Abstract
The fused-type S100 protein profilaggrin and its proteolytic products including filaggrin are important in the formation of a normal epidermal barrier; however, the specific function of the S100 calcium-binding domain in profilaggrin biology is poorly understood. To explore its molecular function, we determined a 2.2Å-resolution crystal structure of the N-terminal fused-type S100 domain of human profilaggrin with bound calcium ions. The profilaggrin S100 domain formed a stable dimer, which contained two hydrophobic pockets that provide a molecular interface for protein interactions. Biochemical and molecular approaches demonstrated that three proteins, annexin II/p36, stratifin/14-3-3 sigma, and Hsp27, bind to the N-terminal domain of human profilaggrin; one protein (stratifin) co-localized with profilaggrin in the differentiating granular cell layer of human skin. Together, these findings suggest a model where the profilaggrin N-terminus uses calcium-dependent and calcium-independent protein-protein interactions to regulate its involvement in keratinocyte terminal differentiation and incorporation into the cornified cell envelope.
Introduction
Profilaggrin is an ∼ 400 kDa human protein critical for normal skin barrier development. It is principally expressed in a differentiation-dependent manner in the stratum granulosum (Presland et al., 2006). Loss-of-function mutations in the profilaggrin (FLG) gene associate it with the skin diseases atopic dermatitis and ichthyosis vulgaris (Akiyama, 2010; Palmer et al., 2006; Sandilands et al., 2006; Smith et al., 2006). Furthermore, variation in copy number of filaggrin monomers within the profilaggrin gene is associated with a dry skin phenotype in the general population (Brown et al., 2012).
In all mammals examined, profilaggrin has four distinct domains: 1) a 92 amino acid N-terminal S100 fused-type calcium-binding domain (CABD); 2) highly basic B domain containing nuclear localization signal (NLS); 3) variable number (species-dependent) of filaggrin units; 4) short C-terminal domain (Presland et al., 1992; Presland et al., 2006). Profilaggrin is highly phosphorylated at Ser/Thr residues within filaggrin units and packaged into keratohyalin granules within the stratum granulosum (Dale et al., 1980; Lonsdale-Eccles et al., 1980). During terminal differentiation of epidermis, when granular cells transition to dead corneocytes, profilaggrin is dephosphorylated and processed by multiple proteases to generate the discrete N-terminal peptide containing the S100 and B domains (PF-NT), and individual filaggrin units (Kam et al., 1993; Presland et al., 2006; Resing et al., 1995). The PF-NT can translocate (NLS within B domain) into the keratinocyte nucleus both in vitro and in vivo (Ishida-Yamamoto et al., 1998; Pearton et al., 2002). It is postulated that PF-NT, once in the nucleus, provides cellular signals directing events of terminal differentiation; it may act as a negative feedback signal controlling epidermal proliferation and homeostasis (Aho et al., 2012).
Profilaggrin, along with six other epidermal proteins, has an S100 domain fused to additional protein sequences, defining a “fused S100-type” subfamily within the larger S100 family (Kizawa et al., 2011). An important unsolved question is why the epidermis expresses several S100 fused-type proteins, whose genes cluster on chromosome 1q21. While isolated S100 proteins have been studied extensively, the functions of fused-type S100 proteins in the epidermis are less clear. Thus, we investigated the structure and binding interactions of the most abundant and best understood member of the S100 fused-type family, profilaggrin. While structures for isolated S100 proteins exist, no atomic resolution structures have been determined for an S100 fused-type calcium-binding protein, nor for any region of profilaggrin.
We report here advances in the molecular structure and biochemical function of the N-terminus of human profilaggrin. First, we determined the 2.2Å resolution x-ray crystal structure of the S100 fused-type CABD of profilaggrin. Second, several biochemical and yeast two-hybrid studies demonstrated the N-terminal fragment of profilaggrin exists both in vitro and in vivo as a homodimer. Third, co-immunoprecipitation and mass spectrometry approaches identified three protein targets for the N-terminus of profilaggrin. This research addresses a specific knowledge gap in human epidermal barrier biology correlating protein structure with molecular function.
