Abstract
Clear cell papillary renal cell carcinoma (ccpRCC) and renal angiomyoadenomatous tumor (RAT) share morphologic similarities with clear cell (ccRCC) and papillary renal cell carcinoma (pRCC). It is a matter of controversy whether their morphologic, immunophenotypic and molecular features allow the definition of a separate renal carcinoma entity. The aim of our project was to investigate specific renal immunohistochemical biomarkers involved in the hypoxia-inducible factor pathway and mutations in the VHL gene to clarify the relationship between ccpRCC and RAT. We investigated 28 ccpRCC and 9 RAT samples by immunohistochemistry using 25 markers. VHL gene mutations and allele losses were investigated by Sanger sequencing and fluorescence in situ hybridization (FISH). Clinical follow-up data were obtained for a subset of the patients. No tumor recurrence or tumor-related death was observed in any of the patients. Immunohistochemistry and molecular analyses led to the reclassification of three tumors as ccRCC and TFE3 translocation carcinomas. The immunohistochemical profile of ccpRCC and RAT samples was very similar but not identical, differing from both ccRCC and pRCC. Especially, the parafibromin and hKIM-1 expression exhibited differences in ccpRCC/RAT compared with ccRCC and pRCC. Genetic analysis revealed VHL mutations in 2/27 (7%) and 1/7 (14%) ccpRCC and RAT samples, respectively. FISH analysis disclosed a 3p loss in 2/20 (10 %) ccpRCC samples. ccpRCC and RAT have a specific morphologic and immunohistochemical profile but they share similarities with the more aggressive renal tumors. Based on our results, we regard ccpRCC/RAT as a distinct entity of renal cell carcinomas.
Keywords: kidney, clear cell papillary renal cell cancer, cytokeratin 7, VHL, renal angiomyoadenomatous tumor, clear cell renal cell carcinoma
Introduction
Clear cell papillary renal cell carcinoma (ccpRCC) has been proposed as a new entity of renal cell cancer by the International Society of Urological Pathology (ISUP) to be included in the next World Health Organization Classification of Renal Tumors (1). It was initially discovered in kidneys with end-stage renal disease in 2006 (2). Since then more than 100 ccpRCC cases have been described and the majority were found in normal functioning kidneys (3–8). They are characterized by tumor cells with clear cytoplasm, linear arrangement of low-grade nuclei located apically distant from the basal membrane and containing varying amounts of tubular, papillary and cystic architecture. Strikingly, the ccpRCC lack mitoses, atypia, pleomorphism, necrosis, hyaline globules, foamy macrophages and vascular invasion. Despite significant morphologic, immunohistochemical and genetic similarities to clear cell renal cell carcinoma (ccRCC) and papillary renal cell carcinoma (pRCC), characteristic genetic differences include VHL gene mutations and 3p losses, found in ccRCC. Gain of the chromosomes 7 and 17 or loss of chromosome Y, are absent or extremely rare in ccpRCC cases (4, 9, 10). No disease defining mutation has been identified to date.
The renal angiomyoadenomatous tumor (RAT) was first reported in the kidney of a 93-year-old male by Michal et al. (11). Nine years later, the same group characterized 5 additional tumors (12). Verine pointed out that ccpRCCs are a major differential diagnosis of RAT and emphasized their morphologic, immunohistochemical, molecular and clinical similarities (13). In the literature many terms have been used to probably describe the same entity, including ccRCC with prominent leiomyomatous proliferation and renal cell carcinoma with smooth muscle stroma (12, 14–17).
The epithelial component of ccpRCC and RAT is composed of cells with abundant clear cytoplasm, strong diffuse CK7 activity and low grade nuclei (Fuhrman grade 1 and 2). Due to their many similarities, several authors regard ccpRCC and RAT to be a variant of the same entity (6, 9, 13, 17, 18).
The aims of our study were to clarify the relationship between ccpRCC and RAT and to identify markers to reliably distinguish ccpRCC and RAT from the biologically more aggressive renal neoplasms.
Materials and Methods
Case Cohort
All tumors were consultation cases from HM and EC and were received from Austria, France, Germany, Italy and Switzerland. Hematoxylin and eosin-stained slides were reviewed for morphologic features of clear cell papillary renal cell carcinoma (ccpRCC) and renal angiomyoadenomatous tumor (RAT) as previously described (3, 5–7, 11, 12, 19). Diagnostic features of ccpRCC include tumor cells with abundant clear cytoplasm, varying papillary, cystic and tubular architecture, low grade nuclei (Fuhrman grade 1 and 2) located apically distant from the basal membrane, and strong diffuse CK7 and CA-IX expression. For diagnosis of RAT, the following criteria were required: cells with clear cytoplasm, low grade nuclei (Fuhrman grade 1 and 2) embedded in a smooth muscle stroma and strong diffuse CK7 staining of the epithelial component. Tumors were staged according to the TNM system (20) and graded according to Fuhrman et al. (21). The morphologic characteristics were scored as previously described (7).
Immunohistochemistry
A total of 25 antibodies were selected as (i) they are involved in the VHL signaling pathway, (ii) they are known to be prognostic biomarkers of ccRCC and (iii) they have been reported as markers of ccpRCCs and RATs in a small group of ccpRCCs described in recent USCAP meetings (2011–2014). TMA sections (2.5 μm) were transferred to glass slides and treated using Ventana Benchmark XT, Bond-max (Leica Microsystems) automated systems, as well as manual protocols. TMA construction was not possible in five of the ccpRCC cases due to absence of tissue. The immunohistochemical staining product was described as nuclear, membranous or cytoplasmic (Table 2). The immunohistochemistry results were interpreted as 0 (negative), 1+ (weak staining), 2+ (moderate staining) and 3+ (strong staining). For statistical analysis all 2+ and 3+ stainings were defined as positive, 0 and 1+ as negative. Antibodies and protocols are listed in Table 2.
Table 2.
