99 | 60 |
| 10c | H |
|
5 | 3.3 | 100 | 0 | 90 |
| 11d | TBS |
|
5 | 99 | 69 | 21 | 100 |
| 10d | H |
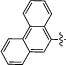
|
5 | 0 | 100 | 0 | 95 |
| 11e | TBS |
|
5 | 100 | 10 | 99 | 47 |
| 10e | H |
|
5 | 23 | 100 | 2.3 | 92 |
| 11f | TBS |
|
5 | 95 | 79 | 95 | 73 |
| 10f | H |
|
5 | 9.0 | 99 | 0 | 100 |
| 11g | TBS |
|
5 | 50 | 100 | 28 | 84 |
| 10g | H |
|
5 | 00 | 96 | 0 | 100 |
| 11h | TBS |
|
5 | 21 | 100 | 6.4 | 93 |
| 10h | H |
|
5 | 00 | 97 | 4.7 | 100 |
| 11i | TBS |
|
5 | 85 | 100 | 75 | 93 |
| 10i | H |
|
5 | 00 | 100 | 10 | 100 |
| 9a | TBS |
|
4 | 92 | 100 | 57 | 96 |
| 9b | TBS |
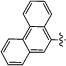
|
4 | 97 | 100 | 77 | 81 |
| 8c | H |
|
4 | 0 | 100 | 8.2 | 100 |
| 8d | H |
|
4 | 14 | 100 | 10 | 100 |
Single concentration assay at 10 μM.
Another important aspect of the current SAR concerns the bioisosterism of the 3′ 1,2,3-triazole ring. Replacing a functional group with its bioisosteres for improved target binding is a common medicinal chemistry practice. Toward this end we synthesized and tested a large number of compounds with 3′ linkers of a few distinct functionalities generally considered as 1,2,3-triazole bioisosteres, including thiazolidinone, amide, sulfonamide, urea/thiourea, and sulfonyl urea. Again the 5′ unprotected analogue for each compound in this series was included in antiviral assays. These efforts resulted in a few notable observations (Table 3). First, bioisosteric replacement by these functionalities did not appear to negatively impact the cytotoxicity profile as the vast majority of compounds in this series remain noncytotoxic. Second, just like the 3′-1,2,3-triazole series, the 5′ TBS protecting group is required for antiviral activity as none of the unprotected analogues were active. Third, many of the TBS protected analogues demonstrated strong inhibition at 10 μM, though some appeared to have noticeably different antiviral potencies between the WNV and DENV assays. For example, thiazolidinones 13b, 13c, and urea 19c showed substantially higher inhibition against WNV than DENV, whereas amides 15f, 15h, urea 19a, 19b, and sulfonylurea 21a were considerably more active against DENV than WNV. This differs from the 3′-triazole series where compounds showed largely similar potencies against WNV and DENV.
Table 3.
Effect of Triazole Bioisosteres on Antiviral Activities against WNV and DENV
| ||||||
|---|---|---|---|---|---|---|
| Compound | R | R′ | WNV | DENV | ||
|
| ||||||
| Inhibition %a | Viability %a | Inhibition %a | Viability %a | |||
| 6 | TBS | NH2 | 0 | 100 | 0 | 100 |
| 13a | TBS |
|
26 | 94 | 22 | 98 |
| 12a | H |
|
0 | 99 | 0 | 100 |
| 13b | TBS |
|
92 | 99 | 49 | 99 |
| 12b | H |
|
9.7 | 100 | 13 | 95 |
| 13c | TBS |
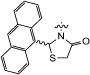
|
85 | 90 | 46 | 97 |
| 12c | H |
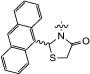
|
12 | 100 | 0 | 96 |
| 15a | TBS |
|
95 | 50 | 92 | 8.0 |
| 14a | H |
|
0 | 100 | 0 | 100 |
| 15b | TBS |
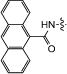
|
0 | 100 | 7.6 | 98 |
| 14b | H |
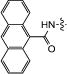
|
0 | 92 | 0 | 95 |
| 15c | TBS |
|
54 | 100 | 28 | 100 |
| 14c | H |
|
00 | 95 | 16 | 94 |
| 15d | TBS |
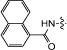
|
37 | 94 | 11 | 99 |
| 14d | H |
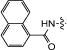
|
00 | 87 | 15 | 98 |
| 15e | TBS |
|
99 | 51 | 98 | 10 |
| 14e | H |
|
00 | 92 | 45 | 97 |
| 15f | TBS |
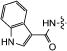
|
19 | 93 | 90 | 27 |
| 14f | H |
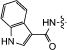
|
0 | 97 | 4.3 | 96 |
| 15g | TBS |
|
24 | 89 | 51 | 98 |
Single concentration assay at 10 μM.
Dose–Response Antiviral Potency
To confirm the observed antiviral potency and gauge activity profile with these scaffolds, we further tested 12 compounds in dose-response fashion. Among selected compounds, 11 were tested against WNV and 7 against DENV using the same assays and control compounds as for single dose testing. The selection of compounds was based on their activity and cell viability against each corresponding replicon cell line at 10 μM. As summarized in Table 4, all compounds tested inhibited WNV and/or DENV in low micromolar range with the majority showing an EC50 of single-digit μM. In addition, all compounds but one (19b) tested against DENV also showed similar level of antiviral activity against WNV, suggesting that our compounds tend to inhibit both flaviviruses. The dose–response inhibition of our compounds against WNV and DENV is further manifested in curves depicted in Figure 2. The challenge with these scaffolds is that, although no or marginal toxicity was observed at 10 μM, most compounds showed only a modest 1–5 fold of selectivity in dose–response testing. The lack of antiviral selectivity represents a common issue in flavivirus antiviral discovery. Nevertheless, our dose–response testing did identify two analogues compound (9a and 15d) that did not exhibit any cytotoxicity at the highest tested concentration (200 μM), suggesting that it is possible to address the toxicity concern of our scaffolds through chemical modifications.
Table 4.
Dose–Response Testing of Selected Compounds against WNV and DENV
| WNV
|
DENV
|
|||
|---|---|---|---|---|
| compound | EC50a (μM) | CC50b (μM) | EC50a (μM) | CC50b (μM) |
| 9a | 7.4 ± 1.3 | >200c | 8.4 ± 0.8 | 21 ± 7.8 |
| 11a | 2.9 ± 2.1 | 13 ± 1.4 | 7.3 ± 1.0 | 7.9 ± 0.7 |
| 11b | 8.4 ± 3.7 | 32 ± 11 | 14 ± 2.8 | 31 ± 1.4 |
| 11i | 7.1 ± 0.4 | 24 ± 3.5 | 7.5 ± 0.7 | 22 ± 7.0 |
| 13b | 3.4 ± 0.7 | 15 ± 3.5 | ||
| 13c | 9.0 ± 0.5 | 27 ± 9.2 | ||
| 15c | 9.9 ± 0.1 | 15 ± 7.1 | ||
| 15d | 33 ± 2.8 | >200c | ||
| 15i | 15 ± 2.1 | 23 ± 2.1 | 9.6 ± 0.6 | 22 ± 4.9 |
| 17b | 8.4 ± 1.6 | 12 ± 2.1 | 7.4 ± 1.4 | 14 ± 4.2 |
| 19b | 11 ± 0 | 23 ± 4.9 | ||
| 19c | 10 ± 0 | 16 ± 1.4 | ||
Concentration inhibiting virus replication by 50%; mean value ± standard deviation from two separate experiments.
Concentration resulting in 50% cell death; mean value ± standard deviation from two separate experiments.
No cytotoxicity observed at the highest dose tested (200 μM).
Figure 2.
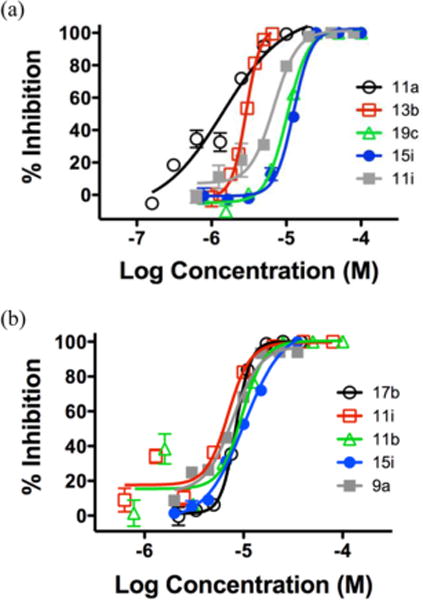
Dose response curves of selected compounds in antiviral assays: (a) WNV assay and (b) DENV assay.
