Abstract
Magnetic resonance imaging (MRI) is an essential tool in the study of muscle anatomy and functional activity in the tongue. Objective assessment of similarities and differences in tongue structure and function has been performed using unnormalized data, but this is biased by the differences in size, shape, and orientation of the structures. To remedy this, we propose a methodology to build a 3D vocal tract atlas based on structural MRI volumes from twenty normal subjects. We first constructed high-resolution volumes from three orthogonal stacks. We then removed extraneous data so that all 3D volumes contained the same anatomy. We used an unbiased diffeomorphic groupwise registration using a cross-correlation similarity metric. Principal component analysis was applied to the deformation fields to create a statistical model from the atlas. Various evaluations and applications were carried out to show the behaviour and utility of the atlas.
Keywords: Vocal tract atlas, Statistical model, MRI, Tongue
1. Introduction
The vocal tract is a complex system that consists of both movable and immovable structures, coordinating numerous functions. It is involved in critical human functions such as breathing, eating, and speaking. The movable structures such as the lips, jaw, tongue, and velum are the primary articulators during speech production. In particular, the human tongue forms remarkably complex shapes. It is a muscular hydrostat [1] with three orthogonal fibre directions and extensive fibre inter-digitations. Any bundle of fibres may contain multiple muscles, including intrinsic and extrinsic muscles, categorized by their function. The understanding of tongue muscle structure and function is essential for the diagnosis and treatment of disease and for the scientific studies of the vocal tract itself. To date, however, frustratingly little is known regarding the relationship between tongue structure and function. This is partly because the complex anatomy of the tongue poses challenges to spatially distinguishing each muscle and characterizing muscle interaction and function [2].
Imaging of the human vocal tract and tongue with magnetic resonance imaging (MRI) has played an essential role in interpreting muscle anatomy and function. MRI is a noninvasive imaging technique which can capture high spatial resolution images (high-resolution MRI), muscle fibre directions (diffusion tensor MRI), and tissue point motion (tagged MRI), leading to functional information about the vocal tract [3–5]. MRI provides quantities that capture features of vocal tract structure and function including surface shapes [6] and the deformation of the internal tongue musculature [4, 5]. MRI-based imaging continues to be a crucial tool for vocal tract research.
Although MRI has played an important role in vocal tract imaging, there are several limiting factors in its use in vocal tract analysis. First, the size and shape of the vocal tract including the tongue and muscles vary from one subject to another as illustrated in Fig. 1. As a result, objective assessment of similarities and differences in tongue structure and function has been performed using unnormalized data; this process, however, is biased by differences in size, shape, and orientation of the structures. Second, there exists no comprehensive framework to assess variability of the vocal tract’s hard and soft tissue structures. Both of these limitations can be addressed by using an atlas, which provides a normalized space in which all subjects from a target population can be mapped and compared. This in turn facilitates quantitative comparisons of anatomical features [7] including their statistical variation [8].
Figure 1.

Examples of high-resolution MR images acquired at rest position. Each subject has different vocal tract proportions. The white rectangle shows the vocal tract area.
Atlases constructed from populations of subject images are widely used computational tools, playing important roles in the diagnosis and treatment of diseases, segmentation and labelling for surgical planning, and provision of a common coordinate space for comparing subjects of all types [9, 10]. Brain atlases, in particular, have been used as references for normalization of groups of individuals, spatial maps for brain delineation, and characterization of tissue distribution [11]. To the best of our knowledge, however, currently there is no atlas constructed from a population of images for the study of vocal tract structure and function.
In this work, we constructed a vocal tract atlas and statistical model using structural MRI. Together, the atlas and the statistical model provide an average description of human vocal tract architecture along with its variability within the training population. The atlas building method is a multi-step procedure as illustrated in Fig. 2. First, twenty normal subjects were imaged using three orthogonal image stacks (i.e., axial, coronal, and sagittal) (Fig. 2A). Second, for each subject a single high-resolution volume was generated via a super-resolution volume reconstruction technique (Fig. 2B) [12]. Third, we processed each volume to remove extraneous image data and bound the vocal tract in order for each stack to have the same anatomical structures (Fig. 2B). Fourth, we applied a state-of-the-art groupwise registration technique to construct an average atlas image volume together with its anatomical correspondences to each subject (Fig. 2C). Manual segmentation of the tongue and a variety of muscle structures was then carried out in the atlas (Fig. 2C). Finally, we performed principal component analysis (PCA) on the deformation fields resulting from atlas building, which yields a statistical model comprising principal modes of the sample covariance on the deformation fields (Figs. 2D and 2E). In this work, we use the term atlas to refer to the average of the warped volumes in the common space. The statistical model refers to the PCA analysis on deformation fields and other quantities computed in the atlas space.
Figure 2.
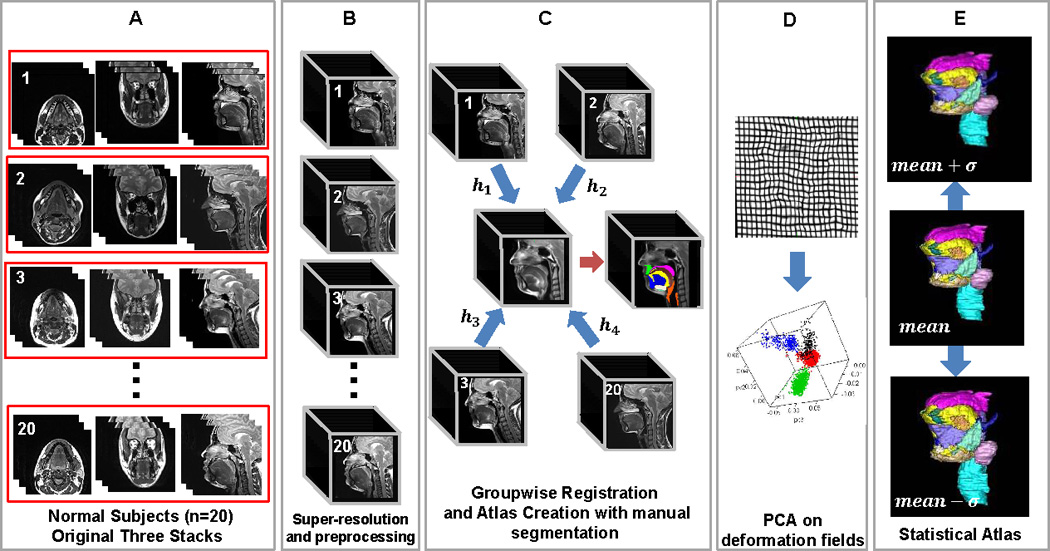
Overview of the atlas construction method.
The remainder of this paper is structured as follows. The atlas building method for the vocal tract is presented in Sec. 2. In Sec. 3, we describe experiments for quantitative and qualitative validations of the atlas using tongue and muscle segmentations. Results and discussion are presented in Secs. 4 and 5, respectively. Finally, the conclusion is given in Sec. 6.
2. Methods and Materials
2.1. Subjects and Data Acquisition
Twenty high-resolution MR datasets were used. All MRI scanning was performed on a Siemens 3.0 T Tim Trio system (Siemens Healthcare Inc., Malvern, PA) with a 12-channel head and a 4-channel neck coil using a segmented gradient echo sequence. In addition, a T2-weighted Turbo Spin Echo sequence with echo train length of 12 and TE/TR of 62ms/2500ms was used. The field-of-view (FOV) of each image was 240 mm×240 mm and was sampled at 256×256 pixels. Each dataset contains a sagittal, coronal, and axial stack of images encompassing the tongue and surrounding structures. The image size for the high-resolution MRI is 256×256×z (where z ranges from 10 to 24) with 0.94 mm×0.94 mm in-plane resolution and 3 mm slice thickness. The datasets were acquired at a rest position and the subjects were required to remain still from 1.5 to 3 minutes for each orientation. Table 1 summarizes the characteristics of the twenty healthy subjects.
Table 1.
