Summary
Enhancers provide critical information directing cell-type specific transcriptional programs, regulated by binding of signal-dependent transcription factors and their associated cofactors. Here we report that the most strongly activated estrogen (E2)-responsive enhancers are characterized by trans-recruitment and in situ assembly of a large 1-2 MDa complex of diverse DNA-binding transcription factors by ERα at ERE-containing enhancers. We refer to enhancers recruiting these factors as mega transcription factor-bound in trans (MegaTrans) enhancers. The MegaTrans complex is a signature of the most potent functional enhancers and is required for activation of enhancer RNA transcription and recruitment of coactivators, including p300 and Med1. The MegaTrans complex functions, in part, by recruiting specific enzymatic machinery, exemplified by DNA-dependent protein kinase. Thus, MegaTrans-containing enhancers represent a cohort of functional enhancers that mediate a broad and important transcriptional program and provide a molecular explanation for transcription factor clustering and hotspots noted in the genome.
Introduction
Functional specialization and precise patterning of different cell and tissue types are vital for all metazoans, which also generate cell- or tissue-specific gene expression patterns. Enhancers, initially defined as DNA elements that act over a distance to positively regulate expression of protein encoding target genes, are the principle regulatory components of the genome that enable such cell-type specific and signal-dependent patterns of gene expression (Banerji et al., 1981; Shlyueva et al., 2014). Each cell type harbors more than 100,000 candidate enhancers in humans, vastly outnumbering protein-coding genes (Bernstein et al., 2012; Heintzman et al., 2009; Shlyueva et al., 2014). This makes it very important to be able to predict and understand which enhancers are actually functionally required for target coding gene transcriptional regulation.
Enhancer activation requires the presence of specific recognition sequences for the cooperative recruitment of DNA-binding transcription factors (TFs) and their cofactors that initially activate gene expression (Rosenfeld et al., 2006). While the role of a large number of coactivator complexes and their associated enzymatic activities is well established (Rosenfeld et al., 2006), the precise biochemical mechanisms by which so many coactivators are recruited and required for the different functional activities at specific enhancer sites remains incompletely understood. Global genomic technologies have uncovered characteristic markers of enhancers and have provided clues as to their activation. Features that have been used to predict enhancers that are likely to be functional include the levels of enhancer RNAs (eRNAs) transcribed from enhancer-like regions in the genome (Li et al., 2013), the presence of the histone acetyltransferase p300/CBP (Visel et al., 2009), the timing of RNA Pol II occupancy (Bonn et al., 2012), and levels of H3K4me2 and H3K27Ac (Chepelev et al., 2012; Heintzman et al., 2009). However, because enhancers identified using these features are not equally functional, additional methods are needed to distinguish the enhancers with different activation potential.
There are ∼2,600 DNA-binding TFs encoded by the human genome (Babu et al., 2004), with about 200-300 TFs being expressed in each cell type (Vaquerizas et al., 2009). A long-standing question is how different TFs collaborate to regulate the enhancer network in a specific cell type. With the large expansion of genome-wide binding data, DNA-binding transcription factors were noted to co-bind to some so-called ‘hotspot’ regions or to cooperatively cluster to some functional enhancers in various organisms or cell lines (Junion et al., 2012; Rada-Iglesias et al., 2012; Siersbaek et al., 2014a; Siersbaek et al., 2014b; Wilson et al., 2010; Yan et al., 2013). However, the underlying mechanism(s) and functional significance of this phenomenon are not well understood.
Recently, the idea of clustered enhancers associated with critical developmental or cancer-associated transcription units has been proposed (Hnisz et al., 2013; Loven et al., 2013; Whyte et al., 2013). The initial definition of this super-enhancer model was described as clusters of enhancers spanning >8-10kb, occupied by critical DNA-binding transcription factors at their cognate binding motifs (Loven et al., 2013; Whyte et al., 2013). These clustered super-enhancers control key coding transcription units in stem cells or various disease states, and exhibit high levels of coactivators, which are suggested to contribute to gene activation. Cancer cells were also noted to acquire super-enhancers regulating oncogene drivers (Hnisz et al., 2013; Loven et al., 2013). While the super-enhancer model can explain the higher expression levels for a small number of genes in some environments, it also highlights the need for exploring the functional activities of single enhancers in the regulation of coding genes critical for development and disease and understanding the phenomenon of TF clustering in short-range genomic regions.
Here we report a new signature of the functionally active estrogen-regulated enhancers, particularly the 1,333 most active ERα enhancers linked to target coding gene activation. This signature is the selective recruitment in trans of an apparent complex of other DNA-binding TFs, including RARα/γ, GATA3, AP2γ, STAT1, AP1, and FoxA1. By gel filtration, we found these TFs migrated with ERα as a 1-2MDa complex(es), referred to as the MegaTrans complex. The MegaTrans complex is almost invariably recruited to functional ERα-bound enhancers, ∼22% of which fit the criteria of being components of super-enhancers. Furthermore, the MegaTrans complex is required for activation of the functional enhancers, apparently based in part on specific recruitment of enzymes. This is exemplified by the functionally important recruitment of the DNA-dependent protein kinase to ERα-regulated enhancers by RARs. The MegaTrans complex, in turn, is also required for activation of eRNA transcription and recruitment of coactivators, including p300 and Med1, and thus exerts critical biological functions, conceptually parallel to what has been proposed for super-enhancers.
Results
Trans-Bound RARs on ERα Active Enhancers Regulate ERα Enhancer Function
ERα functions as a central transcription factor for gene programs that mediate cell growth and proliferation, and it accomplishes this role primarily through enhancer regulation. Amongst the total ∼7,174 ERα-bound enhancers, a subset of 1,333 enhancers that are located in proximity (<200kb) to their regulated coding transcription units have proved to be the most significantly activated upon estrogen stimulation according to levels of H3K27Ac and increased eRNA transcription, and appear to constitute the most potent functional enhancer program (Li et al., 2013).
Our current study was initiated by investigating the possible functional mechanisms by which RARs on retinoic acid response element (RARE)-containing enhancers mediate RA-induced coding gene transcriptional programs, as well as the functional role(s) of RAR at enhancers that accommodate the effects of other signals, such as E2-induced coding gene transcriptional programs (Hua et al, 2009; Ross-Innes et al, 2010). To distinguish the possible binding in cis (the chromatin association of a transcription factor through direct DNA binding at its recognition sites) and in trans (the chromatin association of a transcription factor through protein-protein interaction) functional models of RAR, we engineered MCF7 to express a bacterial biotin ligase (BirA) that can biotinylate a biotin ligase recognition peptide (BLRP)-tagged protein in vivo (Figure S1A). Under control of a Tet-On promoter, wild-type RAR and two DNA-binding domain mutants that cannot bind to RARE DNA sites (Figures S1B and S1C) were expressed at similar levels as the endogenous proteins upon doxycycline induction (Figure S1D). Using these lines, we first performed biotin ChIP-seq for WT and mutant RARα/γ (RARβ is not expressed in MCF7 breast cancer cells) upon RA and E2 stimulation. Comparing wild-type and non-DNA-binding mutants, we found that about 15,000/18,000 of WT RARα/γ-bound sites required the intact RAR DNA-binding ability because binding was lost with mutant RARs, and none of these sites was bound by ERα (Figures S1E, S1F, and S1G). Among these 15,000 sites, 3,540 were enhancers that exhibited RA activation (Figure S1E), exemplified by the ∼700 most active RAR cis-binding enhancers, which showed significant RA-induced eRNA and gene target activation by Global Run-on Sequencing (GRO-seq) (Figure S1H).
However, there were ∼3,000 RARα/γ binding sites that did not depend on RAR DNA-binding ability (Figures S1E and S2A). Remarkably, we found that both RARα and RARγ were recruited to virtually all of the ERα-bound 1,333 active enhancers in response to E2 (Figure 1A). This observation is consistent with previous evidence that RAR can bind to ERα binding sites, although conflicting conclusions were reached regarding its activating or repressive effects (Hua et al, 2009; Ross-Innes et al, 2010). However, ERα did not exhibit co-localization with RARs on ERα non-active enhancers (Figure 1B). By comparing the binding patterns of wild-type and two non-DNA-binding mutants, we found the binding of RARs on the 1,333 ERα active enhancers was in trans (Figures 1C and S2A).
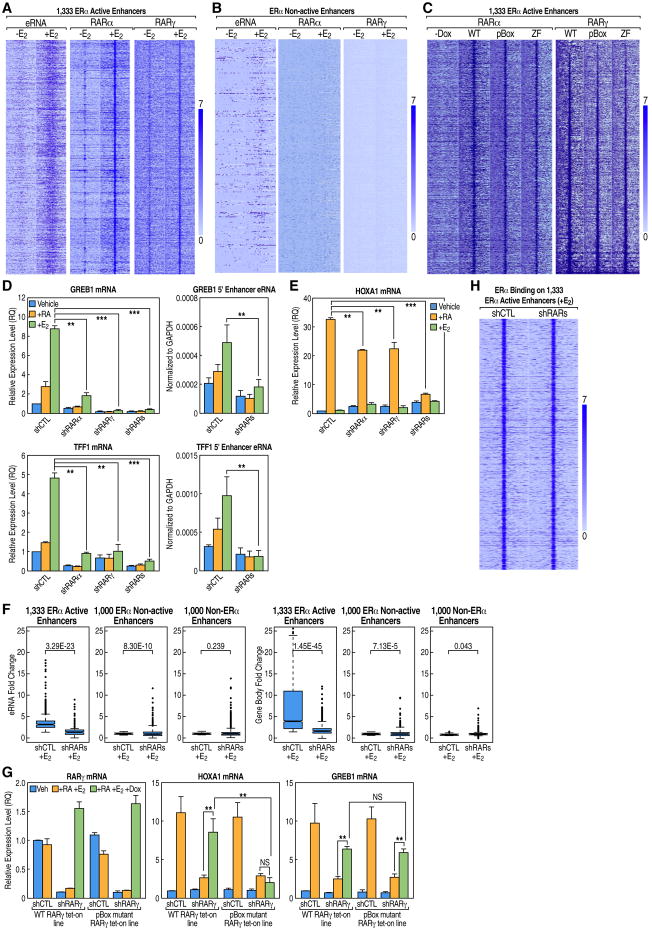
Figure 1. Trans-Bound RARs on ERα Active Enhancers Regulate E2-Liganded Transcription Activation.
