Abstract
BACKGROUND
S-Nitrosothiols have been recognized as biologically-relevant products of nitric oxide that are involved in many of the diverse activities of this free radical.
SCOPE OF REVIEW
This review serves to discuss current methods for the detection and analysis of protein S-nitrosothiols. The major methods of S-nitrosothiol detection include chemiluminescence-based methods and switch-based methods, each of which comes in various flavors with advantages and caveats.
MAJOR CONCLUSIONS
The detection of S-nitrosothiols is challenging and prone to many artifacts. Accurate measurements require an understanding of the underlying chemistry of the methods involved and the use of appropriate controls.
GENERAL SIGNIFICANCE
Nothing is more important to a field of research than robust methodology that is generally trusted. The field of S-Nitrosation has developed such methods but, as S-nitrosothiols are easy to introduce as artifacts, it is vital that current users learn from the lessons of the past.
1. Introduction
Nitric oxide (NO) is a multi-functional ubiquitous free radical molecule generated from the nitric oxide synthase family of enzymes. The most easily understood and widely established mechanisms of NO action involve its interaction with metal centers and other free radicals. For example, NO has a high affinity for ferrous iron and this is exemplified by its ability to bind to the heme iron of soluble guanylyl cyclase in the canonical endothelial-derived relaxing factor pathway. In addition, the reaction of NO with superoxide, which occurs at a diffusion limited rate and results in the formation of peroxynitrite, has been implicated in several pathological conditions.
In addition to these direct and chemically simple mechanisms of NO signaling, it has become clear that NO can also be involved in the post-translational modification of protein amino-acid residues that can lead to protein dysfunction and in some rarer cases, gain in protein function. The two NO-dependent post-translational modifications are tyrosine nitration and protein S-nitrosation. It is worth comparing and contrasting these two modifications. Mechanisms of tyrosine nitration include the indirect reaction of tyrosine with peroxynitrite [1,2], the action of peroxidases in the presence of nitrite (an oxidation production of nitric oxide) [3], and the reaction of NO with tyrosyl radicals [4]. Mechanisms of thiol nitrosation include the oxidation of NO to form nitrosating agents (N2O3), the reaction of NO with a thiyl radical, transition metal ion or metalloprotein assisted nitrosation and direct addition of NO to a thiol followed by oxidation of the resultant radical [5-10]. Both of these modifications are chemically irreversible processes that require oxidation of either the amino acid or NO, and, in most cases, the reduction of oxygen.
The image that is often presented (particularly for S-nitrosation) that NO can reversible associate with a thiol to form an S-nitrosothiol is fundamentally and chemically erroneous, although many figures and schemes published in high quality journals appear to ignore this chemical reality. A major difference between tyrosine nitration and thiol nitrosation is that nitrosation is biologically reversible. Biological systems contain reducing pathways that are able to ‘repair’ this modification at the cost of cellular reducing equivalents, and so processes modified by S-nitrosation are potentially functionally reversible. This is equivalent to kinases and phosphatases forming and reversing protein phosphorylation at the cost of cellular ATP. Although parallels have been drawn between phosphorylation and nitrosation in the literature, the analogy is a poor one, as phosphorylation is rarely limited by ATP supply or thought of as an ATP signaling pathway, whereas S-nitrosation as an NO-dependent signal would need to be responsive to changes in NO formation.
One of the challenges with the study of post-translational modifications is accurate and sensitive detection. This has been a major issue in the field of S-nitrosation and in some cases methodology development has seen published values dramatically reassessed. For example the level of S-nitrosothiol in plasma was initially reported to be 8 μM and more recent reports put this level at tens of nM and some even lower [11]. Accurate determination of levels of modification is clearly important in driving the importance of S-nitrosation, but also important is a full understanding of the range and scope of protein modifications, and a great deal of effort has been put into determining the S-nitrosothiol proteome. In this review we will survey the major methods for the detection of S-nitrosothiols in biological systems.
2. General Considerations
The issue that has caused the highest level of doubt and uncertainty in the field of S-nitrosothiols is the appropriate care that needs to be taken to avoid artifactual S-nitrosothiol formation during sample preparation and analysis. This issue raises doubt and uncertainty in many publications (too many to cite without unfairly singling out those authors) that have used S-nitrosothiol detection methods. The chemical basis of this artifact is that the reaction between nitrous acid and thiols generates S-nitrosothiols [12], and so any method that uses acidification at any point in the detection methodology will inevitably generate S-nitrosothiols in the sample unless the thiols are irreversibly alkylated before acidification. Nitrite removal is also an alternative, but all buffers contain nitrite at some level and nitrite removal strategies invariably include acidification. These issues were recognized early on in the study of S-nitrosothiols and the importance of this issue cannot be stressed enough. Even methods that do not ostensibly use acidification can be compromised. For example, it is easy to overlook the acidic mobile phase in HPLC analysis and it is possible to make S-nitrosothiols on-column from contaminating nitrite. The use of acidic media to remove proteins from beads after immune precipitation may be a routine methodology, but will compromise S-nitrosothiol detection unless thiols are blocked. This is particularly acute if kits are used and the investigator is not fully aware of the pH of the various proprietary solutions.
3. UV-Visible Detection
S-Nitrosothiols are colored and have weak absorbance bands in the UV (330-340 nm) and visible (450-550 nm) range. Depending on the dihedral angle made by the S-N bond, the color of the compounds can vary from green to orange/pink [13]. Although these bands uniquely identify S-nitrosothiols, the extinction coefficients are small and make direct detection virtually impossible in any meaningful biological situation. These absorbance bands have been used to study transnitrosation (the transfer of the nitroso functional group from an S-nitrosothiol to a thiol) and for examining the formation and decay of these compounds in simple systems [14]. These absorbance bands have been utilized in HPLC methods which use UV-Visible detection for accurate quantification of low-molecular weight S-nitrosothiols [15].
One of the simplest methods to detect S-nitrosothiols using spectrophotometry is the Saville reaction [16]. The principal of this reaction is that Hg(II) can decay S-nitrosothiols to form nitrite, and that this nitrite can then be detected using the Griess method [17] for nitrite detection. This involves nitrite dependent diazotization of sulfanilamide, under strongly acidic conditions, followed by coupling a naphthyl ring to produce a strong chromophore (extinction coefficient to about 50 mM−1cm−1). Although this assay can be quite sensitive it has the major drawback of being a difference method. In all biological situations there is significantly more nitrite than S-nitrosothiols. Not only is low-level nitrite a contaminant in most buffers but biological systems with nitric oxide synthase activity generate nitrite. Attempting to use this method to detect S-nitrosothiols on a high background of nitrite is prone to large errors. Generally this method is not suitable for detecting biological nitrosothiols, unless there levels are massively elevated by pharmacological means.
4. Chemiluminescence methods
A wide spread and sensitive detection method for S-nitrosothiols is based on detection of the chemiluminescence generated from the ozone-dependent oxidation of nitric oxide [18]. The chemical basis of this method is the reaction between NO and ozone, which yields nitrogen dioxide in an excited state (NO2*). Upon relaxation to ground state, NO2* emits energy as light (chemiluminescence) in the red and near infrared region (600-3200 nm). The intensity of chemiluminescence is directly proportional with the amount of NO present in the detector and can be recorded by a photomultiplier. This method can be adapted to detect both gaseous samples (as ppb in for example, exhaled breath) and aqueous samples. This method has a rich history in NO research as the first demonstration that NO was generated by biological systems was made using ozone-dependent chemiluminescence [19]
For the detection of S-nitrosothiols, the commonly used apparatus consists of a purge vessel where the S-nitrosothiols are cleaved to generate NO, which is delivered by argon or helium purge to the reaction cell of the detector where ozone-dependent oxidation of NO occurs. The cleavage of the S-NO bond can be completed either with photolysis [20,21] or with chemical reduction. Various reducing mixtures have been developed. The most common of these are acidic tri-iodide [22-26] and copper containing systems, such as Cu(I)/cysteine [27,28], Cu(I)/cysteine/CO [29-31], Cu(II)/ascorbate [32,33]
Typically, a sample consists of an array of NO derivatives, such as nitrite, nitrate, iron nitrosyls, and S-, N- and C-nitroso compounds. Since the detection is based exclusively on oxidation of nitric oxide, and not its metabolites, the selectivity of this method is determined by the cleavage strategy through a combination of reduction of the desired compound and elimination the interfering NO derivatives (Scheme 1).
