Abstract
Response to targeted therapies varies significantly despite shared oncogenic mutations. Nowhere is this more apparent than in BRAF(V600E)-mutated melanomas where initial drug response can be striking and yet relapse is commonplace. Resistance to BRAF inhibitors have been attributed to the activation of various receptor tyrosine kinases (RTKs) though the underlying mechanisms have been largely uncharacterized. Here, we found that EGFR induced vemurafenib resistance is ligand dependent. We then employed whole-genome expression analysis and discovererd that vemurafenib resistance correlated with the loss of MITF, along with its melanocyte lineage program, and with the activation of EGFR signaling. An inverse relationship between MITF, vemurafenib resistance and EGFR was then observed in patient samples of recurrent melanoma and was conserved across melanoma cell lines and patients’ tumor specimens. Functional studies revealed that MITF depletion activated EGFR signaling and consequently recapitulated the resistance phenotype. In contrast, forced expression of MITF in melanoma and colon cancer cells inhibited EGFR and conferred sensitivity to BRAF/MEK inhibitors. These findings indicate that an “autocrine drug resistance loop” is suppressed by melanocyte lineage signal(s), such as MITF. This resistance loop modulates drug response and could explain the unique sensitivity of melanomas to BRAF inhibition.
Introduction
Primary and secondary resistance to molecular therapies remains a cardinal challenge in the clinical setting. For metastatic melanoma, the pace of progress from the benchside discovery of BRAF(V600E) to the bedside delivery of vemurafenib (VEM) has been rapid. As with other targeted agents, however, acquired resistance to selective BRAF inhibitors (SBI) soon followed on the heels of clinical success. COT expression(Johannessen et al., 2010), SOX10 reduction(Sun et al., 2014), BRAF amplification(Shi et al., 2012), splice variation(Poulikakos et al., 2011), NRAS mutagenesis and receptor tyrosine kinase (RTK) activation(Girotti et al., 2013; Nazarian et al., 2010; Villanueva et al., 2010; Yadav et al., 2012), have all been linked to SBI resistance in melanoma. Although these mechanisms all confer a similar phenotype of mediating cell survival and proliferation, the relative contribution of subclonal selection versus epigenetic reprogramming of cell state to the emergence of each mechanism is not known. On the other hand, the lineage dependency of SBI sensitivity suggests that cellular differentiation state might underlie any of the previously described mechanisms of resistance to SBI.
We thus set out to characterize changes in transcriptional programming that occur during the course of in vitro selection for VEM resistance. Using a forward pharmacogenetic screen, we discovered that VEM resistance was associated with different degrees of cellular reprogramming. On the one hand, resistance that emerges from a BRAF splice product is associated with minimal changes in cell state. On the other hand, resistance can also be associated with significant transcriptome changes anchored by the concomitant loss of the master melanocyte lineage regulator, MITF, with the activation of EGFR, a pathway little utilized in these neural crest derived cells. We show that modulating MITF levels alters VEM sensitivity in both melanoma and colon cancer, and that a reciprocal relationship between MITF and EGFR is conserved across melanoma specimens and correlates with VEM response. Our studies point to lineage identity as a major determining factor for SBI sensitivity.
Results
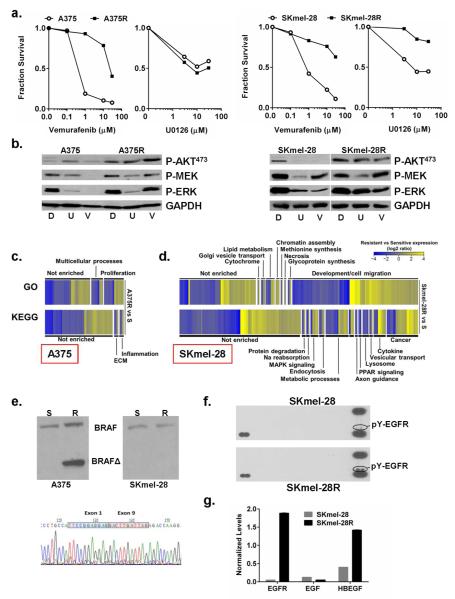
In order to elucidate programmatic changes that occur with VEM resistance, we subjected two melanoma cell lines, A375 and SKmel-28, to escalating doses of VEM in order to generate isogenically-matched pairs of sensitive and resistant cell lines. Dose interrogation showed that both resistant cell lines (i.e. A375R and SKmel-28R) exhibited >10 fold increase in their VEM GI50’s (Fig 1a) compared to their parental counterparts. Both resistant lines showed increased pAKT473 and re engagement of MAPK signaling although the A375R cells retained sensitivity to MEK inhibition while SKmel-28R developed cross-resistance (Fig 1a, 1b). We subsequently subjected all 4 sets of cell lines in triplicate to comparative gene expression analysis.
Figure 1. Characteristics of vemurafenib resistant lines.
(a). A375R and SKmel-28R cells (closed squares) showed >10-fold increase in VEM GI50 compared to their parent cell lines (open circles). SKmel-28R, but not A375R, are also cross-resistant to MEK inhibitor U0126 (5 μM). (b). Kinase signaling responses to DMSO control (labeled D), U0126 (U; 5 μM) and VEM (V; 5 μM) in A375, A375R, SKmel-28 and SKmel-28R lines. Gene Ontology (GO) and KEGG analyses for differentially expressed genes between the A375 and A375R pair (c) and between the SKmel-28 and SKmel-28R pair (d). (e). A375R cells harbor a BRAFΔ2-8 splice variant, which is not present in either the native A375 cells or the SKmel-28/SKmel-28R pair. (f). Phosphotyrosine (pY)-RTK blot analysis shows increased pY-EGFR in the SKmel-28R cells compared to the SKmel-28 cells while qPCR analysis (g) reveals upregulation of HB-EGF and EGFR, but not EGF, in the SKmel-28R line.