(A) Heatmaps of GRO-seq and ChIP-seq data (±E2) data for 1,333 ERα active enhancers showing strong E2-induced eRNA transcription and E2-enhanced binding of both RARα and RARγ, respectively.
(B) Heatmaps of GRO-seq and ChIP-seq for a control group of ERα non-active enhancers exhibiting no RARα/γ binding and no significant E2-induced eRNA transcription.
(C) For the 1,333 ERα active enhancers, heatmaps of ChIP-seq data for the wild-type and two DNA-binding mutants of RARα/γ (+RA and E2) show that their association with these enhancers is DNA binding independent.
(D) Knockdown of either RARα or RARγ by shRNA inhibits ERα target gene induction by E2, as demonstrated by qPCR analysis.
(E) Knockdown of either RARα or RARγ using shRNA inhibits expression of the RAR cis-binding target HoxA1 gene in response to RA, as shown by qPCR analysis.
(F) RARs are required for the E2-liganded activation of ERα active enhancers and their targets, as shown by GRO-seq boxplots. No significant effects were found for either ERα non-active enhancers or non-ERα enhancers.
(G) The pBox mutant RARγ fails to rescue expression of its cis-binding target HoxA1 after knockdown of endogenous RARγ. In contrast, both wild-type and pBox mutant RARγ can rescue expression of the trans-binding target GREB1. For details regarding rescue experiments see Extended Experimental Procedures.
(H) Heatmap showing that knockdown of RARs does not affect ERα binding at the 1,333 active enhancers.
Data are represented as mean ± SEM (NS not significant, **P<0.01, ***P<0.001). See also Figure S1 and S2.
Knockdown of either RARα or RARγ caused a significant decrease in both E2-dependent induction of eRNAs and activation of target coding genes, while knockdown of both caused almost complete inhibition, as assessed by q-PCR of targets such as GREB1 and TFF1 (Figure 1D). The knockdown of RARα and RAR γ, which was confirmed for both RNA and protein levels (Figures S2B and S2C), inhibited RA induction of the HoxA1 gene target as expected (Figure 1E). Boxplot analysis of the GRO-seq experiments showed that the presence of RARs was required for effective induction of both eRNAs and target coding gene transcription units upon E2 treatment (Figures 1F and S2D). RARα/γ knockdown also inhibited classical RAR cis-bound enhancers and their target genes (Figures S1H and S2E). Thus, while RAR binding in cis activates a distinct RA-responsive transcriptional program, its recruitment in trans is also required for effective E2-dependent activation of ERα-bound functional enhancers.
Next, we utilized wild-type and pBox mutant RARγ to test their ability to rescue ERα-regulated enhancer function following endogenous RARγ knockdown (Figure S2F). Intriguingly, the non-DNA-binding mutant receptor continued to be effectively recruited to the ERα-bound regulatory enhancers at the GREB1 gene (Figure S2A) and was capable of restoring full E2-dependent GREB1 gene activation in rescue experiments (Figure 1G). However, as expected, it failed to activate the cis-bound, RAR-regulated HoxA1 gene (Figure 1G).
Administration of ICI 182780 to knockdown ERα caused a loss of RAR binding at the ERα-regulated enhancers (Figures 2D, S3C, and S3D), but did not alter the binding of RARα or RARγ at activated enhancers harboring cis RAR binding sites (data not shown). Knockdown of RAR did not cause down-regulation of ERα RNA or protein levels (Figures S2B and S2C), and did not affect the ERα binding pattern on ERα active enhancers (Figure 1H).
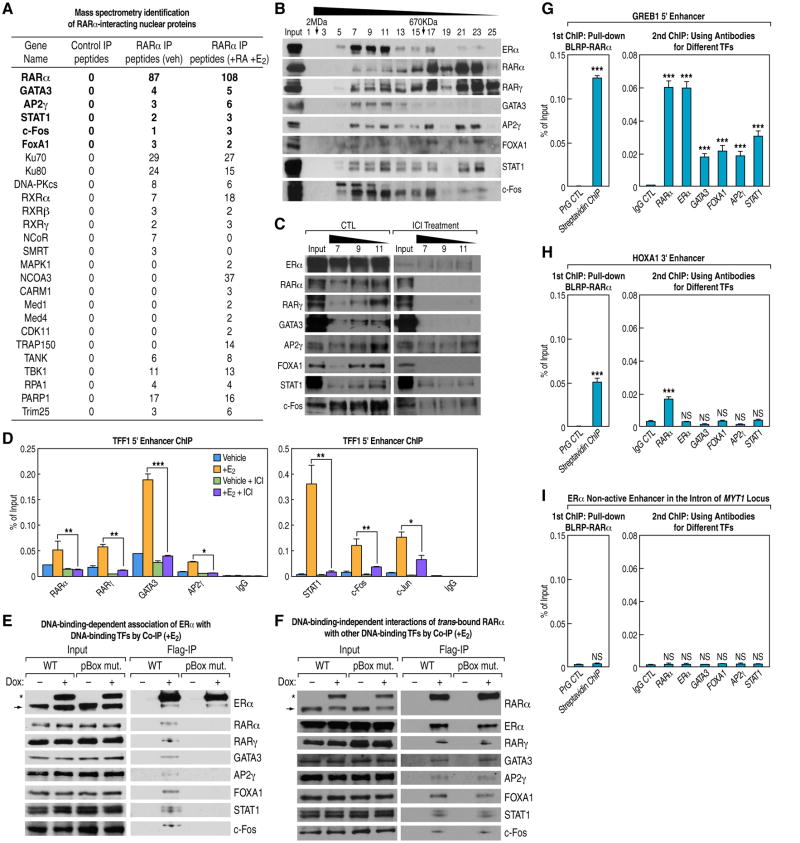
Figure 2. ERα Interacts with a Mega (1-2MDa) Complex of DNA-Binding Transcription Factors at ERE-Containing Active Enhancers.
(A) RARα associates with several DNA-binding TFs, as shown by mass spectrometry analysis after pull down of biotin-tagged RARα and elution with TEV protease digestion. The same inducible BLRP-tagged RARα stable cell line without doxycycline induction was used as a control.
(B) Western blots of gel filtration samples from MCF7 nuclear lysates (+E2) show various DNA-binding TFs associate with ERα in 1-2MDa fractions.
(C) Knockdown of ERα by ICI 182780 causes loss of the DNA-binding TFs in 1-2MDa ERα-containing complex, as revealed by immunoblotting of gel filtration fractions from the 1-2MDa range (fractions 7, 9 and 11).
(D) DNA-binding TFs in the ERα complex bind to an ERα active enhancer at TFF1 locus upon E2 signal, and knockdown of ERα reduces their binding. ChIP signals are presented as percentage of input.
(E) The interaction of ERα with other DNA-binding TFs is dependent on its DNA-binding ability, as shown by co-immunoprecipitation using BLRP-tagged WT or pBox mutant ERα. The asterisk marks BLRP-tagged ERα, and the arrow marks endogenous ERα.
(F) The interaction of RARα with other DNA-binding TFs is independent of its DNA-binding ability, as demonstrated by co-immunoprecipitation of BLRP-tagged WT or pBox mutant RARα and other TFs. The asterisk marks BLRP-tagged RARα, and the arrow marks endogenous RARα.
(G-I) ChIP-reChIP analysis confirms the co-binding of RARα, ERα and other DNA-binding TFs on ERα active enhancers but not on the ERα non-active enhancers or RAR-bound HoxA1 enhancer. ChIP signals are presented as percentage of input and are compared to negative controls.
Data are represented as mean ± SEM (NS not significant, * P<0.05, ** P<0.01, *** P<0.001). See also Figure S3.
Collectively, our data indicate that ERα selectively recruits RARα and RARγ in trans on the functional enhancers regulating the most robustly-activated target coding genes and that this strong activation depends on the ERα-mediated trans-binding of RARs.
ERα Recruits a Mega DNA-Binding Transcription Factor Complex in situ at Functional ERα Enhancers
These findings prompted us to examine the behavior of additional DNA-binding TFs associated with ERα, based on previously reported mass spectrometry analysis of proteins that co-immunoprecipitated with ERα (Mohammed et al, 2013) as well as our own confirmatory data. From these ERα complex data, we noted a number of DNA-binding transcription factors associated with ERα, including RARγ, GATA3, AP2γ, STAT1, and, intriguingly, FoxA1. To complement these observations, we also examined the proteins associated with RAR following pull down from MCF7 cells stably expressing, at physiological levels, biotin-tagged RARα (Figures S1D and S3A). In addition to RARα, RXRs, and many well-known cofactors for nuclear receptors, GATA3 was also detected along with other DNA-binding proteins including AP2γ, STAT1, c-Fos, and FoxA1 (Figure 2A). We then performed gel filtration analysis on nuclear extracts prepared from MCF7 cells in the absence of DNase treatment and analyzed all fractions for ERα, RARα/γ, GATA3, and the other DNA-binding transcription factors identified in the mass spectrometry analysis. This analysis revealed co-elution of ERα, RARα, RARγ, GATA3, AP2γ, FoxA1, STAT1, c-Fos, and other proteins in an estimated 1-2MDa complex(es) (Figure 2B). These components were all present in ERα-immunoprecipitates from nuclear extracts and their association was enhanced upon E2 treatment (Figure S3B). Importantly, knockdown of nuclear ERα by administration of ICI 182780 caused a virtual loss of the entire complex associated with ERα by gel filtration analysis (Figures 2C and S3C) and recruitment of each factor to ERα-bound functional enhancers (Figures 2D and S3D). Thus, the material co-migrating in the gel filtration represented proteins interacting as a complex with ERα rather than artifacts. This complex remained intact in the presence of 250mM NaCl, but was lost under 600mM NaCl high-salt conditions (data not shown).
To further investigate the hypothesis that the ERα-dependent trans-recruitment/assembly of other DNA-binding transcription factors occurs only in situ at ERα active enhancers, we first confirmed that the interactions between ERα and the TFs were dependent on DNA (Figure S3E). Using a non-DNA-binding ERα pBox mutant, which is incapable of binding the estrogen response element (ERE) motif (Stender et al., 2010), we could show that this mutation abolishes the interactions of ERα and these associated TFs (Figure 2E). As a control, a comparable RARα pBox mutant did not affect its interaction with ERα and these TFs (Figure 2F). These data suggest that RARα and other TFs are recruited by ERE-bound ERα to its activated enhancers; thus, the entire complex is assembled in situ on ERα-bound enhancers.