If all the conditions of the analysis are carefully controlled, possible interferences are taken into account, and proper standards in the anticipated concentration ranges are analyzed along with the samples, these methods can be reliably used and detection levels of about 1 picomole can be achieved [34].
Photolysis
The light-induced homolytic cleavage of S-N bond in S-nitrosothiols [20], which was originally performed in a borosilicate glass coil by irradiation with a 200 W mercury vapor lamp, results in NO release with concomitant formation of thiyl radical. In order to prevent further reactions of the products of photolysis, liquid and organic traces are condensed in cold traps (0 °C, and -75 °C, consecutively), and the NO gas is immediately purged into the ozonator chamber.
The method was originally developed for sensitive detection of S-nitrosothiols in mammalian plasma, and later extended for analysis of various low molecular weight S-nitrosothiols, iron nitrosyls, nitroprusside, and L-glycyl-N-nitroso-tryptophan [21]. In that latter study it was shown that metal chelators improve the stability and sensitivity of the assay. Nitrosoproteins were discriminated from circulating low molecular weight S-nitrosotiols by protein precipitation using acid, or alternatively acetone, phenol or ammonium sulfate at neutral pH. The specificity of this assay towards S-nitrosothiols was ensured by analyzing the samples without and with addition of organic or inorganic Hg(II). As mentioned above, Hg(II), which is indicated to be added in slight excess over thiol at pH 7.4 [35], converts S-nitrosothiols to nitrite which is more resistant to photolysis.
S-nitrosothiols show two characteristic absorption maxima, one in 300-350 nm, the other in 530-560 nm range. Typically, the range of shorter wavelength is used for irradiation, although recently visible-wavelength photolysis was also suggested [36] to avoid possible interference with nitrite [21] and nitrate [37]. The light intensity must not be higher than 100 W, otherwise thermal effects might cause signal distortion. NO release from nitrite contaminations also can be prohibited by keeping the pH above 6. The use of a 300 nm cutoff filter is also recommended. The detection limit of this assay is about 5 pmole [20,38,39].
The requirement of mostly custom made photolytic equipment vs. commercially available NO analyzers suitable for chemical reduction-based systems may explain the limited popularity of this method. However, the photolytic properties of S-nitrosothiols have been used as functional diagnostics of their involvement in biological processes [40].
Triiodide-chemiluminescence
The use of tri-iodide to chemically reduce s-nitrosothiols was developed by Samouilov and Zweier [22] . The reduction of S-nitrosothiols is quantitative according to the following chemistry:
In the presence of iodide ions the cycle is catalytic for iodine. Acidic iodide also quantitatively converts nitrite to NO, and the presence of iodine does not change the yield. The nascent NO is immediately purged to the detector. A condenser and a chemical trap containing solution of 0.1 N NaOH placed between the purge vessel and the reaction cell of the NO analyzer protects the detector. Use of glacial acetic acid rather than HCl, and most cases addition of anti-foaming agent help to overcome foaming caused by the protein content of most biological samples. The issue of foaming is least problematic with tri-iodide-based methods compared to copper-based methods, where foaming is a major limitation.
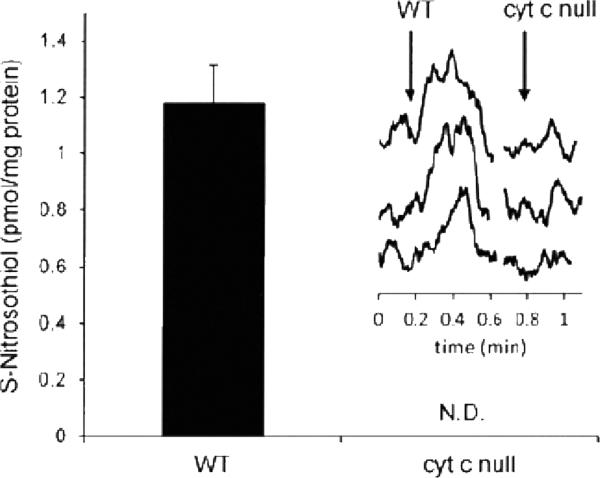
This method is convenient to use, because by sequential addition of a series of chemical scavengers based on the traditional Griess/Saville assay [16,17], nitrite and S-nitrosothiol content of the sample can be determined and discriminated from iron nitrosyls and nitrosamines. The acidic triiodide media reduces all above listed NO derivatives. Nitrite content can be calculated from the difference between the chemiluminescence signal before and after treating the samples with 10 % of 100 mM sulfanilamide/HCl solution, since the nitrite diazotates sulfanilamide, and thus its contribution to the chemiluminescence signal will be removed. S-nitrosothiols can be depleted by subsequent addition of 10% of 50 mM HgCl2, which converts S-nitrosothiols to nitrite (which is removed by sulfanilamide), while nitrosamines and nitrosyl hemoglobin are resistant to mercury treatment [26] and the residual signal can then be subtracted. If iron nitrosyls are expected to be present in large amounts it may be pertinent to remove them using ferricyanide. This has been done in experiments where NO adductions of hemoglobin were quantified [41]. Using this method 1 pmol S-nitrosothiol/mg protein can be quantified in cellular system. An example of using this method to detect very low levels of S-nitrosothiols is shown in Figure 1, taken from [7].
Figure 1.
Low levels of S-nitrosothiols detected in cells using tri-iodide-dependent chemiluminescence (This research was originally published in the Biochemical Journal Broniowska KA, Keszler A, Basu S, Kim-Shapiro DB, Hogg N: Cytochrome c-mediated formation of S-nitrosothiol in cells. Biochem J., 2012, 442191-197 © Portland Press)
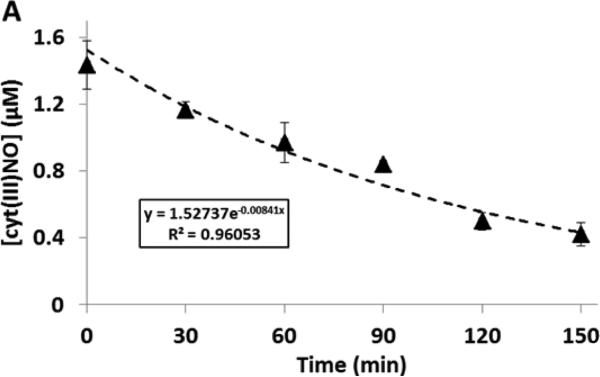
Our experience with this method over many years has brought to light two important issues. 1) Although heme nitrosyls can be subtracted from the S-nitrosothiol signal, by virtue of being Hg(II)-resistant, the level of these compounds may change over the analysis time. For example, the heme nitrosyl of cytochrome c after addition of sulfanilamide and HgCl2 gives a decaying chemiluminescence signal (Figure 2). Consequently, it becomes important to analyze the mercury-treated and non-treated samples in close temporal proximity. This way a possible artifact caused by instability of iron nitrosyl can be eliminated. 2) We also observed that dinitrosyl iron complexes (DNIC), as opposed to iron nitrosyls, are detectable with triiodide chemiluminescence, and are removed by mercury. However, the differential stability of DNIC and S-nitrosothiols makes the specific detection of S-nitrosothiols possible (Keszler A. et al. manuscript in preparation).