Results
Biochemical and structural basis for profilaggrin dimerization
The influence of calcium ions on the aggregation state of the N-terminal CABD of human profilaggrin (residues 1-92, inclusive of N-terminal Met) (PF-CABD) was analyzed using size-exclusion chromatography. In presence of 1 mM EDTA, and absence of calcium ions, recombinant PF-CABD separated into a single major peak (Figure 1a, solid line). In contrast, PF-CABD in 5 mM calcium chloride separated into numerous higher-than-expected molecular weight (MW) aggregates (Figure 1a, dotted line). SDS-PAGE of the collected size-exclusion fractions demonstrated PF-CABD eluted in a narrow range of fractions in the absence of calcium (Figure 1b), but in a broad range of higher-than-expected MW fractions in the presence of 5 mM calcium (Figure 1c). These data suggest PF-CABD undergoes calcium-dependent aggregation.
Figure 1. Calcium-independent dimerization and calcium-dependent self-aggregation of profilaggrin N-terminal S-100 fused-type calcium-binding domain.
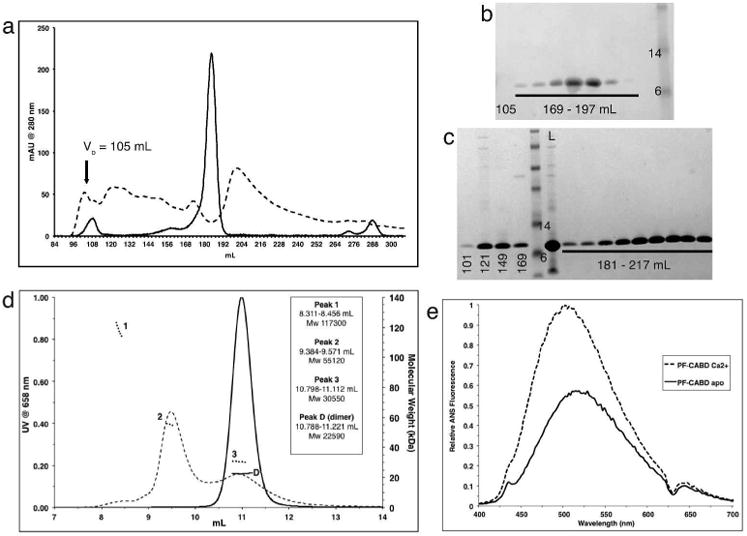
Gel filtration (GF) of PF-CABD with 1mM EDTA (solid line) produced a single peak [180-191mL], but with 5mM CaCl2 (dotted line) produced multiple high MW aggregates [100-186mL]. Void volume (Vo). (b) SDS-PAGE of GF eluted protein (1mM EDTA) within 169-197mL fractions demonstrated PF-CABD protein; none was present in 105mL fraction. (c) SDS-PAGE of GF eluted protein (5mM CaCl2) in 101-169mL fractions demonstrated PF-CABD high MW L = loaded sample. (d) Light scattering of PF-CABD with 1mM EDTA (solid lines) demonstrated a single homogenous peak with MW (22,590 Da) of a PF-CABD dimer (scattering distribution labeled “D”), but with 5mM CaCl2 (dotted lines) demonstrated multiple peaks of higher than expected MW (scattering distributions labeled 1-3). (e) Increased ANS fluorescence emission for PF-CABD (2 μM) in 2mM CaCl2 (dotted line) compared to PF-CABD in 2mM EDTA (solid line) indicated calcium-dependent structural opening of the protein to expose hydrophobic residues.
To further characterize these distinct PF-CABD aggregates, multiple angle light scattering was performed on PF-CABD in the absence and presence of calcium. PF-CABD in 1 mM EDTA without added calcium demonstrated a monodisperse peak of 22,590 Da (Figure 1d, solid lines); this is double the expected calculated MW of 11,194 Da for a PF-CABD monomer. In presence of 5 mM (Figure 1d, dotted lines) or 15 mM (data not shown) calcium chloride, PF-CABD aggregated into multiple peaks corresponding to complexes ranging from 30,550 Da to 117,300 Da. Thus, in absence of calcium, PF-CABD exists as a stable dimer; however, in the 5-15 mM calcium environment tested PF-CABD aggregates in vitro into higher MW species.