Characteristics of the 20 healthy subjects (M: male, F: Female)
| Subject | Age | Gender | Weight (lb) |
Subject | Age | Gender | Weight (lb) |
|---|---|---|---|---|---|---|---|
| 1 | 23 | M | 155 | 11 | 21 | F | 126 |
| 2 | 31 | F | 150 | 12 | 25 | M | 150 |
| 3 | 24 | F | 100 | 13 | 22 | M | 130 |
| 4 | 57 | F | 170 | 14 | 43 | M | 180 |
| 5 | 43 | F | 217 | 15 | 26 | M | 240 |
| 6 | 35 | M | 210 | 16 | 42 | F | 180 |
| 7 | 45 | F | 180 | 17 | 52 | M | 156 |
| 8 | 27 | F | 180 | 18 | 39 | M | 210 |
| 9 | 22 | F | 160 | 19 | 50 | F | 260 |
| 10 | 44 | M | 155 | 20 | 27 | M | 175 |
2.2. Construction of the Atlas and Statistical Models
2.2.1. Super-resolution Volume Reconstruction
It is desirable to start atlas creation from a high-resolution, isotropic 3D MR image volume. However, it was not possible to acquire such a volume directly because the image acquisition time would be longer than most subjects can refrain from swallowing. Therefore, we acquired three separate orthogonal image stacks—axial, sagittal, and coronal—and combined them using a maximum a posteriori-Markov random field (MAP-MRF) super-resolution technique [12]. The resulting volume has an improved signal-to-noise ratio (SNR) and an isotropic spatial resolution (0.94 mm × 0.94 mm × 0.94 mm); and because it uses edge-preserving regularization, the resulting volumes have clearer anatomical details than the source images themselves. A detailed description of the algorithm is found in [12].
2.2.2. Preprocessing
Once super-resolution volumes were generated for each subject, we performed several preprocessing steps prior to atlas construction. These steps include: 1) removing the MRI wraparound artefact, 2) spatially bounding the volumes so that each volume has the same anatomical features, and 3) intensity bias correction to reduce the impact of the intensity inhomogeneities [13]. These steps improve the registration performance in atlas construction (see Figs. 2A vs. 2B).
2.2.1. Groupwise Registration and Atlas Construction
Aligning and combining image data from a group of individuals into a common space allows us to build a model of average vocal tract anatomy, to obtain a volume with increased SNR, and to investigate similarities and differences across subjects [10]. The atlas building procedure involves a groupwise affine registration as an initial transformation followed by a groupwise deformable registration using the Symmetric Normalization (SyN) algorithm [13] to further register the image volumes. We obtained the final atlas by averaging all the registered volumes.
We made use of an unbiased groupwise registration using the SyN algorithm with a cross-correlation (CC) similarity metric [10, 13]. This method has been demonstrated to be among the most accurate intensity-based normalization methods [14]. Let be a group of N subject volumes (N=20 in this work) to be registered using the SyN algorithm and IR be the atlas that is sought. The objective of the atlas construction is to find IR that minimizes
where E is an energy between the atlas and each image. The energy is evaluated by first finding a diffeomorphic registration between the two images and then evaluating a cost criterion based on the integrated CC metric and the two velocity fields describing the symmetric diffeomorphism.
The CC metric is evaluated locally at each spatial position x and is given by [15]
where the local means μIR(x) and μIi(x) are computed in a 5×5×5 square volume around x and the summations are also carried out over the same local volume. The use of a CC metric is particularly important to cope with the inter-subject image intensity inhomogeneity. Next, two optimal diffeomorphic transformations and for each image Ii are computed by minimizing the following functional:
The velocity fields v1i(x, t) and v2i(x, t) are associated with the diffeomorphic transformations as described in [15]. The pairwise energy function is then defined as . The algorithm iterates between finding a new atlas image IR and finding optimal diffeomorphisms and until convergence to a minimum total energy is achieved.
2.2.3. Manual Tongue and Muscle Segmentation
For purposes of validation, a manual segmentation of the tongue surface was performed on each of the twenty subjects and the atlas (the “average” image volume) by one expert observer. With an in-house software based on Lee et al. [16], initial tongue segmentations were obtained and these segmentations were then refined manually using ITK-SNAP software [17]. This approach reduces segmentation time significantly and helps the observer to delineate the tongue regions consistently. These manual segmentations serve as ground truths to validate the mappings used for the atlas building.
In addition, a variety of vocal tract structures (16 in total) in the atlas were manually segmented, including (1) the hyoid bone, (2) the palate and maxillary bone, (3) the mandibular bone, (4) the tongue, (5) the larynx, (6) the soft palate and hard palate mucosa, (7) the geniohyoid muscle, (8) the genioglossus muscle, (9) the hyoglossus muscle, (10) the digastric muscle, (11) the mylohyoid muscle, (12) the styloglossus muscle, (13) the inferior longitudinal muscle, (14) the superior longitudinal muscle, (15) the sublingual gland, and (16) the submandibular gland (see Figs. 3, 4, and 5). These manual segmentations allow construction of the statistical shape model so that it includes these additional muscles and structures in addition to the tongue surface.
Figure 3.
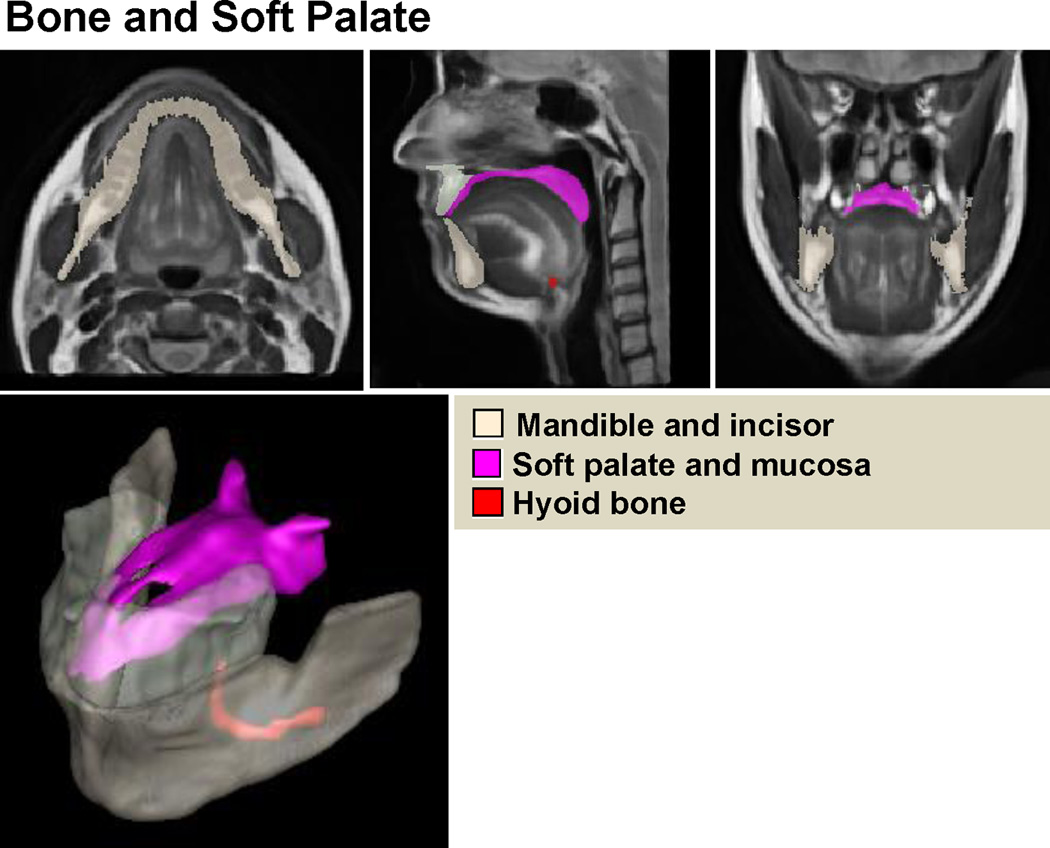
High-resolution vocal tract atlas overlaid with manual segmentation of bone and soft palate. The atlas allows definition of the standard bone and soft palate structures.
Figure 4.