Figure 2.
Decay of heme-nitrosyl signal over time in tri-iodide-dependent chemiluminescence. This signal is due to nitrosyl cytochrome c and illustrates that the Hg(II)-resistant signal is not necessarily stable and so the mercury control sample needs to be injected immediately after the mercury-free sample.
Determination of S-nitrosothiol in blood with reductive chemiluminescence is a complex task since under the anaerobic conditions of the purge vessel the nascent NO is subject to autocapture by the heme to form nitrosylhemoglobin. Although the nitrosylhemoglobin will itself be oxidized to liberate NO, the consequence of autocapture is to broaden the peak of NO release thus reducing sensitivity [42]. Autocapture can be reduced by pre-oxidizing hemoglobin with ferricyanide. S-nitrosothiols in blood samples can be preserved by incubation the sample with a stabilization solution [43-45] containing potassium ferricyanide, which oxidizes the heme to Fe(III), and removes NO from the coordination sphere of heme iron [24,25], and also prevents heme-dependent reduction of S-nitrosohemoglobin [46]; N-ethyl-maleimide (NEM) to bind free thiol; DTPA metal chelator to stabilize S-nitrosothiols, and Nonidet P-40 detergent to facilitate membrane solubilization. Before injecting the sample to the purge vessel the low molecular weight fraction should be separated on a Sephadex G-25 column, and the heme concentration should be determined in the protein fraction. S-nitrosothiol content is established from the difference of chemiluminescence signal of not mercury-treated and mercury-treated (both sulfanilamide supplemented) samples [43].
The reliability of triiodide method has been debated [39,47] by claiming that the NO may react with the reducing medium forming NOI, a nitrosating agent, and that a synergistic effect of the components of stabilization solution might lead to decay of S-nitrosothiols. However, these criticisms are highly speculative and extensive investigations conducted by several laboratories proved the triiodide-base assay to be a highly sensitive and satisfactorily specific method for S-nitrosothiol analysis in biological samples [41,43,48]. The major limitation of the tri-iodide method is that acidification is required and therefore nitrite elimination has to be performed. For this reason investigators have endeavored to develop pH neutral methodologies as discussed below.
Copper/cysteine-based methods
The chemical basis of this method consists of transnitrosation of S-nitrosothiols with cysteine, and reduction of the formed S-nitrosocysteine by Cu(I), which gets oxidized to Cu(II) and subsequently re-reduced by excess cysteine [49]:
At neutral pH this method is specific for S-nitrosothiol when HgCl2 treatment is included, the S-nitrosothiol concentration being calculated from the difference of peak areas with and without Hg(II) treatment. Monitoring the pH during the analysis is critically important, since if it drops below 6, other NO derivatives become detectable decreasing the selectivity of the assay. Injecting nitrite and nitrate as negative control is advisable. Extensive foaming of biological samples can be controlled by addition of antifoaming agent. The assay was modified by sample pretreatment with stabilization solution (potassium ferricyanide/NEM/DTPA) to oxidize heme iron to ferric form, hence impede NO autocapture [44]. Although low molecular weight S-nitrosothiols can be quantified with high efficiency and sensitivity, a major disadvantage of this method is its poor efficacy (35 %) for S-nitrosoproteins,
The original assay (2C) contains two components (cysteine and copper salt), but later it was further developed by supplementation with CO gas (3C) in order to make the technique suitable to handle blood samples [29]. Excess carbon monoxide can prevent autocapture of NO by the heme. The commercially available CO gas should be purified by passing through iodine crystals and charcoal to eliminate metal carbonyls which influence the selectivity. This method requires some specific security protections. First of all CO must be handled with care, the system always should be purged with inert gas before opening to the atmosphere. Moreover, a hopcalite trap is routinely attached to the exhaust of chemiluminescence apparatus to trap ozone from the exhaust, but this must be removed as the reaction of the trap material with CO is strongly exothermic.
Copper/ascorbate chemiluminescence
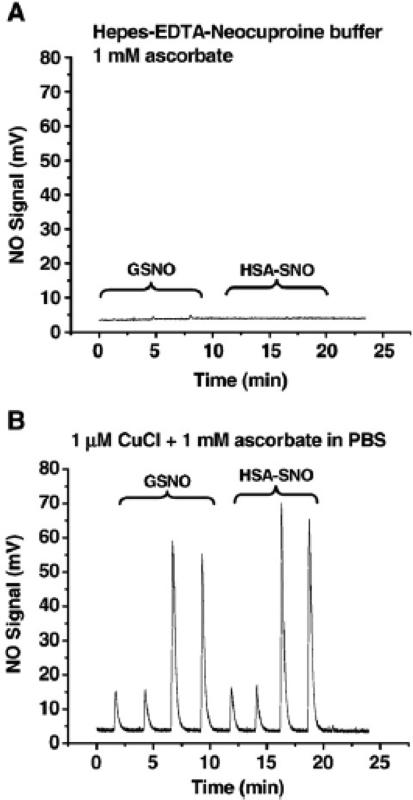
This method was developed to quantify S-nitrosothiols, especially S-nitroso hemoglobin, while avoiding interference with nitrite and nitrosyl hemoglobin [33]. The assay uses Cu(II) salt and ascorbate at neutral pH. Under the anaerobic environment of the purge vessel ascorbate reduces Cu(II) to Cu(I), which reduces S-nitrosothiol to thiol and NO. Cu(I) gets oxidized by reacting with S-nitrosothiol, and recycled by ascorbate. When analyzing S-nitrosoproteins ascorbate should be in excess over copper to ensure the total conversion of Cu(II) to Cu(I). This system unlike the Cu(I)/cysteine based one is efficient not only for low molecular weight S-nitrosothiols but also for S-nitrosoproteins. Data from Cu(I)/Ascorbate-mediate chemiluminescence is shown in Figure 3 [50].
Figure 3.
An example of Cu/Ascorbate-mediated chemiluimesence. Chemilumiesence detection of both protein RSNO and GSNO in the presence of ascorbate with and without Cu(I) addition. (Taken From Wang X, Kettenhofen NJ, Shiva S, Hogg N, Gladwin MT: Copper dependence of the biotin switch assay: Modified assay for measuring cellular and blood nitrosated proteins. Free Radic. Biol. Med. 44(2008)1362-1372 © Elsevier)
A major downside of this method is the heavy foaming even if using antifoaming agent, which makes analysis of high protein containing samples (e.g. plasma) extremely difficult. To overcome this problem the assay was recently modified [32]. The pH was decreased to the acidic region using glacial acetic acid to suppress the foaming, although to some extent, this defeats the object of having a pH neutral method. The interference of nitrite was excluded by addition of sulfanilamide to the sample. The assay is not silent for nitrosamines, so mercury treatment should also be performed.
Alternative reduction-based methods
The above described methods are the most commonly used ones, however, other, less appreciated techniques are also available.
A Cu(I)/I−/I2-mediated assay was developed [23] originally for detection S-nitrosothiols in plasma. The reducing mixture consists of KI in glacial acetic acid and CuSO4, and works at 70°C. This technique is used with combination of Griess/Saville reagents, and addition of NEM prevents artificial S-nitrosothiol generation.
An assay based on quinone-hyroquinone redox system in Tris buffer at pH>10 at 60 °C (Samouilov A. and Zweier J. L. 1998) has the advantage of not detecting nitrite [22]. Acidic samples should be treated with sulfanillamide reagent to remove nitrite.