The calcium-dependent conformational opening of PF-CABD to expose hydrophobic residues was demonstrated by fluorescence spectroscopy (Figure 1e) using the molecular probe 8-anilinonaphthalene-1-sulfonic acid (ANS), which fluoresces greater when bound to hydrophobic surface. Fluorescence results confirm PF-CABD undergoes calcium-dependent conformational/structural change, and suggest non-specific hydrophobic interactions as the mechanism for higher-order (beyond dimer) protein aggregation.
We subsequently determined the 2.2Å resolution x-ray crystal structure of PF-CABD bound to calcium (Table 1). The structure demonstrates PF-CABD is a dimer (Figure 2a). Each PF-CABD monomer forms a four-helix domain similar to other EF-hand calcium-binding proteins and binds two calcium ions (Figures S1-S2). The crystal asymmetric unit (AU) contains two PF-CABD dimers bound together for 4 total copies of PF-CABD per AU; 8 calcium ions (2 per PF-CABD) are bound (Figures 2b, S3). Comparison of PF-CABD inter-helical angles with other select S100 proteins demonstrates unique helical orientations, particularly for helix I/IV and helix II/III interfaces (Table S1); the four PF-CABD subunits in the AU have similar helical orientations (average RMSD 0.82Å).
Table 1. Data collection and refinement statistics.
| Crystal | PF-CABD |
|---|---|
| Diffraction Data | |
|
| |
| Space Group | P212121 |
| Unit Cell Dimensions, | |
| a, b, c (Å) | 40.49, 85.28, 96.90 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution range (outer shell), Å | 50-2.20 (2.24-2.20)* |
| I/σI | 12.7 (1.69) |
| CC(1/2) in outer shell, % | 59.2 |
| Completeness, % | 92.9 (93.3) |
| Rmerge | 0.13 (0.854) |
| Rpim | 0.067 (0.439) |
| Rmeas | 0.147 (0.966) |
| No. of crystals used | 1 |
| No. of unique reflections | 16492 |
| Redundancy (outer shell) | 4.2 (4.1) |
| Wilson B-factor, Å2 | 26.9 |
|
| |
| Refinement | |
|
| |
| Rwork, % | 19.8 (25.2) |
| Rfree, % | 24.2 (32.2) |
| Rfree Test Set Size, % | 5.13 |
| No. of Non-Hydrogen Atoms | |
| Protein | 2978 |
| Ligands/Ions (2PE, Ca) | 64 |
| Waters | 120 |
| R.m.s. Deviations | |
| Bond lengths, (Å) | 0.003 |
| Angles, (°) | 0.719 |
| Chirality | 0.027 |
| Planarity | 0.004 |
| Dihedral, (°) | 16.61 |
| Average B-factor, (overall) Å2 | 46.0 |
| Protein/Ca/2PE/Waters | 47.21/36.70/44.85/42.0 |
Values in parentheses are for highest-resolution shell.
Figure 2. Crystal structure of the N-terminal S100 fused-type calcium-binding domain of human profilaggrin.