DENV Yield Reduction Assay
To verify the observed dose–response antiviral potency, we also tested two selected compounds (9a and 15i) in a DENV yield reduction assay. This assay directly measures the ability of a compound to inhibit viral production. The assay was done by inoculating Vero cells with DENV and adding compound. The titer of the virus produced was determined by plating serial dilutions on fresh Vero cells and counting the corresponding number of plaques. As shown in Figure 3, both compounds tested significantly reduced the titer of DENV at 10 μM, with compound 9a almost completely suppressing viral production (98% inhibition) and 15i inhibiting DENV (67%) as effectively as the lower dosed MPA (1.0 μM). These results strongly indicate that our compounds indeed directly impact viral replication.
Figure 3.
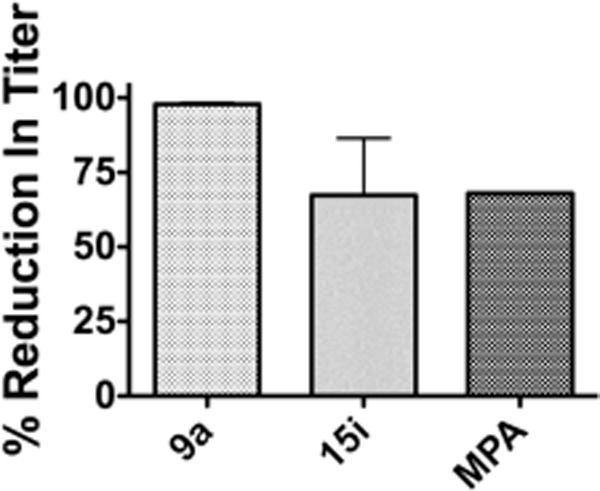
Impact of selected compounds on DENV viral production. Percent reduction in viral titer at 10 μM (average of two separate experiments): 98% for compound 9a and 67% for 15i. Percent reduction at 1.0 μM (one experiment): 68% for MPA.
Antiviral Mechanism of Action
Since the 5′ OH is protected by a silyl group, it is unlikely that these nucleoside derivatives could get phosphorylated in cells and act as chain terminators. While the exact antiviral mechanism of action for our compounds remains unclear, we noticed that another 5′ silylated thymidine-based scaffold as represented by GRL-002 was recently reported as an inhibitor type of WNV MTase.20 Specifically, these analogues were found to mimic the methyl donor SAM and competitively bind to the SAM binding site of the MTase. Since the northern part of our compounds is similar to that of the reported chemotype (Figure 4, a, highlighted), we were prompted to look at the ability of our compounds to bind to MTase. Toward this end, compound 9a was docked into WNV MTase using the reported crystal structure of WNV MTase cocrystallized with Sinefungin (SIN; PDB code 3LKZ).28 In our docking, the predicted binding mode of GRL-002 was found to be identical as reported by Hongmin Li et al.20 The predicted binding mode of compound 9a and its overlay with GRL-002 is shown in Figure 4b. The thymine and sugar moiety in compound 9a were found to be bound identical to that of GRL-002 and SIN. The dimethyl-t-butylsilyl core is predicted to bind in the hydrophobic pocket in which methionine group of SIN occupies which is predicted to confer selective inhibition of viral MTase over human MTase. The 3′ triazole core of compound 9a is found to occupy an additional pocket adjacent to the sugar bound region which is not utilized by earlier reported ligands (GRL-002 and SIN). These additional interactions obtained through the novel designs of the current analogues in the current study would result in the design of potent flavivirus inhibitors. Furthermore, the southern part of 9a, the 3′ triazole substituent, appears to occupy a lot of empty space in the binding groove, which might provide highly beneficial binding interactions absent from GRL-002. Collectively, molecular modeling suggests that it is likely that our novel antiviral compounds target the SAM binding site of the MTase.
Figure 4.
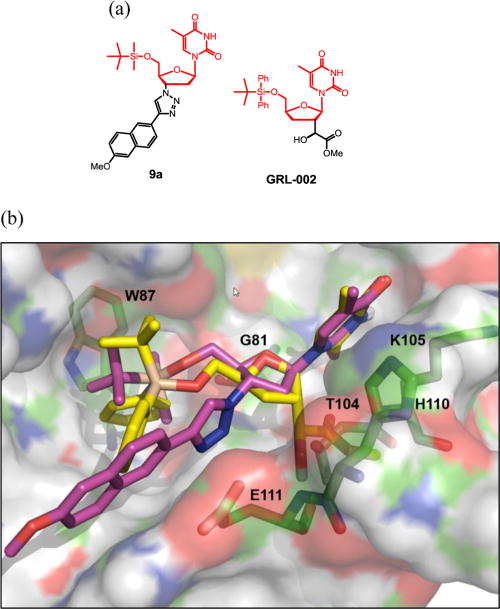
Docking of compound 9a into the crystal structure of WNV MTase (PDB code 3LKZ28). (a) Structure of 9a and GRL-002; (b) overlay with GRL-002 (yellow) and the predicted binding mode of compound 9a (magenta) within the MTase SAM-binding pocket. Residues lining the pocket are highlighted in green sticks.
To confirm the predicted binding mode of our compounds, we tested 12 selected compounds in a previously reported SAM competition assay.29 This assay measures the ability of the compounds to compete against 3H-labeled SAM–MTase complex formation (Figure 5). SIN, a close analogue of SAM and a reported potent inhibitor of SAM binding, was used as a positive control. Remarkably, at 20 μM all tested compounds reduced the formation of the 3H-labeled SAM–MTase complex to the same level as without MTase, suggesting a complete inhibition of SAM binding. Interestingly, the competition of cold SAM or SIN at 20 μM also led to the same level of reduction on the formation of 3H-labeled SAM–MTase complex, implying that our compounds could be competitive inhibitors of SAM binding and that the observed potency against WNV and DENV could contribute to validating viral MTase as a unique antiviral target.
Figure 5.
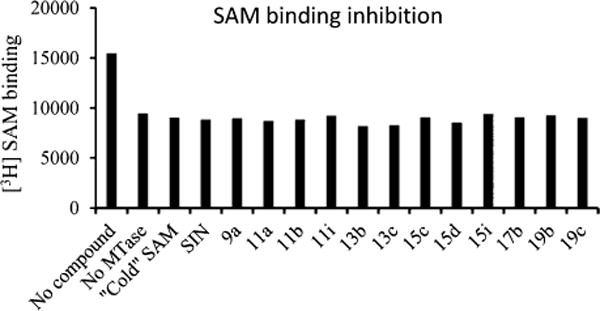
Inhibition of the [3H]-SAM-MTase complex formation by SAM, SIN, and selected compounds at 20 μM concentration. The biotinylated DNV3MTase and 3H-labeled SAM were incubated with or without compounds AdoMet, SIN, and each compound. The reaction mixtures were mixed with the streptavidin-coated SPA beads and quantified using a Microbeta29 scintillation counter.
CONCLUSIONS
5′-Silylated AZT-derived 3′-1,2,3-triazole nucleoside bioisosteric scaffolds were found to consistently inhibit WNV and DENV at low micromolar concentrations without inhibiting HIV or any other viruses tested. SAR showed that both the 5′ silyl protecting group and the 3′ bulky substituent are essential for antiviral activity against WNV and DENV. That none of the 5′-desilylated, potently HIV-inhibiting analogues showed any activity against WNV or DENV indicates that a simple silylation-desilylation process can serve as a switch beween inhibiting WNV/DENV and HIV-1. The antiviral activity in the primary replicon assays was confirmed through a plaque assay where viral titer reduction was observed. Molecular modeling and SAM-binding assay indicate that the observed antiviral activity is likely due to binding to flavivirus MTase.
EXPERIMENTAL SECTION
Chemistry
General Procedures
All commercial chemicals were used as supplied unless otherwise indicated. Dry solvents were either purchased (toluene and MeOH) or dispensed under argon from an anhydrous solvent system with two packed columns of neutral alumina or molecular sieves. Flash chromatography was performed on a Teledyne Combiflash RF-200 with RediSep columns (silica) and indicated mobile phase. All moisture sensitive reactions were performed under an inert atmosphere of ultrapure argon with oven-dried glassware. 1H and 13C NMR spectra were recorded on a Varian 600 MHz spectrometer. Mass data were acquired on an Agilent TOF II TOS/MS spectrometer capable of ESI and APCI ion sources. Analysis of sample purity was performed on a Varian Prepstar SD-1 HPLC system with a Phenomenex Gemini, 5 μm C18 column (250 mm × 4.6 mm). HPLC conditions: solvent A = H2O, solvent B = MeCN; flow rate = 1.0 mL/min; compounds were eluted with a gradient of 20% MeCN/H2O for 5 min then to 100% MeCN for 40 min. Purity was determined by total absorbance at 254 nm. All tested compounds have a purity ≥96.