Summary
All the above discussed methods were developed with the intention to find a relatively easy, fast, and user friendly technique for selective determinations of low levels of S-nitrosothiols in complex biological matrices. The chemiluminescence of ozone-mediated oxidation of nitric oxide seemed to fulfill the criteria. However, a series of various NO metabolites is present in biological milieu, which also gives chemiluminescence signal and makes the specificity for S-nitrosothiol difficult.
The first obstacle is the omnipresent nitrite. The photolysis method uses a 300 nm cutoff filter, while the copper/cysteine-, the copper/iodide-, the original copper/ascorbate, and the quinone-/hydroquinone systems work by keeping the pH neutral or alkaline to avoid the reduction of nitrite. The triiodide-, and modified copper/ascorbate assays eliminate nitrite by using sulfanilamide.
None of the known methods is directly selective for S-nitrosothiol against nitrosamine and most of them neither against heme nitrosyl. All assays use HgCl2 treatment to deplete S-nitrosothiols, and calculate their concentration indirectly from the difference between the signal of not treated and treated samples. Interestingly, our recent findings revealed that certain assays reduce dinitrosyl iron complexes, which are also mercury labile. This fact should not be neglected during the analysis.
A major challenge is the quantification of S-nitrosothiol in blood or in other heme containing samples. Potential source of loss of nascent NO can be its autocapture by the Fe(II) of the heme. In the triiodide- and the modified 2C methods the samples are pretreated with potassium ferricyanide to oxidize the iron to Fe(III) state and hinder the NO autocapture. The 3C technique uses CO to prevent NO coordination to the heme.
All methods are validated by multiple laboratories, and proved to be adequate to measure picomole level of S-nitrosothiols in biological systems. However, the samples are very diverse. Some of them consist of mainly S-nitrosoproteins, others comprise low molecular weight S-nitrosothiols. They may or may not contain blood or erythrocytes. Their protein content is also quite different. If the goal is to determine low concentrations of S-nitrosothiols in complex matrices, it is advisable to try more than one method, and critically evaluate the results.
5. RSNO detection using the biotin-switch technique
While chemiluminsecence assays provide a sensitive and specific method for direct detection of total RSNO, their limitation lies in the inability to differentiate S-nitrosation of specific proteins. As the field of redox signaling has grown in recent years and S-nitrosation has become increasingly recognized as an important nitric oxide-dependent post-translational modification, the ability to clearly identify specific sites on specific protein targets for S-nitrosation is critical to define its contribution to redox-sensitive signaling cascades and subsequent cellular effects. Detection of S-nitrosated proteins has been a major challenge for several reasons including: 1) the nitroso group is not amenable to radio-labeling strategies; 2) reliable, specific antibodies which detect S-nitrosated proteins have not been generated; and 3) chemical probes with specific reactivity with S-nitrosothiols do not exist as they do for other thiol modifications (e.g., dimedone labeling of protein sulfenic acids). Thus, strategies for protein S-nitrosothiol measurement have been driven by indirect detection methods which aim to replace the labile nitroso moiety with a more stable adduct for further analysis using gel-based techniques. A major innovation to this end was the description of the biotin-switch technique by Jaffrey et al in 2001 [51], described in more detail below.
It should be noted that direct antibodies for S-nitrosated proteins are commercially available and have been used for immunohistochemistry [28] and infrequently for western blotting. We have no experience with the use of such antibodies for immunohistochemistry and so can provide no insight; however, we have attempted both immunoprecipitation and western blotting with these products to no avail. While these experiences are necessarily anecdotal, the fact that a great deal of expense and ingenuity has gone into alternative methods to detect and isolate S-nitrosothiols via affinity-based methods speaks volumes as to the lack of confidence placed in direct antibody approaches.
Assay principles
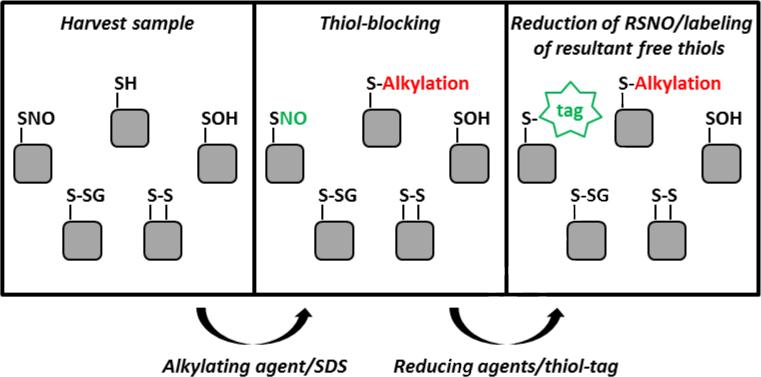
The original biotin-switch technique and all modifications of this protocol have four major steps: 1) thiol-blocking, 2) selective reduction of RSNO, 3) labeling of resultant free thiols, and 4) detection of biotin-labeled thiols (shown schematically in Figure 4). Here, we will discuss the approach employed in the original biotin-switch technique and modifications to this protocol made by other investigators.
Figure 4.
Schematic of Biotin-switch labeling system.
Thiol-blocking
The biotin-switch technique is particularly versatile, as protein samples can be prepared from nearly any source including purified protein [52], cultured cells [53], tissue preparations [54], isolated organelles [55], and plasma [50]. In all cases, care must be taken in sample preparation to retain protein S-nitrosothiols while limiting the introduction of artifacts. Immediately after harvesting the protein sample, free thiols are blocked using an alkylating agent. This is critical, as the efficiency of this step dictates the specificity and sensitivity of this method. Incomplete blocking of thiols will result in false-positive identification of protein S-nitrosothiols. The original biotin-switch technique used MMTS (methyl methanethiosulfonate) as the alkylating agent which forms disulfide bonds with free thiols. Although the kinetics MMTS/thiol reactions are amenable for rapid and robust thiol-blocking, the more labile nature of the disulfide bond has led others to propose the use of N-ethylmaleimide (NEM) and iodoacetamide (IAM) as preferred thiol-blocking reagents. These reagents form thioether instead of disulfide bonds and have become more common recently. Others have expressed concern over the reaction chemistry of MMTS – the disulfide exchange between MMTS and free thiols in the protein sample generates the free thiol form of MMTS, and some have suggested this has the potential to artifactually reduce S-nitrosothiols in the protein sample [56]. Typically, thiol-blocking is expedited by heating samples in the presence of thiol-blocking reagents (e.g., 50°C, 20-60 min), and then the thiol-blocking reagents must be removed from the sample prior to moving on to the reduction step. This can be done using trichloroacetic acid, ethanol, or acetone protein precipitation or small spin columns. Residual thiol-blocking reagents will interfere in the reduction and labeling steps.
Selective reduction of RSNO
Now that the free thiols in the protein sample are alkylated, the goal of the next step is to selectively reduce protein RSNO, so they can be labeled with a biotin- or fluorescent-tag (the ‘switch’). This must occur without reducing disulfides or other thiol modifications, as the technique relies on the assumption that all resultant free thiols are derived from RSNO. The original biotin-switch technique and several other since used 1-30 mM ascorbate (1 h) for selective reduction of RSNO [51,57]; however, from a kinetic perspective with known rate constants for the reaction of ascorbate with S-nitrosoglutathione (12 M−1s−1, pH 7.3) [58], this should be completely inadequate for efficient RSNO reduction[53]. Further studies revealed that trace metal ions present in assay buffers were the likely culprits in facilitating selective RSNO reduction, and ascorbate serves to reduce cupric(II) ions back to the cuprous(I) state. In fact, we examined this in detail and proposed the addition of copper ions and the absence of metal chelators for reduction/labeling assay buffers [50,59]. Under these conditions, RSNO reduction is both rapid and relatively specific. Although reduction of other thiol modifications cannot be completed ruled out, we have shown that at least for the model protein serum albumin, mixed disulfides do not yield false-positives [50].