Antibody Overview
| Antibody | Clone | Species | Vendor | Dilution | Staining Pattern |
|---|---|---|---|---|---|
| β-catenin | 14/Beta-Catenin | Mouse | BD Biosciences | 1:50 | membranous |
| Carbonic anhydrase IX | -- | Rabbit | Abcam Limited | 1:6000 | membranous (partially cytoplasmatic) |
| c-MET | SP44 | Rabbit | Ventana | prediluted | membranous |
| CD10 | SP67 | Mouse | Ventana | prediluted | membranous |
| CD70 | 301731 | Mouse | R&D Systems | 1:75 | membranous |
| CD133 | -- | Rabbit | Abcam Limited | 1:500 | membranous |
| Cytokeratin 7 | OV-TL 12/30 | Mouse | DAKO A/S | 1:100 | membranous |
| Cytokeratin 19 | RCK108 | Mouse | Abcam Limited | 1:200 | membranous |
| E-cadherin | EP700Y | Rabbit | Cell Marque Lifescreen Ltd. | 1:200 | membranous |
| Estrogen receptor | SP1 | Rabbit | Labvision | 1:50 | nuclear |
| GATA-3 | L50-823 | Mouse | Biocare Medical | 1:250 | nuclear |
| GLUT-1 | -- | Rabbit | Millipore Corporation | 1:1000 | membranous |
| hKIM-1 | -- | -- | Bonventre lab | -- | membranous and cytoplasmatic |
| Melanosome | HMB-45 | Mouse | DAKO A/S | 1:50 | cytoplasmic |
| OCT3/4 | N1NK | Mouse | Novocastra Laboratories Ltd | 1:150 | nuclear |
| p16 | JC8 | Mouse | Santa Cruz Biotechnology, Inc. | 1:200 | nuclear |
| p27 | -- | Rabbit | Santa Cruz Biotechnology, Inc. | 1:30 | nuclear |
| p53 | DO-7 | Mouse | DAKO A/S | 1:80 | nuclear |
| Parafibromin | 2H1 | Mouse | Santa Cruz Biotechnology, Inc. | 1:10 | nuclear |
| PAX-2 | -- | Rabbit | ZYMED Laboratories Inc. | 1:100 | nuclear |
| PAX-8 | -- | Rabbit | Protein Tech Group, Inc | 1:200 | nuclear |
| Progesterone receptor | 1E2 | Rabbit | Ventana | prediluted | nuclear |
| TFE3 | MRQ-37 | Rabbit | Cell Marque Lifescreen Ltd. | prediluted | nuclear |
| TFEB | -- | Goat | Santa Cruz Biotechnology, Inc. | 1:100 | nuclear |
| Vimentin | Vim 3B4 | Mouse | DAKO A/S | 1:250 | cytoplasmatic |
Fluorescence In Situ Hybridization
FISH, performed to detect VHL allele losses, was carried out using the ZytoLight ® SPEC VHL/CEN 3 Dual Color Probe (ZytoVision, Bremerhaven, Germany). Tissue sections were cut from FFPE blocks, deparaffinized and hybridized as previously described (22). Sixty non-overlapping tumor nuclei from three different areas were analyzed and the number of VHL and CEN3 signals was recorded for each nucleus. The total number of VHL and CEN3 signals as well as the VHL/CEN3 ratio and the percentage of tumor cells with less than 2 VHL signals were calculated. Tumors were considered VHL deleted if more than 50% of the tumor nuclei displayed less than 2 VHL signals (23). In two cases TFE3 FISH using SPEC TFE3 Dual color break apart probe from ZytoVision were done on whole sections as previously described by our group (24).
VHL Sequencing Analysis
Tumor areas displaying >80% tissue in the epithelial portion of the ccpRCC and RAT were marked on the H&E slides. DNA from FFPE tumor tissue samples was obtained by punching 1–2 tissue cylinders (diameter 0.6mm) from each sample. DNA was extracted from the tumor tissue samples according to the Maxwell 16 FFPE Plus DNA Purification protocol (Promega, Fitchburg, USA) for automated DNA purification. DNA concentrations in the samples were measured using the Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA). PCR of the VHL gene was performed as previously described (25) using approximately 40 ng of DNA for each amplification. DNA sequencing was performed with the dideoxy chain-termination method using the BigDye® Terminator v1.1 Cycle Sequencing kit (Applied Biosystems, Foster City, USA). The same forward and reverse primers were used for the PCR and sequencing. Cycle sequencing products were analyzed using the AbiPrism 3100 Genetic analyzer (Applied Biosystems). The obtained sequences were compared with the NCBI sequence AF010238 using NCBIs Blast 2 Sequences. All VHL point mutations obtained were validated by a second separate PCR and sequencing analysis.
Results
Clinical and Pathologic Findings
The patients with ccpRCC ranged from 29 to 75 years of age (mean age 58 years) and those with RAT from 32 to 68 years of age (mean age 43.3 years) at the time of nephrectomy. Male to female ratio was 1.5:1 in the ccpRCC group (17 men and 11 women) and 3.5:1 in the RAT group (6 men and 1 woman).
Clinical follow-up data was available for 78% (21/27) of the ccpRCC patients and 71% (5/7) of the RAT patients. Mean follow-up time was 29.7 months (range 7 to 84 months) for the ccpRCC patients and 32.3 months (range 25 to 38 months) for the RAT patients. There was no evidence of recurrence or disease-related death in any of the patients. None of the RAT (0/5) patients and 14 % (3/22) of the ccpRCC patients had end-stage renal disease.
In the RAT group, the average diameter of the tumor was 3.1 cm (range 1.8–5.0 cm) compared to 2.6 cm (range 0.5–8 cm) in the ccpRCC group. 67% (4/6) of the RAT patients displayed pathologic stage pT1a and 33% (2/6) stage pT1b. Overall 86% of the tumors (6/7) were Fuhrman nuclear grade 1 and 14% (1/7) were nuclear grade 2. In the ccpRCC cases 77% (20/26) were stage pT1a, 19% (5/26) were pT1b and 4% (1/26) were pT2a. Fuhrman nuclear grade 1 was found in 48% (13/27) and nuclear grade 2 in 52% (14/27) of the tumors. All the ccpRCCs and 6/7 RATs showed at least focal papillary architecture and branched ducts. In contrast to ccpRCC, secretory cells were completely absent in the RAT cases. Both showed variable amounts of cystic areas. All tumors were characterized by absence of mitotic formations, foamy macrophages, calcifications and vascular invasion.
Table 1 summarizes the clinicopathological findings. Morphological characteristics are shown in Table 5.
Table 1.