Profiling of the A375 versus A375R pair revealed strikingly sparse gene expression changes associated with the gain of VEM resistance (Fig S1). Of the 12,466 species surveyed, only 0.37% and 0.52% showed >2-fold induction and suppression, respectively, with maximum changes ranging from a 5.8-fold induction of MAGEA1 to a 6.7-fold suppression of SERPINA3 (Table S1). Gene Ontology (GO) and KEGG categories impacted by these minor expression variations (Fig 1c) included “proliferation” (GO) and “inflammation” and “ECM” (KEGG). Since the A375R cells retained sensitivity to MEK inhibitors (Fig 1a,b), we hypothesized that the resistance lesion was upstream of MEK. Exome sequencing (Table S2) did not detect any acquired mutations in NRAS, HRAS, KRAS, MAP2K1, MAP2K2, CRAF or BRAF, although there was a BRAF(Δ2-8) splice variant that was present in the A375R but not the A375 parent line (Fig 1e). This specific alteration has been reported in a VEM-resistant human tumor specimen (pt #5 from a recent report(Poulikakos et al., 2011)) and therefore is likely driving VEM resistance in A375R cells.
In contrast to the A375 pair, expression analysis of SKmel-28 versus SKmel-28R revealed significant programmatic changes with the emergence of VEM resistance (Fig S1). Overall, 3.4% and 3.0% of the genes exhibited a >2-fold increase or decrease, respectively, with a dynamic range of 114-fold induction (IL1B) and 57-fold suppression (LOC728715, Table S1). GO analysis indicated major shifts in “development/cell migration” genes while KEGG categorization yielded numerous enrichments including “cancer”, “cytokine” and “metabolic processes” (Fig 1d). The SKmel-28R cells lacked putative BRAF splice variants (Fig 1e) and differed from the A375R cells in demonstrating co-resistance to both VEM and MEK inhibition (Fig 1a,b). Comparative phosphotyrosine (pY) RTK blot analysis of the SKmel-28 and SKmel-28R pair uncovered sustained EGFR signaling on SKmel-28R cells (Fig 1f), which has been reported to cause VEM resistance(Corcoran et al., 2012; Girotti et al., 2013). There’s no dramatic alteration of phospho EGFR in A375R (Fig S1C). Additional exome sequencing of SKmel-28 and SKmel-28R did not reveal biologically plausible acquired variants in EGFR, NRAS, HRAS, KRAS, MAP2K1, MAP2K2, CRAF or BRAF (Table S2). Taken together, these results suggest that direct target modification, such as the BRAF splice product in A375R cells, neutralizes drug effects by resetting a specific signaling pathway but leaves few programmatic footprints. In contrast, EGFR activation in SKmel-28R cells appears to be associated with more profound gene expression alterations. We thus set out to clarify the mechanism by which EGFR may have become activated in the SKmel-28R cells.
Since growth factors and cytokines are well known activators of RTK signaling, we first interrogated these genes in the microarray and found that a surprising number was upregulated during the gain-of-resistance in SKmel-28. Among candidate ligand-RTK pairings, HB-EGF-EGFR and GAS6-AXL levels were all increased (Fig S2) though only EGFR appeared to be activated in the phosphotyrosine (pY) RTK blot analysis (Fig 1f). qPCR of Skmel-28R cells confirmed a 39-fold increase in EGFR and a 3.5-fold induction of HB-EGF compared to VEM sensitive Skmel-28 cells (Fig 1g). Thus, an EGFR auto-stimulatory circuit appears to be selectively sustained and mediating resistance in the SKmel-28R cells.
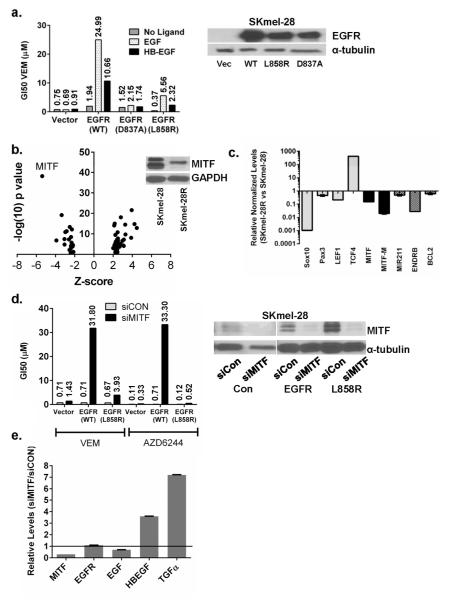
To experimentally validate the EGFR findings, we generated stable SKmel-28 lines expressing wild-type EGFR, oncogenic EGFR(L858R), or kinase-dead EGFR(D837A) (Fig 2a). In the absence of EGFR ligand, there was only a minimal gain in VEM resistance in EGFR overexpression lines, with the gains in VEM GI50’s for SKmel-28EGFR(WT), SKmel-28EGFR(D837A) and SKmel-28EGFR(L858R) cells all less than 3-fold compared to SKmel-28VECTOR (GI50 =0.75 μM). However, upon the addition of EGF or HB-EGF, VEM resistance was dramatically enhanced in wild-type EGFR overexpression lines (Fig 2a). There was a 36-fold and a 12-fold increase in VEM GI50’s when EGF or HB-EGF, respectively, were exogenously added. As expected, the kinase-inactive EGFR(D837A) allele had minimal effects on VEM resistance even in the presence of EGF or HB-EGF. Since both AXL and GAS6 were also upregulated in SKmel-28R compared to SKmel-28 cells in the microarray data, we also transduced AXL into SKmel-28 cells. However, we observed only minimal effects on VEM sensitivity either in the absence or presence of exogenous GAS6 (Fig S3). These results indicate that overexpression of EGFR alone may not be sufficient to induce resistance and that ligand upregulation is a critical component of an “autocrine resistance loop.”