General Procedure 1 for Silylation
To a solution of 5′-hydroxy nucleoside (1.12 mmol, 1.0 equiv) and imidazole (2.24 mmol, 2.0 equiv) in DMF (10 mL) was added appropriate silyl chloride (1.34 mmol, 1.2 equiv), and the mixture was stirred at room temperature for 10–12 h. The reaction progress was monitored by TLC. The solvent was removed in vacuo, diluted with water, and extracted with EtOAc (3 × 20 mL). The organic phase was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted with 2–10% MeOH in CH2Cl2, and yielded the desired compound.
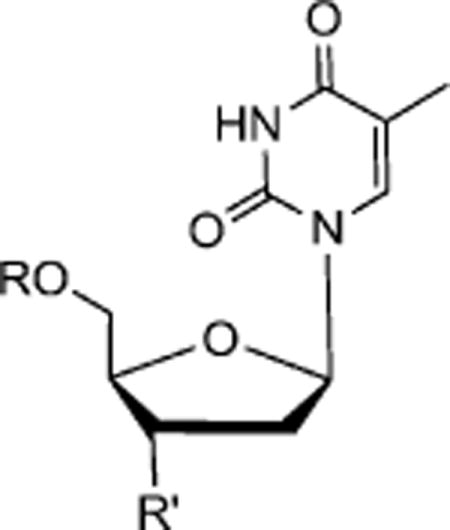
1-((2R,4S,5S)-4-Azido-5-(((tert-butyldimethylsilyl)oxy)methyl)-tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (5)
Yield 91%. 1H NMR (600 MHz, CD3OD) δ 7.55 (s, 1H), 6.15 (t, J = 6.6 Hz, 1H), 4.32–4.31 (m, 1H), 3.95–3.86 (m, 3H), 2.38–2.35 (m, 2H), 1.87 (s, 3H), 0.94 (s, 9H), −0.13 (s, 6H); 13C NMR (150 MHz, CD3OD) δ 164.8, 150.7, 135.9, 110.1, 84.6, 84.5, 62.7, 60.7, 36.8, 25.0, 17.8, 11.2, −6.6; HRMS-ESI(−) m/z calcd for C16H26N5O4Si 380.1754 [M–H]−, found 380.1768.
General Procedure 2 for the Synthesis of 1,4-Triazoles Derivatives via CuACC (8a–e)
To the mixture of AZT (0.375 mmol, 1.0 equiv) and alkyne (0.375 mmol, 1.0 equiv) in 4.0 mL of THF/H2O (3:1) was added freshly prepared 1 M solution of sodium ascorbate (0.1 equiv) in water, followed by the addition of freshly prepared 1 M solution of CuSO4·5H2O (0.06 equiv) in water. The heterogeneous reaction mixture was stirred at room temperature for 12 h and monitored by TLC and MS. After the completion, the reaction was evaporated to dryness. The crude product was purified by column chromatography, eluted with 2–10% MeOH in CH2Cl2, yielded desired 1,4-triazole.
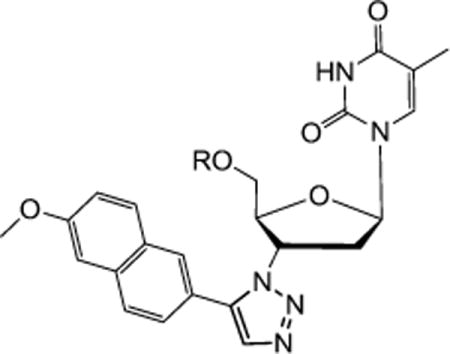
1-((2R,4S,5S)-5-(Hydroxymethyl)-4-(4-(6-methoxynaphthalen-2-yl)-1H-1,2,3-triazol-1-yl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (8a)
Yield 91%. 1H NMR (600 MHz, DMSO-d6) δ 11.35 (s, 1H, 3-NH), 8.83 (s, 1H), 8.31 (s, 1H), 7.94 (d, J = 8.4 Hz, 1H), 7.88 (t, J = 7.2 Hz, 2H), 7.83 (s, 1H), 7.33 (s, 1H), 7.18 (dd, J = 2.6 Hz, J = 9.0 Hz, 1H), 6.45 (t, J = 6.6 Hz, 1H), 5.40–5.42 (m, 1H), 5.30 (t, J = 5.0 Hz, 1H, 5′–OH), 4.28–4.29 (m, 1H), 3.87 (s, 3H, OMe), 3.66–3.75 (m, 2H), 2.69–2.83 (m, 2H), 1.81 (s, 3H, CH3); C NMR (150 MHz, DMSO-d6) δ 164.6, 157.9, 150.9, 147.2, 136.8, 134.4, 130.0, 128.9, 127.9, 125.9, 124.0, 121.3, 119.6, 110.3, 106.4, 84.8, 84.5, 61.4, 61.1, 55.6, 37.5, 12.5; HRMS-ESI(+) m/z calcd for C23H24N5O5 450.1777 [M + H]+, found 450.1775.
General Procedure 3 for the Synthesis of 1,5-Triazoles Derivatives via RuACC (10a–i)
To the mixture of AZT (0.5 mmol, 1.0 equiv) and alkyne (0.75 mmol, 1.5 equiv) in dry THF (5.0 mL) was added catalytic amount of Cp*RuCl(PPh3)2 (0.05 equiv) and stirred at 60°C for 1–2 days. The reaction was monitored by TLC and MS. The reaction mixture was evaporated to dryness, and the crude product was purified by column chromatography, eluted with 2–10% MeOH in CH2Cl2, yielded desired 1,5-triazole.
1-((2R,4S,5S)-5-(Hydroxymethyl)-4-(5-(6-methoxynaphthalen-2-yl)-1H-1,2,3-triazol-1-yl) tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (10a)
Yield 39%. The reaction of AZT (150 mg, 0.56 mmol) with alkyne (143 mg, 0.83 mmol) yielded compound 18e (98 mg, 39%) as a yellow solid. mp 134–136°C; 1H NMR (600 MHz, DMSO-d6) δ 11.36 (s, 1H, 3-NH), 8.03 (s, 1H), 7.95–7.96 (m, 2H), 7.89 (d, J = 8.4 Hz, 1H), 7.75 (s, 1H), 7.56 (d, J = 8.4 Hz, 1H), 7.41 (s, 1H), 7.25 (dd, J = 2.4 Hz, J = 8.8 Hz, 1H), 6.57 (t, J = 6.8 Hz, 1H), 5.21–5.23 (m, 2H), 4.38–4.39 (m, 1H), 3.88 (s, 3H, OMe), 3.56 (dd, J = 1.8 Hz, J = 12.0 Hz, 1H), 3.46 (dd, J = 2.4 Hz, J = 12.0 Hz, 1H), 2.58–2.64 (m, 2H), 1.73 (s, 3H, CH3); 13C NMR (150 MHz, DMSO-d6) δ 164.1, 158.7, 150.9, 138.6, 136.5, 134.8, 133.4, 130.3, 129.1, 128.5, 128.0, 127.2, 121.6, 120.1, 110.1, 106.3, 85.4, 85.0, 61.8, 58.7, 55.8, 38.2, 12.7; HRMS-ESI(+) m/z calcd for C23H24N5O5 450.1777 [M + H]+, found 450.1811.
General Procedure 4 for the Synthesis of 4-Thiazolidinone Derivatives (13a–c)
The mixture amine 6 (0.14 mmol, 1.0 equiv) and appropriate aldehyde (0.28 mmol, 2.0 equiv) in MeOH (10 mL) was heated at 50°C for 2–3 h, and the solvent was evaporated under reduced pressure. The solid obtained was dissolved in toluene (10 mL), and dropwise thioglycolic acid (0.72 mmol, 5.0 equiv) was added and the reaction was carried out at reflux temperature for 12 h. The reaction progress was monitored by TLC. The solvent was removed in vacuo, diluted with water and extracted with EtOAc (3 × 15 mL), and washed with an aqueous solution of NaHCO3. The combined organic phase was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted with CH2Cl2 and 4–10% MeOH in CH2Cl2, and yielded the desired 4-thiazolidinone.