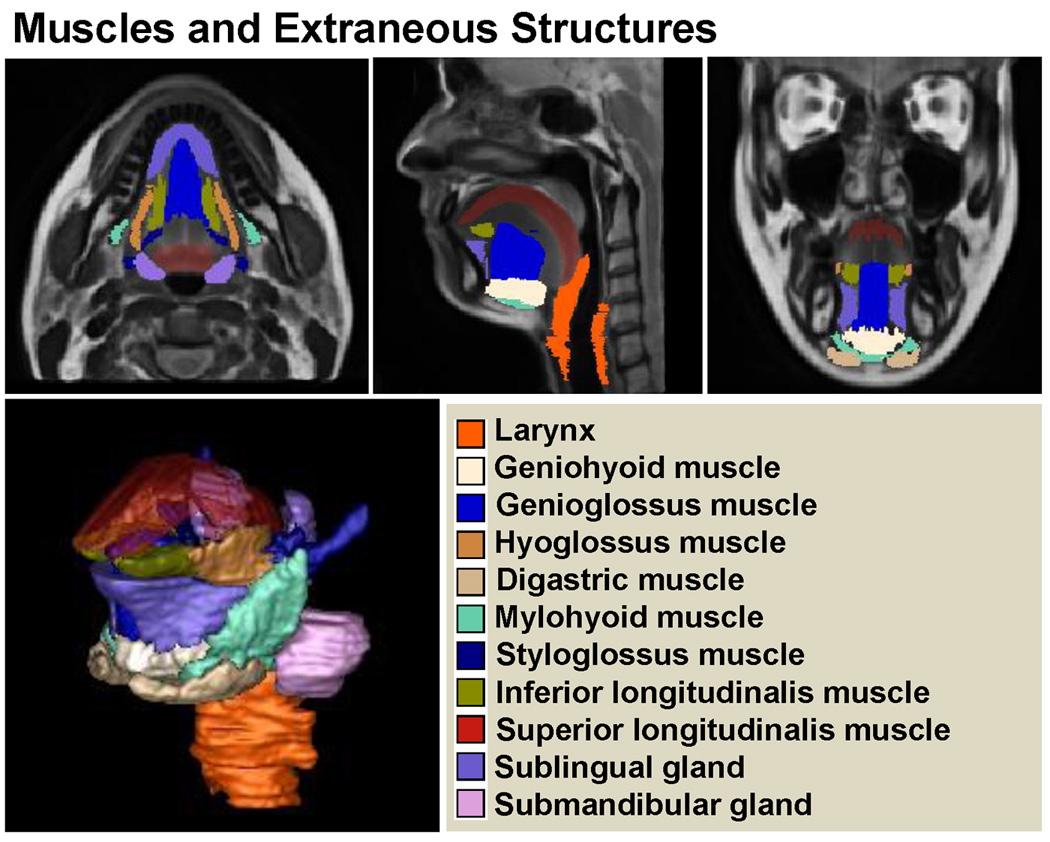
High-resolution vocal tract atlas overlaid with manual segmentation of muscles and extraneous structures. The atlas allows definition of the standard muscular structures.
Figure 5.
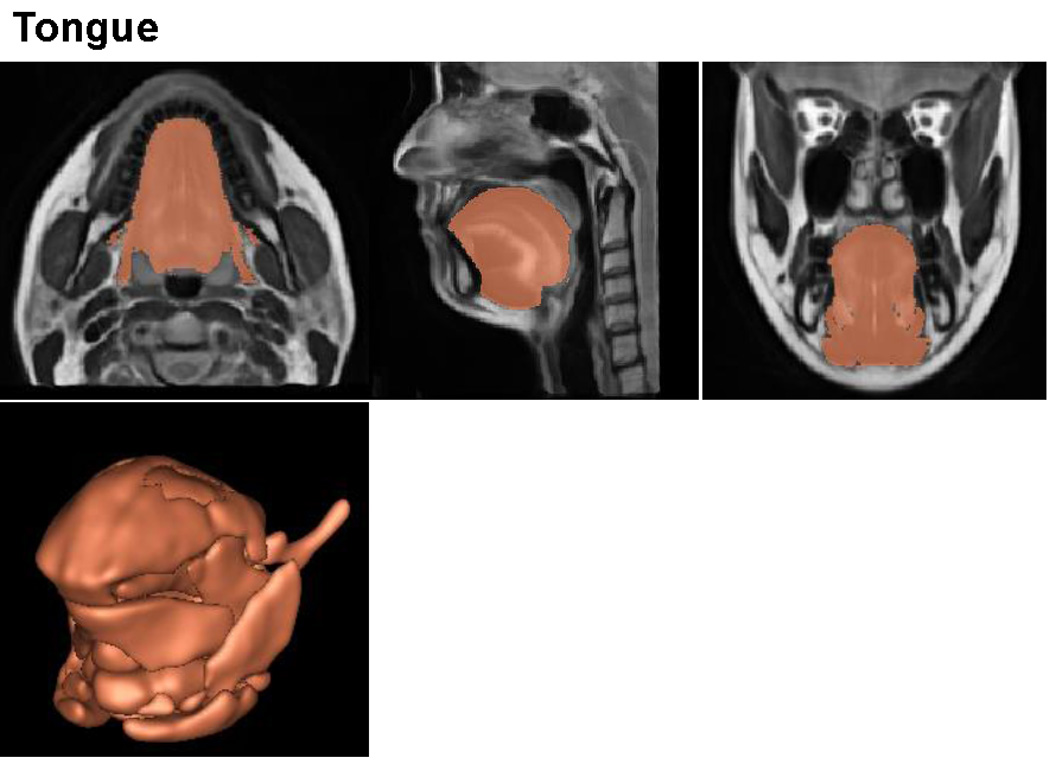
High-resolution vocal tract atlas overlaid with manual segmentation of the tongue. The atlas allows definition of the standard tongue.
2.2.4. Statistical Models Using PCA
In this section, we describe a PCA method to create the statistical models in atlas space. PCA is used to analyze the variability of the entire volume (see Fig. 6 below) as well as shape variability of the segmented structures within the volume (see Fig. 7 below). We apply PCA to the deformation fields required to map individual anatomies to the atlas, which provides second-order statistics by finding the principal modes of the sample covariance on the deformation fields. The PCA used here is a global PCA for the entire anatomy, which is different from multi-object statistical models such as in [18–21]. Let us first denote the deformation fields, {h1, h2, …, hN}, N = 20, defined by the mappings from an individual anatomy to the atlas. In this work, we are particularly interested in two deformations. They are Deformation I, which is a global transformation that represents the homogeneous plus local volume changes achieved by removing the rigid transformations from the cascade of the affine transformation and local deformations (SyN), and Deformation II, which represents the local deformations provided by SyN only. The average deformation field at every point is then given by
where N=20. The sample covariance of the deformation fields is constructed by
where the mean offset map, hi−mh, is a column vector.
Figure 6.
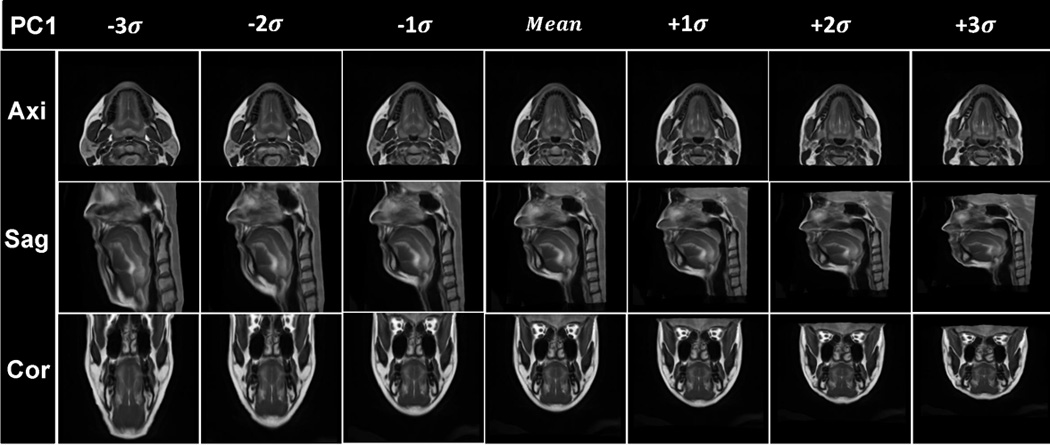
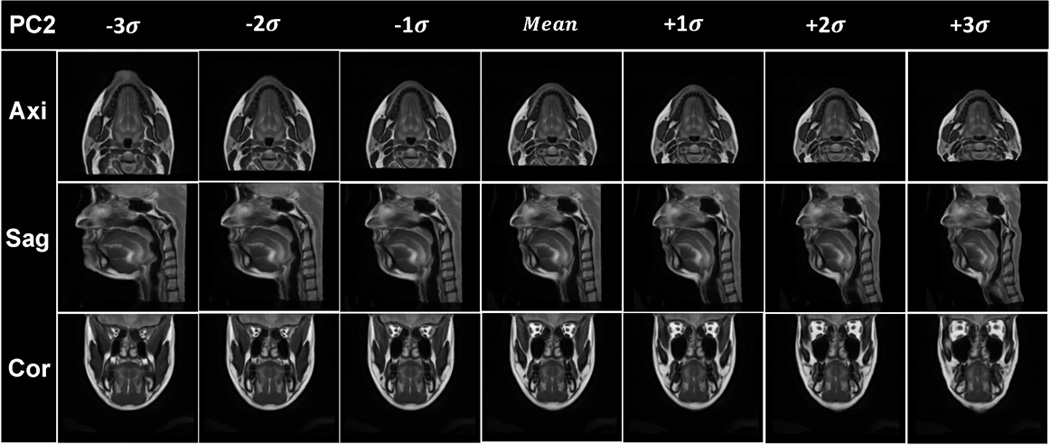
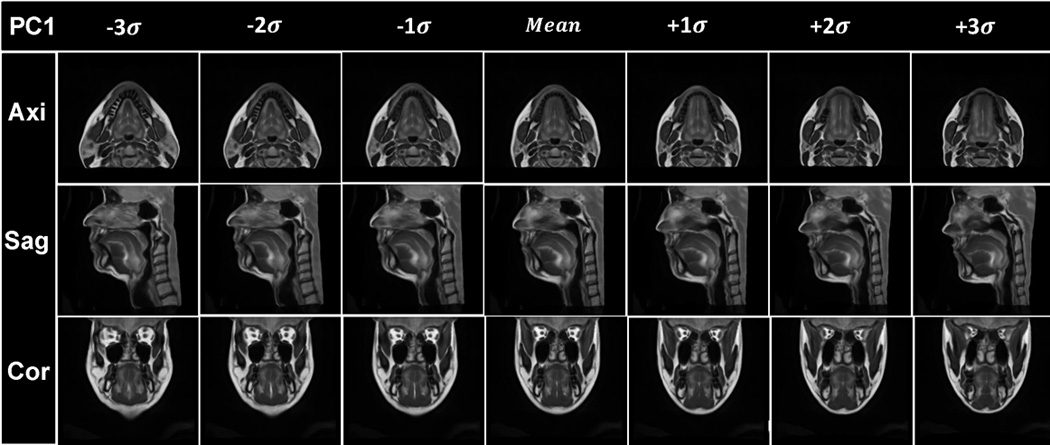
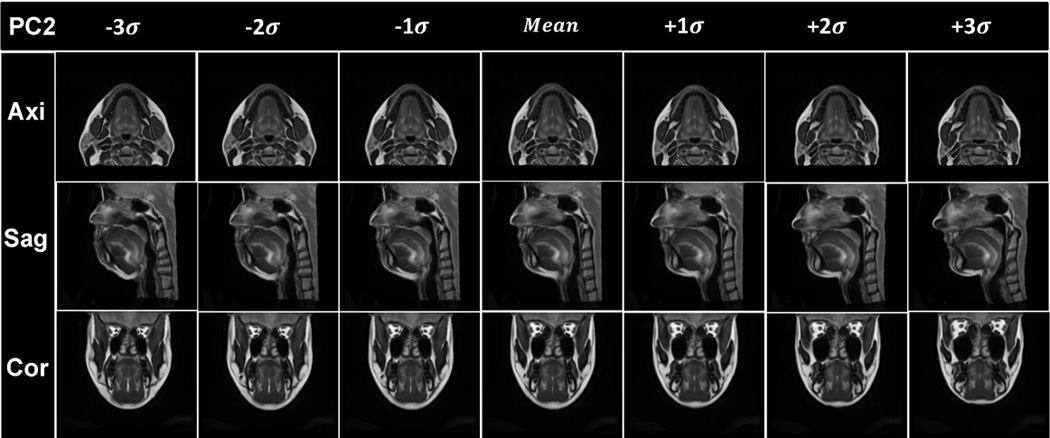
Visualization of the results of PCA analysis. The vocal tract atlas was warped by the first two PC’s using Deformations I and II. (a) PC1 of Deformation I shows variance in vertical length changes. (b) Atlas warping by PC2 of Deformation I shows variance in AP width. (c) Atlas warping by PC1 of Deformation II shows variance in the LR width. (d) Atlas warping by PC2 of Deformation II represents vertical differences in the posterior oral cavity. Deformation I represents the homogeneous plus local volume changes by removing the rigid transformations from the cascade of the affine transformation and local deformations (SyN) and Deformation II represents the local deformations only (SyN).
Figure 7.
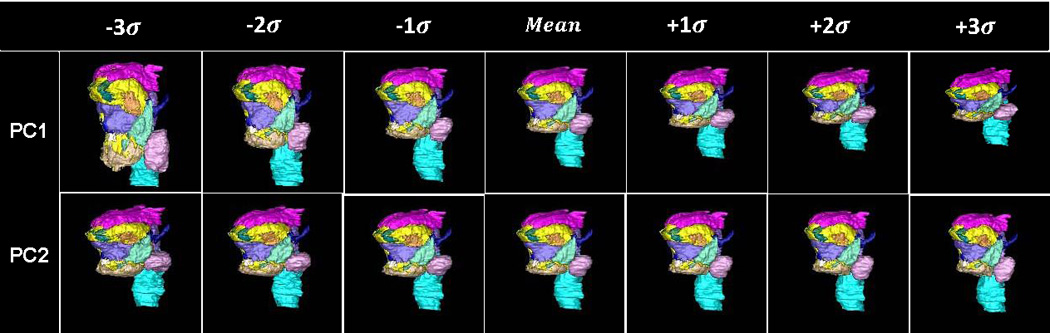
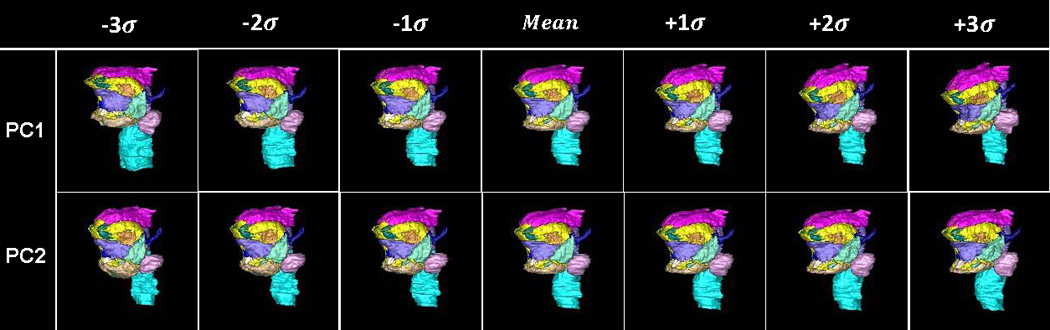
The surface renderings of segmented structures of the vocal tract atlas. (a) The surface of the substructures of the vocal tract atlas was warped by PC1 and PC2 of Deformation I, respectively. (b) The surface of the substructures of the vocal tract atlas was warped by the PC1 and PC2 of Deformation II, respectively.