To further confirm that these factors were, indeed, co-recruited to the same transcription units, rather than the consequence of differential recruitment behavior in different cell populations, we performed serial pairwise two-step ChIP analyses to assess the co-recruitment of RARα with ERα, GATA3, FoxA1, AP2γ, and STAT1 on the same ERα-bound enhancers. Using a BLRP-tagged RARα stable cell line, two step ChIP was performed with biotin-streptavidin pull-down of RARα in the first round followed by immunoprecipitation with antibodies for RARα (as positive control), ERα, GATA3, FoxA1, AP2γ, and STAT1. In each case, we found that these proteins were present on the interrogated active enhancers, including the GREB1 enhancer (Figure 2G). In contrast, as a control, this was not the case of the RAR cis-bound enhancer regulating the HoxA1 transcription unit (Figure 2H). Thus, the MegaTrans complex was co-recruited to ERα-bound active enhancers but not to functional enhancers that directly bind RARα in cis. RARα and the other TFs also were not present at ERα-bound, non-active enhancers (Figure 2I). Double-ChIP experiments performed with a BLRP-tagged GATA3 stable line similarly demonstrated the co-binding of GATA3 with ERα and all of the other TFs at ERα active enhancers but not at either the HoxA1 enhancer or ERα non-active enhancers (Figure S3F). Together, these data indicate that a new feature of the active, regulatory ERα-bound enhancers, in addition to their increased levels of eRNA transcription, is the selective recruitment of this “MegaTrans complex”.
Trans-Bound GATA3 Also Regulates Functional ERα Enhancers
To explore the possible functional consequences of the additional ERα-interacting transcription factors, we next explored the potential recruitment and function of GATA3 on ERα active and non-active enhancers. ChIP-seq experiments revealed, as in the case of RARα and RARγ, that GATA3 was recruited in an E2-dependent fashion to active enhancers (Figure 3A), but not inactive enhancers (Figure 3B). Because we found the presence of GATA3 on functional ERα-bound enhancers that did not harbor apparent GATA3 cis-binding elements by motif analysis, we again assessed the possibility that GATA3 was recruited in trans to these ERα-bound active enhancers. Knockdown of ERα by administration of ICI 182780 inhibited GATA3 recruitment to ERα active enhancers (Figures 2D and S3D). Because direct or indirect ERα and GATA3 interactions were suggested by immunoprecipitation experiments (Figure S3B), we investigated the consequences of disrupting the ability of GATA3 to bind to cognate DNA sites by two different mutations of the second zinc finger that is required for cis-binding of GATA3 (Nesbit et al, 2004) (Figure S4A). We generated inducible BLRP-tagged stable lines expressing wild-type and the two DNA-binding mutants at physiological levels (Figure S4B), and biotin ChIP-seq revealed they were equally well recruited, apparently in trans, to these ERα-bound active enhancers (Figures 3B and S4C). By comparing the ChIP-seq data for wild-type and DNA-binding mutants, we found that amongst ∼18,000 wild-type GATA3 binding peaks about 5,000 were retained in the two GATA3 mutants, and these trans-binding sites featured ERE as the top motif by Homer analysis (Figure S4D). For the ∼13,000 cis-binding peaks, GATA motifs were enriched and a heatmap of the non-ERα enhancers containing a GATA motif was used to confirm a total loss of binding of the two non-DNA-binding GATA3 mutants (Figures 3C and S4D).
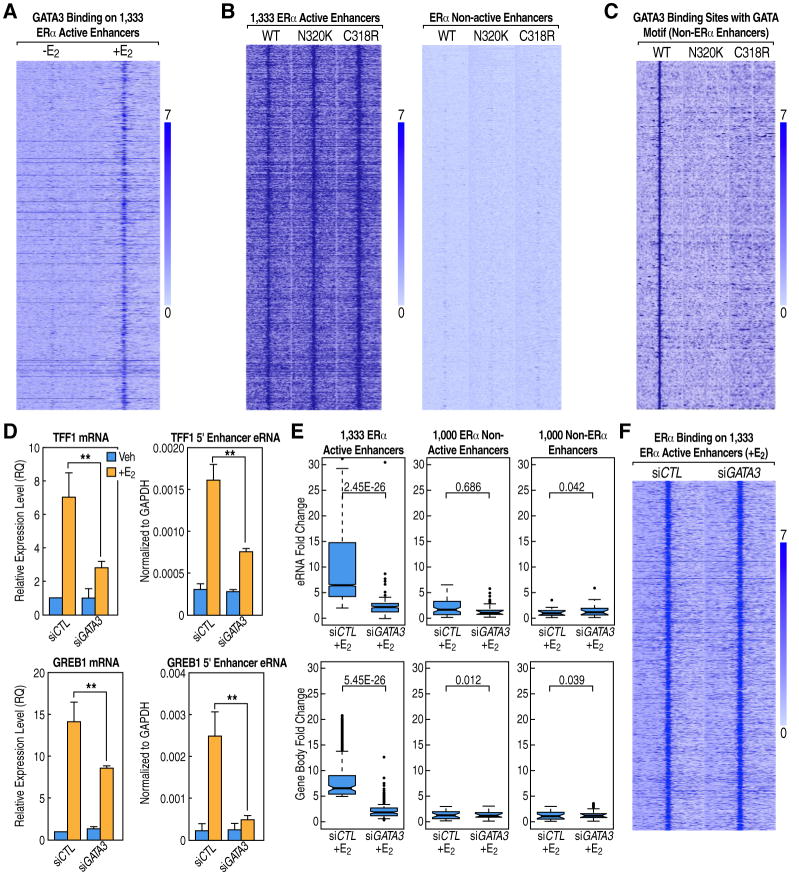
Figure 3. Trans-Bound GATA3 on ERα Active Enhancers Regulates ERα E2-Liganded Transcription Activation.
(A) Heatmap displaying GATA3 binding at the 1,333 ERα active enhancers is enhanced by E2.
(B) Heatmaps of ChIP-seq data for wild-type and two DNA-binding mutants of GATA3 (+E2) show the binding of GATA3 to these ERα active enhancers is not dependent on its DNA-binding ability. There is no binding of either wild-type or mutants GATA3 to ERα non-active enhancers.
(C) Heatmap of ChIP-seq data for wild-type and two DNA-binding mutants of GATA3 (+E2) shows the binding of GATA3 to these non-ERα enhancers that contain the GATA motif requires its DNA-binding ability.
(D) Knockdown GATA3 affects ERα-dependent activation of eRNA transcription and coding gene expression for GREB1 and TFF1 genes. Mean ± SEM based on three independent qPCR experiments (** P<0.01).
(E) GRO-seq boxplots showing that GATA3 is required for the E2-liganded activation of ERα active enhancers and their coding gene targets.
(F) Heatmap showing that knockdown of GATA3 does not affect ERα binding at the 1,333 active enhancers.
See also Figure S4.
Using qPCRs or GRO-seq analysis, we explored the consequences of specific siRNA-mediated knockdown of GATA3 on E2-dependent induction of eRNAs. We found a dramatic inhibition of the eRNA activation events on active enhancers (Figures 3D, 3E, and S4E) but no effect on ERα-bound non-activated enhancers or non-ERα-bound enhancers (Figure 3E). The same inhibition effects were also found for gene body expression of the targets of these 1,333 ERα active enhancers (Figures 3D, 3E, and S4E). Knockdown of GATA3 did not affect ERα gene expression at either the RNA or protein level (Figures S4F and S4G) or ERα binding at active enhancers (Figure 3F). Thus, GATA3 and RARs, as components of a complex of DNA-binding TFs associated in trans with ERα on active enhancers, are required for E2-dependent enhancer activation.
ERα Active Enhancers Are Regulated by the MegaTrans Complex
We next investigated whether other DNA-binding transcription factors present in the 1-2MDa “complex” (MegaTrans) co-migrating with ERα were also recruited to E2-actived enhancers even in the absence of their cognate DNA-binding elements. We reviewed our own and published ChIP-seq data from MCF7 cells for other DNA-binding TFs present in the MegaTrans complex (Joseph et al., 2010; Theodorou et al., 2013). E2-regulated active enhancers were found to harbor AP2γ, FoxA1, c-Jun, and c-Fos, along with RARα/y and GATA3 (Figures 4A and 4B), but these TFs were not present on non-active enhancers (Figures 4A and S5A). Similar to RARα/γ and GATA3, the recruitment of the other TFs was also increased by E2 and abolished by knockdown of nuclear ERα using ICI 182780 (Figures 4C, 2D, and S3D)
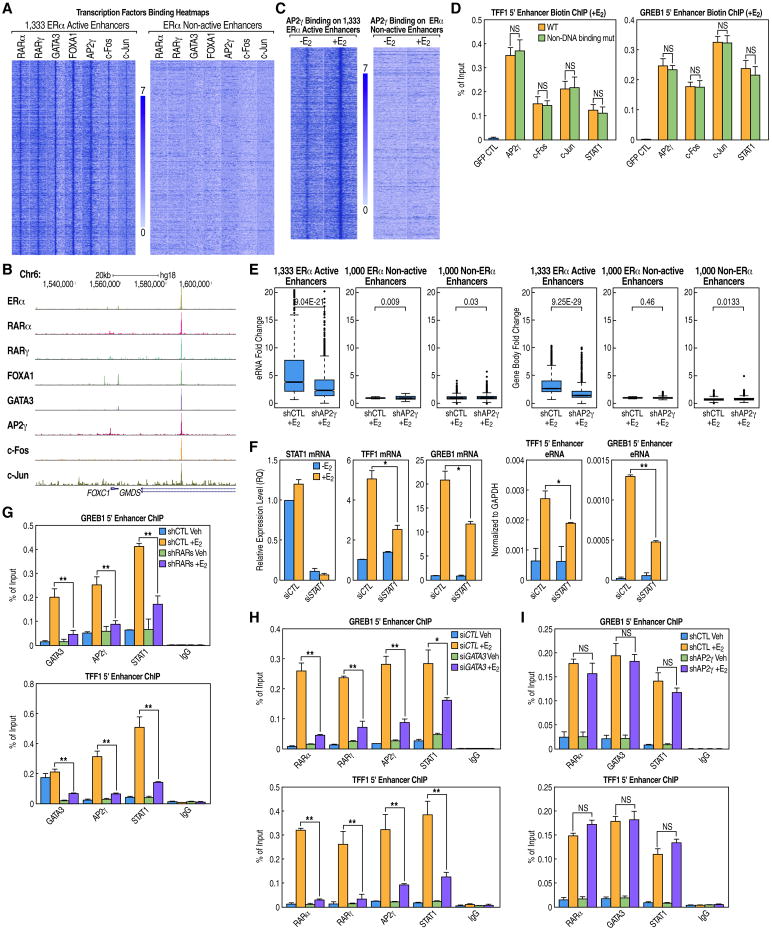
Figure 4. ERα Active Enhancers Are MegaTrans Enhancers Regulated by DNA-Binding TFs.