Figure 2. Loss of MITF contributes to an EGFR autocrine resistance loop in SKmel-28R cells.
(a). Stable expression in SKmel-28 cells of EGFR(WT) or EGFR(L858R), but not kinase-dead EGFR(D837A), leads to VEM resistance in the presence of exogenously added EGF (10 ng/mL) or HB-EGF (10 ng/mL), but not in the absence of ligand. Expression of EGFR alone does not significantly increase VEM resistance. (b). Using the Ingenuity software, transcription factor analysis of genes differentially expressed between SKmel-28 and SKmel-28R indicate a strong suppression of MITF target genes. Western blotting confirms the loss of MITF protein in SKmel-28R cells (inset). (c) qPCR shows loss of MITF expression in SKmel-28R cells along with that of other melanocytic lineage regulators (SOX10, PAX3, LEF1) and various MITF targets (MITF M, MiR211, ENDRB, BCL2). Levels of TCF4 (ITF2), which is known to suppress MITF(Furumura et al., 2001), is greatly elevated. (d). siRNA-mediated depletion of MITF (denoted siMITF) in stable SKmel-28EGFR(WT) and SKmel-28EGFR(L858R) cells leads to enhanced VEM and AZD6244 resistance even in the absence of EGFR ligand. (e). RNA levels of HB-EGF and TGFα, two known ligands of EGFR, are increased in SKmel-28 cells where MITF has been depleted by siRNA. Thus, loss of MITF closes the “autocrine resistance loop” in the face of higher EGFR expression. Numbers above columns represent actual GI50 values.
To elucidate determinants of this resistance loop, we next performed transcriptional factor analysis on differentially-expressed genes in SKmel-28R versus SKmel-28 cells (Table S3). As shown in Fig 2b, MITF suppression was the leading transcriptional footprint with a Z-score of −5.391 (p=6.37×10−39); there was also inhibition of SOX10 activity (Z-score, −2.153, p=2.59×10−5). These findings are consonant with the categorical change of “development/cell migration” genes, as recovered by GO classification of the microarray data. Validation of the microarray analysis by qPCR and western blotting confirmed dramatic reductions in MITF and MITF-M along with several downstream MITF targets: BCL2, EDNRB, and miR-211 (Fig 2c). Overall, the acquisition of VEM resistance in SKmel-28 cells appears to have silenced the entire melanocytic program as positive upstream MITF regulators (LEF1, PAX3 and SOX10) were all diminished while the negative MITF regulator TCF4 was increased by 420-fold (Fig 2c).
To determine if MITF loss cooperates with EGFR activation in mediating resistance, we depleted MITF in the stable EGFR SKmel-28 lines (Fig 2d) using siRNA and observed the acquisition of strong resistance against both VEM (45-fold increase in GI50) and AZD6244 (300-fold increase in GI50) in the SKmel-28EGFR(WT) cells even in the absence of EGFR ligand (Fig 2d). siMITF had no acute effects on the amount of EGFR and EGF but did immediately increase the levels of HB-EGF (3.6-fold) and TGFα (7.2-fold), both of which are known ligands for EGFR (Fig 2e). In addition, neuregulin 1, (NRG1), an indirect activator of EGFR through ERBB4 or ERBB3 binding, was also increased 5.1-fold after MITF knockdown (data not shown). These findings indicate that MITF suppression may create an autocrine ligand-rich environment which synergizes with EGFR upregulation to mediate resistance.
In order to independently replicate the MITF/EGFR observation, we generated additional pairs of VEM sensitive and resistant melanoma cell lines and found that the emergence of VEM resistance in another line, MGH-CH1, was also correlated with MITF loss and EGFR activation (Fig S4a,b). Using the aforementioned strategy, we stably introduced EGFR(WT), EGFR(L858R) and EGFR(D837A) alleles into MGH-CH1 cells and assessed VEM sensitivity. As shown in Fig S4c, both MGH-CH1EGFR(WT) and MGH-CH1EGFR(L858R) cells exhibited ligand-dependent VEM resistance compared to MGH-CH1vector and MGH-CH1EGFR(D837A) cells, which was consistent with the SKmel-28 results. Next, suppression of MITF in MGH-CH1EGFR(L858R) cells also led to a gain of resistance to both VEM and AZD6244 in the absence of ligand (Fig S4d). In contrast to SKmel-28 cells, however, the EGFR(L858R) allele seemed to play a stronger role than wildtype EGFR in the MGH-CH1 cells.