Significant concerns have arisen regarding the use of high levels of ascorbate in some assay conditions. Ascorbate was shown to reduce disulfides of microtubule proteins [60] and Giustarini et al reported that 1-50 mM ascorbate readily reduces the disulfide bonds of DTNB (Ellman's reagent) and the more physiologic disulfides glutathione disulfide, cystine, and cystinylglycine in a concentration- and time-dependent manner [61]. Huang and Chen also described an ascorbate-dependent false-positive signal when examining S-nitrosation of bovine serum albumin [62]. These concerns can be largely circumvented with additional positive- and negative-controls in each experiment. For example, samples can be processed in parallel in the absence of reducing agents (often termed ‘no reduction’ controls). Signal obtained from these samples represents background labeling from incomplete thiol-blocking in the first step and is not derived from selective reduction of modified thiols. In addition, we find that the inclusion of a positive-control dithiothreitol (DTT) reduced sample can provide additional insight into the differential reduction of S-nitrosothiols versus disulfides in the experimental system [59].
The realization that protein sulfenic acids are an important biological thiol modification brings these species to the table as possible artifacts in switch methods. Although some attempts have been made to rule these out (see [57]), it is hard to see a chemical reason why the reductive conditions of switch assays would not pick up these species. This is an area where a more thorough investigation is required for select proteins that exhibit stable sulfenic acid modifications.
More recently, alternative reducing agents have been used for selective reduction of RSNO in hopes of overcoming the selectivity and sensitivity issues of ascorbate-based biotin-switch techniques. A series of triphenylphosphine ester derivatives have been developed and applied in the biotin-switch technique. Although the number of publications using this approach is limited, the early reports indicate phosphine-mediated selective reduction is specific for S-nitrosothiols in complex biological samples, does not reduce protein disulfides or hydrogen peroxide-dependent thiol oxidation, and is sensitive enough to detect changes in protein S-nitrosothiols from inducible nitric oxide synthase (iNOS)-derived nitric oxide [63]. Earlier this year, methylhydrazine was also suggested to be a new and improved selective-reductant for the biotin-switch technique [64]. Wiesweg et al characterized in detail the reaction of methylhydrazine with model nitroso compounds including S-nitrosoglutathione (GSNO), S-nitroso-N-acetyl-penicillamine (SNAP), N-acetyl-N-nitrosotryptophan (NANT), and S-nitrosoalbumin. They also compared detection of S-nitrosoalbumin using the biotin-switch technique with methylhydrazine- or ascorbate only-reduction (not copper/ascorbate) and demonstrated improved sensitivity using methylhydrazin [64]. Whether these new strategies for selective reduction of RSNO in the biotin-switch technique become established remains to be seen.
Labeling of resultant free thiols
Once protein RSNO are reduced, the resultant free thiol is labeled with a biotin- or fluorescent-tag. Here, we have discussed the reduction and labeling steps separately, but in practice, they typically occur together in a single incubation period. The reduction takes place in the presence of thiol-labeling reagents, so as S-nitrosothiols are reduced, they can be immediately tagged. The original biotin-switch technique used Biotin (bt)-HPDP (N-[6-(Biotinamido)hexyl]-3’-(2’-pyridyldithio)propionamide) to label resultant free thiols [51]. Bt-HPDP forms disulfide bonds with free thiols, leaving them biotin-tagged. These newly biotinylated proteins are often detected after affinity purification using avidin beads (discussed in more detail below), and the fact that bt-HPDP-labeled proteins can be easily released from avidin beads by reducing off the tag with DTT or β-mercaptoethanol makes it a preferred thiol label for many investigators. The downside to this approach is the biotin-tag is not retained in sample processing and thus its presence on a protein of interest cannot be confirmed in later steps. For this reason, biotin-conjugated maleimido and iodoacetamide linking agents (e.g., biotin-IAM; BIAM) have also been applied to the biotin-switch technique with success. In addition, fluorophore-conjugated thiol labels have come into favor as 2D proteomic and fluorescence difference gel electrophoresis (DIGE) techniques have become more accessible.
Detection of labeled thiols
The final step in the biotin-switch technique is the detection of the biotinylated proteins. The most widely applied detection method is affinity purification using avidin beads and Western blot analysis using antibodies specific for a protein of interest. This approach is particularly useful for investigations in which a candidate protein(s) has been previously identified and was used by Jaffrey et al in the original biotin-switch paper to detect neuronal NOS-dependent S-nitrosation of the N-methyl-D-aspartate receptor [51]. For those more interested in cataloging more global changes in the S-nitroso proteome, 2D proteomic approaches coupled to Western blot analysis is a more appropriate detection approach; however, for this application it is critical that the biotin tag be retained in sample processing (discussed above), as avidin-based detection is required [Huang, Journal of Proteome Research 2009]. More recently, SNO-DIGE techniques have been described by several groups [55,59] which couple the principles of the biotin-switch technique with fluorescent thiol tags (e.g., maleimide-conjugated cyanine dyes) and 2DDIGE analysis. The advantages of this approach are two-fold. Direct in-gel detection eliminates the need for transfer to membranes and immuno-detection and registration between protein spots on the gel and membrane. The ability to multiplex with different spectrally resolvable fluorescent tags also allows for the analysis of more than one sample per gel and limits gel-to-gel variability. This has led to the identification and functional characterization of novel targets of S-nitrosation in a more quantitative manner. Finally, efforts have also been made to identify protein S-nitrosation in situ from prepared and mounted tissue slices by adapting the biotin-derivatization for immunohistochemical detection. The benefit of this approach is it allows for localization of RSNO to specific regions in cells or whole-tissue preparations [65,66], and it has been used, for example, to define S-nitrosated proteins (e.g., Ran GTPase) in the lung [65].
Caveats and conclusions
It is clear the biotin-switch technique has been a critical innovation in the field of nitric oxide biology which has significantly furthered our understanding of the (patho)physiological role of protein S-nitrosation in health and disease. However, significant caveats exist in analysis and interpretation of biotin-switch results. Notably, issues related to incomplete thiol-blocking prior to reduction/labeling, the selectivity of reducing agents for RSNO versus other thiol modifications, artifactual detection of false-positives due to assay conditions, and (anecdotal) unreliable replication of results between laboratories must be considered. There remains a need for new approaches to address these issues and the development of reagents that allow for direct detection and/or direct derivatization of the nitroso moiety in the future.