Clinicopathological findings
| # | Age | Gender | Laterality | Grade (Fuhrman) |
Size (cm) |
Number of Tumors |
Stage | ESRD | Tumor- related death |
Follow-up (months) |
Tumor Recurrence |
Metastasis | Special remarks |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAT | |||||||||||||
| 1 | 32 | M | right | 2 | 1.8 | 1 | pT1a | no | na | na | no | no | |
| 2 | 32 | M | left | 2 | 4.7 | 2 | pT1b | na | na | na | na | na | IBD, papillary RCC type 1, kidney adenomas |
| 3 | 34 | M | right | 2 | 1.8 | 2 | pT1a | na | no | 35 | na | na | |
| 4 | 45 | F | right | 2 | na | 1 | na | na | na | na | na | na | |
| 5 | 56 | M | left | 2 | 2 | 1 | pT1a | no | no | 25 | no | no | |
| 6 | 68 | M | na | 1 | 3.5 | 1 | pT1a | na | na | na | na | na | |
| 7 | 36 | M | left | 2 | 5 | 1 | pT1b | no | no | 19 | no | no | |
| CCPRCC | |||||||||||||
| 1 | 55 | M | na | 2 | na | na | na | na | na | na | na | na | |
| 2 | 58 | M | right | 2 | 0.9 | 1 | pT1a | no | no | 39 | no | no | |
| 3 | 57 | F | right | 1 | 4 | 1 | pT1b | na | no | 25 | na | na | |
| 4 | 50 | M | na | 2 | 3 | 1 | pT1a | na | na | na | na | na | |
| 5 | 38 | M | left | 2 | 2 | 1 | pT1a | yes | na | na | na | na | IgA nephritis |
| 6 | 63 | M | right | 2 | 2 | na | pT1a | na | na | na | na | na | Large cell b-cell lymphoma |
| 7 | 62 | M | left | 2 | 3 | 2 | pT1a | no | no | 12 | no | no | papillary RCC type 2 |
| 8 | 31 | F | left | 2 | 8 | 1 | pT2a | na | na | na | na | na | |
| 9 | 55 | M | right | 1 | 4.5 | 1 | pT1b | no | no | 24 | no | no | |
| 10 | 68 | F | na | 1 | 3.8 | pT1a | na | na | na | na | na | ||
| 11 | 51 | M | left | 2 | 1.3 | 1 | pT1a | no | no | 11 | no | no | |
| 12 | 75 | F | left | 2 | 5.1 | 1 | pT1b | no | no | 7 | no | no | |
| 13 | 38 | M | left | 1 | 2.2 | 1 | pT1a | no | no | 8 | no | no | |
| 14 | 53 | F | right | 1 | 2 | 1 | pT1a | no | no | 84 | no | no | |
| 15 | 51 | M | left | 2 | 1 | 8 | pT1a | no | no | 71 | no | no | |
| 16 | 62 | M | left | 1 | 1.3 | 1 | pT1a | yes | no | 67 | no | no | |
| 17 | 52 | M | left | 1 | 5 | 1 | pT1b | no | no | 60 | no | no | |
| 18 | 71 | F | right | 2 | 2.5 | 1 | pT1a | no | no | 39 | no | no | |
| 19 | 57 | F | left | 1 | 1.5 | 1 | pT1a | no | no | 37 | no | no | |
| 20 | 72 | M | na | 1 | 0.5 | 2 | pT1a | no | no | 29 | no | no | |
| 21 | 61 | F | left | 2 | 5 | 1 | pT1b | no | no | 22 | no | no | |
| 22 | 71 | M | right | 1 | 2 | 1 | pT1a | no | no | 21 | no | no | |
| 23 | 53 | F | right | 1 | 1 | 3 | pT1a | no | no | 17 | no | no | |
| 24 | 70 | F | left | 1 | 2.8 | 1 | pT1a | no | no | 15 | no | no | |
| 25 | 65 | F | na | 1 | 0.5 | 1 | pT1a | no | no | 14 | no | no | |
| 26 | 74 | m | left | 2 | 1.8 | 1 | pT1a | yes | no | 12 | no | no | |
| 27 | 54 | m | left | 2 | 2 | 1 | pT1a | no | no | 10 | no | no | |
Abbreviations: ESRD: end-stage renal disease; n. a.: not available; RAT: renal angiomyoadenomatous tumor; IBD: inflammatory bowel disease; ccpRCC: clear cell papillary renal cell carcinoma
Table 5.
Morphological characteristics of RAT and ccpRCC
| # | Papillary Architecture | Branched Ducts | Secretory Cells | % Cystic |
|---|---|---|---|---|
| RAT | ||||
| 1 | 1f | yes | no | 15 |
| 2 | 0 | no | no | 0 |
| 3 | 2f | yes | no | 65 |
| 4 | 1f | yes | no | 5 |
| 5 | 2f | yes | no | 0 |
| 6 | 1f | yes | no | 55 |
| 7 | 2f | yes | no | 15 |
| ccpRCC | ||||
| 1 | 1 | yes | yes | 85 |
| 2 | 2 | yes | no | 10 |
| 3 | 3 | yes | yes | 10 |
| 4 | 3 | yes | no | 10 |
| 5 | 2 | yes | no | 15 |
| 6 | 3 | yes | yes | 55 |
| 7 | 1 | yes | yes | 0 |
| 8 | 3 | yes | no | 0 |
| 9 | 3 | yes | no | 0 |
| 10 | 2 | yes | no | 5 |
| 11 | 3 | yes | no | 0 |
| 12 | 3 | yes | yes | 40 |
| 13 | 1 | yes | yes | 15 |
| 14 | 1 | yes | no | 5 |
| 15 | 2 | yes | no | 20 |
| 16 | 2 | yes | no | 10 |
| 17 | 1 | yes | yes | 15 |
| 18 | 3 | yes | yes | 45 |
| 19 | 2 | yes | no | 30 |
| 20 | 2 | yes | yes | 20 |
| 21 | 2 | yes | no | 5 |
| 22 | 1 | yes | no | 10 |
| 23 | 3 | yes | yes | 55 |
| 24 | 1 | yes | yes | 10 |
| 25 | 3 | yes | no | 35 |
| 26 | 2 | yes | yes | 5 |
| 27 | 3 | yes | yes | 30 |
f = focal
Scored as previously described (7)
Immunohistochemical Findings
The immunohistochemical findings are detailed in Table 3. ccpRCC and RAT were strongly positive for CK7, CK19, CA-IX, GLUT-1, E-cadherin, vimentin, β-catenin, parafibromin, PAX-2, PAX-8, p27, p53 and c-MET. Staining for GLUT-1 (p = 0.0572), CD 70 (p = 0.1499) and p16 (p = 0.3702) differed slightly in the RAT samples compared with ccpRCCs, although differences did not show statistical significance. Following the recent results by Cui et al. (26), Aron et al. (53) and Schwartz et al. (56), we tested parafibromin, hKIM-1 (27) and CD133 expression to distinguish ccpRCC/RAT from ccRCC/pRCC. As shown in Table 4, the expression difference reached statistical significance (p <0.0001). The biomarkers CD70 (28), MET (29) and E-cadherin (30) were able to distinguish between ccpRCC/RAT and ccRCC (p <0.0001). Furthermore, the hKIM-1 and parafibromin were able to distinguish between ccpRCC/RAT and pRCC. All ccpRCC cases exhibited a characteristic CA-IX “cup-like” distribution, sparing the luminal border as it has been described in the literature before (6, 31). In contrast, the RAT tumors and the ccpRCC-like tumor with the VHL mutation showed a circumferential membranous staining pattern. Two RAT samples stained weakly positive for TFE3 and were, therefore, further analyzed by HMB45 and TFEB. Both stainings revealed a negative result. Additionally, TFE3 FISH was performed (see below).
Table 3.
Results of Immunohistochemistry. Percentage relates to number of interpretable cases.
| Antibody | ccpRCC | RAT |
|---|---|---|
| β-catenin | 95.5 | 85.7 |
| Carbonic anhydrase IX | 95.5 | 85.7 |
| CD10 | 31.8 | 66.7 |
| CD70 | 22.7 | 0 |
| CD133 | 81.8 | 100 |
| c-MET | 91.3 | 100 |
| Cytokeratin 7 | 100 | 100 |
| Cytokeratin 19 | 88.9 | 100 |
| E-cadherin | 100 | 100 |
| Estrogen receptor | 4.3 | 0 |
| GATA-3 | 31.8 | 42.9 |
| GLUT-1 | 95.5 | 85.7 |
| hKIM-1 | 23.8 | 0 |
| OCT3/4 | 8.7 | 0 |
| p16 | 18.2 | 42.9 |
| p27 | 100 | 100 |
| p53 | 72.7 | 71.4 |
| Parafibromin | 95.5 | 100 |
| PAX-2 | 63.6 | 100 |
| PAX-8 | 95.5 | 100 |
| Progesterone receptor | 0 | 0 |
| Vimentin | 95.5 | 100 |
Table 4.