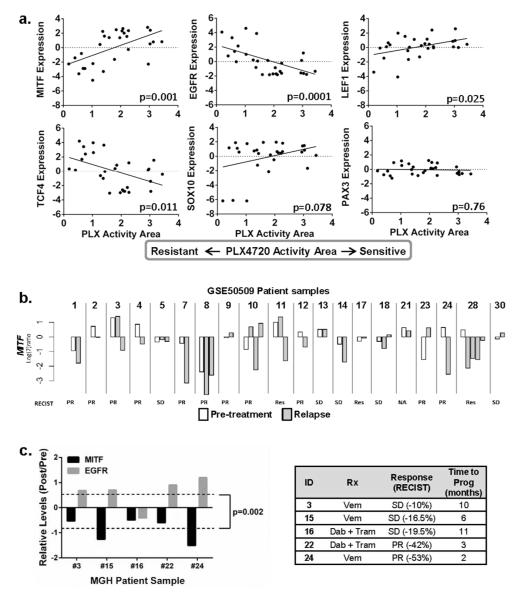
We next sought broader evidence of an interaction between levels of MITF, EGFR and therapeutic sensitivity. First, we examined the Cancer Cell Line Encyclopedia (CCLE) database(Barretina et al., 2012) and found significant correlations (Fig 3a) between PLX4720 resistance and low MITF (p=0.001), high EGFR (p=0.0001), low LEF1 (p=0.025) and high TCF4 (p=0.011); PLX4720 insensitivity was marginally related to low SOX10 (p=0.078) but not to PAX3 (p=0.76). In addition, copy number analysis using a set of BRAF(V600E) melanoma lines(Ji et al., 2012) revealed that MITF amplification is associated with increased sensitivity to both VEM and the MEK inhibitor U0126 (Fig S5), further strengthening MITF’s role in melanoma’s response to MAPK pathway inhibition. Thus, the pattern of lineage silencing, high EGFR, and SBI insensitivity appears to be preserved across a panel of melanoma lines. We next set out to confirm the relationship between acquired VEM resistance and MITF levels in tumor specimens. We first examined changes in MITF expression using a publicly available microarray data set (Gene Expression Omnibus (GEO) GSE50509) which captured gene expression data for 20 pre-treatment and 30 relapse melanoma specimens from 20 patients. As shown in Fig 3b, the majority of relapsed tumors showed evidence of MITF loss. Since EGFR probes in GSE50509 did not pass our quality filter, we also verified trends in MITF and EGFR expression by qPCR using an in-house collection of BRAF(V600E)-mutated tumors from patients treated with BRAF±MEK inhibitors. The average relative log2-fold change between pre- and post-relapse specimens was −0.88 for MITF and 0.61 for EGFR (Fig 3c; p=0.002). In 4/5 tumor pairs, the relapse specimen had lower MITF levels but higher EGFR levels compared to the pre-treatment samples.
Figure 3. Expression of the melanocyte lineage program and EGFR correlates with VEM sensitivity.
(a). PLX4720 sensitivity (i.e. increased PLX activity area) is positively correlated with higher MITF, LEF1 and SOX10 levels and negatively associated with higher EGFR and TCF4 levels. (b). Data from GSE50509 showing MITF levels in pre-treatment (white columns) and post-relapse (gray columns) specimens. Results are shown as log2 ratios normalized to the mean intensity of pre-treatment samples. (c). Relative (post-relapse to pre-treatment) and normalized (to GUSB) levels of MITF and EGFR in clinical specimens from patients on BRAF/MEK inhibitor trials. RNA levels were determined by qPCR. There is a significant trend towards decreased MITF expression and increased EGFR expression in the post-relapse samples compared to pre-treatment samples.
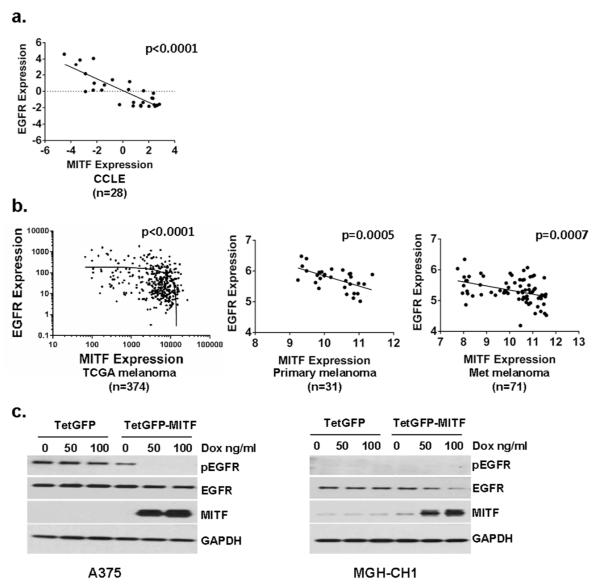
The overall expression patterns that relate both MITF and EGFR to VEM response also suggest that the two molecules may exhibit innate reciprocity. First, a significant negative correlation between MITF and EGFR was identified in the 28 CCLE melanoma lines (Fig 4a; p<0.0001). Turning to melanoma specimens rather than cell lines, significant reciprocal relationships were also observed in the 374 tumors (p<0.0001) available through The Cancer Genome Atlas (TCGA; Fig 4b, Table S4) and 31 primary melanomas (p=0.0005) and 71 metastatic melanoma specimens (p=0.0007) available in the GEO (GSE46517; Fig 4b, Table S4). Implicit in these findings is the possibility that MITF may directly counter-regulate EGFR signaling. To test this hypothesis, we induced MITF expression short-term in A375 and MGH-CH1 cells using the Tet-on system (Fig 4c). In the A375 cells, there was a modest reduction in EGFR but a complete abrogation of pEGFR. In the MGH-CH1 cells, there was a dramatic loss of EGFR though basal levels of pEGFR were undetectable. Interestingly, with the acute elevation of MITF levels, there appeared to be a dose-dependent growth retardation in both cell lines (Fig S6).
Figure 4. Reciprocity between MITF and EGFR in melanoma.
(a). An inverse relationship between levels of MITF and EGFR can be observed across 28 CCLE melanoma samples and (b) 374 melanoma tumor specimens in the TCGA and 31 primary melanoma tumors and 71 metastatic melanoma specimens from GSE46517. Linear regression indicated within dot plot; of note, the TCGA data are plotted in log-log scale and thus the linear regression line appears downwardly curved. (c). Induction of MITF in A375 and MGH-CH1 cells using a Tet-on promoter leads to a complete suppression of pEGFR and a slight decrease in EGFR in the A375 cells and a significant decrease in EGFR in the MGH-CH1 cells.