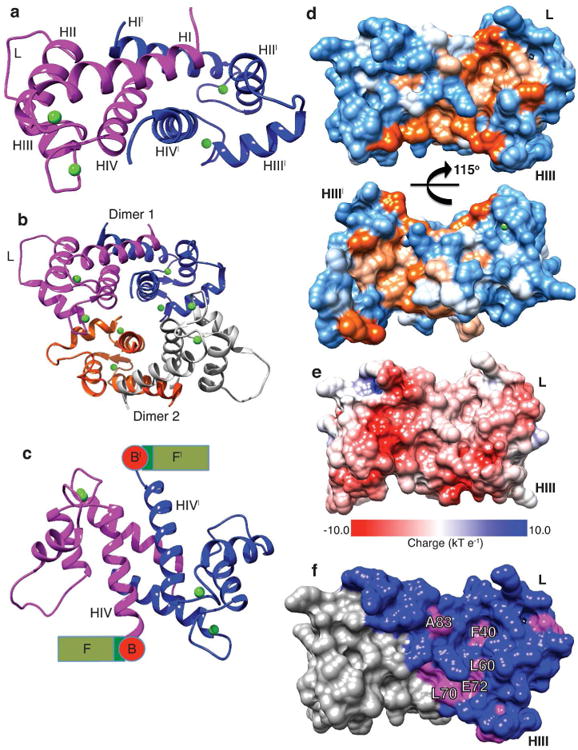
(a) PF-CABD is biologically a dimer (protein 1, magenta; protein 2, blue with prime symbol). PF-CABD monomer forms a four-helix bundle. (b) The crystal AU contained two PF-CABD dimers (dimer 1, magenta/blue; dimer 2, orange/gray). The tetramer core is composed of four helix IV helices. (c) PF-CABD dimer rotated forward 120° about x-axis compared to panel 2a to display the antiparallel helix IV plane connected to a schematic of remaining profilaggrin sequence. B = B domain; F = filaggrin units. (d) Molecular surface of hydrophobic pocket in PF-CABD dimer illustrating two hydrophobic pockets (orange color gradient based on degree of hydrophobicity); polar residues = blue; orientation 40° backward rotation about x-axis compared to Figure 2a. (e) Electrostatic surface potential of PF-CABD dimer, demonstrating acidic calcium-binding loops (red) and uncharged pocket (white). Basic residues = blue. (f) Only 5 residues are conserved in the hydrophobic pocket (labeled, magenta) across S100 fused-type protein family. Non-conserved residues colored blue. Calcium ions = green. L = interhelical linker; H = helix.
The dimerization interface is formed primarily by α-helices I and IV from interdigitating monomers. Helices I and IV from one monomer form a V-shaped groove that is occupied by helix I from the dimerization partner. Monomer interdigitation creates two α-helical planes: “helix I” plane (two antiparallel N-terminal helices of homodimer) and “helix IV” plane (two antiparallel C-terminal helices of homodimer). This antiparallel dual-plane helical architecture structurally positions the remainder of profilaggrin on opposite sides of the homodimer in a sterically feasible state (Figure 2c). Residues stabilizing the interhelical interfaces are mainly hydrophobic or aromatic (Table S1).
Stabilizing interactions exist for the N-terminus (Leu3, Leu4) and C-terminus (Tyr85) (Table S1). Tyr85 is stabilized by a 4 aromatic ring cluster; Tyr85 of one monomer stacks against several phenylalanine residues derived from the opposing dimer molecule: Phe14, Phe70, and Phe73 (Figure S4). The total combined area of the molecular surface buried between dimerizing subunits is ∼3000Å2.
Yeast-two hybrid (Y2H) experiments examined the ability of the human profilaggrin N-terminus (PF-NT) [CABD (residues 1-92) plus B domain (residues 93-293)] to form homodimers in vivo. PF-NT homodimerization was observed only when both bait and prey constructs contained full-length PF-NT (Figures S5-S6); a similar positive interaction was seen using full PF-NT and a slightly shorter profilaggrin peptide (259 residues; AB-259). No Y2H interaction was seen with shorter constructs containing only part of the B-domain. PF-CABD was unable to form homodimers in yeast, under conditions where S100A2 could form a functional protein interaction. Western analysis with PF-NT antibodies showed all B domain constructs were abundantly expressed in yeast cells; however, PF-CABD constructs were less well expressed, but detectable on immunoblots (data not shown). The in vivo Y2H experiments complement the in vitro biochemical data and together reveal a dimerization capacity for human PF-NT.
Profilaggrin N-terminal S100 domain contains a unique hydrophobic pocket
The molecular surface of the PF-CABD dimer demonstrates two distinct, symmetric hydrophobic pockets arranged ∼115° opposite each other (Figures 2d-e). Each pocket (one per PF-CABD subunit) is formed by accessible surface area (ASA) contributions from 24 residues, 14 of which are hydrophobic and account for 47.4% of the pocket ASA (Table S2). In addition, there are 5 polar (31.8% ASA), 2 basic (14.4% ASA), and 3 acidic (6.4% ASA) residues contributing to the molecular surface properties of the pocket. The two pockets are connected by a small hydrophobic surface patch (∼226Å2) formed by adjacent Phe 78 residues. Previous investigations on target selectivity in S100 proteins showed, despite sharing a common EF-hand structural motif and moderate sequence similarity, S100 proteins have unique surface properties at their target protein binding sites, enabling functional diversity (Bhattacharya et al., 2004). One dimer subunit contained a PEG 400 molecule bound in the hydrophobic pocket (Figure S7).