6. Mass Spectrophotometric and Proteomic Analysis of Protein S-Nitrosation
Direct mass spectrometric analysis
S-Nitrosation of a thiol adds 29 Da and so is potentially easily detectable by mass spectrometry. Electrospray Ionization Mass Spectrometry (ESI-MS) was used as early as 1995 to examine peptide and protein S-nitrosation [67]. This clearly demonstrates the ability of ESI-MS to directly detect S-nitrosation of purified peptides and proteins. However, even in this case, higher temperature of the capillary transfer tube resulted in significant degradation of S-nitrosothiol indicating that more aggressive ionization strategies were unlikely to be successful. However, this study also illustrates a methodological issue that affected much of the early work on MS detection of S-nitrosothiols which is worth highlighting. Acidification of nitrite in the presence of thiols is a very efficient way to make S-nitrosothiols, and so if peptides/proteins are prepared for analysis in acetic acid (or any acidified solution) without prior thiol block, it is not possible to distinguish NO-dependent S-nitrosation from nitrous acid-dependent nitrosation. As nitrite is the major product of oxygen-dependent NO oxidation it will always be present in solutions of NO, and so these methods likely detected far higher levels of S-nitrosothiols than were generated by the chemistry of NO. Another early example of this is the detection by ESI-MS of a nitrosated fragment of NfκB p50, nicely showing the S-nitrosation of a fragment of this protein but most likely generated from acidification of nitrite-containing buffers [68]. Hemoglobin, being by far the most abundant protein in the red cell, was a good target for the use of direct MS analysis in a biological matrix. Hemolysate was incubated with S-nitrosocysteine (which will modify thiols via a NO-independent transnitrosation reaction) and direct LC-ESI-MS analysis showed a 29 Da shifts of the α- and β-globin subunits, but showing selective S-nitrosation of a single β-chain cysteine at low concentration of nitrosating agent. This study observed the S-nitroso peptides could be recovered after tryptic digest of hemoglobin, however, it should be again pointed out acidic conditions were used in many parts of this study, including the digestion, with no apparent thiol blocking step. Mass accuracy is clearly important in these studies, as a 30 Da shift is indicative of a sulfinate formation, and this has been previously observed in the oxidation of cathepsin K by NO donors [69]
Mass spectrometric analysis of S-nitrosothiols has mainly been used to analyze protease digests in proteomic studies after switch-type methods. The SNOSID method of Hao et al [70] was one of the first to attempt this by performing trypsinolysis after biotin switch, and pulling down labeled peptides. In this method biotin-HPDP was used and the label was reduced off before MS analysis. Consequently it was not possible to confirm the exact site of labeling (if the peptide contained multiple cysteines) or if the peptide was not a target of nitrosation but was disulfide-bonded to the biotin-labeled peptide. This limitation was partially overcome by Greco et al [71] who used acidic conditions to release the biotinylated peptides from the affinity matrix and so retained the biotin on the peptide where its additional mass could be used to identify sites of modification. The use of irreversible biotinylation reagents has recently been suggested so that reducing conditions can be employed to disrupt disulfide bonds without loss of biotin label [72]. More recently, direct affinity capture of S-nitrosated proteins has been accomplished with organo-mercury affinity resin allowing the circumvention of reduction issues [73]. This method has recently been used to perform in vivo proteomic assessment of nitrosated proteins [74].
7. Conclusions
Any field of scientific endeavor will only be as good as its methods, and methods take time to be fully explored and rigorously assessed. The S-nitrosothiol field has matured to where the methods may not be perfect, but the issues and caveats are well known. It is however, that newcomers to this field fully understand the limitations of the methods they use and do not repeat the mistakes of the past or use these methods without a proper understanding of their limitations. This issue is most pertinent in switch methods. Although commercial kits are available, use of these kits does not mitigate any of the responsibility of the investigator to assess the limitations of the method and perform appropriate controls. Finally it must be stressed that knowing that a protein is S-nitrosated is not enough to assign this modification a biological role. Many modifications may be functionally silent either because the target thiol medication plays no role in protein function, or because the extent of thiol modification is insufficient to impact that particular metabolic or signaling pathway. This latter point is of particular concern as methods become increasingly sensitive. It is likely that some low level of protein S-nitrosation occurs as an inevitable consequence of using NO as a signaling molecule and we should be careful not to assign specific functions to this this baseline noise. It is not clear how many of the proteins present in proteomic screen of S-nitrosation represent functionally important modifications. Nevertheless, these screens are invaluable for providing targets for hypotheses that can tested by additional more specific methods. A recent example of this is the identification of VLCAD (very long chain acyl co-enzyme A dehydrogenase) from a proteomic screen which was later shown to be activated by S-nitrosation [74] implicated a role of S-nitrosation in β-oxidation.
S-Nitrosothiols Detection Methods are described
We discuss and compare chemiluminescence Detection methods
We describe switch-type methods for S-nitrosothiol detection
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin JC, Smith CD, Beckman JS. Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase. Arch. Biochem. Biophys. 1992;298:431. doi: 10.1016/0003-9861(92)90431-u. [DOI] [PubMed] [Google Scholar]
- 2.Radi R. Peroxynitrite, a stealthy biological oxidant. J. Biol. Chem. 2013 doi: 10.1074/jbc.R113.472936. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Vliet A, Eiserich JP, Halliwell B, Cross CE. Formation of reactive nitrogen species during peroxidase-catalyzed oxidation of nitrite. A potential additional mechanism of nitric oxide-dependent toxicity. J. Biol. Chem. 1997;272:7617. doi: 10.1074/jbc.272.12.7617. [DOI] [PubMed] [Google Scholar]
- 4.Gunther MR, Hsi LC, Curtis JF, Gierse JK, Marnett LJ, Eling TE, Mason RP. Nitric oxide trapping of the tyrosyl radical of prostaglandin H synthase-2 leads to tyrosine iminoxyl radical and nitrotyrosine formation. J. Biol. Chem. 1997;272:17086. doi: 10.1074/jbc.272.27.17086. [DOI] [PubMed] [Google Scholar]
- 5.Madej E, Folkes LK, Wardman P, Czapski G, Goldstein S. Thiyl radicals react with nitric oxide to form S-nitrosothiols with rate constants near the diffusion-controlled limit. Free Radic. Biol. Med. 2008;44:2013. doi: 10.1016/j.freeradbiomed.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein S, Czapski G. Mechanism of the nitrosation of thiols and amines by oxygenated NO solutions: the nature of the nitrosating intermediates. J. Am. Chem. Soc. 1996;118:3419. [Google Scholar]