Using singular value decomposition, the covariance matrix is decomposed into the set of orthogonal modes of variation and a diagonal matrix of corresponding singular values. The principal components (PC) of variation are the eigenvectors ei of the covariance matrix C, and a deformation field h can be approximated using the first k eigenvectors ei corresponding to the largest eigenvalues λi by
where ωi ∈ ℝ are weights for the different PCs. The instances of the atlas and segmented structures can then be computed by projection onto the PCs as given by
where V and M denote the atlas and the segmented structures in the atlas space and VS and MS represent the volume and shapes with different modes of variation. The mean shape, the PCs, and the associated eigenvalues compose the statistical model.
3. Evaluations and Applications
In this section, we describe a series of experiments to assess the vocal tract atlas and statistical models both qualitatively and quantitatively and to demonstrate potential applications of the atlas. The experiments include PCA analysis on the deformation fields, segmentation of the tongue using transformations used in building the atlas, and deformation-based voxel analysis.
3.1. PCA analysis
PCA’s are computed on two deformations (Deformations I and II) of the data to assess the contribution of homogeneous and local deformations. We create statistical models of the volume and shapes of different muscles using the two deformations and describe the effect of the PCs computed using each deformation.
3.2. Segmentation of the Tongue Surface Using the Atlas
We perform leave-one-out experiments using twenty-one subjects by creating twenty-one atlases, each made up of twenty subjects. We then use an atlas-based segmentation to delineate the tongue of each excluded subject in all twenty-one experiments. We use the same registration method used in our atlas building, which includes an affine registration, followed by a deformable registration using the SyN algorithm with the CC similarity measure. Two measurements are used to gauge the accuracy of the segmentation results. The first measurement is the dice similarity coefficient (DSC) defined as
where AS and Ag are the areas enclosed by the segmented contour registered to the excluded subject and the manual segmentation in the excluded subject, respectively. The second measurement is the intraclass correlation coefficient (ICC) [22, 23], demonstrating the reliability of the volume measurements obtained from different methods. The mean and standard deviation of the measurements are computed to evaluate the variation of the segmentation results. In our study, statistical significance is evaluated using a paired t-test (p<0.05).