(A) Heatmaps of ChIP-seq data for different TFs (+E2) displaying strong binding of these DNA-binding TFs at the 1,333 ERα active enhancers but not at ERα non-active enhancers.
(B) UCSC browser snapshot image of an ERα active enhancer for FoxC1, which exemplifies a MegaTrans-bound enhancer (+E2).
(C) Heatmap showing AP2γ binding at ERα active enhancers, but not at ERα non-active enhancers, in response to E2.
(D) WT and non-DNA-binding mutants of MegaTrans TF components bind equivalently to two ERα active enhancers of TFF1 and GREB1, as demonstrated by biotin ChIP using BLRP-tagged TFs (GFP served as control). For details regarding DNA-binding domain mutagenesis see Extended Experimental Procedures. ChIP signals are presented as percentage of input.
(E) GRO-seq boxplots showing that AP2γ is required for ligand-dependent activation of both eRNA and target gene body transcription for ERα active enhancers.
(F) STAT1 is required for the activation of ERα active enhancers and coding gene expression by E2 for GREB1 and TFF1 genes, as demonstrated by knockdown and qPCR analysis.
(G-I) Knockdown of RARs or GATA3, but not AP2γ, greatly reduces the E2-enhanced occupancy of DNA-binding TFs on two ERα active enhancers of TFF1 and GREB1. ChIP signals are presented as percentage of input.
Data are represented as mean ± SEM (NS not significant, *P<0.05, **P<0.01). See also Figure S5.
In order to investigate whether, in fact, all DNA-binding transcription factors present in the MegaTrans complex were recruited in trans to ERα functional enhancers, a series of DNA-binding domain mutations were generated for AP2γ, c-Fos, c-Jun, and STAT1. ChIP-qPCR data on the GREB1 and TFF1 enhancers showed that the binding of the non-DNA-binding mutants at these two ERα active enhancers was comparable to that of the wild-type proteins (Figure 4D), which confirms the trans-recruitment of these TFs by ERα.
Based on the roles of RARs and GATA3 on ERα active enhancers, we evaluated the functional effects of other recruited transcription factors. Beginning with AP2γ, we found that, in addition to its recruitment in response to E2 on ERα regulatory enhancers (Figure 4C), knockdown of AP2γ caused a dramatic inhibition of eRNA and target coding gene expression assayed by both qPCR and GRO-seq (Figures 4E, S5B, and S5C). Similarly, as STAT1 was also recruited to ERα-bound enhancers (Figures 2D and S3D), we evaluated its effect on two well-described ERα bound/regulated enhancers. Again, we found a functional contribution to the outcome of E2-induced activation of enhancer transcription and target coding gene expression (Figure 4F). The same regulatory effects were also demonstrated upon knockdown of two AP1 components, c-Jun and c-Fos (Figures S5D and S5E), that were present in the MegaTrans complex (Figures 4A and 4B)
To begin to assess the interdependency of the components of the MegaTrans complex on recruitment to ERα-bound functional enhancers, we tested the consequences of knockdown of RARα/γ, GATA3, and AP2γ on GREB1 and TFF1 enhancer occupancy, We found a marked inhibition of recruitment of other MegaTrans components upon knockdown of RARα/γ and GATA3 (Figures 4G and 4H) but not by knockdown of AP2γ (Figure 4I), consistent with interdependency of at least some components of the complex for recruitment of various other components. RARα/γ and GATA3 may serve as key functional components, along with ERα, in recruitment/assembly of the MegaTrans complex on functional ERα-bound enhancers.
FoxA1 Is Required for ERα Recruitment and MegaTrans Complex Stabilization on ERα Active Enhancers
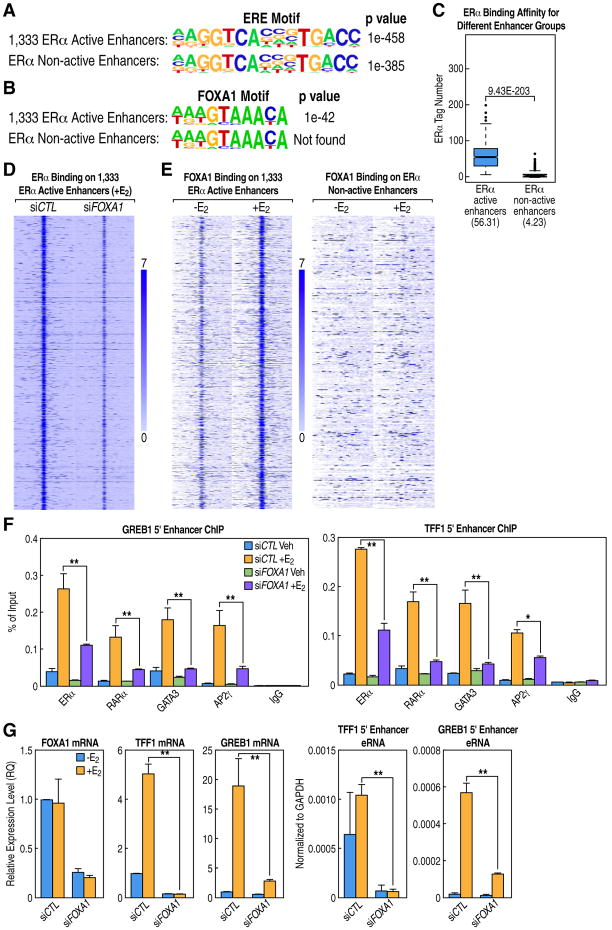
These experiments raised a question about potential differences in DNA sequence features between ERα active enhancers that bound the MegaTrans complex and ERα non-active enhancers that did not. Comparison of the EREs between these two groups revealed that its frequency and the primary consensus sequences were essentially identical (Figure 5A). In contrast, and in accord with the well-known importance of FoxA1 as a pioneer factor for ERα binding (Hurtado et al, 2011), we noted that the functional, MegaTrans-bound 1,333 ERα active enhancers generally harbor a FoxA1 binding motif within 200 bp of the ERE sites, while the FoxA1 motif was virtually absent on the non-functional, non-MegaTrans-bound ERα enhancers (Figure 5B). Indeed, the affinity for ERα is >90% lower on the non-functional than functional ERα-bound enhancers (Figure 5C). Consistent with FoxA1 functioning as a key determinant of ERα binding (Hurtado et al, 2011), our data showed greatly reduced binding of ERα at the 1,333 ERα-bound active enhancers upon FoxA1 knockdown (Figure 5D)
Figure 5. FoxA1 Performs Dual Roles on ERα Active Enhancers.
(A-B) ERα and FoxA1 motif analyses using Homer program for 1,333 ERα active enhancers and ERα non-active enhancers (see Extended Experimental Procedures for analysis details).
(C) Boxplot based on ERα ChIP-seq data (+E2) showing higher binding affinity of ERα at 1,333 ERα active enhancers than at ERα non-active enhancers.
(D) Heatmap showing that knockdown of FoxA1 greatly reduces ERα binding at the 1,333 active enhancers.
(E) Heatmap showing FoxA1 binding at 1,333 ERα active, but not at ERα non-active enhancers, is enhanced in response to E2.
(F) Conventional ChIP assays for TFF1 and GREB1 enhancers showing knockdown of FoxA1 substantially reduced binding of ERα and the MegaTrans components following E2 treatment. ChIP signals are presented as percentage of input.
(G) FoxA1 is required for the activation of ERα active enhancers in response to E2, as exemplified by the effects of FoxA1 knockdown on coding gene expression and eRNA transcription for GREB1 and TFF1 genes.
Data are represented as mean ± SEM (* P<0.05, ** P<0.01).
Because FoxA1 appears to be a component of the MegaTrans complex based on gel filtration and co-IP data (Figures 2B and S3B), and also exhibits E2-induced binding at the 1,333 ERα active enhancers (Figure 5E), we speculate that FoxA1 potentially plays dual roles in the binding of ERα to functional enhancers and in ERα-dependent recruitment of the MegaTrans complex. Indeed, knockdown of FoxA1 caused a dramatic impairment of ERα binding on the functional ERα enhancers (Figure 5D), which was accompanied by a loss of recruitment of the MegaTrans complex on this functional enhancer cohort (Figure 5F) and inhibition of both eRNA and gene body activation (Figure 5G). Thus FoxA1 is distinct from the other DNA-binding TFs in the MegaTrans complex that apparently do not affect ERα binding upon knockdown (Figures 1H and 3F).
Roles of MegaTrans Complex in Co-Activator Recruitment and in Super-Enhancer Function
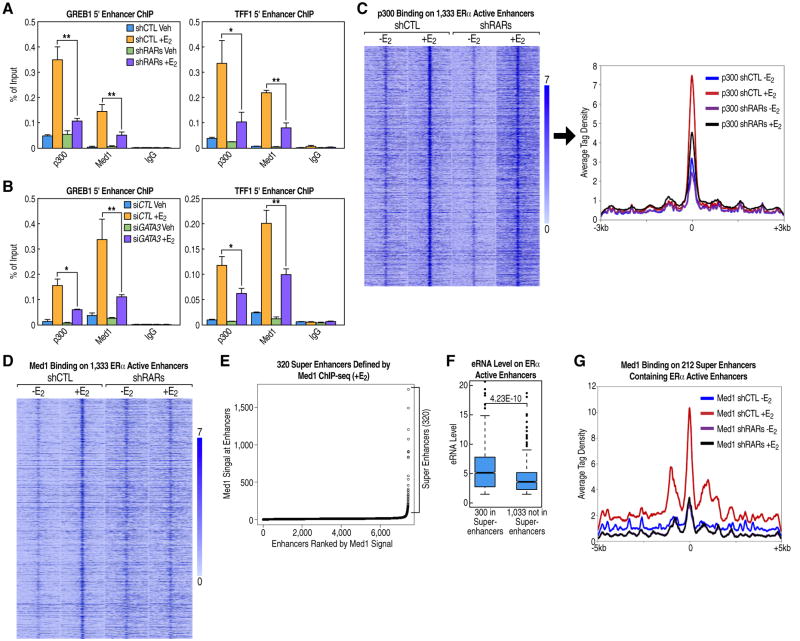
A basic aspect of the mechanism by which MegaTrans components function is their requirement for effective activation of E2-induced eRNAs on the functional enhancers. Accordingly, we assessed the recruitment of the coactivator p300 by qPCR and ChIP-seq upon knockdown of RARα/γ or GATA3. All of these knockdowns inhibited the E2-induced accumulation of p300 on activated enhancers (Figures 6A, 6B, 6C, and S6A), consistent with a previous report of a role for RARα in p300 recruitment (Ross-Innes et al, 2010). Based on the importance of Mediator complex for enhancer function, putatively due to its roles in enhancer:promoter looping events (Kagey et al, 2010), we also evaluated the effects of RARα/γ and GATA3 knockdown on E2-dependent recruitment of Med1 to functional enhancers by qPCR, finding a dramatic inhibition following these knockdowns (Figures 6A and 6B). This result was confirmed genome-wide by ChIP-seq (Figures 6D and S6B)
Figure 6. Trans-Bound TFs on MegaTrans Enhancers Are Required for Recruitment of ERα Co-Activators and Super-Enhancer Function.