Different expression patterns of hKIM-1, Parafibromin, CD70, CD133, MET and E-cadherin in ccPRCC, ccRCC and pRCC (own data and literature).
| Antibody | ccpRCC | ccRCC | Fisher Exact (P) | pRCC | Fisher Exact (P) |
|---|---|---|---|---|---|
| hKIM-1 | 27.3% (6/22) | 74% (54/73)* | <0.0001 | 93% (28/30)* | <0.0001 |
| Parafibromin | 91.3% (21/23) | 7% (4/61)# | <0.0001 | 19% (7/37)# | <0.0001 |
| CD70 | 26.1% (6/23) | 78% (197/252)§ | <0.0001 | 32% (113/348)§ | 0.6475 |
| CD133 | 78.3% (18/23) | 14.3% (3/21)x | <0.0001 | 53% (8/15) x | 0.16 |
| MET | 91.3% (21/23) | 0% (0/96)y | <0.0001 | 90% (18/20)y | 1 |
| E-cadherin | 100% (22/22) | 31.9% (22/69)z | <0.0001 | n. a. | n. a. |
Fluorescence In Situ Hybridization Findings
Three deletions of the short arm of chromosome 3 were identified. All of them occurred in the ccpRCC cases (3/21, 14%) and no deletion was found in the RAT cases (0/7, 0%). The presence of the 3p deletions in the two ccRCC controls were correctly identified. In 9 of the cases FISH was not performed as there was not sufficient tissue after VHL mutation analysis and immunohistochemistry.
TFE3 FISH was performed with the two above-mentioned RAT-like cases that showed weak TFE3 expression. One case showed the typical break apart pattern in >85% of the cells, while the second case was negative. Both cases were reclassified as translocation carcinomas due to immunohistochemical TFE3 positivity.
VHL Gene Mutation Analysis
Three VHL mutations were detected in the ccpRCC group (3/27, 11%) in exon 2 (c.351G>C/p.Trp117Cys, c.461C>T/p.Pro154Leu, c.388G>C/p.Val130Leu) and one in the RAT group (1/7, 14%) in exon 1 (c.174_177delGCCG /p.Pro59GlyfsX7). We identified two cases, harboring both a VHL mutation and 3p loss. One case showed a 3p loss but no VHL mutation and two cases with a VHL mutation showed no 3p loss.
Discussion
In the present study we have sequenced the largest number of ccpRCC (27) and RAT (7) cases to date. We found a VHL mutation rate of 11% in ccpRCC and 14% in RAT. Furthermore, we analyzed hypoxia-inducible factor pathway-related proteins to compare these findings with recent findings.
ccpRCC and RAT are currently underrecognized. Recent studies have revealed that they are not rare (7, 32, 33) and that among all RCC the ccpRCC have a prevalence rate between 1.2% and 4.1%, thus representing up to 4,500 new cases of renal cancer in the United States annually (7, 32, 34). Awareness of its morphologic and immunohistochemical features is imperative for a correct classification. In a recent publication Gill et al. underscored the necessity of reclassifying low grade and low stage ccRCC as up to 7% of the cases are in fact ccpRCC (33).
Morphologically, ccpRCC and RAT share many features. Their epithelial component is composed of cells with clear cytoplasm and low grade nuclei. Both tumors have various amounts of smooth muscle stroma, and their epithelial component is characterized by either cystic or papillary architecture. In our cohort the majority of the RAT samples had focal papillary features of the epithelial component, which are typically diffuse CK7 and CA-IX positive. The most relevant differential diagnoses include ccRCC that exhibit papillary features, pRCC exhibiting clear cell characteristics and Xp11 translocation carcinoma. In our cohort, two cases initially classified as RAT had to be re-classified as Xp11 translocation carcinomas after immunohistochemistry and TFE3 FISH analysis. The translocation carcinomas were identified by nuclear TFE3 protein expression. Only one case showed a positive TFE3 FISH result. It is controversial whether TFE3 positivity is sufficient to diagnose TFE3 translocation carcinoma (24, 35), but, from these 2 cases, we concluded that TFE3 translocation cancer falls within the differential diagnostic spectrum of ccpRCC/RAT. Another differential diagnosis for the case with weak TFE3 staining and negative FISH is TFEB-associated RCC. Those tumors can overlap tremendously with the TFE3 rearranged RCC (36, 37). To rule out this differential diagnosis we did two additional immunohistochemical stainings (HMB45 and TFEB). Both stainings showed a negative result making that differential diagnosis unlikely.
One ccpRCC case was reclassified as ccRCC. That case exhibited typical ccpRCC morphology but was completely negative for CK7 and strongly positive for hKIM-1. This case also revealed a mutation in the VHL gene and a 3p loss in the FISH analysis. These findings highlight the importance of molecular testing and should raise awareness of ccpRCC mimicking ccRCC (38).
VHL gene mutations are the genetic hallmark of ccRCC. Initially, it was reported that VHL alterations are absent in ccpRCC. However, three groups have recently identified VHL mutations in ccpRCC at frequencies varying from 15% to 27% (39–41). In concordance with these studies, we also identified VHL gene alterations in ccpRCC, but the prevalence of VHL gene mutations is significantly lower than in ccRCC (42–44). The discrepancy between the number of mutations found in our ccpRCC cases and that reported may be explained by the different detection methods employed, including single nucleotide polymorphism (SNPs) genotyping array, Sanger sequencing, and by the limited number of cases in previous studies. Alternatively, cases with VHL mutations could represent ccRCCs with morphology and immunoprofile which closely mimics that of clear ccpRCC and RAT tumor. Currently, ccpRCC are diagnosed on the basis of morphology and diffuse strong CK7 expression. The absence of VHL mutations/3p deletions is not diagnostic for ccpRCC. Therefore we suggest to diagnose tumors with diffuse CK7 expression combined with the typical morphology as ccpRCC. In previous studies CCRCC with diffuse CK7 profile have had a completely different prognosis than CCRCC without that CK expression pattern (45). These previous findings justify such an approach. VHL inactivation leads to a HIF-dependent CA-IX and GLUT-1 up-regulation. We only found few VHL mutations, but in combination with CA-IX and GLUT-1 immunoreactivity in both ccpRCC and RAT. This clearly sets the ccpRCC and RAT apart from ccRCC, which shows VHL mutations in up to 80% of the cases (44, 46). Therefore, we believe that the HIF pathway may be activated in a VHL-independent manner in most ccpRCCs and RATs, also hypothesized by Rohan et al. (6).
Recently, Lawrie et al. found various mutations in ccpRCC by using Next Generation Sequencing (NGS), including a non-synonymous T992I mutation in the MET proto-oncogene (47). This gene was originally described as causing hereditary pRCC (48). Interestingly, Lawrie et al. detected no VHL mutation, but found overexpression in all five members of the miR-200 family. The miR-200 family plays an essential role in tumor suppression by inhibiting epithelial-mesenchymal transition (49). To support Lawrie’s results we also noted immunoreactivity for E-cadherin and β-catenin. These findings suggest that epithelial-to-mesenchymal transition (EMT) may be incomplete or blocked in ccpRCC contributing to their indolent course (47).