- 7.Broniowska KA, Keszler A, Basu S, Kim-Shapiro DB, Hogg N. Cytochrome c-mediated formation of S-nitrosothiol in cells. Biochem. J. 2012;442:191. doi: 10.1042/BJ20111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keszler A, Zhang Y, Hogg N. The Reaction between Nitric Oxide, Glutathione and Oxygen in the Presence and Absence of Protein: How are S-Nitrosothiols Formed? Free Radic. Biol. Med. 2009;48:55. doi: 10.1016/j.freeradbiomed.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue K, Akaike T, Miyamoto Y, Okamoto T, Sawa T, Otagiri M, Suzuki S, Yoshimura T, Maeda H. Nitrosothiol Formation Catalyzed by Ceruloplasmin. Implication for cytoprotective mechanism in vivo. J. Biol. Chem. 1999;274:27069. doi: 10.1074/jbc.274.38.27069. [DOI] [PubMed] [Google Scholar]
- 10.Forman HJ, Fukuto JM, Torres M. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am. J. Physiol. Cell Physiol. 2004;287:C246–C256. doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- 11.Gladwin MT, Wang X, Hogg N. Methodological vexation about thiol oxidation versus S-nitrosation--A commentary on “An ascorbate-dependent artifact that interferes with the interpretation of the biotin-switch assay”. Free Radic. Biol. Med. 2006;41:557. doi: 10.1016/j.freeradbiomed.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 12.Field L, Dilts RV, Ravichandran R, Lanhert PG, Carnahan PG. An unusually stable thionitrite from N-acetyl-DL-penicillamine. X-Ray, crystal and molecular structure of 2-(acetylamino)-2-carboxy-1,1-dimethylethyl thionitrite. J. Chem. Soc. Chem. Commun. 1978:249. [Google Scholar]
- 13.Bartberger MD, Houk KN, Powell SC, Mannion JD, Lo KY, Stamler J, Toone EJ. Theory, Spectroscopy and crystolographic analysis of S-nitrosothiols: Conformational distribution dictates spectroscopic behavior. J. Am. Chem. Soc. 2000;122:5889. [Google Scholar]
- 14.Singh RJ, Hogg N, Joseph J, Kalyanaraman B. Mechanism of nitric oxide release from S-nitrosothiols. J. Biol. Chem. 1996;271:18596. doi: 10.1074/jbc.271.31.18596. [DOI] [PubMed] [Google Scholar]
- 15.Hogg N. The kinetics of S-Transnitrosation - a reversible second-order reaction. Anal. Biochem. 1999;272:257. doi: 10.1006/abio.1999.4199. [DOI] [PubMed] [Google Scholar]
- 16.Saville B. A scheme for the colorimetric determination of microgram amounts of thiols. Analyst. 1958;83:670. [Google Scholar]
- 17.Griess P. Bemerkungen zu der Abhandlung der HH. Weselky und Benedikt Ueber einige Azoverbindungen. Chemische Berichte. 1879;12:426. [Google Scholar]
- 18.Clyne MAA, Thrush BA, Wayne RP. Kinetics of chemiluminescent reaction between nitric oxide and ozone. Trans. Faraday Soc. 1964;60:359. [Google Scholar]
- 19.Palmer RM, Ferrige AG, Moncada S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature. 1987;327:524. doi: 10.1038/327524a0. [DOI] [PubMed] [Google Scholar]
- 20.Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel D, Valeri CR, Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc. Natl. Acad. Sci. USA. 1992;89:7674. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alpert C, Ramdev N, George D, Loscalzo J. Detection of S-nitrosothiols and other nitric oxide derivatives by photolysis-chemiluminescence spectrometry. Anal. Biochem. 1997;245:1. doi: 10.1006/abio.1996.9947. [DOI] [PubMed] [Google Scholar]
- 22.Samouilov A, Zweier JL. Development of chemiluminescence-based methods for specific quantitation of nitrosylated thiols. Anal. Biochem. 1998;258:322. doi: 10.1006/abio.1998.2609. [DOI] [PubMed] [Google Scholar]
- 23.Marley R, Feelisch M, Holt S, Moore KP. A chemiluminescence based assay for S-nitrosoalbumin and other plasma S-nitrosothiols. Free Radic. Res. 2000;32:1. doi: 10.1080/10715760000300011. [DOI] [PubMed] [Google Scholar]
- 24.Gladwin MT, Shelhamer JH, Schechter AN, Pease-Fye ME, Waclawiw MA, Panza JA, Ognibene FP, Cannon RO., III Role of circulating nitrite and S-nitrosohemoglobin in the regulation of regional blood flow in humans. Proc. Natl. Acad. Sci. USA. 2000;97:11482. doi: 10.1073/pnas.97.21.11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gladwin MT, Wang X, Reiter CD, Yang BK, Vivas EX, Bonaventura C, Schechter AN. S-Nitrosohemoglobin is unstable in the reductive erythrocyte environment and lacks O2/NO-linked allosteric function. J. Biol. Chem. 2002;277:27818. doi: 10.1074/jbc.M203236200. [DOI] [PubMed] [Google Scholar]
- 26.Feelisch M, Rassaf T, Mnaimneh S, Singh N, Bryan NS, Jourd'heuil D, Kelm M. Concomitant S-, N-, and heme-nitros(yl)ation in biological tissues and fluids: implications for the fate of NO in vivo. FASEB J. 2002;16:1775. doi: 10.1096/fj.02-0363com. [DOI] [PubMed] [Google Scholar]
- 27.Fang K, Ragsdale NV, Carey RM, Macdonald T, Gaston B. Reductive Assays for S-Nitrosothiols: Implications for Measurements in Biological Systems. Biochem. Biophys. Res. Commun. 1998;252:535. doi: 10.1006/bbrc.1998.9688. [DOI] [PubMed] [Google Scholar]
- 28.Gow AJ, Chen Q, Hess DT, Day BJ, Ischiropoulos H, Stamler JS. Basal and Stimulated Protein S-Nitrosylation in Multiple Cell Types and Tissues. J. Biol. Chem. 2002;277:9637. doi: 10.1074/jbc.C100746200. [DOI] [PubMed] [Google Scholar]
- 29.Doctor A, Platt R, Sheram ML, Eischeid A, McMahon T, Maxey T, Doherty J, Axelrod M, Kline J, Gurka M, Gow A, Gaston B. Hemoglobin conformation couples erythrocyte S-nitrosothiol content to O2 gradients. Proc. Natl. Acad. Sci. USA. 2005;102:5709. doi: 10.1073/pnas.0407490102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgard DA, Abraham J, Allen A, Craft J, Foley W, Robinson J, Wells B, Xu C, Stedman DH. Chemiluminescent reactions of nickel, iron, and cobalt carbonyls with ozone. Appl. Spectrosc. 2006;60:99. doi: 10.1366/000370206775382730. [DOI] [PubMed] [Google Scholar]
- 31.Palmer LA, Doctor A, Chhabra P, Sheram ML, Laubach VE, Karlinsey MZ, Forbes MS, Macdonald T, Gaston B. S-nitrosothiols signal hypoxia-mimetic vascular pathology. J Clin. Invest. 2007;117:2592. doi: 10.1172/JCI29444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagababu E, Rifkind JM. Determination of s-nitrosothiols in biological fluids by chemiluminescence. Methods Mol. Biol. 2011;704:27. doi: 10.1007/978-1-61737-964-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagababu E, Ramasamy S, Rifkind JM. S-Nitrosohemoglobin: A mechanism for its formation in conjunction with nitrite reduction by deoxyhemoglobin. Nitric Oxide. 2006;15:20. doi: 10.1016/j.niox.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Basu S, Azarova NA, Font MD, King SB, Hogg N, Gladwin MT, Shiva S, Kim-Shapiro DB. Nitrite Reductase Activity of Cytochrome c. J. Biol. Chem. 2008;283:32590. doi: 10.1074/jbc.M806934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon TJ, Stamler JS. Concerted nitric oxide/oxygen delivery by hemoglobin. Methods Enzymol. 1999;301:99. doi: 10.1016/s0076-6879(99)01073-3. [DOI] [PubMed] [Google Scholar]
- 36.Riccio DA, Nutz ST, Schoenfisch MH. Visible photolysis and amperometric detection of S-nitrosothiols. Anal. Chem. 2012;84:851. doi: 10.1021/ac2031805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dejam A, Kleinbongard P, Rassaf T, Hamada S, Gharini P, Rodriguez J, Feelisch M, Kelm M. Thiols enhance NO formation from nitrate photolysis. Free Radic. Biol. Med. 2003;35:1551. doi: 10.1016/j.freeradbiomed.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Gow A, Doctor A, Mannick J, Gaston B. S-Nitrosothiol measurements in biological systems. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007;851:140. doi: 10.1016/j.jchromb.2007.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer LA, Gaston B. S-Nitrosothiol Assays That Avoid the Use of Iodine. In: Enrique Cadenas and Lester Packer, editor. Methods in Enzymology Nitric Oxide, Part F. Academic Press; 2008. pp. 157–176. [DOI] [PubMed] [Google Scholar]
- 40.Clementi E, Brown GC, Feelisch M, Moncada S. Persistant inhibition of cell respiration by nitric oxide: Crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc. Natl. Acad. Sci. USA. 1998;95:7631. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Bryan NS, MacArthur PH, Rodriguez J, Gladwin MT, Feelisch M. Measurement of Nitric Oxide Levels in the Red Cell: Validation of tri-iodide-based chemiluminescence with acid-sulfanilamide pretreatment. J. Biol. Chem. 2006;281:26994. doi: 10.1074/jbc.M603953200. [DOI] [PubMed] [Google Scholar]