(A-B) Knockdown of RARs or GATA3 greatly reduces the E2-enhanced binding of p300 and Med1 to ERα active enhancers. ChIP signals are presented as percentage of input.
(C) Heatmap and tag density plot of p300 ChIP-seq data for four different conditions demonstrating that knockdown of RARs by shRNA reduces E2-enhanced p300 recruitment on 1,333 ERα active enhancers.
(D) Trans-bound RARs are required for E2-enhanced recruitment of the co-activator Med1 to ERα active enhancers, as shown by a heatmap of Med1 ChIP-seq data on 1,333 ERα active enhancers.
(E) A Med1 tag density plot based on Med1 ChIP-seq (+E2) data and clustering of enhancers identifies ∼320 super-enhancers in MCF7 cells (see Extended Experimental Procedures for analysis details).
(F) A boxplot analysis based on GRO-seq data (+E2) of eRNA expression levels for two groups of ERα active enhancers: the 300 ERα active enhancers located in super-enhancers (median: 5.14) and 1,033 ERα active enhancers that are not located in super-enhancers (median: 3.59).
(G) Tag density plot showing knockdown of trans-bound RARs, which affects the function of ERα active enhancers, reduces the E2-enhanced Med1 signal at 212 super-enhancers that contain ERα active enhancers.
Data are represented as mean ± SEM (*P<0.05, ** P<0.01). See also Figure S6.
Based on the criteria developed in the initial description of super-enhancers (Hnisz et al, 2013; Whyte et al, 2013), we assessed the number of super-enhancers in MCF7 cells by Med1 ChIP-seq under both − and + E2 conditions. While there are only ∼122 super-enhancers under the −E2 condition, E2 treatment increases the total to ∼320 such enhancers (Figure 6E), of which ∼212 contained at least one ERα-bound functional enhancer, including one at the c-Myc gene locus (Figures S6C). Thus, only ∼300 of the 1,333 ERα-bound functional enhancers characterized by MegaTrans complex fulfill the current definition of being located in super-enhancers. The efficacy of this subset of 300 ERα-bound active enhancers was only slightly better than the other 1,033 ERα-bound active enhancers with respect to eRNA induction (Figure 6F). Thus, the functional strength of the ERα- bound enhancers, irrespective of their presence in a super-enhancer, is predicted by the presence of the MegaTrans complex.
Actually, for the 212 super-enhancers that contain ERα active enhancers, their Med1 levels were also dependent on the E2 signal (Figure S6C). Interestingly, we observed greatly reduced levels of Med1 at these 212 super-enhancers following knockdown of RARs (Figure 6G), suggesting that MegaTrans enhancers are important constituents in the function of these clustered super-enhancers.
DNA-Binding TFs of the MegaTrans Complex Might Recruit Specific Functionally Required Components for Enhancer Activation
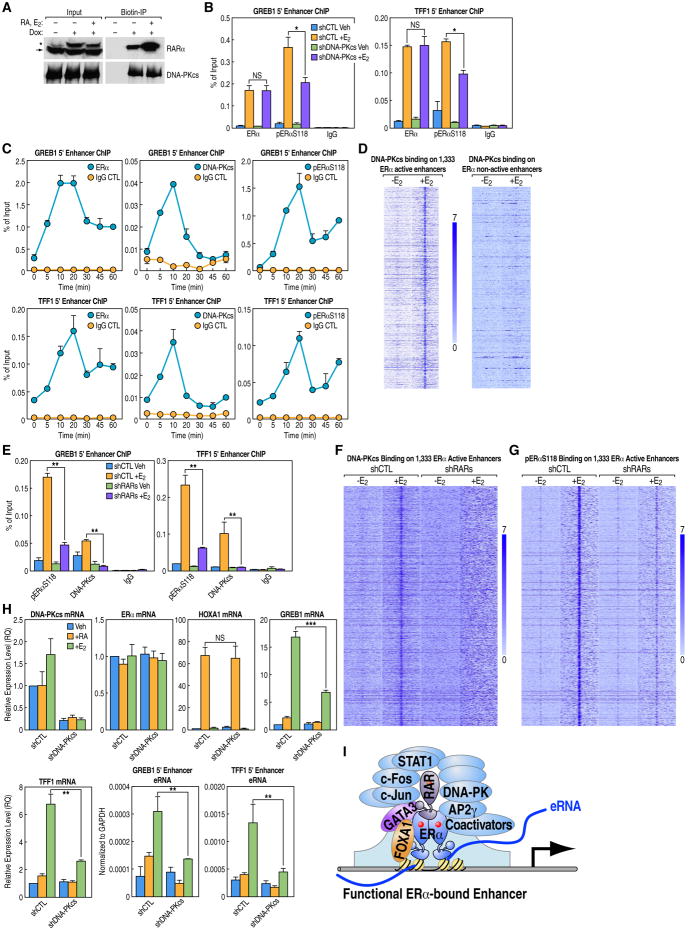
Based on the presence of specific non-transcription factor components in the mass spectrometry analysis of RARα-associated proteins (Figure 2A), we evaluated the functional significance of these additional proteins. We elected to focus on DNA-dependent protein kinase (DNA-PK), comprising the catalytic subunit DNA-PKcs, Ku70, and Ku80, since all three DNA-PK subunits were present in the RARα pull down as revealed by mass spectrometry. We confirmed these associations by co-IP and Western blot analysis (Figure 7A). DNA-PKcs has previously been reported as a component of the ERα complex that directly phosphorylates S118 of ERα (Foulds et al, 2013), and we confirmed that knockdown of DNA-PKcs partially impacted phosphorylation of ERα S1 18 without affecting ERα binding at ERα active enhancers (Figure 7B). Using a specific antibody against DNA-PKcs for ChIP analysis, we first evaluated the temporal kinetics of its potential recruitment on the GREB1 and TFF1 enhancers, finding recruitment at approximately 10 minutes following E2 treatment of MCF7 cells (Figure 7C). In addition, a specific antibody against phosphorylated ERα S1 18 revealed strong enrichment that peaked at 20 minutes, slightly after the recruitment of ERα and DNA-PKcs (Figure 7C). According to these observations, we conducted ChIP-seq analysis of DNA-PKcs in MCF7 cells after 10 minutes of E2 treatment, which revealed 12,629 peaks that mostly located in intergenic regions (Figure S7A). Of the detected peaks, 971 were on the ERα-bound, MegaTrans-containing active enhancers but few were present on non-active enhancers (Figures 7D and S7B). A second antibody for DNA-PKcs yielded similar ChIP-seq results, confirming the specificity of the signal (data not shown).
Figure 7. Trans-Bound RAR May Contribute to the Recruitment of DNA-PK Kinase as a Co-Activator for ERα Active Enhancers.
(A) Western blots demonstrating interaction of doxycycline-induced BLRP-RARα protein with DNA-PKcs after pull down by streptavidin magnetic beads. The asterisk marks BLRP-tagged RARα, and the arrow marks endogenous RARα.
(B) Conventional ChIP assays for TFF1 and GREB1 enhancers showing DNA-PKcs is not required for the occupancy of ERα but is partially required for the presence of S1 18-phosphorylated ERα (pERαS1 18). ChIP signals are presented as percentage of input.
(C) The kinetics of ERα, DNA-PKcs, and pERαS118 occupancy at ERα active enhancers. ChIP signals are presented as percentage of input.
(D) Heatmaps, based on ChIP-seq data, showing DNA-PKcs binding at 1,333 ERα active enhancers is enhanced by E2, while its binding is not apparent at ERα non-active enhancers.
(E) Knockdown of RARs by shRNA greatly reduces DNA-PKcs binding to ERα active enhancers and affects enrichment of pERαS118. ChIP signals are presented as percentage of input.
(F) Heatmap of DNA-PKcs ChIP-seq data showing loss of E2-enhanced DNA-PKcs binding to ERα active enhancers upon knockdown of both RARs.
(G) Heatmap of pERαS1 18 ChIP-seq data demonstrating partial reduction of E2-enhanced pERαS118 binding to ERα active enhancers upon knockdown of both RARs.
(H) Knockdown of DNA-PKcs by shRNA affects E2-liganded activation of gene body and eRNA transcription for GREB1 and TFF1 genes but does not affect ERα levels or RA induction of the HoxA1 gene, as demonstrated by qPCR.
(I) Working model of a MegaTrans enhancer. At ERα active enhancers that contain ERE and FoxA1 motifs, DNA-bound ERα and FoxA1 dynamically recruit in situ the functionally required MegaTrans complex of DNA-binding TFs, including RAR, GATA3, AP2γ, STAT1, and AP1. The trans-bound components of the MegaTrans complex may recruit specific, functional enzymatic machinery, exemplified by the recruitment of DNA-PK.
Data are represented as mean ± SEM (NS not significant, * P<0.05, ** P<0.01, ***P<0.001). See also Figure S7.