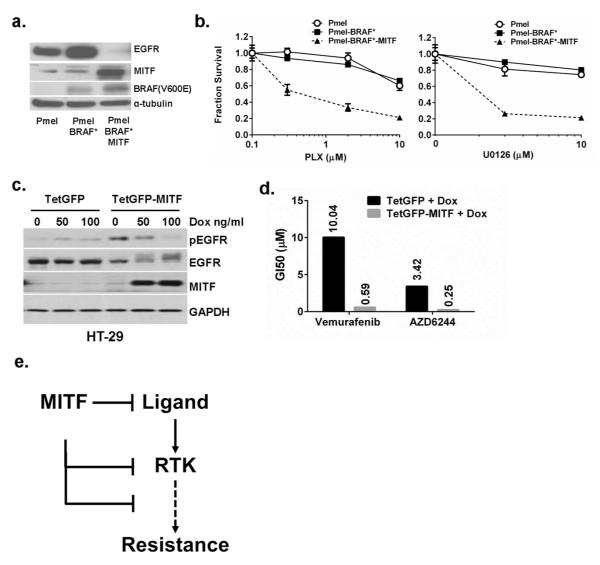
In order to directly assess the drug sensitizing effects of MITF, we used a set of immortalized primary melanocyte lines (Pmel; Fig 5a) which have stably incorporated BRAF(V600E) alone (Pmel-BRAF*) or both BRAF(V600E) and MITF (Pmel-BRAF*-MITF) together(Garraway et al., 2005). These cells were selected because they harbor the minimal essential elements for defining drug response. As shown in Fig 4a, the native Pmel line had low MITF expression and no BRAF(V600E) as assessed by the VE-1 antibody. EGFR was clearly expressed in both the Pmel and Pmel-BRAF* cells. On the other hand, Pmel-BRAF*-MITF cells had near abolition of EGFR expression. The Pmel-BRAF*-MITF cells were also significantly more sensitive to the selective BRAF inhibitor PLX4720 and MEK inhibitor U0126 (Fig 5b) compared to both Pmel and Pmel-BRAF* cells suggesting that the addition of MITF conferred enhanced drug sensitivity even in cells with extant BRAF(V600E). We next determined if MITF can have a similar impact on drug response in non-pigment cells by using a Tet-on MITF system that was engineered into the BRAF(V600E)-mutated HT29 colon cancer line. Forced expression of MITF reduced the levels of pEGFR (Fig 5c) and engendered a >10-fold increase in sensitivity (as measured by GI50’s) to both VEM and AZD6244 (Fig 5d). These results support a role for MITF in regulating EGFR and in modulating drug response for both pigment cells and non-pigment cells.
Figure 5. Overexpression of MITF confers therapeutic sensitivity.
(a). Immortalized primary melanocytes (Pmel) with stable expression of BRAF(V600E) and MITF (Pmel-BRAF*-MITF) have lower EGFR levels than either control Pmel cells or Pmel cells with only BRAF(V600E) (Pmel-BRAF*). (b). Pmel-BRAF*-MITF (solid triangle) cells are more sensitive to both PLX and U0126 than Pmel (open circle) and Pmel-BRAF* (solid squares) cells. (c). The colon cancer cell line HT29, which harbors a BRAF(V600E) mutation, was transduced with a Tet-on vector (TetGFP) or Tet-on MITF (TetGFP-MITF) and induced to express MITF using the indicated doses of doxycycline. (d). Induced expression of MITF in HT29 leads to increased sensitivity to vemurafenib and AZD6244, as determined by a >10-fold reduction in the GI50. (e). One possible model suggests that MITF, or other lineage determinants, may influence therapeutic resistance by modulating levels of ligand (e.g. HB-EGF), RTKs (such as EGFR) or other downstream components of RTK signaling.
Discussion
Acquired therapeutic resistance in melanoma has been ascribed to various mechanisms. However, the relationship between these acquired lesions and underlying transcriptional programs are not well defined. Our studies suggest that a balance between lineage identity and RTK activation modulates drug sensitivity. More specifically, loss of MITF potentiates an EGFR “autocrine resistance loop” that is not normally utilized by the melanocyte lineage, which then mediates therapeutic resistance.
Our analysis of the CCLE data(Barretina et al., 2012), and those of others(Konieczkowski et al., 2014), supports the notion that melanomas with weak lineage identity (low MITF, LEF1 and SOX10) appear to be more resistant to PLX. This hypothesis would harmonize with the rapid ability of colorectal cancers to utilize a lineage-appropriate expression of EGFR to undermine the effects of SBIs in tumors with BRAF mutations(Corcoran et al., 2012; Prahallad et al., 2012). Thus, cells with nominal MITF may predominate during the course of selection. Alternatively, MITF may directly, or indirectly, suppress the EGFR signaling system (Fig 5e) as suggested by our experiments modulating MITF in immortalized melanocytes, melanoma and colon cancer cells. The use of Pmel cells with stable and genetically-defined elements (i.e. BRAF(V600E) and MITF) provides perhaps the most precise and direct evidence that MITF-enriched cells adopt a low-EGFR state that is more drug sensitive. With forced MITF expression in A375 and MGH-CH1 melanomas and HT29 colon cancer cells, there was a direct reduction of EGFR and/or pEGFR.