- 42.Rogers SC, Khalatbari A, Gapper PW, Frenneaux MP, James PE. Detection of human haemoglobin-bound nitric oxide. J. Biol. Chem. 2005:M501179200. doi: 10.1074/jbc.M501179200. [DOI] [PubMed] [Google Scholar]
- 43.Macarthur PH, Shiva S, Gladwin MT. Measurement of circulating nitrite and S-nitrosothiols by reductive chemiluminescence. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007 doi: 10.1016/j.jchromb.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Basu S, Wang X, Gladwin MT, Kim-Shapiro DB. Chemiluminescent detection of S-nitrosated proteins: comparison of tri-iodide, copper/CO/cysteine, and modified copper/cysteine methods. Methods Enzymol. 2008;440:137. doi: 10.1016/S0076-6879(07)00808-7. [DOI] [PubMed] [Google Scholar]
- 45.Piknova B, Schechter AN. Measurement of nitrite in blood samples using the ferricyanide-based hemoglobin oxidation assay. Methods Mol. Biol. 2011;704:39. doi: 10.1007/978-1-61737-964-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spencer NY, Zeng H, Patel RP, Hogg N. Reaction of S-nitrosoglutathione with the heme group of deoxyhemoglobin. J. Biol. Chem. 2000;275:36562. doi: 10.1074/jbc.M005347200. [DOI] [PubMed] [Google Scholar]
- 47.Hausladen A, Rafikov R, Angelo M, Singel DJ, Nudler E, Stamler JS. Assessment of nitric oxide signals by triiodide chemiluminescence. Proc. Natl. Acad. Sci. USA. 2007;104:2157. doi: 10.1073/pnas.0611191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu S, Wang X, Gladwin MT, Kim-Shapiro DB. Chemiluminescent detection of S-nitrosated proteins: comparison of tri-iodide, copper/CO/cysteine, and modified copper/cysteine methods. Methods Enzymol. 2008;440:137. doi: 10.1016/S0076-6879(07)00808-7. [DOI] [PubMed] [Google Scholar]
- 49.Rogers SC, Gibbons LB, Griffin S, Doctor A. Analysis of S-nitrosothiols via copper cysteine (2C) and copper cysteine - Carbon monoxide (3C) methods. Methods. 2012 doi: 10.1016/j.ymeth.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Kettenhofen NJ, Hogg N, Gladwin MT. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic. Biol. Med. 2008;44:1362. doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2001;3:193. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 52.Sinha V, Wijewickrama GT, Chandrasena RE, Xu H, Edirisinghe PD, Schiefer IT, Thatcher GR. Proteomic and mass spectroscopic quantitation of protein S-nitrosation differentiates NO-donors. ACS Chem. Biol. 2010;5:667. doi: 10.1021/cb100054m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Keszler A, Broniowska KA, Hogg N. Characterization and application of the biotin-switch assay for the identification of S-nitrosated proteins. Free Radic. Biol. Med. 2005;38:874. doi: 10.1016/j.freeradbiomed.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 54.Cutler MJ, Plummer BN, Wan X, Sun QA, Hess D, Liu H, Deschenes I, Rosenbaum DS, Stamler JS, Laurita KR. Aberrant S-nitrosylation mediates calcium-triggered ventricular arrhythmia in the intact heart. Proc. Natl. Acad. Sci. U. S. A. 2012;109:18186. doi: 10.1073/pnas.1210565109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chouchani ET, Hurd TR, Nadtochiy SM, Brookes PS, Fearnley IM, Lilley KS, Smith RA, Murphy MP. Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem. J. 2010;430:49. doi: 10.1042/BJ20100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgoyne JR, Eaton P. A rapid approach for the detection, quantification, and discovery of novel sulfenic acid or S-nitrosothiol modified proteins using a biotin-switch method. Methods Enzymol. 2010;473:281. doi: 10.1016/S0076-6879(10)73015-9. [DOI] [PubMed] [Google Scholar]
- 57.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic. Biol. Med. 2009;46:119. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holmes AJ, Williams DLH. Reaction of ascorbic acid with S-nitrosothiols: clear evidence for two distinct reaction pathways. J. Chem. Soc. Perkin Trans. 2000;2:1639. [Google Scholar]
- 59.Kettenhofen NJ, Wang X, Gladwin MT, Hogg N. In-gel detection of S-nitrosated proteins using fluorescent methods. Methods Enzymol. 2008;441:53. doi: 10.1016/S0076-6879(08)01204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Landino LM, Koumas MT, Mason C, Alston JA. Ascorbic acid reduction of microtubule protein disulfides and its relevance to protein S-nitrosylation assays. Biochem. Biophys. Res. Commun. 2006;340:347. doi: 10.1016/j.bbrc.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 61.Giustarini D, le-Donne I, Colombo R, Milzani A, Rossi R. Is ascorbate able to reduce disulfide bridges? A cautionary note. Nitric. Oxide. 2008;19:252. doi: 10.1016/j.niox.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 62.Huang B, Chen C. An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radic. Biol. Med. 2006;41:562. doi: 10.1016/j.freeradbiomed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 63.Li S, Wang H, Xian M, Whorton AR. Identification of protein nitrosothiols using phosphine-mediated selective reduction. Nitric. Oxide. 2012;26:20. doi: 10.1016/j.niox.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiesweg M, Berchner-Pfannschmidt U, Fandrey J, Petrat F, de GH, Kirsch M. Rocket fuel for the quantification of S-nitrosothiols. Highly specific reduction of S-nitrosothiols to thiols by methylhydrazine. Free Radic. Res. 2013;47:104. doi: 10.3109/10715762.2012.744836. [DOI] [PubMed] [Google Scholar]
- 65.Ckless K, Reynaert NL, Taatjes DJ, Lounsbury KM, van der Vliet A, Janssen-Heininger Y. In situ detection and visualization of S-nitrosylated proteins following chemical derivatization: identification of Ran GTPase as a target for S-nitrosylation. Nitric Oxide. 2004;11:216. doi: 10.1016/j.niox.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 66.Aesif SW, Janssen-Heininger YM, Reynaert NL. Protocols for the detection of s-glutathionylated and s-nitrosylated proteins in situ. Methods Enzymol. 2010;474:289. doi: 10.1016/S0076-6879(10)74017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mirza UA, Chait BT, Lander HM. Monitoring reactions of nitric oxide with peptides and proteins by electrospray ionization-mass spectrometry. J. Biol. Chem. 1995;270:17185. doi: 10.1074/jbc.270.29.17185. [DOI] [PubMed] [Google Scholar]
- 68.Matthews JR, Botting CH, Panico M, Morris HR, Hay RT. Inhibition of NF-kappaB DNA binding by nitric oxide. Nucleic Acids Res. 1996;24:2236. doi: 10.1093/nar/24.12.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Percival MD, Ouellet M, Campagnolo C, Claveau D, Li C. Inhibition of cathepsin K by nitric oxide donors: evidence for the formation of mixed disulfides and a sulfenic acid. Biochemistry. 1999;38:13574. doi: 10.1021/bi991028u. [DOI] [PubMed] [Google Scholar]
- 70.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. USA. 2006;103:1012. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Greco TM, Hodara R, Parastatidis I, Heijnen HFG, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc. Natl. Acad. Sci. USA. 2006;103:7420. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang B, Chen C. Detection of protein S-nitrosation using irreversible biotinylation procedures (IBP) Free Radic. Biol. Med. 2010;49:447. doi: 10.1016/j.freeradbiomed.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 73.Griffith OW, Stuehr DJ. Nitric oxide synthases: properties and catalytic mechanism. Annu. Rev. Physiol. 1995;57:707. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 74.Doulias PT, Tenopoulou M, Greene JL, Raju K, Ischiropoulos H. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci. Signal. 2013;6:rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]