In order to determine whether trans-bound RAR is required for the recruitment of DNA-PKcs at ERα active enhancers, we performed ChIPs for both DNA-PKcs and pERαS118 after knockdown of RARα/γ. We found that RARα/γ knockdown substantially reduced the levels of both DNA-PKcs and pERαS118 at ERα active enhancers (Figures 7E, 7F, 7G, S7C, and S7D), suggesting that trans-bound RARs may be required for the functionally relevant recruitment of DNA-PKcs at these ERα enhancers.
Knockdown of DNA-PKcs significantly inhibited E2-induced activation of ERα-bound functional enhancers and their target coding gene expression but did not affect RA-induced HoxA1 activation (Figure 7H). Consistently, the treatment of MCF7 cells with the DNA-PK kinase inhibitor NU7441 also inhibited ERα-dependent target activation (Figure S7E). Thus, at least one role of RARs that are recruited to ERα-bound functional enhancers may be to facilitate the concomitant recruitment of a specific protein kinase. It is possible that, analogous to this role of RARs role in recruitment of DNA-PK, other DNA-binding TFs components in the MegaTrans complex also contribute to the recruitment of additional enzymatic factors that are required for functional enhancer activation.
Discussion
The MegaTrans Complex Is a Signature of ERα Functional Enhancers
Here, we suggest that, in addition to the critical recruitment of an ever-increasing number of well-characterized coactivator complexes, many with specific enzymatic functions, activation of the most robust subset of ERα enhancers by E2 is dependent upon, and can be predicted by, their ability to recruit a complex of established DNA-binding transcription factors, referred to as the MegaTrans complex (Figure 7I). This complex appears to be recruited/assembled in trans on ERα-bound functional enhancers and requires the presence of ERα. In addition to the requirement for ERα, certain other components of the complex appear to be necessary for its assembly on functional enhancers; for example, knockdown of RARα/γ and GATA3 abolishes recruitment of other components of the complex and inhibits enhancer/target coding gene activation. Although the precise biochemical interactions that underlie the formation of the MegaTrans complex remain incompletely defined, our data on the effects of DNase I treatment and DNA-binding domain mutation suggest that the MegaTrans complex assembles in situ at ERα-bound, ERE-containing enhancers, which also harbor nearby FoxA1 cis-binding sites.
While the idea that DNA-binding transcription factors can be recruited in trans to either activate or repress specific target coding genes is well established (Langlais et al, 2012; Pascual et al, 2005; Reichardt et al, 1998), this study provides an initial description of a ligand-dependent recruitment in trans of a complex of DNA-binding transcription factors that proves important for ERα function. Using the published criteria for defining super-enhancers (Hnisz et al, 2013; Whyte et al, 2013), only ∼22% of the MegaTrans functional enhancers can be classified as components of super-enhancers, and we note that there is only a very slight distinction in the levels of eRNA induction in response to E2 on the functional MegaTrans enhancers associated with super-enhancers compared to those not associated with the super-enhancers. Thus, recruitment of the MegaTrans complex serves as a mark that distinguishes the most active enhancers of the estrogen-regulated transcriptional program.
These observations raise several corollary questions. First, does this MegaTrans complex serve on all active or activated enhancers, irrespective of the DNA-binding transcription factors bound in cis to those enhancers? It appears that the RARE-containing functional enhancers, which recruit RARα/γ in cis, do not recruit this complex or ERα (Figure S1G). Therefore, we speculate that there may be a number of distinct MegaTrans complexes that are recruited only by certain regulatory DNA-binding factors, and these complexes, analogous to events for ERα-regulated enhancers, serve to mark and initiate other specific enhancer activation events. Second, how is the MegaTrans complex selectively recruited only to the functional ERα-bound enhancers? Based on our initial data, we suggest that the answer likely involves the apparent dual roles of the “pioneer factor” FoxA1, which is selectively recruited to the functional, MegaTrans-dependent enhancers at <200bp from the ERE but is also required for the binding of ERα to these enhancers. In addition to its established pioneering role, FoxA1 may also make an important contribution to the recruitment/stabilization of the MegaTrans complex. We are tempted to speculate that, in addition to promoting cooperative binding of ERα to enhancers, FoxA1 may cause a conformational alteration in the ERα receptor, either directly or via altered enhancer DNA architecture, that facilitates the recruitment of the MegaTrans complex; however, it is formally possible that the increased affinity of the ERα for the enhancer alone determines binding of the MegaTrans complex. These questions and other undefined aspects of the MegaTrans complex represent fascinating issues for future investigation.
The MegaTrans Complex as a Platform for Regulatory Enzymes
In light of the already large number of important coactivator complexes, why would these additional DNA-binding transcription factors, most of which are recruited to the active enhancers by the ERE-bound ERα, be required? First, we have found that they play important “early” roles in enhancer function as they are important for eRNA induction and the ligand-dependent increase of p300 and Med1 occupancy on the enhancers. Thus, components of the MegaTrans complex are required to license the recruitment of well-known, important coactivators, as exemplified by p300 and Mediator subunits. In this regard, the DNA-binding transcription factors summoned to bind in trans through ERα are subserving functions that are quite analogous to those of the recognized coactivator complexes, many of which feature associated/intrinsic enzymatic activities. Similarly, we note that RARs are capable of interacting with many known or potential coactivators, and we have focused on one such potential regulator. The enzyme DNA-PKcs binds to RARs and is recruited with rapid temporal kinetics to ERα-bound functional enhancers. Additionally, knockdown of DNA-PKcs partially phenocopies the functional consequences of RARα/γ knockdown in MCF7 cells. Therefore, we are tempted to speculate that components of the MegaTrans complex individually recruit various enzymes/factors that collectively are important mechanisms in initial activation of the functional enhancer program. DNA-PK is a kinase with multiple targets, including ERα on Ser118 (Foulds et al, 2013), which we find occurs on the active ERα-bound enhancers, dependent of the presence of RARs on these functional enhancers. It is particularly intriguing that DNA-PKcs is associated with the Ku80 complex, classically considered to be involved in DNA damage repair (Hartley et al, 1995; Jin and Weaver, 1997), which may in fact be pertinent to its functions in transcriptional control events. The rapid appearance of DNA-PKcs on the ligand-regulated enhancers is analogous to other examples of recruited protein kinases in gene regulation events (Perissi et al., 2008; Tee et al, 2014).
Thus, investigation of the ERα-regulated enhancers has revealed that an additional and critical component of the most active enhancers is the ERα-dependent recruitment of the MegaTrans complex, which combinatorially promotes recruitment of additional coactivators/enzymes that increase enhancer activation and target coding gene transcription. Analogous to the hypothesis that super-enhancers regulate critical developmental or disease-associated coding gene transcriptional programs, MegaTrans complex recruitment appears to serve as a mechanism of marking/empowering enhancers to control key aspects of the regulatory transcriptional programs in a specific cell type. The super-enhancer model defines the combinatorial effects of multiple, clustered enhancers spanning >8-10kb, while the MegaTrans enhancer model explains the different functional activity of single enhancers.
MegaTrans Enhancers as a Commonly Utilized Strategy?
The uncovering of another layer of machinery involved in the effective activation of ERα-regulated enhancers raises the possibility that distinct MegaTrans enhancers exist for other classes of DNA-binding TFs that are responsible for activation of unique transcriptional programs. We note that ChIP-seq analyses for many established DNA-binding TFs have revealed their binding on enhancers that do not harbor any known cognate binding sequences. This raises the possibility that these TFs might exert roles, in trans, on other transcription programs analogous to the effects of the MegaTrans complex on the ERα-regulated functional enhancers. The ‘hotspot’ or ‘clustering’ phenomenon of DNA-binding TFs has recently been reported in several different organisms (Junion et al., 2012; Rada-Iglesias et al, 2012; Siersbaek et al, 2014a; Siersbaek et al., 2014b; Wilson et al, 2010; Yan et al, 2013). However, the underlying molecular mechanism(s) and functional significance are not well understood. Our results provide a functional model to explain at least many cases of the clustering phenomena. Specifically, our data suggest that the DNA-dependent binding of ERα and FoxA1 at ERα functional enhancers establishes a platform for recruiting a MegaTrans complex of other DNA-binding TFs by protein-protein interactions (in trans). MegaTrans complex-bound enhancers function as more robust enhancers by recruiting certain unique factors and enzymes, such as DNA-PK. Thus, our study provides new insights into the understanding of the TF clustering phenomenon. Our data simultaneously help to explain why ChIP-seq analyses reveal roughly 50% of the regions occupied by many of the DNA-binding TFs assayed in the ENCODE project do not harbor cognate DNA-binding motifs.
Experimental Procedures
A detailed description of all methods and any associated references is provided in the Extended Experimental Procedures, which is included in the supplemental information section.
Cell Culture and BLRP-Tagged Stable Cell Lines
MCF7 cells, initially obtained from ATCC, were maintained in culture and treated as described (Li et al., 2013). To study binding patterns for wild-type and non-DNA-binding mutants of RARα/γ, GATA3, ERα and other TFs, we first established a parental MCF7 stable cell line that expressed BirA enzyme and Tet-Repressor. We then used this parental cell line to make doxycycline-inducible stable cell lines expressing BLRP-tagged proteins at close to endogenous levels. BLRP-tagged proteins were biotinylated in vivo by BirA enzyme, allowing for pull downs to be performed with NanoLink™ streptavidin magnetic beads (Solulink) under very stringent washing conditions.
Chromatin Immunoprecipitation (ChIP) and Global Run-on Sequencing (GRO-seq)
ChIP-qPCRs, ChIP-seqs and GRO-seqs were performed as previously reported (Li et al, 2013). Immunoprecipitated DNA was recovered by purification on QIAquick spin columns (Qiagen) after decrosslinking and then analyzed by qPCR using primers listed in Table S1. The qPCR-validated DNA samples were used to make libraries for deep sequencing. The details of ChIP-seq and GRO-seq data analysis are included in the Extended Experimental Procedures.
Supplementary Material
Figure S1. RARs Have Cis- and Trans-Binding Functional Models in MCF7, Related to Figure 1
(A) Schematic diagram depicting the in vivo biotinylation BirA-BLRP system, in which BirA biotin ligase can biotinylate a lysine residue in the BLRP tag, and BLRP-tagged proteins but not non-specific biotin labeled proteins can be eluted by TEV protease digestion.
(B) Diagram of two different DNA-binding domain mutations for RARα and RARγ. Arrows point out the mutation sites.
(C) Both zinc finger and pBox mutations abolish RAR ability to activate RARE luciferase reporter. Mean ± SEM based on three independent experiments (***P<0.001).