We also observed an increase in pEGFR with siMITF depletion but only when cells were stably endowed with ectopic EGFR. Both SKmel-28 and MGH-CH1 lines express robust levels of MITF but exquisitely low levels of EGFR. The prevalence of this phenotype across the melanoma population may explain the markedly higher response rate to SBIs compared to BRAF mutant colorectal cancer. The cooperativity between MITF depletion and overexpression of EGFR and its ligands suggests that MITF loss may contribute to resistance by augmenting levels of ligands, such as HB-EGF and TGF-α, neuregulin 1 or IL-8, which has reported to be negatively regulated by MITF(Hari Kishore et al., 2012) and which is known to transactivate EGFR(Itoh et al., 2005). Interestingly, a recent report shows that IL8 signaling can induce chemo-resistance by maintaining melanoma initiating cells(Wilson et al., 2014). Lastly, low MITF and high EGFR (and perhaps AXL(Konieczkowski et al., 2014)) could mark exclusive cellular states that harbor distinct therapeutic susceptibilities unrelated to the direct function of these proteins. This static view is supported by the innate reciprocity between MITF and EGFR expression which is preserved across multiple collections of melanoma tumors for which expression data are available (Fig 4a, b). However, the precise mechanism(s) by which MITF interacts with the EGFR signaling system remains the subject of ongoing investigation.
Although there is burgeoning appreciation for a link between melanocyte lineage identity and RTK signaling, there have been several reports examining the role of MITF in dictating drug response. On the one hand, Smith et al. found that when A375 cells were induced into resistance against MEK inhibition, emergent subclones exhibited higher MITF and SMURF2 levels(Smith et al., 2013). Along these lines, Johannessen et al. used A375 cells in a reverse screen and found that genes in the cAMP pathway induced MITF, strengthened lineage identity and conferred resistance to RAF, MEK and ERK inhibitors(Johannessen et al., 2013); it is worth noting, however, that one of the patient tumors reported in the paper clearly showed loss of MITF with relapse (#16; extended data Fig 10d ref(Johannessen et al., 2013)). On the other hand, Konieczkowski et al. found that intrinsic PLX resistance is associated with low MITF along with high NF-κB levels(Konieczkowski et al., 2014). In a related study, Sun et al. also showed that depletion of lineage gene SOX10, but not MITF, increased EGFR and VEM resistance(Sun et al., 2014). In that report, sh(MITF) did not alter VEM sensitivity though it may be related to the cell line used; in our hands, SKmel-28 cells have 11-fold more MITF than A375 cells (data not shown) and thus the A375 cells may have come to depend on SOX10 signaling to a greater extent. Instead of inducing resistance by SOX10 loss only, our study indicates that MITF loss only confers resistance in the presence of EGFR expression.
Our data indicates that RTK induced resistance is dependent on ligand, which broadens the picture of growth factors in drug resistance, which was initially suggested in paracrine HGF induced primary BRAFi resistance(Straussman et al., 2012; Wilson et al., 2012). Our data is also consonant with a recent report that high EGFR expressing cells often lack EGFR activation, presumably due to lack of ligands, and are still sensitive to BRAFi; thus, EGFR levels alone cannot be used as a biomarker of vemurafenib resistance(Gross et al., 2014). Our results also suggest that ligand measurement in blood or other body fluid might be an eventual method to predict drug resistance clinically. It is intriguing that stable EGFR phosphorylation by constitutively active L858R mutant does not induce resistance, insight from which might offer a more detailed understanding of EGFR-dependent resistance and strategies to overcome resistance.
In summary, lineage reprogramming appears to be directly coupled to RTK activation in the setting of therapeutic resistance. These comparative molecular studies provide a framework for understanding shifts in transcriptional states as resistance lesions emerge under drug selection. Efforts are now underway to recover useful therapeutic agendas to overcome these programs.
Materials and Methods
Compounds, antibodies and reagents
Vemurafenib, PLX4720, AZD6244 and U0126 were from Selleck Chemicals (Houston, TX). EGF, HB-EGF and p-RTK arrays were purchased from R&D (Minneapolis, MN). Doxycycline was from Clontech (Mountain View, CA). Antibodies against p-EGFR, EGFR, BRAF, p-MEK. MEK, p-ERK, ERK, p-AKT, AKT, GAPDH and α-tubulin were from Cell Signaling (Danvers, MA). CellTiter-Glo cell viability assay was from Promega (Madison, WI). AlamarBlue cell viability reagent was from Life Technologies (Grand Island, NY). Human phospho-RTK array kit was from R&D systems (Minneapolis, MN). siRNA against MITF and non-targeting control siRNA were from GE Dharmacon (Lafayette, CO).
Long-term cell proliferation assays
Cells were seeded into 6-well plates (1 × 104 cells per well) and cultured both in the absence and presence of drugs as indicated. 10-15 days later, cells were fixed with paraldehyde and stained with 1% crystal violet for 10min. The plates were then washed with ddH2O and dried in air before photographed.
Drug treatment and cell viability assay
Cell viability was determined by AlamarBlue (Life Technologies) fluorescence assay and CellTiter-Glo (Promega) luminescence assay. Approximately 12 hours before drug treatment, cells were seeded at a density of 3,000 cells (100 μL) per well in a 96-well plate. The plates were incubated with drugs for 48 hours. Ten microliters of AlamarBlue was added to each well and incubated for 3 hours at 37°C. AlamarBlue is fluorescent substrates reduced by mitochondrial enzyme activity in viable cells. Alternatively, 30 μL of CellTiter-Glo was added to each well and incubated, protected from light on an orbital shaker, for 10 minutes. CellTiter-Glo contains luciferase, which catalyzes the oxygenation of luciferin (creating light) according to the amount of ATP present. Fluorescence or luminescence intensity was determined using a Molecular Devices plate reader with an excitation filter centered on 540 nm and an emission filter centered on 590 nm or with an integration time of 500 milliseconds and measuring total light emitted, respectively. GI50’s were calculated using the software CompuSyn (http://www.combosyn.com).