To analyze primary sequence and molecular surface conservation across all 7 members of the S100 fused-type protein family, a multiple sequence alignment was generated (Figure S8) and conserved and non-conserved residues mapped onto the PF-CABD structure (Figure 2f). Excluding the N-terminal Met, S100 fused-type proteins conserve 26 residues, defining “conserved” as at least 6 out of 7 family members contain the same amino acid at a given position. Of those 26, only 5 are within the hydrophobic pocket; thus, of 24 residues comprising the hydrophobic pocket (Table S2), only 21% are conserved. This illustrates how S100 fused-type proteins utilize sequence diversity within the hydrophobic pocket to generate unique binding interfaces for potential protein targets in skin.
Identification of human profilaggrin N-terminus-associated proteins
To identify proteins that associate with the 32 kDa human PF-NT, we performed immunoprecipitation of human foreskin cell lysates with profilaggrin N-terminal B1 antibody and fractionated the total precipitate using SDS-PAGE. Western analysis of immunoprecipitate with profilaggrin B1 antibody confirmed the presence of this peptide in the mixture. Eight bands visible by SDS-PAGE after immunoprecipitation with profilaggrin B1 antibody were selected for trypsin digestion and mass spectrometry. Three proteins (in addition to profilaggrin N-terminus itself) were identified by mass spectrometry: annexin II (p36 subunit), stratifin (14-3-3sigma), and hsp27. Each protein was represented by three or more peptides encompassing 10.6-21% of the identified polypeptide (Table S3). PF-NT was represented by 19 peptides spanning 128/293 residues (43.7%).
Stratifin and annexin II directly interact with human profilaggrin N-terminus
Yeast-two hybrid (Y2H) studies were conducted to further dissect interactions of PF-NT with stratifin and annexin II in vivo (Table 2; Figures 3 and S9). Human stratifin associated with both PF-NT and prey proteins containing the CABD plus varying amounts of the B domain (residues 120-293); however, stratifin did not interact with human PF-CABD alone. Stratifin additionally interacted with mouse PF-AB (Table 2; Figure S9). Stratifin also forms homodimers in vivo (Yaffe et al., 1997), but in yeast this interaction is relatively weak compared to the association with human or mouse PF-NT. Association of human profilaggrin B domain with stratifin in a calcium-independent manner is consistent with the presence of several potential 14-3-3 binding sites in profilaggrin (Table S4).
Table 2. Summary of Yeast-two Hybrid Data: Interaction of Profilaggrin N-terminus with Human Annexin II/p36 and Stratifin/14-3-3σ.
| Bait Protein (pOBD construct) | Prey Protein (pOAD construct) | Strength of interaction a |
|---|---|---|
| Annexin II, 1-339 | Human AB | +++ |
| Annexin II, 1-339 | Human AB (S100 mutant)b | +/-c |
| Annexin II, 1-339 | Human A | ++ |
| Annexin II, 1-339 | Mouse AB | ++ |
| Annexin II, 1-339 | Mouse A | ++ |
| Annexin II, 1-44 | Human AB | ++ |
| Annexin II, 1-44 | Mouse A | ++ |
| Annexin II, 15-339 | Human AB | - |
| Annexin II, 15-339 | Mouse A | - |
| Stratifin | Human AB d | ++ |
| Stratifin | Human AB (S100 mutant)b | ++ |
| Stratifin | Human A | - |
| Stratifin | Human B | + |
| Stratifin | Mouse AB | ++ |
| Stratifin | Mouse A | + |
| Stratifin | Mouse B | + |
| Stratifin | Stratifin | +/-c |
+++, robust growth on 10 mM 3-AT, ++, weaker growth (and less colonies) on 10 mM 3-AT; +, weak growth on 10 mM 3-AT.