Other genetic alterations characteristic for pRCC include gain of the chromosome 7 and loss of the chromosome Y. However, in ccpRCC, gain of the chromosome 7 have very rarely been reported (4, 5, 9, 10) and no loss of the chromosome Y has been observed to date. Fisher et al. found a unique gene expression profile of ccpRCC when investigating 8 different genes, with only some expression levels comparable with those observed in ccRCC and pRCC (50).
In our FISH analysis, we identified three chromosome 3p deletions in 20 ccpRCC and in 7 RAT samples. All 3p deletions occurred in ccpRCC with a frequency of 14.3 %, but none was detected in RAT. To date, only 4 cases with a 3p loss have been reported in ccpRCC (34, 41). Interestingly, the single case described by Martignoni et al. (41) concurrently harbored a VHL mutation like two of our three cases with a 3p loss. Shi et al. (34) also used FISH and observed monosomy of chromosome 3 in three cases in a series of 11 ccpRCC all lacking mutations in the VHL gene. In 2009, Shannon et al. (14) published a study on 5 ccRCC with smooth muscle stroma and found loss of the entire chromosome 3 in two cases and a 3p loss in one case using FISH. In contrast, Martignoni found no 3p loss in a series of three cases of ccRCC with smooth muscle stroma (17). Given these molecular findings, it has been suggested that RAT and ccRCC with smooth muscle stroma are interchangeable terms (51). However, some of the cases of ccRCC with smooth muscle stroma, particularly those that showed 3p loss, might represent ccRCCs with exuberant, infiltrative smooth muscle, whereas the others might in fact be RAT tumors, particularly the ones that do not show 3p loss (15). Additionally, recent data shows that some tumors with RAT morphology and immunophenotype share a common mutation in the TCEB1 gene which inactivated the VHL pathway and upregulated proteins along the hypoxia-inducible pathway (52). Twenty-five different antibodies were used to characterize ccpRCC and RAT. We were particularly interested in hypoxia-inducible factor pathway-related proteins as well as other antibodies, which were reportedly used in small series of ccpRCC cases in the 2011 and 2014 USCAP meetings. This gave us the opportunity to compare immunohistochemical findings in ccpRCC and RAT to clarify their interrelationship. Remarkably, there were no statistically significant differences in the staining properties in any of the antibodies in ccpRCC compared to RAT.
Parafibromin and hKIM-1 expression levels differed significantly between ccpRCC/RAT and ccRCC/pRCC. Cui et al. (26) recently demonstrated that parafibromin can be very helpful in differentiating ccpRCC from ccRCC and pRCC. In a study by Aron et al. (53), the difference in the staining positivity rate of ccpRCC and ccRCC was even more striking compared with our study. In addition to parafibromin and hKIM-1 expression, CD70 also proved to be a useful marker in differentiating ccpRCC from ccRCC, since CD70 expression is rare in ccpRCC and very frequent in ccRCC. CD70 was used for immunohistochemistry because we have previously demonstrated that CD70 is a potential biomarker for ccRCC (28, 54). The importance of immunohistochemical stainings in the correct identification of true ccpRCC was also highlighted by Williamson et al. (55). They studied 14 ccpRCC-like tumors, which could not be distinguished from ccpRCC morphologically, but which showed a high 3p deletion frequency (82%) and showed a different immunohistochemical profile, with negative or localized CK7 staining as the most striking feature. These characteristics also led to a reclassification of one of our tumors, primarily diagnosed as ccpRCC.
Recently, Schwartz et al. studied different stem cell markers in renal cancers. They reported a 90 % positivity rate for OCT 3/4 in a series of 10 ccpRCC samples (56). This finding is discrepant to our positivity rate of 8.7%, which may be due to the use of different antibodies or immunohistochemical protocols. However, similarly to Schwartz et al. (56), we also detected a high positivity rate of stem cell marker CD133 (81.8% and 100%, respectively) in ccpRCC. Interestingly, Schwartz et al. reported a CD133 positivity rate of only 14% in ccRCC. It can therefore be concluded that CD133 is an additional tool to distinguish ccRCC from ccpRCC/RAT.
In concordance with Munari et al. (57), we found that about one third of ccpRCC are positive for GATA3, a protein crucial for the regulation of Th2 development and function. However, given that only a moderate staining intensity was seen in no more than 10 % of the tumor cells, we do not consider OCT3/4 and GATA3 as diagnostic tools to differentiate ccpRCC from ccRCC.
No previous studies have reported cancer-related death, vascular invasion or metastasis in ccpRCC (4–7, 51, 58), suggesting that the disease follows an indolent course. Benign biologic behavior was also observed in all RAT cases (12, 14, 59). This is comparable to multilocular cystic RCC, which has an excellent prognosis with no disease recurrence after surgery (7, 12, 59). Specific molecular alterations may account for the indolent course of multilocular cystic RCC. Proposals have been put forward to rename multilocular cystic RCC as multilocular cystic renal cell neoplasm of low malignant potential to underscore this specific biologic behavior (1). Our group has reported that the expression of p27, CA-IX, CK7 and CK19 is associated with a better prognosis in sporadic RCC (45, 60). Interestingly, our ccpRCC/RAT cases stained strongly positive for all of these markers. Hence, the indolent clinical course of ccpRCC/RAT might in part be due to this specific signaling pathway. However, some of our low grade ccRCC included in our previous publications may in fact be unrecognized ccpRCC (33, 45).
In summary, we have demonstrated that ccpRCC and RAT cannot be distinguished from one another by immunohistochemistry and molecular analyses and both follow a benign clinical course. We regard them as a spectrum of a distinct tumor entity. Precise diagnosis is crucial since it has an excellent prognosis. Given the reliability of TFE3 immunohistochemistry, TFE3 FISH should be performed in cases with equivocal TFE3 immunohistochemistry (35). Taking into account the controversial relevance of the VHL mutation analysis in this differential diagnosis, direct VHL sequencing is not helpful in separation of ccRCC with prominent smooth muscle stroma from RAT. Our results suggest that a panel of antibodies against CK7, parafibromin and MET are a helpful tool to differentiate most ccpRCC/RAT from other renal tumors (Table 4). In some difficult cases VHL mutation testing and TFE3 FISH analysis are helpful tools to distinguish ccRCC and TFE3 translocation carcinoma from ccpRCC/RAT (Figure 5).
Figure 5.
Proposed diagnostic workflow for renal cell carcinoma showing H&E features indicative for ccpRCC/RAT.