Stable infections
Lentiviral transduction was used to alter the levels of MITF. Lentiviral supernatant was produced by transient transfection of HEK293T cells (ATCC) using lipofectamine (Life Technologies, Inc.), according to the manufacturer’s instructions. The viral-containing supernatants were harvested 48 hours after transfection and filtered through a 0.45 μm filter unit. To transduce melanoma cells with lentivirus, logarithmically growing melanoma cells were seeded at a density of 2×105 cells per well in 6-well plates. A total of 0.5 mL of lentivirus suspension and 8 μg/mL of polybrene were added to DMEM with 10% FBS in a total volume of 1 mL. Cells were incubated at 37°C for 12 hours before removing the medium and replacing with 2 mL of fresh DMEM for expansion of the transductants. Cells were selected with puromycin at 1.5 μmol/L for another 5 days before further experiments.
Protein lysate preparation and immunoblots
Cells were seeded in medium containing 10% fetal bovine serum (FBS) for 24 h, and then washed with PBS and lysed with RIPA buffer supplemented with Halt protease inhibitor cocktail (Pierce). Equal amounts of protein (5–20 μg) were loaded onto 4%–20% SDS polyacrylamide mini-gels (Bio-Rad), transferred to polyvinylidene difluoride membranes. After being blocked in 5% milk in TBS-Tween for 1 hour, blots were incubated with primary antibodies overnight, followed by horseradish peroxidase-conjugated secondary antibody (1:5,000) for 45 minutes. Antigen–antibody complexes were detected by enhanced chemiluminescence. Human p-RTK arrays were performed according to the manufacturer’s instructions.
Melanoma patient tumor samples
Human melanoma specimens (all BRAF V600 mutated) were obtained from patients undergoing treatment with vemurafenib, dabrafenib or trametinib in accordance with a protocol approved by the MGH Institutional Review Board (IRB). All patients provided written informed consent for analysis, as approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board (DF/HCC Protocol 11-181).
RNA isolation and RT–PCR
RNA isolation from cell lines harvested with TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. cDNA synthesis was performed with Maxima Universal First Strand cDNA Synthesis Kit (#K1661, Thermo scientific) according to manufacturer’s instruction. cDNA was obtained by reverse transcription using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, AB) according to the manufacturer’s instructions. Levels of individual genes were quantified using a TaqMan Gene Expression Assays (Life Technologies) as previously described(Yang et al., 2006): EGFR (Hs01076078_m1), EGF(Hs01099999_m1), HB-EGF(Hs00181813_m1), MITF(Hs01117294_m1), TCF4(Hs00162613_m1), MIR211 (Hs04231471_s1), TGFA (Hs00608187_m1), SOX10(Hs00366918_m1), BCL2(Hs00608023_m1), PAX3(Hs00240950_m1), LEF1(Hs01548150_m1), CREB1(Hs00231713_m1), NRG1(Hs00247620_m1). Human GUSB (# 4333767T, Life Technologies) was used as an endogenous control. Primer sequences for MITF-M were from Dynek et al(Dynek et al., 2008 Dynek et al., 2008). Real time PCR was performed using the LightCycler480 (Roche), 95°C 10 min for denature and then 95°C × 10sec, 60°C to 62°C × 50sec for 45 cycles. The normalized, relative levels of the genes between samples were expressed as the log2 ratio (e.g., −10 = 2−10 2 = 1,024-fold reduction in expression in sample 1 vs sample 2).
Supplementary Material
Fig S1. Differences in log(2) expression levels of genes in A375 and A375R (A) and SKmel-28 and SKmel-28R (B). (C). p-EGFR expression in A375 and A375R cells. (D). Morphology of Skmel-28 and Skmel-28R. The pictures were taken with light microscope (10X). VEM sensitive SKmel-28 melanoma cells were more bipolar and stellate in appearance c/w pigment cells while VEM resistant SKmel-28R cells were more polygonal and epithelioid in appearance.
Fig S2. Relative levels (SKmel-28R vs. SKmel-28) of growth-factor related genes in samples 1-3.
Fig S3. Overexpression of AXL with or without GAS6 does not significantly increase resistance to VEM.
Fig S4. Functional analysis of resistance in MGH-CH1 cell line. (A). VEM inhibition curves for MGH-CH1/MGH-CH1R (B) phosphotyrosine-RTK blot shows increased pY-EGFR and decreased MITF in MGH-CH1 resistant (MGH-CH1R) cells compared to native MGH-CH1 cells. (C). Addition of EGF and HBEGF increases VEM resistance in MGH-CH1EGFR(WT) and MGH-CH1EGFR(L858R) lines. (D). Depletion of MITF in MGH-CH1EGFR(L858R) cells leads to enhanced resistance to both VEM and AZD6244.
Fig S5. MITF amplification is associated with increased VEM and U0126 sensitivity.
Fig S6. Induction of MITF leads to proliferative arrest. A375 and MGH-CH1 cells were transduced with (A) TetGFP-vector or (B) TetGFP-MITF. (C) Addition of doxycycline shows increase in GFP fluorescence in both TetGFP-vector and TetGFP-MITF. (C,D). Increasing doxycycline leads to proliferative arrest only in the TetGFP-MITF cells (D) but not the TetGFP-vector controls (C).
Table S1. RNA expression profiles of A375, SKmel-28 cells and their resistant counterparts.
Table S2. Exome sequencing data for major MAPK signaling genes in A375, SKmel-28 cells and their resistant counterparts.