This protein contains mutations in key amino acids involved in calcium binding (see text for details).
Very weak but detectable growth on histidine-deficient media containing 10 mM 3-AT.
Stratifin exhibited a positive interaction with all human profilaggrin constructs containing varying lengths of the B domain, i.e. PF-AB 120, PF-AB 140, PF-AB 160, PF-AB 218, PF-AB 259 and PF-AB 293. Growth was similar (++) for all bait-prey combinations on histidine-deficient media containing 10 mM 3-AT.
Figure 3. The profilaggrin S100 domain interacts with the N-terminus of annexin II.
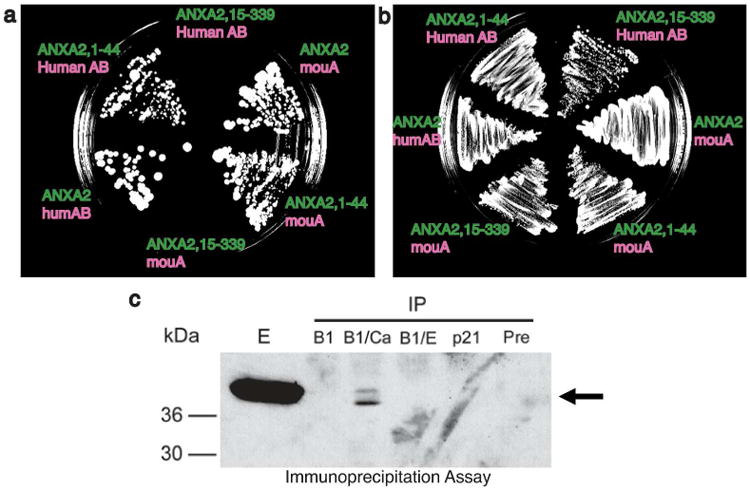
Yeast two-hybrid analysis (panels a and b) demonstrated the A (S100) domain of profilaggrin interacts with the N-terminus of annexin II. (a) Bait (green) and prey (pink) plasmid combinations were plated on HLT-deficient media containing 10 mM 3-amino-1,2,4-triazole. Both full length ANXA2 and a truncated N-terminal protein (ANXA2, 1-44) interacted with human and mouse profilaggrin N-terminus, but an ANXA2 protein lacking the first 14 amino acids (ANXA2, 15-339) showed no Y2H signal. (b) Control (LT-deficient) plate showing confluent growth of bait/prey combinations. (c) Association of profilaggrin N-terminus and annexin II in vitro is calcium-dependent. Epidermal proteins were immunoprecipitated with either profilaggrin B domain (B1) antibody, a p21/WAF1 antibody, or a pre-immune rabbit control. Immunoprecipitated proteins were separated on SDS/polyacrylamide gels and immunblotted with annexin II antibody. The lanes show immunoprecipitation with: B1 antibody, with no additions; B1/Ca, B1 antibody with the addition of 5 mM CaCl2; B1/E, B1 antibody with the addition of 5 mM EDTA; p21/WAF1 antibody or pre-immune serum, with no additions. E represents a control epidermal extract to show annexin II.
Annexin II/p36 interacts with human and mouse PF-CABD domain in a calcium-dependent manner as well as the corresponding PF-AB regions (Table 2; Figure 3; Supplemental Discussion). Removal of the N-terminal 14 amino acids from annexin II, which is the domain that binds S100A10 (Bharadwaj et al., 2013), prevented its interaction with human PF-NT (Figure 3). These studies demonstrate PF-NT and S100A10 bind the same N-terminal domain of annexin II. Immunoprecipitation experiments using human epidermal lysates confirmed annexin II associates with PF-NT; however, detectable annexin II could only be pulled down with profilaggrin N-terminal antibody in the presence of added calcium (Figure 3c).
To investigate the role of calcium coordination on annexin II binding and homodimerization, six mutations were made to key calcium coordination residues in PF-NT [Two in N-terminal pseudo/non-canonical EF-hand (D21A, E31V) and four in C-terminal canonical EF-hand (D61A, I62T, D63A, E72V)]. Mutant PF-NT showed diminished binding affinity with annexin II in yeast (Table 2); by contrast, alteration of calcium binding did not diminish the strength of interaction with stratifin (Figure S9). Homodimerization of mutant human PF-NT protein in yeast was not affected (data not shown), consistent with our biochemical data showing stable homodimer formation in the absence of calcium.