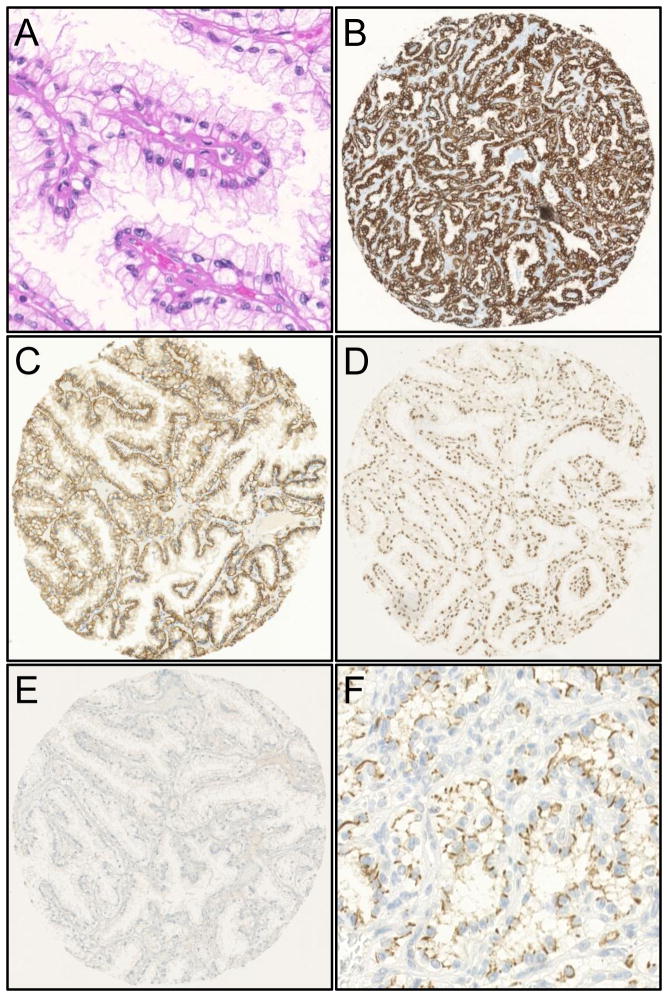
Figure 1.
Clear cell papillary renal cell carcinoma. H&E stain (A) and typical immunohistochemical profile with diffuse membranous CK7 positivity (B), membranous “cup-like” CA-IX positivity (C), nuclear parafibromin positivity (D), hKIM-1 negativity (E) and CD133 positivity (F).
Figure 2.
H&E morphology of a ccpRCC-like (A-C) and a RAT-like (D-F) case. Diagnostic features of ccpRCC include tumor cells with abundant clear cytoplasm, varying papillary, cystic and tubular architecture and low grade nuclei (Fuhrman grade 1 and 2) located apically distant from the basal membrane. The epithelial part of RAT tumors is composed of cells with clear cytoplasm, low grade nuclei (Fuhrman grade 1 and 2) embedded in a smooth muscle stroma.
Figure 3.
Molecular features of a ccpRCC-like (A) and a RAT-like (C) case both exhibiting a circumferential CA-IX staining pattern, harboring a VHL mutation (D: c.174_177delGCCG/p.Pro59GlyfsX7; E: c.351G>C/p.Trp117Cys) and a 3p deletion detected by fluorescence in situ hybridization (B). The mutation sites are denoted by an arrow. The boundaries between exon and intron are indicated. The upper base pair letter sequence shows the wild type (WT) sequence (D, E). Tumor cells harbor only one VHL (green) signal and two CEN3 copies (orange) (B).
Figure 4.
ccpRCC look-alike showing classic ccpRCC morphology on the HE stain (A), however with a typical ccRCC immunohistochemical profile showing CK7 negativity (B), CA-IX positivity (C), hKIM-1 positivity (D) and proof of VHL mutation (E).
Acknowledgments
We thank Susanne Dettwiler, André Fitsche, and Martina Storz for their outstanding technical assistance and Dorothee Pflueger for helping to interpret the TFE3 FISH data. We thank the sequencing service at the Institute of Surgical Pathology for performing numerous sequencing reactions.
The project was supported by the Swiss National Science Foundation to HM (3238BO-103145) and the Zurich Cancer League to HM. JVB received grants from the NIH (R37DK39773, RO1DK072381).
Footnotes
Disclosure/conflict of interest
JVB is co-inventor on KIM-1 patents, which have been assigned to Partners Healthcare and licensed by Partners Healthcare to a number of companies. All other authors have no conflict of interest.
References
- 1.Srigley JR, Delahunt B, Eble JN, et al. The International Society of Urological Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am J Surg Pathol. 2013;37:1469–1489. doi: 10.1097/PAS.0b013e318299f2d1. [DOI] [PubMed] [Google Scholar]
- 2.Tickoo SK, de Peralta-Venturina MN, Harik LR, et al. Spectrum of epithelial neoplasms in end-stage renal disease: an experience from 66 tumor-bearing kidneys with emphasis on histologic patterns distinct from those in sporadic adult renal neoplasia. Am J Surg Pathol. 2006;30:141–153. doi: 10.1097/01.pas.0000185382.80844.b1. [DOI] [PubMed] [Google Scholar]
- 3.Adam J, Couturier J, Molinie V, et al. Clear-cell papillary renal cell carcinoma: 24 cases of a distinct low-grade renal tumour and a comparative genomic hybridization array study of seven cases. Histopathology. 2011;58:1064–1071. doi: 10.1111/j.1365-2559.2011.03857.x. [DOI] [PubMed] [Google Scholar]
- 4.Aydin H, Chen L, Cheng L, et al. Clear cell tubulopapillary renal cell carcinoma: a study of 36 distinctive low-grade epithelial tumors of the kidney. Am J Surg Pathol. 2010;34:1608–1621. doi: 10.1097/PAS.0b013e3181f2ee0b. [DOI] [PubMed] [Google Scholar]
- 5.Gobbo S, Eble JN, Grignon DJ, et al. Clear cell papillary renal cell carcinoma: a distinct histopathologic and molecular genetic entity. Am J Surg Pathol. 2008;32:1239–1245. doi: 10.1097/PAS.0b013e318164bcbb. [DOI] [PubMed] [Google Scholar]
- 6.Rohan SM, Xiao Y, Liang Y, et al. Clear-cell papillary renal cell carcinoma: molecular and immunohistochemical analysis with emphasis on the von Hippel-Lindau gene and hypoxia-inducible factor pathway-related proteins. Mod Pathol. 2011;24:1207–1220. doi: 10.1038/modpathol.2011.80. [DOI] [PubMed] [Google Scholar]
- 7.Williamson SR, Eble JN, Cheng L, et al. Clear cell papillary renal cell carcinoma: differential diagnosis and extended immunohistochemical profile. Mod Pathol. 2013;26:697–708. doi: 10.1038/modpathol.2012.204. [DOI] [PubMed] [Google Scholar]
- 8.Herrera LP, Hirsch M, Comperat E, et al. Clear Cell-Papillary Renal Cell Carcinoma (CP-RCC) Not Associated with End Stage Renal Disease: Clinicopathologic Analysis of 50 Tumors Confirming a Novel Subtype of Renal Cell Carcinoma (RCC) Occurring in a Sporadic Setting. Mod Pathol. 2011;24(Suppl):197A. [Google Scholar]
- 9.Wolfe A, Dobin SM, Grossmann P, et al. Clonal trisomies 7, 10 and 12, normal 3p and absence of VHL gene mutation in a clear cell tubulopapillary carcinoma of the kidney. Virchows Arch. 2011;459:457–463. doi: 10.1007/s00428-011-1137-3. [DOI] [PubMed] [Google Scholar]
- 10.Kuroda N, Shiotsu T, Kawada C, et al. Clear cell papillary renal cell carcinoma and clear cell renal cell carcinoma arising in acquired cystic disease of the kidney: an immunohistochemical and genetic study. Ann Diagn Pathol. 2011;15:282–285. doi: 10.1016/j.anndiagpath.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Michal M, Hes O, Havlicek F. Benign renal angiomyoadenomatous tumor: a previously unreported renal tumor. Ann Diagn Pathol. 2000;4:311–315. doi: 10.1053/adpa.2000.17890. [DOI] [PubMed] [Google Scholar]
- 12.Michal M, Hes O, Nemcova J, et al. Renal angiomyoadenomatous tumor: morphologic, immunohistochemical, and molecular genetic study of a distinct entity. Virchows Arch. 2009;454:89–99. doi: 10.1007/s00428-008-0697-3. [DOI] [PubMed] [Google Scholar]
- 13.Verine J. Renal angiomyoadenomatous tumor: morphologic, immunohistochemical, and molecular genetic study of a distinct entity. Virchows Arch. 2009;454:479–480. doi: 10.1007/s00428-009-0741-y. [DOI] [PubMed] [Google Scholar]