(D) Western blots showing that the doxycycline-induced expression levels of BLRP-tagged RARα/γ (marked by asterisk) are similar to their endogenous protein (marked by arrow) levels. The blots with streptavidin-HRP show the in vivo biotinylation levels for tagged-RARα/γ.
(E) cis and trans binding sites of RAR in MCF7 cells. The average tag density plots show the differential binding of wild-type and mutant RAR on these sites.
(F) ChIP-seq heatmap of RARα and ERα for 15,000 RAR cis-binding sites, showing no binding of ERα at these sites.
(G) UCSC browser snapshot of ChIP-seq showing the cis-binding of RARα to the HoxA cluster depends on its DNA-binding ability.
(H) GRO-seq boxplots showing that RARs are required for RA-liganded activation of both eRNA and gene body transcription.
Figure S2. Both Cis- and Trans-Binding of RARs Regulates Ligand-Dependent Transcription Activation, Related to Figure 1
(A) UCSC browser snapshot of ChIP-seq showing the trans-binding of RARα at the GREB1 locus does not depend on its DNA-binding ability.
(B) qPCR results show robust shRAR knockdown efficiency and that ERα RNA levels are not affected by knockdown of RAR. Data are represented as mean ± SEM based on three independent experiments.
(C) Western blots show the protein knockdown efficiency of two different shRNA constructs for both RARα/γ. Knockdown of RARα/γ does not affect ERα protein level. Histone H3 was used as loading control.
(D) UCSC browser image of GRO-seq showing knockdown of RARs affects E2-induced activation of TFF1 gene. The arrow points to an ERα active enhancer, which also has lower E2 induction of eRNA upon knockdown of RARs.
(E) UCSC browser snapshot of GRO-seq showing knockdown of RARs affects RA-induced activation of HoxA1 gene.
(F) A Tet-On inducible system was used to express either wild-type or pBox mutant RARγ in rescue experiments. Western blot was used to check the knockdown efficiency of endogenous RARγ (marked by arrow) by a shRNA that targets to 3′UTR of RARγ mRNA and the induced expression of wild-type or pBox-mutant RARγ protein (marked by asterisk). GAPDH blot was used as an internal loading control.
Figure S3. ERα In Situ Recruits a DNA-Binding TF Complex to ERE-Containing Active Enhancers, Related to Figure 2
(A) Silver staining of RARα complex to detect the pull-down of BLRP-RARα and its associated nuclear proteins. The same cell line without doxycycline treatment was used as a control to filter background proteins after mass spectrometry.
(B) The E2-dependent interaction between ERα and other DNA-binding TFs was confirmed by coimmunoprecipitation of BLRP-tagged ERα and the other TFs.
(C) Test of ERα protein levels after ICI 182780 knockdown showing that a treatment of 3 hours yielded the best ERα protein knockdown. GAPDH blot was used as an internal loading control.
(D) DNA-binding TFs in the ERα complex bind to an ERα active enhancer located 5′ of GREB1 upon E2 signal and ERα knockdown reduces their binding. ChIP signals are presented as percentage of input.
(E) The DNA-dependent interactions between ERα and other DNA-binding TFs were confirmed by co-immunoprecipitation of BLRP-tagged ERα and the other TFs.
(F) ChIP-reChIP analyses were performed to confirm the co-binding of GATA3, ERα, and other DNAbinding TFs to ERα active enhancers but not HoxA1 enhancer or ERα non-active enhancers. ChIP signals are presented as percentage of input.
In (B) and (E), the asterisk marks BLRP-tagged ERα and the arrow marks endogenous ERα. Data are represented as mean ± SEM (NS not significant, *P<0.05, **P<0.01, ***P<0.001).
Figure S4. Dual-Binding Models for GATA3 and Trans-bound GATA3 Also Regulates the Function of ERα Active Enhancers, Related to Figure 3
(A) Schematic diagram showing two different mutagenesis approaches for disrupting the GATA3 DNA-binding domain.
(B) Western blots show the expression levels of BLRP-tagged GATA3 (marked by asterisk) are similar to their endogenous protein (marked by arrow) levels. The blots with streptavidin-HRP show the in vivo biotinylation level for GATA3.
(C) UCSC browser snapshot of ChIP-seq illustrating the cis-binding and trans-binding of GATA3 at the FoxC1 gene locus. In the snapshot, the trans-binding of GATA3 at one ERα active enhancer (marked by arrow) is evident.
(D) The cis and trans dual binding models of GATA3 in MCF7.
(E) UCSC browser snapshot of GRO-seq showing knockdown of GATA3 affects the E2-induced activation of P2RY2 gene at both the enhancer and the gene body. The arrow points to an ERα active enhancer.
(F) Western blots confirm the effective siRNA knockdown of GATA3 and that its knockdown does not affect ERα protein level. GAPDH was used as a loading control.
(G) qPCR results showing robust siGATA3 knockdown efficiency of RNA levels and no concomitant change in ERα RNA level. Data are represented as mean ± SEM based on three independent experiments.
Figure S5. ERα Active Enhancers Are MegaTrans Enhancers Regulated by DNA-Binding TFs, Related to Figure 4
(A) UCSC browser snapshot of ChIP-seq showing an ERα non-active enhancer (pointed by arrow) at the TNIP1 locus without the binding of MegaTrans complex.
(B) Knockdown of AP2γ by shRNA affects ligand-dependent activation of both eRNA transcription and coding gene expression for GREB1 and TFF1 genes.
(C) UCSC browser image of GRO-seq showing knockdown of AP2γ affects the E2-induced activation of P2RY2 gene at both the enhancer and the gene body. The arrow points to an ERα active enhancer.
(D-E) Knockdown of c-Jun or c-Fos by lentivirus shRNA affects E2-dependent ERα activation of the coding gene body and eRNA transcription for GREB1 and TFF1 genes, as demonstrated by qPCR.
Data are represented as mean ± SEM (*P<0.05, **P<0.01).
Figure S6. Trans-Bound TFs on ERα MegaTrans Enhancers Are Required for Recruitment of ERα Co-Activators and Super-Enhancer Function, Related to Figure 6
(A) UCSC browser image of ChIP-seq showing the ERα active enhancer (marked by arrow) at the Smad7 locus exhibits a lower level of p300 recruitment upon E2 signal after RARα and RARγ shRNA knockdown.
(B) UCSC browser snapshot of ChIP-seq showing the ERα active enhancer (marked by arrow) at the TFF1 locus has a lower level of E2-induced Med1 recruitment after RARα and RARγ shRNA knockdown.
(C) UCSC browser image of ChIP-seq showing one regular ERα active enhancer (marked by arrow) and one super-enhancer containing several ERα active enhancers at the Myc gene locus. Both regions have stronger Med1 ChIP-seq signals upon E2 treatment, suggesting that a small percentage of ERα MegaTrans enhancers locate in super-enhancers and regulate their function.
Figure S7. Trans-Bound RAR May Contribute to Recruit DNA-PK Kinase as a Co-Activator for ERα Active Enhancers, Related to Figure 7
(A) The distribution of 12,629 DNA-PKcs ChIP-seq (+E2) peaks.
(B) UCSC browser snapshot of DNA-PKcs ChIP-seq data (-E2 and +E2) showing E2 enhanced binding of DNA-PKcs to an ERα active enhancer (pointed by arrow) at the TFF1 locus.
(C) UCSC browser snapshot of ChIP-seq showing the ERα active enhancer (marked by arrow) at the TFF1 locus loses the recruitment of DNA-PKcs upon E2 stimulation after RARα and RARγ knockdown by shRNAs.
(D) UCSC browser snapshot of ChIP-seq showing the ERα active enhancer (marked by arrow) at the TFF1 locus has a lower level of S118-phosphorylated ERα upon E2 stimulation after RARα and RARγ shRNA knockdown.
(E) Knockdown of DNA-PKcs by siRNA or inhibition of DNA-PKcs kinase activity using NU7441 affects the activation of TFF1 by E2. Knockdown of DNA-Pkcs did not affect ERα levels. Data are represented mean ± SEM based on three independent qPCR experiments (**P<0.01, ***P<0.001).
Table S1. All qPCR Primers Used in this Study Related to Experimental Procedures Section
Table S2. siRNAs and shRNAs Used in this Study Related to Experimental Procedures Section
Table S3. Oligos Used in the GRO-seq Protocol Related to Experimental Procedures Section
Highlights.
ERα-regulated enhancers exhibit trans-recruitment of a DNA-binding factor complex.
The DNA-binding transcription factors are assembled in a 1-2MDa complex by ERα.
The MegaTrans complex recruits specific enzymatic machinery to enhancers.
The MegaTrans complex serves as a functional signature of most regulated enhancers.
Acknowledgments
The authors are grateful to Janet Hightower for assistance with figure preparation, to Rachel Pardee for proofreading and to Dr. Majid Ghassemian (UCSD) for assistance in mass spectrometry. We thank Dr. Dimple Notani (UCSD) for critically reading the manuscript, and Drs. Namit Singh and Kevin Corbett (UCSD) for their experimental contributions on gel filtration studies. We thank Dorota Skowronska-Krawczyk and Bogdan Tanasa for critical comments and discussions on this paper. M.G.R. is an investigator with the Howard Hughes Medical Institute. This work was supported by grants from NIH and NCI to M.G.R.
Footnotes
Accession Numbers: The GEO accession number for all deep sequencing data reported in this paper is GSE60272, which includes ChIP-seq data set (GSE60270) and GRO-seq data set (GSE60271).
Supplemental Information: Supplemental information includes Extended Experimental Procedures, seven figures, and three tables and can be found with this article online at XXX
Author Contributions: Z.L. and M.G.R. conceived the original ideas, designed the project and wrote the paper. Z.L. preformed the majority of the experiments with participation from F.Y., W.L., S.O., M.J.F., X.S., F.Z., K.O. and A.K.. D.M. performed most bioinformatic analyses with the assistance from Q.M..