1-((2R,4S,5S)-5-(((tert-Butyldimethylsilyl)oxy)methyl)-4-(4-oxo-2-phenylthiazolidin-3-yl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (13a)
Yield 65%. Diastereomeric mixture (1:1.5), 1H NMR (600 MHz, CD3OD) δ 7.65–7.48 (m, 6H), 6.52 (t, J = 6.6 Hz, 1H), 5.97–5.95 (m, 1H), 4.46–4.44 (m, 1H), 4.11–4.05 (m, 1H), 3.94–3.84 (m, 2H), 3.71 (dd, J = 1.8 Hz, J = 11.6 Hz, 1H), 3.16 (dd, J = 2.4 Hz, J = 11.6 Hz, 1H), 2.22–2.20 (m, 2H), 1.92 (s, 3H), 0.94 (s, 9H), 0.06 (s, 3H), −0.00 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 172.6, 164.8, 150.8, 139.9, 136.0, 129.2, 127.3, 127.1, 110.0, 86.3, 81.0, 65.0, 63.0, 55.4, 37.5, 32.2, 25.0, 17.7, 11.2, −6.5, −6.6; HRMS-ESI(−) m/z calcd for C25H34N3O5SSi 516.1988 [M–H]−, found 516.1975.
General Procedure 5 for Amide Coupling (15a–i)
To the mixture of acid (0.21 mmol, 1.0 equiv), EDCI(0.23 mmol, 1.1 equiv) and HOBt(0.23 mmol, 1.1 equiv) in CH2Cl2:DMF (4:1, 10 mL) was added amine 6 (0.21 mmol, 1.0 equiv) and stirred for 6–10 h at room temperature under nitrogen atmosphere. The reaction progress was monitored by TLC. The solvent was removed in vacuo and extracted with EtOAc (3 × 20 mL). The organic phase was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted with CH2Cl2 and 2–10% MeOH in CH2Cl2, and yielded the desired compound.
N-((2S,3S,5R)-2-(((tert-Butyldimethylsilyl)oxy)methyl)-5-(5-meth-yl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-6-methoxy-2-naphthamide (15a)
Yield 72%. mp 125–129°C; 1H NMR (600 MHz, CD3OD) δ 8.31 (s, 1H), 7.86–7.84 (m, 3H), 7.70 (s, 1H), 7.29 (s, 1H), 7.21–7.19 (m, 1H), 6.33 (t, J = 6.6 Hz, 1H), 4.78–4.76 (m, 1H), 4.14–4.13 (m, 1H), 4.03–4.01 (m, 1H), 3.96 (dd, J = 1.2 Hz, J = 11.4 Hz, 1H), 3.93 (s, 3H), 2.47–2.41 (m, 2H), 1.91 (s, 3H), 0.95 (s, 9H), 0.14 (s, 3H), 0.14 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 169.0, 159.4, 150.8, 136.6, 136.0, 130.0, 128.7, 127.3, 126.7, 124.0, 119.3, 110.0, 105.2, 85.0, 84.6, 63.0, 54.4, 49.9, 37.6, 33.3, 25.0, 17.9, 11.2, −6.5. −6.6; HRMS-ESI(−) m/z calcd for C28H36N3O6Si 538.2373 [M–H]−, found 538.2358.
General Procedure 6 for Synthesis of Sulfonamide (17a–c)
To a solution of amine 6 (0.17 mmol, 1.0 equiv) and triethyl amine (0.34 mmol, 2.0 equiv) in CH2Cl2 (10 mL) was added sulfonyl chloride (0.21 mmol, 1.2 equiv) at 0°C and slowly warmed to room temperature and stirred under a nitrogen atmosphere for 12 h. The reaction progress was monitored by TLC. The reaction was stopped by adding water and extracted with CH2Cl2 (3 × 20 mL). The organic layer was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted with 1–10% MeOH in CH2Cl2, and yielded the desired sulfonamide.
N-((2S,3S,5R)-2-(((tert-Butyldimethylsilyl)oxy)methyl)-5-(5-meth-yl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-4-chlorobenzenesulfonamide (17a)
Yield 84%. mp 179–180°C; 1H NMR (600 MHz, CD3OD) δ 7.85 (d, J = 8.4 Hz, 2H), 7.59 (d, J = 9.0 Hz, 2H), 7.46 (s, 1H), 6.11 (t, J = 6.6 Hz, 1H), 3.92–3.89 (m, 1H), 3.88–3.86 (m, 1H), 3.83 (d, J = 12.0 Hz, 1H), 3.65 (dd, J = 3.0 Hz, J = 12.0 Hz, 1H), 2.15–2.13 (m, 1H), 2.10–2.07 (m, 1H), 1.84 (s, 3H), 0.88 (s, 9H), 0.06 (s, 6H); 13C NMR (150 MHz, CD3OD) δ 162.4, 150.7, 139.6, 138.7, 135.8, 129.2, 128.3, 110.1, 84.9, 84.4, 62.1, 52.3, 37.8, 25.0, 17.8, 11.2, −6.6; HRMS-ESI(−) m/z calcd for C22H31N3O6SSiCl 528.1391 [M–H]−, found 528.1398.
General Procedure 7 for Synthesis of Urea and Thiourea (19a–d)
To a solution of amine 6 (0.17 mmol, 1.0 equiv) in CH2Cl2 (10 mL) was added isocyanate/thioisocyanate (0.25 mmol, 1.5 equiv) at 0°C and stirred for 1 h and then slowly warmed to room temperature and stirred under a nitrogen atmosphere for overnight. The reaction progress was monitored by TLC and MS. The reaction was stopped by adding water and extracted with CH2Cl2 (3 × 20 mL). The organic layer was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted with 1–10% MeOH in CH2Cl2, and yielded the desired urea or thiourea.
N-((2S,3S,5R)-2-(((tert-Butyldimethylsilyl)oxy)methyl)-5-(5-meth-yl-2,4-dioxo-3,4-1-((2S,3S,5R)-2-(((tert-Butyldimethylsilyl)oxy)-methyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-tetrahydrofuran-3-yl)-3-phenylurea (19a)
Yield 66%. mp 112–115°C; 1H NMR (600 MHz, CD3OD) δ 7.65 (s, 1H), 7.33 (d, J = 8.4 Hz, 2H), 7.24 (t, J = 7.8 Hz, 2H), 6.97 (t, J = 7.8 Hz, 1H), 6.23 (t, J = 6.6 Hz, 1H), 4.42−4.40 (m, 1H), 3.98–3.95 (m, 2H), 3.90–3.88 (m, 1H), 2.34–2.29 (m, 2H), 1.89 (s, 3H), 0.93 (s, 9H), −0.14 (s, 3H), −0.13 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 164.9, 156.2, 150.8, 139.1, 136.0, 128.4, 122.2, 118.9, 110.0, 85.5, 84.5, 63.0, 50.0, 38.2, 25.0, 17.9, 11.3, −6.5, −6.6; HRMS-ESI(−) m/z calcd for C23H33N4O5Si 473.2220 [M–H]−, found 473.2236.
General Procedure 8 for Synthesis of Sulfonylurea (21a–b)
To a solution of amine 6 (0.17 mmol, 1.0 equiv) and triethyl amine (0.25 mmol, 1.5 equiv) in CH2Cl2 (10 mL) was added sulfonyl isocyanate (0.25 mmol, 1.5 equiv) and heated at 40°C for 1 h and then stirred at room temperature for 10 h. The reaction progress was monitored by TLC and MS. The reaction was stopped by adding water and extracted with CH2Cl2 (3 × 20 mL) and washed with 0.1 N HCl. The combined organic layer was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted with 1–10% MeOH in CH2Cl2, and yielded the desired sulfonylurea.
N-(((2S,3S,5R)-2-(((tert-Butyldimethylsilyl)oxy)methyl)-5-(5-meth-yl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-carbamoyl)-4-methylbenzenesulfonamide (21a)
Yield 60%. 1H NMR (600 MHz, CD3OD) δ 7.87 (d, J = 7.8 Hz, 2H), 7.57 (m, 1H), 7.39 (d, J = 8.4 Hz, 2H), 6.17 (t, J = 6.6 Hz, 1H), 4.36–4.32 (m, 1H), 3.88–3.86 (m, 2H), 3.70–3.69 (m, 1H), 2.43 (s, 3H), 2.28–2.27 (m, 2H), 1.87 (s, 3H), 0.88 (s, 9H), 0.04 (s, 3H), 0.03 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 164.8, 150.7, 144.2, 136.0, 129.2, 127.3, 110.0, 84.7, 84.3, 62.4, 49.5, 37.5, 25.0, 20.1, 17.9, 11.3, −6.6, −6.7; HRMS-ESI(−) m/z calcd for C24H35N4O7SSi 551.1996 [M–H]−, found 551.1987.