3.3. Deformation-based Voxel Analysis
The deformation fields (e.g., Deformations I and II) obtained in atlas construction carry information about global differences in the local volume when the Jacobian of the deformation field is applied. In addition, the deformation fields reveal the degree and location where the tongue volume varies during the atlas building. In this analysis, we derive the volume changes using Deformations I and II. We first compute the Jacobian of Deformation I to characterize volume changes that indicate anatomical changes. In order to observe volume changes in each muscle, a mask obtained from the muscle segmentation is used to restrict the muscle region in the atlas space. The Jacobian of the deformation field at each voxel is defined as follows:
The Jacobian determinant is then computed to calculate the volume changes at each voxel. The volume changes [24] in each muscle is given by
where |J(x)| denotes the Jacobian determinant and M(x):Ω ⊂ ℝ3 → (0,1) denotes the segmented mask region defined in the atlas space Ω.
4. Results
The computations were performed on a Dell R415 server running Fedora Linux 14, the system has 12 processors with a clock speed of 2.73GHz and 47.25GB of RAM. The computation time to create the atlas was 196 hours. The final vocal tract atlas and manual tongue and muscle segmentations are illustrated in Figs. 3, 4, and 5. The vocal tract atlas allows us to define the standard anatomy. The bone structure and palate are shown in Fig. 3, various muscles and other structures are shown in Fig. 4, and the tongue is shown in Fig. 5, respectively.
4.1. PCA Analysis
Figs. 6(a) and 6(b) show the statistical volume atlas using PC1 and PC2 of Deformation I, respectively. PC1 and PC2 appear to provide horizontal and vertical stretch and represent 63% and 19% of the statistical variability, respectively. This indicates that the greatest transformation between subjects is affine (i.e., global stretching, shearing, and scaling), accounting for 82% of the variance.
Figs. 6(c) and 6(d) show the statistical volume atlas using PC1 and PC2, respectively, of Deformation II. PC1 appears to represent local changes in the distribution of tissue in the lower half of the face and tongue so that positive PC1 shows a thin face from left-to-right and front-to-back and is elongated vertically, whereas negative PC1 shows a wide face front-to-back and left-to-right and is shortened vertically. These local effects account for 31% of the variance. In particular, for PC2, the most salient effect is the change from a very sloped chin (see images in Fig. 6(d), column −3σ) to a more horizontal chin (see images in Fig. 6(d), column +3σ).
Figs. 7(a) and 7(b) depict the 3D statistical shape atlas using PC1 and PC2 of Deformations I and II, respectively, applied to the segmented structures in the atlas. The shape changes are the same as those shown in Fig. 6, but here one can better appreciate the 3D characteristics of the shape changes and also observe a differentiation of effects on separate parts of the anatomy. Table 2 lists the variances accounting for each PC of Deformations I and II.
Table 2.
PC loadings for each deformation
| PC | Deformation I | Deformation II |
|---|---|---|
| PC1 | 63% | 21% |
| PC2 | 19% | 11% |
| PC3 | 7% | 9% |
| PC4 | 5% | 8% |
| PC5 | 1% | 7% |
4.2. Segmentation of the Tongue Surface Using the Atlas
In this section, we describe the results of tongue segmentation using an atlas-based segmentation. Fig. 8 depicts an example of manual segmentation of the tongue (first row) and the segmentation obtained using the atlas-based segmentation (second row). Fig. 9 shows an example of the tongue volume comparison between the atlas-based segmentation and manual segmentation. The mean values of the tongue volume calculated from twenty-one subjects by using the atlas-based segmentation and manual segmentation of the subject were 118.1±30.9 ml and 120.9±29.3 ml, respectively. There was excellent correlation between the two measurements (r=0.9, p=0.23), with a best-fit linear relationship of y=1.0207x. The ICC was found to be 0.85.
Figure 8.
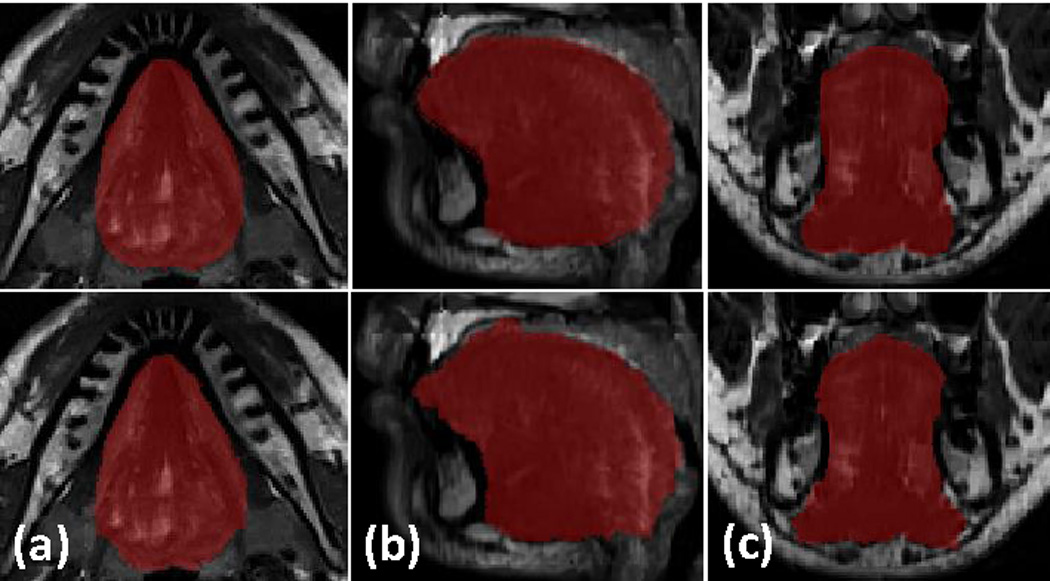
The first row shows one resulting volume from super-resolution volume reconstruction overlaid with a tongue manual segmentation. The second row shows segmentation using the atlas and the inverse mapping. Axial, sagittal, and coronal directions are shown in (a), (b), and (c), respectively. The DSC is 0.938 in this case.
Figure 9.
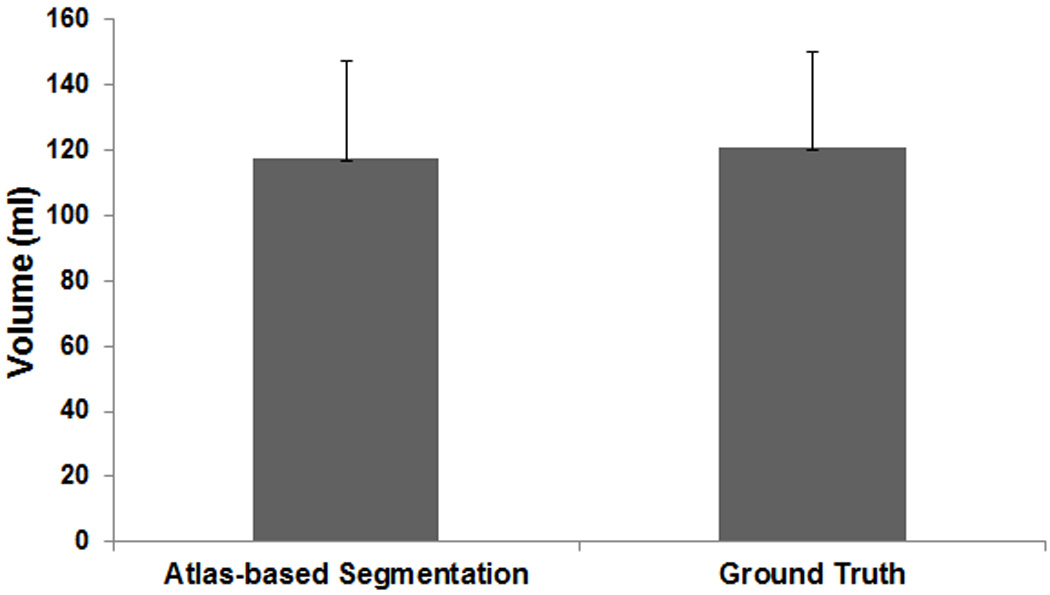
A plot of average volume of the original tongue region and segmentation by using the atlas and the inverse mapping.