Table S3. Ingenuity Transcription Factor analysis to identify alterations in acquired resistant melanoma cells.
Table S4. Data retrieved from TCGA and GSE46517 for EGFR and MITF analysis.
Acknowledgements
This work was supported in part by the National Institutes of Health (K24 CA149202, P01CA163222, 5T32CA071345, 5T32AR007098), the American Skin Association and the generous donors to the MGH Millennium Melanoma Fund. The work was further supported by the Swedish Cancer Society, Swedish Research Council, Berta Kamprad Foundation, Gunnar Nilsson Cancer Foundation and Gustav Vth Jubilee Foundation.
Footnotes
Conflict of Interest None
REFERENCES
- Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–35. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynek JN, Chan SM, Liu J, et al. Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas. Cancer Res. 2008;68:3124–32. doi: 10.1158/0008-5472.CAN-07-6622. [DOI] [PubMed] [Google Scholar]
- Furumura M, Potterf SB, Toyofuku K, et al. Involvement of ITF2 in the transcriptional regulation of melanogenic genes. J Biol Chem. 2001;276:28147–54. doi: 10.1074/jbc.M101626200. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Girotti MR, Pedersen M, Sanchez-Laorden B, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov. 2013;3:158–67. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A, Niemetz-Rahn A, Nonnenmacher A, et al. Expression and activity of EGFR in human cutaneous melanoma cell lines and influence of vemurafenib on the EGFR pathway. Targeted oncology. 2014 doi: 10.1007/s11523-014-0318-9. [DOI] [PubMed] [Google Scholar]
- Hari Kishore A, Li XH, Word RA. Hypoxia and PGE(2) regulate MiTF-CX during cervical ripening. Molecular endocrinology. 2012;26:2031–45. doi: 10.1210/me.2012-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Joh T, Tanida S, et al. IL-8 promotes cell proliferation and migration through metalloproteinase-cleavage proHB-EGF in human colon carcinoma cells. Cytokine. 2005;29:275–82. doi: 10.1016/j.cyto.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Ji Z, Njauw CN, Taylor M, et al. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J Invest Dermatol. 2012;132:356–64. doi: 10.1038/jid.2011.313. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Johnson LA, Piccioni F, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–42. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczkowski DJ, Johannessen CM, Abudayyeh O, et al. A Melanoma Cell State Distinction Influences Sensitivity to MAPK Pathway Inhibitors. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Shi H, Moriceau G, Kong X, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature communications. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MP, Ferguson J, Arozarena I, et al. Effect of SMURF2 targeting on susceptibility to MEK inhibitors in melanoma. J Natl Cancer Inst. 2013;105:33–46. doi: 10.1093/jnci/djs471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R, Morikawa T, Shee K, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Wang L, Huang S, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–22. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Lee JT, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BJ, Saab KR, Ma J, et al. ABCB5 maintains-melanoma initiating cells through a proinflammatory cytokine signaling circuit. Cancer Res. 2014;74:4196–207. doi: 10.1158/0008-5472.CAN-14-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Zhang X, Liu J, et al. Reactivation of mitogen-activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. J Biol Chem. 2012;287:28087–98. doi: 10.1074/jbc.M112.377218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Zhang G, Pittelkow MR, et al. Expression profiling of UVB response in melanocytes identifies a set of p53-target genes. J Invest Dermatol. 2006;126:2490–506. doi: 10.1038/sj.jid.5700470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Differences in log(2) expression levels of genes in A375 and A375R (A) and SKmel-28 and SKmel-28R (B). (C). p-EGFR expression in A375 and A375R cells. (D). Morphology of Skmel-28 and Skmel-28R. The pictures were taken with light microscope (10X). VEM sensitive SKmel-28 melanoma cells were more bipolar and stellate in appearance c/w pigment cells while VEM resistant SKmel-28R cells were more polygonal and epithelioid in appearance.
Fig S2. Relative levels (SKmel-28R vs. SKmel-28) of growth-factor related genes in samples 1-3.
Fig S3. Overexpression of AXL with or without GAS6 does not significantly increase resistance to VEM.
Fig S4. Functional analysis of resistance in MGH-CH1 cell line. (A). VEM inhibition curves for MGH-CH1/MGH-CH1R (B) phosphotyrosine-RTK blot shows increased pY-EGFR and decreased MITF in MGH-CH1 resistant (MGH-CH1R) cells compared to native MGH-CH1 cells. (C). Addition of EGF and HBEGF increases VEM resistance in MGH-CH1EGFR(WT) and MGH-CH1EGFR(L858R) lines. (D). Depletion of MITF in MGH-CH1EGFR(L858R) cells leads to enhanced resistance to both VEM and AZD6244.
Fig S5. MITF amplification is associated with increased VEM and U0126 sensitivity.
Fig S6. Induction of MITF leads to proliferative arrest. A375 and MGH-CH1 cells were transduced with (A) TetGFP-vector or (B) TetGFP-MITF. (C) Addition of doxycycline shows increase in GFP fluorescence in both TetGFP-vector and TetGFP-MITF. (C,D). Increasing doxycycline leads to proliferative arrest only in the TetGFP-MITF cells (D) but not the TetGFP-vector controls (C).
Table S1. RNA expression profiles of A375, SKmel-28 cells and their resistant counterparts.
Table S2. Exome sequencing data for major MAPK signaling genes in A375, SKmel-28 cells and their resistant counterparts.
Table S3. Ingenuity Transcription Factor analysis to identify alterations in acquired resistant melanoma cells.
Table S4. Data retrieved from TCGA and GSE46517 for EGFR and MITF analysis.