General Procedure 9 for Deprotection of Silyl Ether
To a solution of silyl ether (1.0 equiv) in THF was added dropwise 1 N solution of TBAF in THF (1.5 equiv) and stirred at rt for 5–10 h. The progress of the reaction was monitored by TLC and MS. The reaction was stopped by adding water and extracted with EtOAc (3 × 20 mL), washed with saturated solution of NaCl. The combined organic layer was dried over Na2SO4, filtered and concentrated. The crude product was purified by column chromatography, eluted with 1–10% MeOH in CH2Cl2, yielded the desired compound.
N-((2S,3S,5R)-2-(Hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-6-methoxy-2-naphthamide (14a)
Yield 85%. mp >250°C; 1H NMR (600 MHz, CD3OD) δ 8.32 (s, 1H), 7.94 (s, 1H), 7.87–7.84 (m, 3H), 7.29 (s, 1H), 7.20–7.19 (m, 1H), 6.33 (t, J = 6.6 Hz, 1H), 4.78–4.76 (m, 1H), 4.07–4.06 (m, 1H), 3.93 (s, 3H), 3.92–3.84 (m, 2H), 2.50–2.46 (m, 2H), 1.91 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 168.8, 157.7, 147.3, 139.5, 136.7, 130.1, 127.4, 126.7, 124.0, 119.3, 107.4, 85.0, 84.6, 63.9, 59.1, 50.3, 37.3, 11.0; HRMS-ESI(–) m/z calcd for C22H22N3O6Si 424.1509 [M–H]−, found 424.1515.
1-((2R,4S,5S)-4-Amino-5-(((tert-butyldimethylsilyl)oxy)methyl)-tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (6)
To a solution of azide 5 (1.0 g, 2.62 mmol) in MeOH (20 mL) was added 10% Pd/C (0.1 g) under hydrogen atmosphere at 1 atm pressure and stirred overnight at room temperature. The suspension was filtered over Celite and washed with excess MeOH. The solvent was removed under reduced pressure, and the crude product was triturated with ethyl acetate and hexane yielded pure amine 6 as a white solid (0.72 g, 2.02 mmol, 78%). 1H NMR (600 MHz, CD3OD) δ 7.85 (s, 1H), 6.19 (t, J = 6.6 Hz, 1H), 3.92 (dd, J = 2.4 Hz, J = 11.4 Hz, 1H), 3.85 (dd, J = 2.4 Hz, J = 11.4 Hz, 1H), 3.75–3.74 (m, 1H), 3.55–3.54 (m, 1H), 2.23–2.19 (m, 2H), 1.86 (s, 3H), 0.92 (s, 9H), 0.15 (s, 6H); 13C NMR (150 MHz, CD3OD) δ 164.9, 136.2, 109.8, 87.1, 84.6, 62.8, 51.0, 48.1, 40.2, 25.0, 11.2, −6.6, −6.7; HRMS-ESI(−) m/z calcd for C16H28N3O4Si 354.1849 [M–H]−, found 354.1836.
Synthesis of ((2S,3S,5R)-3-(5-(6-Methoxynaphthalen-2-yl)-1H-1,2,3-triazol-1-yl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl)methyl acetate (24)
To a solution of triazole 10a (30 mg, 0.07 mmol) in pyridine (1.0 mL) was added Ac2O (0.08 mg, 0.073 mmol) and stirred at room temperature for 5 h. The reaction progress was monitored by TLC and MS. The reaction was stopped by adding water and pyridine was removed in vacco. The residue was extracted with CH2Cl2 (3 × 15 mL) and washed with 0.1 N HCl. The combined organic layer was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted with CH2Cl2 and then 1–5% MeOH in CH2Cl2, and yielded the desired compound 24 (28 mg, 0.06 mmol, 87%) as a white solid. 1H NMR (600 MHz, CD3OD) δ 7.96–7.94 (m, 2H), 7.88–7.86 (m, 2H), 7.50–7.48 (m, 2H), 7.33 (s, 1H), 7.23 (d, J = 12.0 Hz, 1H), 6.47 (t, J = 6.6 Hz, 1H), 5.43–5.42 (m, 1H), 4.65–4.62 (m, 1H), 4.17 (dd, J = 4.2 Hz, J = 12.6 Hz, 1H), 4.14 (dd, J = 2.4 Hz, J = 12.6 Hz, 1H), 3.94 (s, 3H), 3.07–3.04 (m, 1H), 2.84–2.82 (m, 1H), 1.84 (s, 3H), 1.65 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 171.5, 170.2, 159.1, 156.7, 150.6, 136.9, 135.1, 129.5, 128.6, 127.7, 126.2, 119.7, 110.1, 105.3, 86.6, 82.0, 62.5, 58.7, 48.1, 37.2, 18.8, 13.0; HRMS-ESI(−) m/z calcd for C25H24N5O6 490.1727 [M–H]−, found 490.1739.
1-((2R,4S,5S)-5-(Methoxymethyl)-4-(5-(6-methoxynaphthalen-2-yl)-1H-1,2,3-triazol-1-yl)tetrahydrofuran-2-yl)-3,5-dimethylpyrimidine-2,4(1H,3H)-dione (25)
To a solution of triazole 10a (50 mg, 0.11 mmol) in DMF (2.0 mL) was added NaH (9 mg, 0.22 mmol) at 0°C and stirred for 5 min. CH3I (0.08 mg, 0.073 mmol) was then added dropwise to the mixture at 0°C and slowly warmed to room temperature and stirred for 6 h. The reaction progress was monitored by TLC and MS. The reaction was stopped by adding water, and DMF was removed in vacco. The residue was extracted with CH2Cl2 (3 × 20 mL) and washed with brine. The combined organic layer was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted with CH2Cl2 and then 1–5% MeOH in CH2Cl2, and yielded the desired compound 25 (42 mg, 0.088 mmol, 78%) as a white solid. 1H NMR (600 MHz, CD3OD) δ 7.91−7.88 (m, 2H), 7.82–7.80 (m, 2H), 7.69 (s, 1H), 7.44 (d, J = 9.0 Hz, 1H), 7.28 (s, 1H), 7.19–7.17 (m, 1H), 6.56 (t, J = 6.6 Hz, 1H), 5.34–5.33 (m, 1H), 4.51–4.50 (m, 1H), 3.89 (s, 3H), 3.57 (dd, J = 3.0 Hz, J = 10.2 Hz, 1H), 3.34 (dd, J = 3.0 Hz, J = 10.8 Hz, 1H), 3.21 (s, 3H), 3.13 (s, 3H), 2.95–2.92 (m, 1H), 2.62–2.59 (m, 1H), 1.81 (s, 3H); HRMS-ESI(−) m/z calcd for C25H27N5O5 477.2012 [M–H]−, found 477.2024.
Synthesis of 1-((2R,4S,5S)-5-((Ethoxymethoxy)methyl)-4-(5-(6-methoxynaphthalen-2-yl)-1H-1,2,3-triazol-1-yl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (26)
To a solution of triazole 10a (25 mg, 0.055 mmol) and N,N-diisopropylethylamine (14 mg, 0.11 mmol) in CH2Cl2 (3 mL) was added chloromethyl ethyl ether (8.0 mg, 0.083 mmol) and stirred at room temperature for 2 h. The reaction progress was monitored by TLC and MS. The reaction was stopped by adding water, extracted with CH2Cl2 (3 × 20 mL), and washed with 0.1 N HCl. The combined organic layer was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted with CH2Cl2 and then 1–5% MeOH in CH2Cl2, and yielded the desired compound 26 (18 mg, 0.037 mmol, 66%) as a white solid. 1H NMR (600 MHz, CD3OD) δ 7.95–7.93 (m, 2H), 7.84–7.85 (m, 3H), 7.51–7.49 (m, 1H), 7.32 (s, 1H), 7.22–7.21 (m, 1H), 6.65 (t, J = 6.6 Hz, 1H), 5.54–5.39 (m, 1H), 5.36–5.35 (m, 2H), 4.57–4.56 (m, 1H), 3.93 (s, 3H), 3.78 (dd, J = 3.0 Hz, J = 12.6 Hz, 1H), 3.61–3.57 (m, 3H), 2.87–2.84 (m, 1H), 2.65–2.62 (m, 1H), 1.86 (s, 3H), 1.14 (t, J = 7.2 Hz, 3H); 13C NMR (150 MHz, CD3OD) δ 163.8, 159.0, 151.0, 139.0, 135.8, 135.1, 132.4, 129.4, 128.6, 127.6, 126.2, 120.8, 119.6, 109.5, 105.3, 86.6, 85.4, 70.2, 65.1, 61.0, 58.0, 54.5, 38.3, 13.9, 11.6; HRMS-ESI(−) m/z calcd for C26H28N5O6 506.2040 [M–H]−, found 506.2054.