A Bland-Altman analysis for manual segmentation and segmentation using the atlas is shown in Fig. 10; this indicates a satisfactory result with only one outlier. A plot of all DSC results is provided in Fig. 11. Across all twenty-one atlases, the mean and standard deviation of the DSC is 0.91 and 0.03, respectively. We found that two subjects had low DSCs (0.83 and 0.84) and their heads were quite rotated backward and forward. Rigid body registration, followed by deformable registration improved the dice coefficient to 0.84 and 0.86, respectively. The above results show that the atlas-based segmentation of the whole tongue from the atlas to the individuals accurately characterizes the geometry and volume provided by manual delineation.
Figure 10.
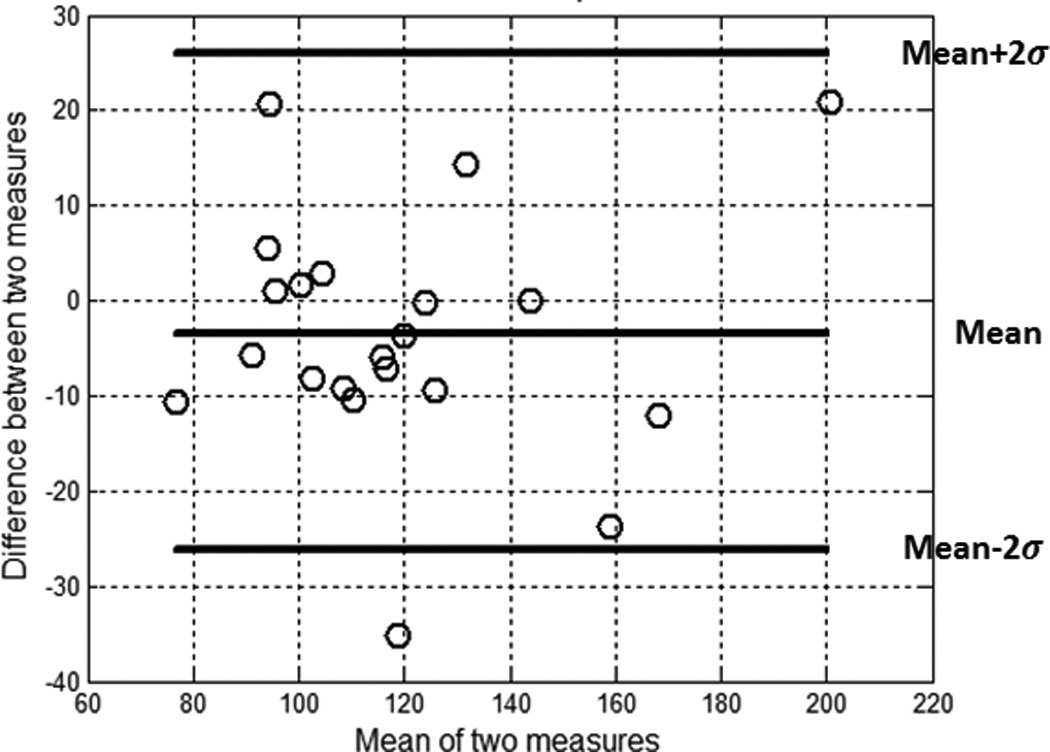
The Bland-Altman plot characterizing agreement between manual segmentation and segmentation using atlas.
Figure 11.
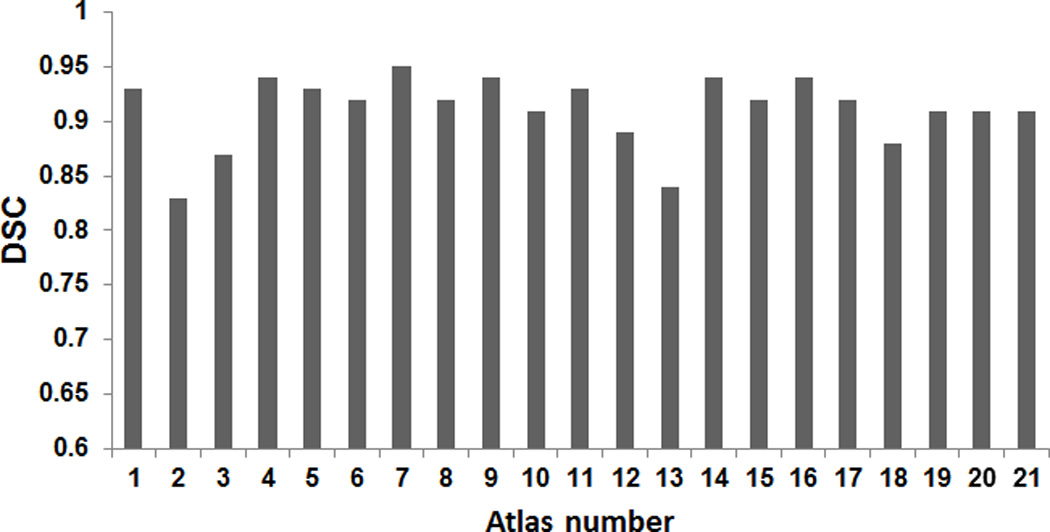
The dice similarity coefficient for all subjects used in the atlas building. The mean and standard deviation is 0.92±0.05.
4.3. Deformation-based Voxel Analysis
We present here a deformation-based voxel analysis including the Jacobian map and volume ratio using the Jacobian determinant. The 16 manually segmented structures shown in Figs. 3, 4, and 5 identify the regions of interest in the tongue and vocal tract. By analysing the Jacobian map derived from the deformation fields used in atlas-building or a pair-wise registration between the atlas and a subject, one can observe the volume and anatomical differences of each muscle. Fig. 12 shows an example of the log Jacobian map of Deformation I of the whole tongue in one subject. The red and blue colors indicate tissue expansion and shrinkage, respectively, relative to the atlas. Figs. 13(a) and 13(b) illustrate the statistics of the volume ratio for each muscle between the twenty subjects and the atlas using Deformations I and II, respectively. The mean and standard deviation of the volume ratios for the combination of the muscle volumes in the whole tongue using Deformations I and II is 1.0±0.2 and 1.0±0.1, respectively. The volume ratios for each of the 16 structures varied more, indicating that the distribution of muscle volume is not uniform across subjects.
Figure 12.
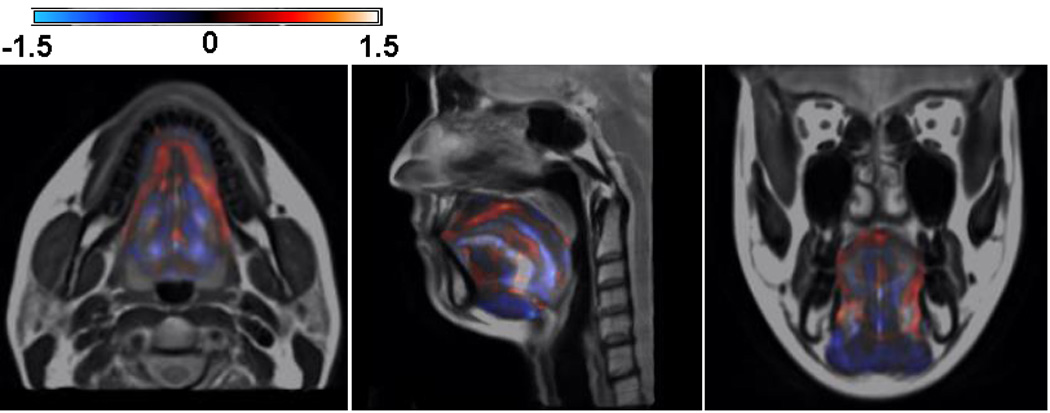
Illustration of the log Jacobian map of Deformation I. The blue shows the tissue expansion and the red indicates the volume shrinkage relative to the atlas. It is clearly shown that log Jacobian map exhibits the volume and anatomical changes of each muscle during the atlas building.
Figure 13.