- 14.Shannon BA, Cohen RJ, Segal A, et al. Clear cell renal cell carcinoma with smooth muscle stroma. Hum Pathol. 2009;40:425–429. doi: 10.1016/j.humpath.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Kuhn E, De Anda J, Manoni S, et al. Renal cell carcinoma associated with prominent angioleiomyoma-like proliferation: Report of 5 cases and review of the literature. Am J Surg Pathol. 2006;30:1372–1381. doi: 10.1097/01.pas.0000213277.45715.82. [DOI] [PubMed] [Google Scholar]
- 16.Singh C, Kendi AT, Manivel JC, et al. Renal angiomyoadenomatous tumor. Ann Diagn Pathol. 2012;16:470–476. doi: 10.1016/j.anndiagpath.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Martignoni G, Brunelli M, Segala D, et al. Renal cell carcinoma with smooth muscle stroma lacks chromosome 3p and VHL alterations. Mod Pathol. 2014 May;27(5):765–74. doi: 10.1038/modpathol.2013.180. [DOI] [PubMed] [Google Scholar]
- 18.Behdad A, Monzon FA, Hes O, et al. Relationship between sporadic clear cell-papillary renal cell carcinoma (CP-RCC) and renal angiomyoadenomatous tumor (RAT) of the kidney: analysis by virtualkaryotyping, fluorescent in situ analysis and immunohistochemistry (IHC) Mod Pathol. 2011;24(Suppl):179A. [Google Scholar]
- 19.Michal M, Hes O, Kuroda N, et al. Difference between RAT and clear cell papillary renal cell carcinoma/clear renal cell carcinoma. Virchows Arch. 2009;454:719. doi: 10.1007/s00428-009-0788-9. [DOI] [PubMed] [Google Scholar]
- 20.Edge SB, Byrd DR, Compton CC, et al. AJCC cancer staging manual. New York: Springer-Verlag; 2010. [Google Scholar]
- 21.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–663. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 22.Schildhaus HU, Deml KF, Schmitz K, et al. Chromogenic in situ hybridization is a reliable assay for detection of ALK rearrangements in adenocarcinomas of the lung. Mod Pathol. 2013;26:1468–1477. doi: 10.1038/modpathol.2013.95. [DOI] [PubMed] [Google Scholar]
- 23.Sanjmyatav J, Hauke S, Gajda M, et al. Establishment of a multicolour fluorescence in situ hybridisation-based assay for subtyping of renal cell tumours. Eur Urol. 2013;64:689–691. doi: 10.1016/j.eururo.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Pflueger D, Sboner A, Storz M, et al. Identification of molecular tumor markers in renal cell carcinomas with TFE3 protein expression by RNA sequencing. Neoplasia. 2013;15:1231–1240. doi: 10.1593/neo.131544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Teichman A, Comperat E, Behnke S, et al. VHL mutations and dysregulation of pVHL- and PTEN-controlled pathways in multilocular cystic renal cell carcinoma. Mod Pathol. 2011;24:571–578. doi: 10.1038/modpathol.2010.222. [DOI] [PubMed] [Google Scholar]
- 26.Cui C, Lal P, Master S, et al. Expression of parafibromin in major renal cell tumors. Eur J Histochem. 2012;56:e39. doi: 10.4081/ejh.2012.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin F, Zhang PL, Yang XJ, et al. Human kidney injury molecule-1 (hKIM-1): a useful immunohistochemical marker for diagnosing renal cell carcinoma and ovarian clear cell carcinoma. Am J Surg Pathol. 2007;31:371–381. doi: 10.1097/01.pas.0000213353.95508.67. [DOI] [PubMed] [Google Scholar]
- 28.Ruf M, Mittmann C, Nowicka AM, et al. pVHL/HIF-Regulated CD70 Expression Is Associated with Infiltration of CD27+ Lymphocytes and Increased Serum Levels of Soluble CD27 in Clear Cell Renal Cell Carcinoma. Clin Cancer Res. 2015 Feb 15;21(4):889–98. doi: 10.1158/1078-0432.CCR-14-1425. [DOI] [PubMed] [Google Scholar]
- 29.Choi JS, Kim MK, Seo JW, et al. MET expression in sporadic renal cell carcinomas. J Korean Med Sci. 2006;21:672–677. doi: 10.3346/jkms.2006.21.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai J. Roles of transcriptional factor Snail and adhesion factor E-cadherin in clear cell renal cell carcinoma. Exp Ther Med. 2013;6:1489–1493. doi: 10.3892/etm.2013.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tickoo SK, Reuter VE. Differential diagnosis of renal tumors with papillary architecture. Adv Anat Pathol. 2011;18:120–132. doi: 10.1097/PAP.0b013e31820cb3dd. [DOI] [PubMed] [Google Scholar]
- 32.Zhou H, Zheng S, Truong LD, et al. Clear cell papillary renal cell carcinoma is the fourth most common histologic type of renal cell carcinoma in 290 consecutive nephrectomies for renal cell carcinoma. Hum Pathol. 2014;45:59–64. doi: 10.1016/j.humpath.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Gill S, Kandel S, Xu B. Frequency of Clear Cell Papillary Renal Cell Carcinoma in Cases of Low Grade Clear Cell Renal Cell Carcinoma: A 12 Year Retrospective Study from a Single Cancer Center. Mod Pathol. 2013;26(Suppl):212A. doi: 10.1177/1066896915613432. [DOI] [PubMed] [Google Scholar]
- 34.Shi SS, Shen Q, Xia QY, et al. Clear cell papillary renal cell carcinoma: a clinicopathological study emphasizing ultrastructural features and cytogenetic heterogeneity. Int J Clin Exp Pathol. 2013;6:2936–2942. [PMC free article] [PubMed] [Google Scholar]
- 35.Green WM, Yonescu R, Morsberger L, et al. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am J Surg Pathol. 2013;37:1150–1163. doi: 10.1097/PAS.0b013e31828a69ae. [DOI] [PubMed] [Google Scholar]