Synthesis of 1-((2R,4S,5S)-4-(5-(6-Methoxynaphthalen-2-yl)-1H-1,2,3-triazol-1-yl)-5-(((tetrahydro-2H-pyran-2-yl)oxy)methyl)-tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (27)
To a solution of triazole 10a (20 mg, 0.044 mmol) and dihydropyran (5.0 mg, 0.053 mmol) in CH2Cl2 (3 mL) was added a catalytic amount of pTSA (10 mol %) and stirred at room temperature for 8 h. The reaction progress was monitored by TLC and MS. The reaction was stopped by adding water and extracted with CH2Cl2 (3 × 20 mL). The combined organic layer was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted with CH2Cl2 and then 1–5% MeOH in CH2Cl2, and yielded the desired compound 27 (17 mg, 0.032 mmol, 72%) as a white solid. 1H NMR (600 MHz, CD3OD) δ 7.95–7.93 (m, 2H), 7.87–7.85 (m, 2H), 7.65–7.64 (m, 1H), 7.51–7.50 (m, 1H), 7.33–7.32 (m, 1H), 7.23–7.21 (m, 1H), 6.70 (t, J = 6.6 Hz, 1H), 5.53–5.47 (m, 1H), 4.60–4.58 (m, 1H), 4.03–4.02 (m, 1H), 3.93 (s, 3H), 3.91 (dd, J = 3.6 Hz, J = 12.0 Hz, 1H), 3.65–3.64 (m, 2H), 3.09–3.07 (m, 1H), 3.03–3.00 (m, 1H), 2.73–2.68 (m, 1H), 1.82 (s, 3H), 1.57–1.17 (m, 6H); 13C NMR (150 MHz, CD3OD) δ 164.8, 159.1, 150.7, 136.3, 135.1, 129.5, 128.6, 127.7, 126.3, 120.9, 119.7, 110.2, 105.3, 99.6, 85.8, 84.2, 66.2, 62.6, 58.3, 54.5, 38.5, 30.1, 24.7, 19.4, 13.0, 11.1; HRMS-ESI(–) m/z calcd for C28H30N5O6 532.2196 [M–H]−, found 532.2184.
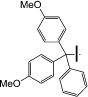
Synthesis of 1-((2R,4S,5S)-5-((Bis(4-methoxyphenyl)(phenyl)-methoxy)methyl)-4-(5-(6-methoxynaphthalen-2-yl)-1H-1,2,3-triazol-1-yl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (28)
To a solution of triazole 10a (30 mg, 0.07 mmol) in pyridine (1.0 mL) was added DMTrCl (0.27 mmol, 0.08 equiv) and stirred at 60°C for 6 h. The reaction progress was monitored by TLC and MS. The reaction was stopped by adding water and pyridine was removed in vacco. The residue was extracted with CH2Cl2 (3 × 10 mL) and washed with 0.1 N HCl. The combined organic layer was dried over Na2SO4, filtered, and concentrated. The crude product was purified by column chromatography, eluted CH2Cl2 and then with 1–5% MeOH in CH2Cl2, and yielded the desired compound 28 (39 mg, 0.05 mmol, 78%) as a white solid. 1H NMR (600 MHz, CD3OD) δ 7.83–7.81 (m, 2H), 7.79 (s, 1H), 7.72 (d, J = 9.0 Hz, 1H), 7.60 (d, J = 1.2 Hz, 1H), 7.36–7.34 (m, 1H), 7.32 (d, J = 3.2 Hz, 1H), 7.21–7.19 (m, 1H), 7.10–7.06 (m, 5H), 6.90–6.88 (m, 4H), 6.59 (t, J = 6.4 Hz, 1H), 6.56–6.52 (m, 4H), 5.60–5.58 (m, 1H), 4.58–4.57 (m, 1H), 3.94 (s, 3H), 3.67 (s, 3H), 3.66 (s, 3H), 3.29–3.27(m, 1H), 3.06–3.02 (m, 2H), 2.92–2.90 (m, 1H), 1.36 (s, 3H); 13C NMR (150 MHz, CD3OD) δ 164.8, 159.0, 158.7, 158.6, 150.6, 144.0, 139.2, 136.4, 135.1, 134.9, 132.3, 129.7, 129.6, 129.5, 128.6, 128.4, 127.8, 127.7, 127.3, 126.5, 126.1, 120.7, 119.7, 112.6, 112.6, 110.3, 105.5, 86.5, 85.6, 83.6, 62.3, 57.7, 54.5, 54.3, 54.2, 38.4, 10.5; HRMS-ESI(−) m/z calcd for C44H40N5O7 750.2928 [M–H]−, found 750.2937.
Biology
Replicon Assays
Compounds were evaluated for antiviral properties using viral subgenomic replicon-containing cells. For WNV, the baby hamster kidney replicon cell line BHK-WII RepRen1B (obtained from Dr. T. Pierson, NIH/NIAID) was used and for DENV, the baby hamster kidney replicon cell line BHK pD2-hRucPac-2ATG30 (obtained from Dr. M. Diamond, Washington University, School of Medicine) was used. The level of replicon RNA produced by the respective viral proteins can be monitored by measuring the activity of the renilla luciferase that is embedded and expressed within each of the WNV and DENV subgenomic replicons. Both replicon lines were maintained in Dulbecco’s modified Eagle’s (DME) medium supplemented with 10% fetal bovine serum (FBS), 100 IU streptomycin/penicillin per ml and 10 μg/mL plasmocin (InvivoGen). The medium for the BHK-WII RepRen1B cells was supplemented with 5 μg/mL blasticidin (Life Technologies), and the medium for the BHK pD2-hRucPac-2ATG was supplemented with 3 μg/mL puromycin (Life Technologies). Three thousand WNV replicon-containing cells per well or 1.5 × 103 DENV replicon-containing cells per well were plated in white opaque 96-well plates in the absence of antibiotic selection, and the next day, compounds dissolved in DMSO were added to triplicate wells in culture medium. The compounds were initially tested at 10 μM final concentration and each plate also contained DMSO alone, medium alone, and a control inhibitory compound. Lycorine, a natural product and published WNV inhibitor, was used at 1 μM for experiments with the WNV replicon-containing cells. MPA, a published DENV inhibitor, at 1 μM was used as a control inhibitory compound for experiments with the DENV replicon-containing cells. Three days later, medium was replaced with a 1:1000 dilution of ViVi-Ren Live Cell Substrate (Promega) in DME minus phenol red and 10% FBS. Luminescence was measured with a Molecular Devices M5e plate reader. Mean values of triplicate wells were determined and compared to the mean value for the wells that received DMSO alone. For compounds selected for dose–response experiments, the concentration of compound that reduced luciferase activity by 50% was defined as the 50% effective concentration (EC50). The EC50 was determined by comparing luciferase activity for eight serial dilutions of the compound and vehicle treated cells using GraphPad Prism software.
Cell Viability Assay
Approximately 3 × 103 WNV replicon-containing cells per well or 1.5 × 103 DENV replicon-containing cells per well were plated in a clear 96-well tissue culture plate (Corning) in the absence of antibiotic selection. The next day, the cells were exposed to culture medium containing compound dissolved in DMSO, DMSO alone, or nothing added and incubated at 37°C and 5% CO2 for 3 days. CellTiter 96 AQueous One Solution Cell Proliferation reagent (Promega) was added according to manufacturer’s instructions and the level of the bioconverted product measured by spectrometry at 450 nm with a SpectraMax E5 (Molecular Devices) as an indication of cell viability. Initial screening of compounds was performed at 10 μM final concentration. All samples were performed in triplicate and mean values for triplicate wells were compared to mean values of triplicate wells receiving DMSO. For compounds that were selected for dose–response experiments, the concentration of compound that reduced cell proliferation by 50% was defined as the 50% cytotoxic concentration (CC50). The CC50 was determined by comparing absorbance readings from eight serial dilutions of compound and vehicle treated cells using GraphPad Prism software.