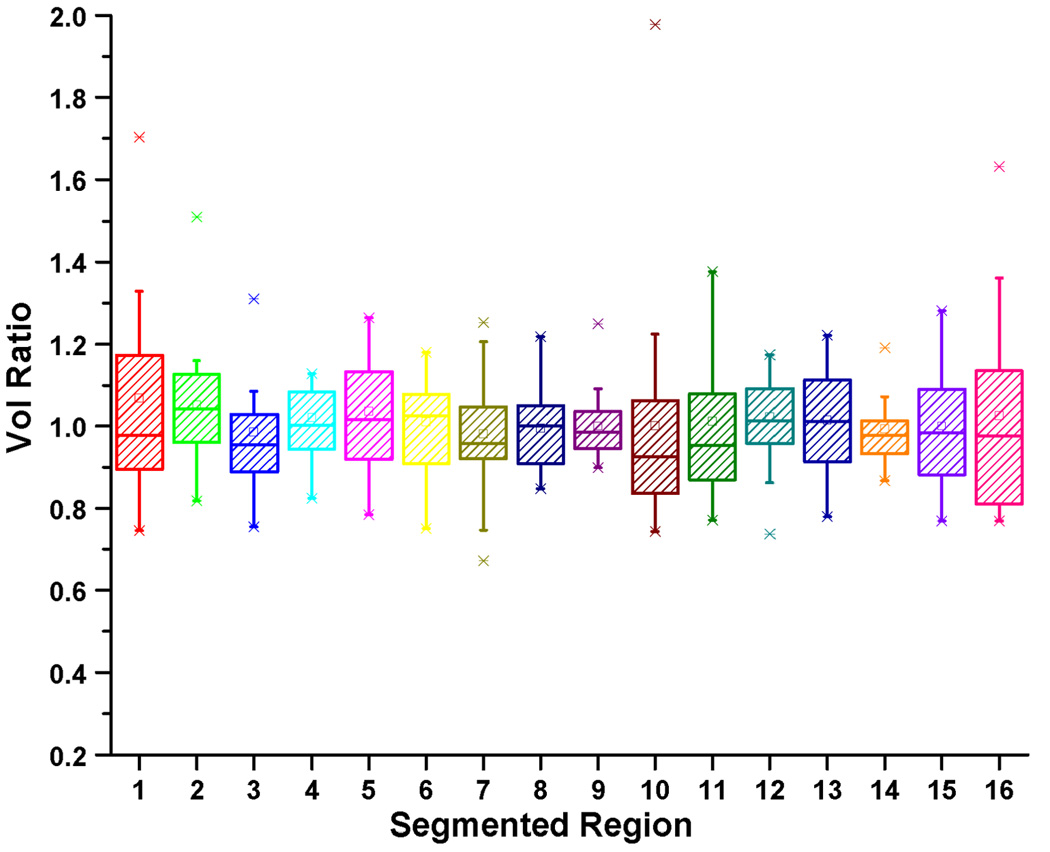
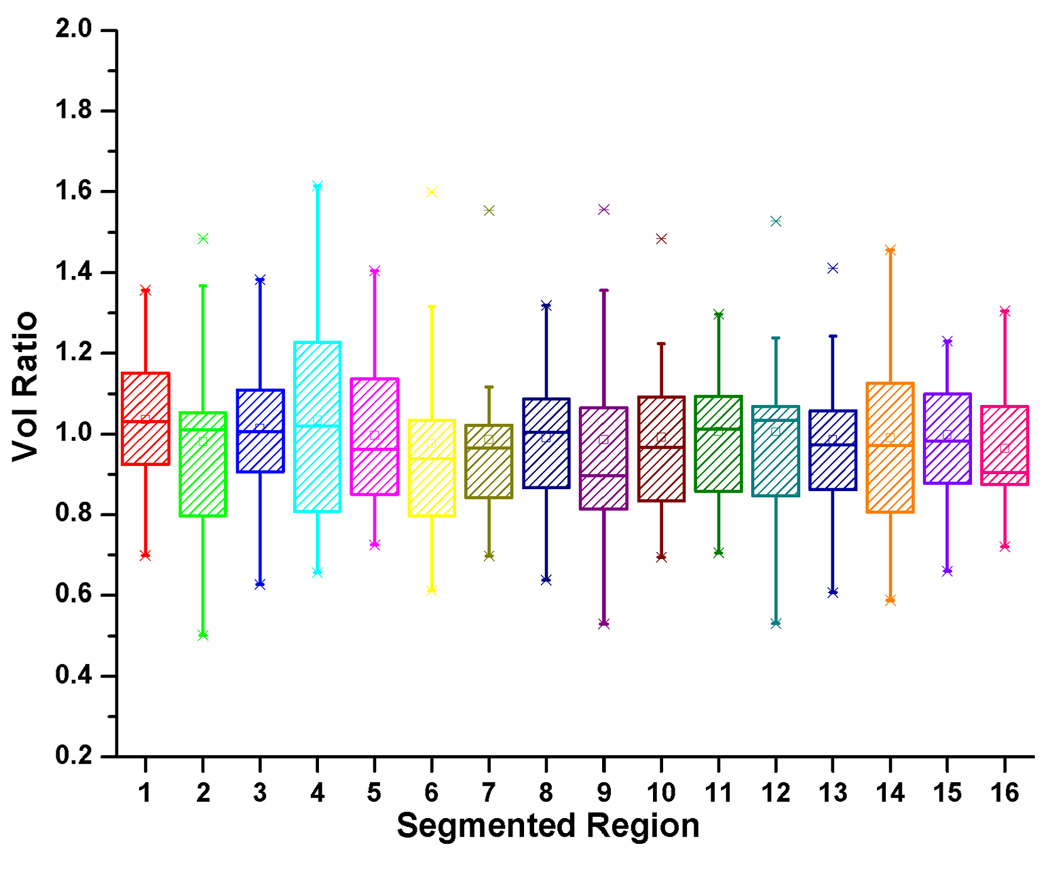
The box-and-whisker diagram for the volume ratio in each muscle using (a) Deformation I and (b) Deformation II. Please note that the combined volume ratio was preserved across subjects.
5. Discussion
An anatomical vocal tract atlas was created using a symmetric and diffeomorphic groupwise registration from the super-resolution volumes of twenty subjects. PCA was used to construct statistical models for the whole volume and for the segmented vocal tract structures.
There are several important characteristics and potential uses of this vocal tract atlas. First, the atlas represents an average, not a specific subject, and therefore any use of it is not biased by a specific individual's anatomical features. Second, the entire volume and all the segmented structures are normalized at the same time, saving computation time in this and future atlases. Third, information from the atlas can be incorporated into the segmentation or registration process as statistical prior information. Fourth, the atlas allows the capture of anatomical variability by providing a coordinate space for motion analyses using PCA or cluster analysis of velocity fields as in [25]. In addition, statistical analyses can be applied to individual structures or the entire volume.
The transformations needed to deform subjects to the atlas were analysed by PCA. Affine transformation is adequate when the difference between images involves global rotation, scaling, and shearing. Deformable registration is used to capture the local shearing or scaling as shown in Figs. 6(c) and 6(d). In the present study, Deformation I was sufficient to capture the most of the variance between subjects as these are all normal adults. However, when creating atlases or transforming other populations into this atlas, such as patients with structural malformations or children, then the local deformation may yield a larger percentage of variance.
For different reasons, the literature indicates the lack of consensus on how to measure volume differences across subjects. For example, there has been some literature questioning the relative sizes of muscles volume and relative sizes of muscles consistent across subjects with different total muscle volumes [26]. In their work, these questions were addressed by measuring muscle volumes in healthy subjects from MRI. In the present work, however, deformation fields were used to measure volume differences of muscles and extraneous structures relative to the atlas.
We see a few directions for improvement in the present work. First, we will characterize the muscle interaction and function using both muscle segmentation and motion information obtained from the speech data. Second, we will compare muscle volume differences from (1) the atlas results, (2) those results transformed back into subject’s space, and (3) unnormalized results to further determine the quality of the atlas results.
Following from analogous work in the brain [27, 28], we anticipate several applications of the atlas to the analysis of tongue image data. The atlas may be used to register functional data from different subjects to a common anatomical space, thereby allowing group-level inferences [29]. Individual differences in tongue and muscle structures as assessed by different imaging modalities, such as diffusion tensor imaging, could be related to normal variations of behaviour or one of many disease states. Thus far, it has been difficult to accurately characterize the relationship between structure (i.e., anatomy) and function (i.e., speech) due to the varied sizes and shapes of the tongue and its muscles. With future studies, this atlas has the potential to accurately inform how the interactions among different muscles impact overall speech production, opening a new window for the investigation of dynamic speech production and speech disorders.
6. Conclusion
In this work, we presented a vocal tract atlas construction method and statistical models of vocal tract variation from twenty normal subjects. Super-resolution volumes were constructed and unbiased groupwise registration was used to create the atlas and symmetric and diffeomorphic deformation fields from the atlas to the subjects. Principal component analysis applied to the deformation fields provided a statistical model of anatomic variation. PC1 and PC2 of Deformations I and II accounted for 82% and 33% of the variance, respectively. The single atlas-based segmentation of the whole tongue from the atlas to the individuals yielded accurate characterization of the geometry and volume (the mean of DSC was 0.9). This vocal tract provides an integrative framework in which individual subjects can be mapped and compared, thus opening new vistas for structural and functional studies of the vocal tract.