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22:490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds CE, Feng Q, Ding C, Bailey S, Hunsaker TL, Malovannaya A, Hamilton RA, Gates LA, Zhang Z, Li C, et al. Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Mol Cell. 2013;51:185–199. doi: 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Weaver DT. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. The EMBO J. 1997;16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L, Pan YF, Li G, Lim M, Thomsen JS, et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-alpha. Mol Syst Biol. 2010;6:456. doi: 10.1038/msb.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EE. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148:473–486. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell. 2012;47:38–49. doi: 10.1016/j.molcel.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, Rueda OM, Holmes KA, Theodorou V, Robinson JL, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbit MA, Bowl MR, Harding B, Ali A, Ayala A, Crowe C, Dobbie A, Hampson G, Holdaway I, Levine MA, et al. Characterization of GATA3 mutations in the hypoparathyroidism, deafness, and renal dysplasia (HDR) syndrome. J Biol Chem. 2004;279:22624–22634. doi: 10.1074/jbc.M401797200. [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Scafoglio C, Zhang J, Ohgi KA, Rose DW, Glass CK, Rosenfeld MG. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol Cell. 2008;29:755–766. doi: 10.1016/j.molcel.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11:633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Siersbaek R, Baek S, Rabiee A, Nielsen R, Traynor S, Clark N, Sandelin A, Jensen ON, Sung MH, Hager GL, et al. Molecular architecture of transcription factor hotspots in early adipogenesis. Cell Rep. 2014a;7:1434–1442. doi: 10.1016/j.celrep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, Poulsen LL, Rogowska-Wrzesinska A, Jensen ON, Mandrup S. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014b;7:1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, Benner C, Glass CK, Katzenellenbogen BS. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol. 2010;30:3943–3955. doi: 10.1128/MCB.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee WW, Shen SS, Oksuz O, Narendra V, Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2013;23:12–22. doi: 10.1101/gr.139469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013;154:801–813. doi: 10.1016/j.cell.2013.07.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. RARs Have Cis- and Trans-Binding Functional Models in MCF7, Related to Figure 1
(A) Schematic diagram depicting the in vivo biotinylation BirA-BLRP system, in which BirA biotin ligase can biotinylate a lysine residue in the BLRP tag, and BLRP-tagged proteins but not non-specific biotin labeled proteins can be eluted by TEV protease digestion.
(B) Diagram of two different DNA-binding domain mutations for RARα and RARγ. Arrows point out the mutation sites.
(C) Both zinc finger and pBox mutations abolish RAR ability to activate RARE luciferase reporter. Mean ± SEM based on three independent experiments (***P<0.001).
(D) Western blots showing that the doxycycline-induced expression levels of BLRP-tagged RARα/γ (marked by asterisk) are similar to their endogenous protein (marked by arrow) levels. The blots with streptavidin-HRP show the in vivo biotinylation levels for tagged-RARα/γ.
(E) cis and trans binding sites of RAR in MCF7 cells. The average tag density plots show the differential binding of wild-type and mutant RAR on these sites.
(F) ChIP-seq heatmap of RARα and ERα for 15,000 RAR cis-binding sites, showing no binding of ERα at these sites.
(G) UCSC browser snapshot of ChIP-seq showing the cis-binding of RARα to the HoxA cluster depends on its DNA-binding ability.
(H) GRO-seq boxplots showing that RARs are required for RA-liganded activation of both eRNA and gene body transcription.
Figure S2. Both Cis- and Trans-Binding of RARs Regulates Ligand-Dependent Transcription Activation, Related to Figure 1
(A) UCSC browser snapshot of ChIP-seq showing the trans-binding of RARα at the GREB1 locus does not depend on its DNA-binding ability.
(B) qPCR results show robust shRAR knockdown efficiency and that ERα RNA levels are not affected by knockdown of RAR. Data are represented as mean ± SEM based on three independent experiments.
(C) Western blots show the protein knockdown efficiency of two different shRNA constructs for both RARα/γ. Knockdown of RARα/γ does not affect ERα protein level. Histone H3 was used as loading control.
(D) UCSC browser image of GRO-seq showing knockdown of RARs affects E2-induced activation of TFF1 gene. The arrow points to an ERα active enhancer, which also has lower E2 induction of eRNA upon knockdown of RARs.
(E) UCSC browser snapshot of GRO-seq showing knockdown of RARs affects RA-induced activation of HoxA1 gene.
(F) A Tet-On inducible system was used to express either wild-type or pBox mutant RARγ in rescue experiments. Western blot was used to check the knockdown efficiency of endogenous RARγ (marked by arrow) by a shRNA that targets to 3′UTR of RARγ mRNA and the induced expression of wild-type or pBox-mutant RARγ protein (marked by asterisk). GAPDH blot was used as an internal loading control.
Figure S3. ERα In Situ Recruits a DNA-Binding TF Complex to ERE-Containing Active Enhancers, Related to Figure 2
(A) Silver staining of RARα complex to detect the pull-down of BLRP-RARα and its associated nuclear proteins. The same cell line without doxycycline treatment was used as a control to filter background proteins after mass spectrometry.
(B) The E2-dependent interaction between ERα and other DNA-binding TFs was confirmed by coimmunoprecipitation of BLRP-tagged ERα and the other TFs.
(C) Test of ERα protein levels after ICI 182780 knockdown showing that a treatment of 3 hours yielded the best ERα protein knockdown. GAPDH blot was used as an internal loading control.
(D) DNA-binding TFs in the ERα complex bind to an ERα active enhancer located 5′ of GREB1 upon E2 signal and ERα knockdown reduces their binding. ChIP signals are presented as percentage of input.
(E) The DNA-dependent interactions between ERα and other DNA-binding TFs were confirmed by co-immunoprecipitation of BLRP-tagged ERα and the other TFs.
(F) ChIP-reChIP analyses were performed to confirm the co-binding of GATA3, ERα, and other DNAbinding TFs to ERα active enhancers but not HoxA1 enhancer or ERα non-active enhancers. ChIP signals are presented as percentage of input.
In (B) and (E), the asterisk marks BLRP-tagged ERα and the arrow marks endogenous ERα. Data are represented as mean ± SEM (NS not significant, *P<0.05, **P<0.01, ***P<0.001).
Figure S4. Dual-Binding Models for GATA3 and Trans-bound GATA3 Also Regulates the Function of ERα Active Enhancers, Related to Figure 3
(A) Schematic diagram showing two different mutagenesis approaches for disrupting the GATA3 DNA-binding domain.
(B) Western blots show the expression levels of BLRP-tagged GATA3 (marked by asterisk) are similar to their endogenous protein (marked by arrow) levels. The blots with streptavidin-HRP show the in vivo biotinylation level for GATA3.
(C) UCSC browser snapshot of ChIP-seq illustrating the cis-binding and trans-binding of GATA3 at the FoxC1 gene locus. In the snapshot, the trans-binding of GATA3 at one ERα active enhancer (marked by arrow) is evident.
(D) The cis and trans dual binding models of GATA3 in MCF7.
(E) UCSC browser snapshot of GRO-seq showing knockdown of GATA3 affects the E2-induced activation of P2RY2 gene at both the enhancer and the gene body. The arrow points to an ERα active enhancer.
(F) Western blots confirm the effective siRNA knockdown of GATA3 and that its knockdown does not affect ERα protein level. GAPDH was used as a loading control.
(G) qPCR results showing robust siGATA3 knockdown efficiency of RNA levels and no concomitant change in ERα RNA level. Data are represented as mean ± SEM based on three independent experiments.
Figure S5. ERα Active Enhancers Are MegaTrans Enhancers Regulated by DNA-Binding TFs, Related to Figure 4
(A) UCSC browser snapshot of ChIP-seq showing an ERα non-active enhancer (pointed by arrow) at the TNIP1 locus without the binding of MegaTrans complex.
(B) Knockdown of AP2γ by shRNA affects ligand-dependent activation of both eRNA transcription and coding gene expression for GREB1 and TFF1 genes.
(C) UCSC browser image of GRO-seq showing knockdown of AP2γ affects the E2-induced activation of P2RY2 gene at both the enhancer and the gene body. The arrow points to an ERα active enhancer.
(D-E) Knockdown of c-Jun or c-Fos by lentivirus shRNA affects E2-dependent ERα activation of the coding gene body and eRNA transcription for GREB1 and TFF1 genes, as demonstrated by qPCR.
Data are represented as mean ± SEM (*P<0.05, **P<0.01).
Figure S6. Trans-Bound TFs on ERα MegaTrans Enhancers Are Required for Recruitment of ERα Co-Activators and Super-Enhancer Function, Related to Figure 6
(A) UCSC browser image of ChIP-seq showing the ERα active enhancer (marked by arrow) at the Smad7 locus exhibits a lower level of p300 recruitment upon E2 signal after RARα and RARγ shRNA knockdown.
(B) UCSC browser snapshot of ChIP-seq showing the ERα active enhancer (marked by arrow) at the TFF1 locus has a lower level of E2-induced Med1 recruitment after RARα and RARγ shRNA knockdown.
(C) UCSC browser image of ChIP-seq showing one regular ERα active enhancer (marked by arrow) and one super-enhancer containing several ERα active enhancers at the Myc gene locus. Both regions have stronger Med1 ChIP-seq signals upon E2 treatment, suggesting that a small percentage of ERα MegaTrans enhancers locate in super-enhancers and regulate their function.
Figure S7. Trans-Bound RAR May Contribute to Recruit DNA-PK Kinase as a Co-Activator for ERα Active Enhancers, Related to Figure 7
(A) The distribution of 12,629 DNA-PKcs ChIP-seq (+E2) peaks.
(B) UCSC browser snapshot of DNA-PKcs ChIP-seq data (-E2 and +E2) showing E2 enhanced binding of DNA-PKcs to an ERα active enhancer (pointed by arrow) at the TFF1 locus.
(C) UCSC browser snapshot of ChIP-seq showing the ERα active enhancer (marked by arrow) at the TFF1 locus loses the recruitment of DNA-PKcs upon E2 stimulation after RARα and RARγ knockdown by shRNAs.
(D) UCSC browser snapshot of ChIP-seq showing the ERα active enhancer (marked by arrow) at the TFF1 locus has a lower level of S118-phosphorylated ERα upon E2 stimulation after RARα and RARγ shRNA knockdown.
(E) Knockdown of DNA-PKcs by siRNA or inhibition of DNA-PKcs kinase activity using NU7441 affects the activation of TFF1 by E2. Knockdown of DNA-Pkcs did not affect ERα levels. Data are represented mean ± SEM based on three independent qPCR experiments (**P<0.01, ***P<0.001).
Table S1. All qPCR Primers Used in this Study Related to Experimental Procedures Section
Table S2. siRNAs and shRNAs Used in this Study Related to Experimental Procedures Section
Table S3. Oligos Used in the GRO-seq Protocol Related to Experimental Procedures Section