DENV Yield Reduction Assay
This assay measures the ability of a compound to inhibit virus production. Vero cells (maintained in DME with 10% FBS and streptomycin/penicillin) were plated in 12-well dishes at 4 × 105 cells per well. The next day, those cells were inoculated with DENV Type 2 New Guinea C strain (ATCC #VR-1584) in infection medium (DMEM, 2% FBS and 10 mM HEPES, pH 7.5) at a multiplicity of infection of 0.2 for 2 h at 37°C and 5% CO2 with gentle rocking every 15 min. The cells were then washed once in Vero culture medium and compounds were added at indicated final concentrations in Vero culture medium. Two days later, the supernatant was harvested and subjected to a low-speed spin to remove any cells. 10-fold serial dilutions of the clarified supernatant were performed and 0.25 mL of each dilution plated onto 1.5 × 105 Vero cells per well in 24-well plates. After 2 h incubation at 37°C and 5% CO2, the inoculum was removed, cells washed two times in PBS and the cells were overlaid in plaque medium (MEM, 5% FBS, 1.3% w/v methyl cellulose and 10 mM HEPES, pH 7.5). After 5 days at 37 °C and 5% CO2, the plaque medium was removed, and the cells were washed twice in PBS and fixed in methanol:acetic acid (3:1) solution (30 min at room temperature). The cells were then stained with Giemsa (0.05% Giemsa w/v, 5% methanol, 5% glycerol) for 20 min at room temperature, washed five times with water, and dried, and the plaques were counted. The titer of virus produced from cells in the presence of the compound was calculated as the number of plaques multiplied by the dilution factor and then converted to plaque forming units per mL.
Expression, denaturation, refolding, and purification of the DNV3MTase
The DNV3MTase was expressed, denatured, refolded, and purified as described.31 Briefly, the E. coli cells were lysed in a denaturing buffer containing 50 mM Tris, pH 8.0, 500 mM NaCl, 10 mM β-Me, 10% glycerol, and 8 M urea. The denatured MTase cell lysate was loaded to the Ni-NTA affinity column under denaturing condition and extensively washed (>30 column volume) with the lysis buffer in the presence of 10 mM imidazole. The MTase-bound Ni-NTA beads were transferred to a dialysis bag and dialyzed overnight at 4°C against a buffer containing 25 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10 mM β–ercaptoethanol (β-Me), and 10% glycerol. Homemade 3C protease was added to the dialysis bag, and the mixture continued to dialyze overnight at 4°C. The protease treated mixture was collected in an empty Bio-Rad Econo column. Flow-through was collected, and the beads were washed with the dialysis buffer for 3–6 column volumes or until the OD280 less than 0.1. The wash fractions and the flow-through were combined and concentrated to 5 mL and was subjected to gel filtration chromatography in a buffer containing 25 mM Tris-HCl, pH 7.5, 200 mM NaCl, 10% glycerol, 2 mM DTT, using a Superdex S-200 column (GE HealthCare). The MTase fractions were collected and concentrated to 10 mg/mL and flash-frozen in liquid nitrogen for crystallization and functional analysis.
Biotinylation of MTase
Biotin was conjugated to the DNV3MTase using the EZ-Link NHS-biotin Kit (Pierce), as described.29 Specifically, the MTases of WNV (30 μM) and DENV3 (65 μM) were dialyzed into phosphate buffered saline (PBS), and mixed with the biotin reagent at a final concentration of 1 mM at 23°C overnight. Unconjugated biotin was removed by FPLC over an HiTrap desalting column (GE HealthCare), and the ratio of conjugated biotin to the DENV3MTase (13:1) was determined using a Biotin Quantitation kit (Pierce).
SAM Binding Inhibition Assay
Biotinylated DNV3MTase (580 nM) was mixed with the polyvinyltoluene (PVT) scintillation proximity assay (SPA) beads (1.5 mg/mL, PerkinElmer) and 20 μM of SAM, sinefungin (SIN), or each compound in the SAM binding buffer (20 mM Tris pH 7.5, 50 mM NaCl, 10 mM KCl, 2 mM MgCl2, 2 mM MnCl2, 0.05% CHAPS) in a white 96-well clear-bottom plate. The samples were mixed by gentle rocking for 20 min at 23°C, followed by the addition of 1.65 μCi of 3H-SAM (425 nM) to a final sample volume of 50 μL. After mixing for another 15 min at 23°C, samples were then centrifuged for 2 min at 500g and analyzed on a Microbeta2 2450 plate counter (PerkinElmer) using the default 3H-Scintillation Proximity Assay protocol within the manufactory software.
Modeling and Docking
Molecular modeling was performed using the Schrodinger small molecule drug discovery suite 2013–2. The crystal structure of West Nile Virus (WNV methyltransferase cocrystallized with Sinefungin (SIN), an Adomet analogue (PDB code 3LKZ) was obtained from protein data bank32 as reported by Hongmin Li et al.28 The above structure was subjected to analysis and found that the native ligand SIN was bound to the hydrophobic pocket, adjacent to the Adomet-binding site of WNV methyltransferase. This pocket is highly conserved among flaviviruses and the residues within this hydrophobic pocket are found to be highly critical in virus replication and cap methylations.
This model was subjected to protein preparation wizard33,34 (Schrodinger Inc.) in which missing hydrogens atoms were added and zero-order bonds to metals were created followed by the generation of metal binding states. The structure of protein was minimized using the OPLS 2005 force field35 to optimize the hydrogen bonding network and converge heavy atoms to the RMSD of 0.3 Å. The receptor grid generation tool in Maestro (Schrodinger Inc.)36 was used to define an active site around the SIN ligand to cover all the residues within 12 Å. The ligands GRL-002 and 9a were drawn using Maestro and subjected to Lig Prep37 to generate conformers, possible protonation at pH of 7 ± 3 and metal binding states which serves as an input for the docking process. All the dockings were performed using Glide XP38 (Glide, version 6.0) mode with the van der Waals radii of nonpolar atoms for each of the ligands were scaled by a factor of 0.8. All the ligands within the hydrophobic pocket of WNV were further refined post docking by minimizing under implicit solvent to account for the local protein flexibility.
Supplementary Material
Acknowledgments
This research was supported partially by the Research Development and Seed Grant Program of the Center for Drug Design, University of Minnesota, and partially by grants (AI094335) from the National Institute of Health (NIH; to H.L.).
ABBREVIATIONS USED
- WNV
West Nile virus
- DENV
Dengue virus
- AZT
3′-azidothymidine
- SAR
structure–activity relationship
- HIV
human immunodeficiency virus
- SAM
S-adenosyl-L-methionine
- MTase
methyltransferase
- MPA
mycophenolic acid
- NNRTI
non-nucleoside reverse transcriptase inhibitor
- CuAAC
copper(I)-catalyzed azide–alkyne cycloaddition
- RhAAC
ruthenium(II)-catalyzed azide–alkyne cycloaddition
- TBS
tertbutyldimethylsilyl
- SIN
sinefungin
Footnotes
Supporting Information
Spectral characterization 1H, 13C and HRMS of new compounds (8b–e, 9a–b, 10b–i, 11a–i, 12a–c, 13b–c, 15b–i, 14b–i, 16a–c, 17b–c, 18a–d, 19b–d, 21b, 20a, 22, and 23). The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.5b00327.
Notes
The authors declare no competing financial interest.