Acknowledgments
This work was supported by NIH grants: R01CA133015, K99/R00DC009279, and K99DC012575. We would like to thank Dr. Ian Stavness for valuable discussion on the volume ratio of the muscles.
Contributor Information
Jonghye Woo, Department of Neural and Pain Sciences, University of Maryland, Baltimore, MD, 21201, USA. telephone: 410-706-1269, fax: 410-706-0865, jschant@gmail.com.
Junghoon Lee, Radiation Oncology and Molecular Radiation Sciences, Johns Hopkins University, School of Medicine, Baltimore MD 21231, USA, telephone: 410-502-1477, fax: 410-516-5566, junghoon@jhu.edu.
Emi Z. Murano, Otolaryngology-Head and Neck Surgery, Johns Hopkins University, School of Medicine, Baltimore, MD, 21218, telephone: 410-706-780, emurano1@jhmi.edu
Fangxu Xing, Electrical and Computer Engineering, Johns Hopkins University, Baltimore, MD 21218; telephone: 410-516-5192, fxing1@jhu.edu.
Meena Al-Talib, Department of Neural and Pain Science, University of Maryland, Baltimore, MD, 21201, USA. telephone: 410-706-1269, fax: 410-706-0865, maltali10@gmail.com.
Maureen Stone, Department of Neural and Pain Science, Department of Orthodontics, University of Maryland, Baltimore, MD, USA. telephone: 410-706-1269, fax: 410-706-0865, mstone@umaryland.edu.
Jerry L. Prince, Electrical and Computer Engineering, Johns Hopkins University, Baltimore, MD 21218; telephone: 410-516-5192, prince@jhu.edu
References
- 1.Kier WM, Smith KK. Tongues, tentacles and trunks: the biomechanics of movement in muscular-hydrostats. Zoological Journal of the Linnean Society. 1985;83:307–324. [Google Scholar]
- 2.Gaige TA, Benner T, Wang R, Wedeen VJ, Gilbert RJ. Three dimensional myoarchitecture of the human tongue determined in vivo by diffusion tensor imaging with tractography. J Magn Reson Imaging. 2007 Sep;26:654–661. doi: 10.1002/jmri.21022. [DOI] [PubMed] [Google Scholar]
- 3.Narayanan S, Nayak K, Lee S, Sethy A, Byrd D. An approach to real-time magnetic resonance imaging for speech production. J Acoust Soc Am. 2004 Apr;115:1771–1776. doi: 10.1121/1.1652588. [DOI] [PubMed] [Google Scholar]
- 4.Parthasarathy V, Prince JL, Stone M, Murano EZ, Nessaiver M. Measuring tongue motion from tagged cine-MRI using harmonic phase (HARP) processing. J Acoust Soc Am. 2007 Jan;121:491–504. doi: 10.1121/1.2363926. [DOI] [PubMed] [Google Scholar]
- 5.Stone M, Davis EP, Douglas AS, NessAiver M, Gullapalli R, Levine WS, Lundberg A. Modeling the motion of the internal tongue from tagged cine-MRI images. J Acoust Soc Am. 2001 Jun;109:2974–2982. doi: 10.1121/1.1344163. [DOI] [PubMed] [Google Scholar]
- 6.Narayanan SS, Alwan AA, Haker K. An articulatory study of fricative consonants using magnetic resonance imaging. The Journal of the Acoustical Society of America. 1995;98:1325–1347. [Google Scholar]
- 7.Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC, Snyder AZ. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. Neuroimage. 2004 Oct;23:724–738. doi: 10.1016/j.neuroimage.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Thompson PM, Woods RP, Mega MS, Toga AW. Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain. Hum Brain Mapp. 2000 Feb;9:81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toga AW, Thompson PM, Mori S, Amunts K, Zilles K. Towards multimodal atlases of the human brain. Nat Rev Neurosci. 2006 Dec;7:952–966. doi: 10.1038/nrn2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yushkevich PA, Avants BB, Pluta J, Das S, Minkoff D, Mechanic-Hamilton D, Glynn S, Pickup S, Liu W, Gee JC, Grossman M, Detre JA. A high-resolution computational atlas of the human hippocampus from postmortem magnetic resonance imaging at 9.4 T. Neuroimage. 2009 Jan 15;44:385–398. doi: 10.1016/j.neuroimage.2008.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi F, Yap PT, Wu G, Jia H, Gilmore JH, Lin W, Shen D. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One. 2011;6:e18746. doi: 10.1371/journal.pone.0018746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo J, Murano EZ, Stone M, Prince JL. Reconstruction of high-resolution tongue volumes from MRI. IEEE Trans Biomed Eng. 2012 Dec;59:3511–3524. doi: 10.1109/TBME.2012.2218246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008 Feb;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009 Jul 1;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011 Feb 1;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Woo J, Xing F, Murano EZ, Stone M, Prince JL. Semi-Automatic Segmentation of Tongue for 3D Motion Analysis with Dynamic MRI; IEEE International Symposium on Biomedical Imaging (ISBI); San Francisco. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yushkevich PA, Piven J, Cody H, Ho S, Gee JC, Gerig G. User-guided level set segmentation of anatomical structures with ITK-SNAP. Insight Jounral. 2005;1 [Google Scholar]
- 18.Cerrolaza JJ, Villanueva A, Cabeza R. Hierarchical statistical shape models of multiobject anatomical structures: application to brain MRI. IEEE Trans Med Imaging. 2012 Mar;31:713–724. doi: 10.1109/TMI.2011.2175940. [DOI] [PubMed] [Google Scholar]
- 19.Duta N, Sonka M. Segmentation and interpretation of MR brain images: an improved active shape model. IEEE Trans Med Imaging. 1998 Dec;17:1049–1062. doi: 10.1109/42.746716. [DOI] [PubMed] [Google Scholar]
- 20.Lu C, Pizer SM, Joshi S, Jeong J-Y. Statistical multi-object shape models. International Journal of Computer Vision. 2007;75:387–404. [Google Scholar]
- 21.Tsai A, Wells W, Tempany C, Grimson E, Willsky A. Coupled multi-shape model and mutual information for medical image segmentation. Inf Process Med Imaging. 2003 Jul;18:185–197. doi: 10.1007/978-3-540-45087-0_16. [DOI] [PubMed] [Google Scholar]
- 22.Bogovic JA, Jedynak B, Rigg R, Du A, Landman BA, Prince JL, Ying SH. Approaching expert results using a hierarchical cerebellum parcellation protocol for multiple inexpert human raters. Neuroimage. 2013;64:616–629. doi: 10.1016/j.neuroimage.2012.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGraw KO, Wong SP. Forming inferences about some intraclass correlation coefficients. Psychological methods. 1996;1:30–46. [Google Scholar]
- 24.Woo J, Slomka P, Nakazato R, Balaji T, Germano G, Berman D, Dey D. SPIE Medical Imaging. San Diego: 2012. Feasibility of Determining Myocardial Transient Ischemic Dilation from Cardiac CT by Automated Stress/Rest Registration. [Google Scholar]
- 25.Stone M, Liu X, Chen H, Prince JL. A preliminary application of principal components and cluster analysis to internal tongue deformation patterns. Comput Methods Biomech Biomed Engin. 2010 Aug;13:493–503. doi: 10.1080/10255842.2010.484809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holzbaur KR, Murray WM, Gold GE, Delp SL. Upper limb muscle volumes in adult subjects. J Biomech. 2007;40:742–749. doi: 10.1016/j.jbiomech.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Gholipour A, Akhondi-Asl A, Estroff JA, Warfield SK. Multi-atlas multi-shape segmentation of fetal brain MRI for volumetric and morphometric analysis of ventriculomegaly. NeuroImage. 2012 Apr 15;60:1819–1831. doi: 10.1016/j.neuroimage.2012.01.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasan KM, Walimuni IS, Abid H, Datta S, Wolinsky JS, Narayana PA. Human brain atlas-based multimodal MRI analysis of volumetry, diffusimetry, relaxometry and lesion distribution in multiple sclerosis patients and healthy adult controls: implications for understanding the pathogenesis of multiple sclerosis and consolidation of quantitative MRI results in MS. J Neurol Sci. 2012 Feb 15;313:99–109. doi: 10.1016/j.jns.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datta R, Lee J, Duda J, Avants BB, Vite CH, Tseng B, Gee JC, Aguirre GD, Aguirre GK. A digital atlas of the dog brain. PLoS One. 2012;7:e52140. doi: 10.1371/journal.pone.0052140. [DOI] [PMC free article] [PubMed] [Google Scholar]