- 36.Argani P, Yonescu R, Morsberger L, et al. Molecular confirmation of t(6;11)(p21;q12) renal cell carcinoma in archival paraffin-embedded material using a break-apart TFEB FISH assay expands its clinicopathologic spectrum. Am J Surg Pathol. 2012;36:1516–1526. doi: 10.1097/PAS.0b013e3182613d8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith NE, Illei PB, Allaf M, et al. t(6;11) renal cell carcinoma (RCC): expanded immunohistochemical profile emphasizing novel RCC markers and report of 10 new genetically confirmed cases. Am J Surg Pathol. 2014;38:604–614. doi: 10.1097/PAS.0000000000000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersson F, Grossmann P, Hora M, et al. Renal cell carcinoma with areas mimicking renal angiomyoadenomatous tumor/clear cell papillary renal cell carcinoma. Hum Pathol. 2013;44:1412–1420. doi: 10.1016/j.humpath.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 39.Xu W, Deng F-M, Melamed J, et al. Incidence and Genetic Characteristics of Clear Cell Tububopapillary Renal Cell Carcinoma. Mod Pathol. 2014;27(Suppl):270A. [Google Scholar]
- 40.Behdad A, Monzon FA, Hes O, et al. Relationship between Sporadic Clear Cell-Papillary Renal Cell Carcinoma (CP-RCC) and Renal Angiomyoadenomatous Tumor (RAT) of the Kidney: Analysis by Virtual-Karyotyping, Fluorescent In Situ Analysis and Immunohistochemistry (IHC) Mod Pathol. 2011;24(Suppl):179A. [Google Scholar]
- 41.Martignoni G, Segala D, Borze I, et al. VHL Mutation, VHL Methylation, Chromosome 3p and Whole Genomic Status in Clear Cell Papillary Renal Cell Carcinoma. Mod Pathol. 2013;26(Suppl):233A. [Google Scholar]
- 42.Young AC, Craven RA, Cohen D, et al. Analysis of VHL Gene Alterations and their Relationship to Clinical Parameters in Sporadic Conventional Renal Cell Carcinoma. Clin Cancer Res. 2009;15:7582–7592. doi: 10.1158/1078-0432.CCR-09-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halat S, Eble JN, Grignon DJ, et al. Multilocular cystic renal cell carcinoma is a subtype of clear cell renal cell carcinoma. Mod Pathol. 2010;23:931–936. doi: 10.1038/modpathol.2010.78. [DOI] [PubMed] [Google Scholar]
- 44.Rechsteiner MP, von Teichman A, Nowicka A, et al. VHL gene mutations and their effects on hypoxia inducible factor HIFalpha: identification of potential driver and passenger mutations. Cancer Res. 2011;71:5500–5511. doi: 10.1158/0008-5472.CAN-11-0757. [DOI] [PubMed] [Google Scholar]
- 45.Mertz KD, Demichelis F, Sboner A, et al. Association of cytokeratin 7 and 19 expression with genomic stability and favorable prognosis in clear cell renal cell cancer. Int J Cancer. 2008;123:569–576. doi: 10.1002/ijc.23565. [DOI] [PubMed] [Google Scholar]
- 46.Nickerson ML, Jaeger E, Shi Y, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrie CH, Larrea E, Larrinaga G, et al. Targeted next-generation sequencing and non-coding RNA expression analysis of clear cell papillary renal cell carcinoma suggests distinct pathological mechanisms from other renal tumour subtypes. J Pathol. 2014;232:32–42. doi: 10.1002/path.4296. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 49.Korpal M, Lee ES, Hu G, et al. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisher KE, Yin-Goen Q, Alexis D, et al. Gene expression profiling of clear cell papillary renal cell carcinoma: comparison with clear cell renal cell carcinoma and papillary renal cell carcinoma. Mod Pathol. 2014;27:222–230. doi: 10.1038/modpathol.2013.140. [DOI] [PubMed] [Google Scholar]
- 51.Alexiev BA, Drachenberg CB. Clear cell papillary renal cell carcinoma: Incidence, morphological features, immunohistochemical profile, and biologic behavior: A single institution study. Pathol Res Pract. 2014 Apr;210(4):234–41. doi: 10.1016/j.prp.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 52.Guo J, Tretiakova MS, Troxell ML, et al. Tuberous sclerosis-associated renal cell carcinoma: a clinicopathologic study of 57 separate carcinomas in 18 patients. Am J Surg Pathol. 2014;38:1457–1467. doi: 10.1097/PAS.0000000000000248. [DOI] [PubMed] [Google Scholar]
- 53.Aron M, Zhang P, De Peralta-Venturina M, et al. Expression of Novel Markers Human Kidney Injury Molecule-1 (Hkim-1), S100A1 and Napsin A in the Differential Diagnosis of Renal Cell Carcinomas (RCC) with Clear and Papillary Features. Mod Pathol. 2012;25(Suppl):190A. [Google Scholar]
- 54.Boysen G, Bausch-Fluck D, Thoma CR, et al. Identification and functional characterization of pVHL-dependent cell surface proteins in renal cell carcinoma. Neoplasia. 2012;14:535–546. doi: 10.1596/neo.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williamson WSR, Zhang S, Eble JN, et al. Clear cell papillary renal cell carcinoma-like tumors in patients with von Hippel-Lindau disease are unrelated to sporadic clear cell papillary renal cell carcinoma. Am J Surg Pathol. 2013;37:1131–1139. doi: 10.1097/PAS.0b013e318282dab8. [DOI] [PubMed] [Google Scholar]
- 56.Schwartz JD, Amin MB, Zhang PL. Immunohistochemical Profile of Stem/Progenitor Cell Marker CD133 in Variants of Renal Tumors. Mod Pathol. 2012;25(Suppl):240A. [Google Scholar]
- 57.Munari E, Segala D, Gobbo S, et al. GATA3 Expression in Clear Cell Papillary Renal Cell Carcinoma and Renal Cell Carcinoma with Prominent Leiomyomatous Proliferation Is a Further Evidence of the Relationship between These Two Entities. Mod Pathol. 2014;27(Suppl):250A. [Google Scholar]
- 58.Leroy X, Camparo P, Gnemmi V, et al. Clear cell papillary renal cell carcinoma is an indolent and low-grade neoplasm with overexpression of cyclin-D1. Histopathology. 2014;64:1032–1036. doi: 10.1111/his.12359. [DOI] [PubMed] [Google Scholar]
- 59.Williamson SR, Halat S, Eble JN, et al. Multilocular cystic renal cell carcinoma: similarities and differences in immunoprofile compared with clear cell renal cell carcinoma. Am J Surg Pathol. 2012;36:1425–1433. doi: 10.1097/PAS.0b013e31825b37f0. [DOI] [PubMed] [Google Scholar]
- 60.Dahinden C, Ingold B, Wild P, et al. Mining tissue microarray data to uncover combinations of biomarker expression patterns that improve intermediate staging and grading of clear cell renal cell cancer. Clin Cancer Res. 2010;16:88–98. doi: 10.1158/1078-0432.CCR-09-0260. [DOI] [PubMed] [Google Scholar]