References
- 1.Takhampunya R, Ubol S, Houng HS, Cameron CE, Padmanabhan R. Inhibition of dengue virus replication by mycophenolic acid and ribavirin. J Gen Virol. 2006;87:1947–1952. doi: 10.1099/vir.0.81655-0. [DOI] [PubMed] [Google Scholar]
- 2.Diamond MS, Zachariah M, Harris E. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology. 2002;304:211–221. doi: 10.1006/viro.2002.1685. [DOI] [PubMed] [Google Scholar]
- 3.Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: Direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 2001;98:6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrey JD, Smee DF, Sidwell RW, Tseng C. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antiviral Res. 2002;55:107–116. doi: 10.1016/s0166-3542(02)00013-x. [DOI] [PubMed] [Google Scholar]
- 5.Borowski P, Lang M, Haag A, Schmitz H, Choe J, Chen HM, Hosmane RS. Characterization of imidazo[4,5-d]pyridazine nucleosides as modulators of unwinding reaction mediated by West Nile virus nucleoside triphosphatase/helicase: Evidence for activity on the level of substrate and/or enzyme. Antimicrob. Agents Chemother. 2002;46:1231–1239. doi: 10.1128/AAC.46.5.1231-1239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang N, Chen HM, Koch V, Schmitz H, Minczuk M, Stepien P, Fattom AI, Naso RB, Kalicharran K, Borowski P, Hosmane RS. Potent inhibition of NTPase/helicase of the West Nile Virus by ring-expanded (“fat”) nucleoside analogues. J Med Chem. 2003;46:4776–4789. doi: 10.1021/jm030277k. [DOI] [PubMed] [Google Scholar]
- 7.Zhou GC, Weng Z, Shao X, Liu F, Nie X, Liu J, Wang D, Wang C, Guo K. Discovery and SAR studies of methionine-proline anilides as dengue virus NS2B-NS3 protease inhibitors. Bioorg Med Chem Lett. 2013;23:6549–6554. doi: 10.1016/j.bmcl.2013.10.071. [DOI] [PubMed] [Google Scholar]
- 8.Samanta S, Lim TL, Lam Y. Synthesis and in vitro evaluation of West Nile virus protease inhibitors based on the 2-{6-[2-(5-phenyl-4H-{1,2,4]triazol-3-ylsulfanyl)acetylamino]benzothiazol-2-ylsul fanyl}-acetamide scaffold. ChemMedChem. 2013;8:994–1001. doi: 10.1002/cmdc.201300114. [DOI] [PubMed] [Google Scholar]
- 9.Nitsche C, Schreier VN, Behnam MA, Kumar A, Bartenschlager R, Klein CD. Thiazolidinone-peptide hybrids as dengue virus protease inhibitors with antiviral activity in cell culture. J Med Chem. 2013;56:8389–8403. doi: 10.1021/jm400828u. [DOI] [PubMed] [Google Scholar]
- 10.Lim HA, Ang MJ, Joy J, Poulsen A, Wu W, Ching SC, Hill J, Chia CS. Novel agmatine dipeptide inhibitors against the West Nile virus NS2B/NS3 protease: a P3 and N-cap optimization study. Eur J Med Chem. 2013;62:199–205. doi: 10.1016/j.ejmech.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 11.Lai H, Dou D, Aravapalli S, Teramoto T, Lushington GH, Mwania TM, Alliston KR, Eichhorn DM, Padmanabhan R, Groutas WC. Design, synthesis and characterization of novel 1,2-benzisothiazol-3(2H)-one and 1,3,4-oxadiazole hybrid derivatives: potent inhibitors of Dengue and West Nile virus NS2B/NS3 proteases. Bioorg Med Chem. 2013;21:102–113. doi: 10.1016/j.bmc.2012.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammamy MZ, Haase C, Hammami M, Hilgenfeld R, Steinmetzer T. Development and characterization of new peptidomimetic inhibitors of the West Nile virus NS2B-NS3 protease. ChemMedChem. 2013;8:231–241. doi: 10.1002/cmdc.201200497. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, Samanta S, Cui T, Lam Y. Synthesis and in vitro evaluation of West Nile virus protease inhibitors based on the 1,3,4,5-tetrasubstituted 1H-pyrrol-2(5H)-one scaffold. ChemMedChem. 2013;8:1554–1560. doi: 10.1002/cmdc.201300244. [DOI] [PubMed] [Google Scholar]
- 14.Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R. Identification and biochemical characterization of small-molecule inhibitors of west nile virus serine protease by a high-throughput screen. Antimicrob. Agents Chemother. 2008;52:3385–3393. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knox JE, Ma NL, Yin Z, Patel SJ, Wang WL, Chan WL, Ranga Rao KR, Wang G, Ngew X, Patel V, Beer D, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of West Nile NS3 protease: SAR study of tetrapeptide aldehyde inhibitors. J Med Chem. 2006;49:6585–6590. doi: 10.1021/jm0607606. [DOI] [PubMed] [Google Scholar]
- 16.Caillet-Saguy C, Lim SP, Shi PY, Lescar J, Bressanelli S. Polymerases of hepatitis C viruses and flaviviruses: structural and mechanistic insights and drug development. Antiviral Res. 2014;105:8–16. doi: 10.1016/j.antiviral.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Gong EY, Kenens H, Ivens T, Dockx K, Vermeiren K, Vandercruyssen G, Devogelaere B, Lory P, Kraus G. Expression and purification of dengue virus NS5 polymerase and development of a high-throughput enzymatic assay for screening inhibitors of dengue polymerase. Methods Mol Biol. 2013;1030:237–247. doi: 10.1007/978-1-62703-484-5_19. [DOI] [PubMed] [Google Scholar]
- 18.Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malet H, Masse N, Selisko B, Romette JL, Alvarez K, Guillemot JC, Tolou H, Yap TL, Vasudevan S, Lescar J, Canard B. The flavivirus polymerase as a target for drug discovery. Antiviral Res. 2008;80:23–35. doi: 10.1016/j.antiviral.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Chen H, Liu L, Jones SA, Banavali N, Kass J, Li Z, Zhang J, Kramer LD, Ghosh AK, Li H. Selective inhibition of the West Nile virus methyltransferase by nucleoside analogs. Antiviral Res. 2013;97:232–239. doi: 10.1016/j.antiviral.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, Monaghan P, Chung KY, Dong H, Liu B, Bodenreider C, Lee G, Ding M, Chan WL, Wang G, Jian YL, Chao AT, Lescar J, Yin Z, Vedananda TR, Keller TH, Shi PY. Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem. 2011;286:6233–6240. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong H, Liu L, Zou G, Zhao Y, Li Z, Lim SP, Shi PY, Li H. Structural and functional analyses of a conserved hydrophobic pocket of flavivirus methyltransferase. J Biol Chem. 2010;285:32586–32595. doi: 10.1074/jbc.M110.129197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sirivolu VR, Vernekar SKV, Ilina T, Myshakina NS, Parniak MA, Wang ZQ. Clicking 3′-Azidothymidine into Novel Potent Inhibitors of Human Immunodeficiency Virus. J Med Chem. 2013;56:8765–8780. doi: 10.1021/jm401232v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem, Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Chen X, Xue P, Sun HH, Williams ID, Sharpless KB, Fokin VV, Jia G. Ruthenium-catalyzed cycloaddition of alkynes and organic azides. J Am Chem Soc. 2005;127:15998–15999. doi: 10.1021/ja054114s. [DOI] [PubMed] [Google Scholar]
- 26.Bolognese A, Correale G, Manfra M, Lavecchia A, Novellino E, Barone V. Thiazolidin-4-one formation. Mechanistic and synthetic aspects of the reaction of imines and mercaptoacetic acid under microwave and conventional heating. Org Biomol Chem. 2004;2:2809–2813. doi: 10.1039/B405400H. [DOI] [PubMed] [Google Scholar]
- 27.Zou G, Puig-Basagoiti F, Zhang B, Qing M, Chen L, Pankiewicz KW, Felczak K, Yuan Z, Shi PY. A single-amino acid substitution in West Nile virus 2K peptide between NS4A and NS4B confers resistance to lycorine, a flavivirus inhibitor. Virology. 2009;384:242–252. doi: 10.1016/j.virol.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong HP, Liu LH, Zou G, Zhao YW, Li Z, Lim SP, Shi PY, Li HM. Structural and Functional Analyses of a Conserved Hydrophobic Pocket of Flavivirus Methyltransferase. J Biol Chem. 2010;285:32586–32595. doi: 10.1074/jbc.M110.129197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Zhou B, Brecher M, Banavali N, Jones SA, Li Z, Zhang J, Nag D, Kramer LD, Ghosh AK, Li HM. S-Adenosyl-Homocysteine Is a Weakly Bound Inhibitor for a Flaviviral Methyltransferase. PLoS One. 2013;8:e76900. doi: 10.1371/journal.pone.0076900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitby K, Pierson TC, Geiss B, Lane K, Engle M, Zhou Y, Doms RW, Diamond MS. Castanospermine, a potent inhibitor of dengue virus infection in vitro and in vivo. J Virol. 2005;79:8698–8706. doi: 10.1128/JVI.79.14.8698-8706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brecher MB, Li Z, Zhang J, Chen H, Lin Q, Liu B, Li H. Refolding of a fully functional flavivirus methyltransferase revealed that S-adenosyl methionine but not S-adenosyl homocysteine is copurified with flavivirus methyltransferase. Protein Sci. 2015;24:117–128. doi: 10.1002/pro.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.http://www.rcsb.org/pdb/explore.do?structureId=3lkz.
- 33.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 34.Schrödinger . Schrödinger Release 2013–2: Schrödinger Suite 2013 Protein Preparation Wizard; Epik version 2.5. Schrödinger, LLC; New York, NY: 2013. Impact version 6.0, Schrödinger, LLC, New York, NY, 2013; Prime version 3.3, Schrödinger, LLC, New York, NY, 2013. [Google Scholar]
- 35.Jorgensen WL, Maxwell DS, TiradoRives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 36.Schrödinger . Schrödinger Release 2013–2: Maestro, version 9.5. Schrödinger, LLC; New York, NY: 2013. [Google Scholar]
- 37.Schrödinger . Schrödinger Release Release 2013–2: LigPrep, version 2.7. Schrödinger, LLC; New York, NY: 2013. [Google Scholar]
- 38.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