Stratifin and human profilaggrin N-terminus co-localize in the epidermal granular layer
To determine if stratifin and profilaggrin N-terminus co-localize in human epithelial cells, we performed double-label immunofluorescence microscopy on paraformaldehyde-fixed human skin (Figure 4a-b). The results confirm PF-NT and stratifin are co-expressed in the stratum granulosum of human skin and co-localize in vivo, primarily at the cell periphery (Figure 4b).
Figure 4. Profilaggrin N-terminus and stratifin co-localize in human epidermal granular cells.
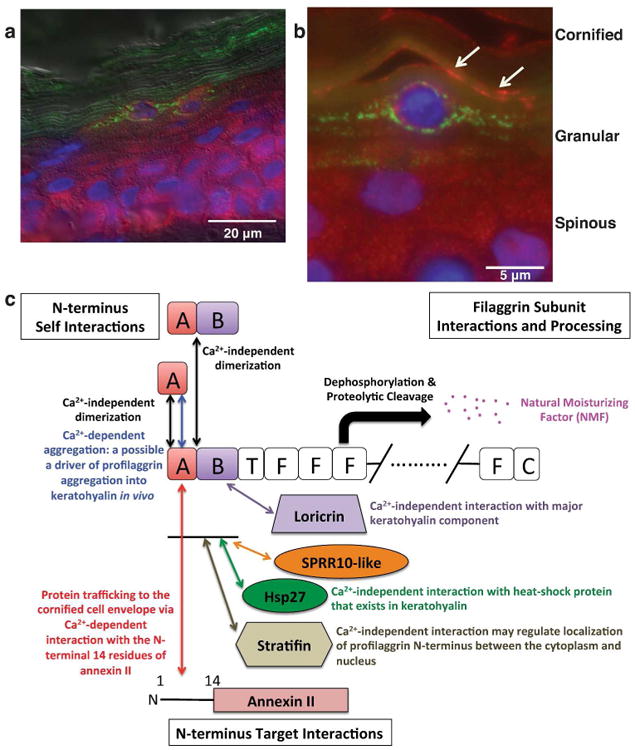
Double label immunofluorescence was performed on fixed adult human skin using antibodies directed against the profilaggrin N-terminus (green) and stratifin (red). Panel (a) shows that stratifin is expressed throughout the epidermis, while profilaggrin is restricted to the granular layer and anuclear stratum corneum. (b) Shown is a vertical section of adult human skin immunolabeled with stratifin (red) and PNT (green) antibodies, with nuclei counterstained with DAPI. Stratifin is localized through the cytoplasm in spinous cells, but in the upper granular layer it is concentrated at the cell periphery where it co-localizes with PNT (orange labeling, arrows). Profilaggrin is also present in KHGs in the characteristic granular pattern, but shows little or no association with stratifin when present in the granular (profilaggrin) form. (c) Proposed biological functions of a calcium-dependent and calcium-independent protein interaction network for profilaggrin in human epidermis (based on the current study and the previous study by Yoneda et al., 2012).
Discussion
Few atomic resolution structures support the biology currently known about terminal epidermal differentiation or the stratum corneum. X-ray crystal structures of the major proteins known to exist in the stratum corneum (e.g., involucrin, keratins 1/10, loricrin, profilaggrin) to our knowledge have not been previously reported. This study correlates profilaggrin structure with its function and establishes an important foundation for future structural studies in epidermal biology. Moreover, it shows at least one member of the fused-type S100 family follows the paradigm of S100 proteins, i.e. calcium binding, peptide homodimerization and physical association with other intracellular proteins. This is important because the function of fused-type S100 domains in other epidermal proteins is unknown.
Advances presented here establish several principles for epidermal barrier biology: 1) profilaggrin's N-terminus undergoes calcium-independent homodimerization and calcium-dependent non-specific hydrophobic aggregation; 2) the S100 fused-type CABD contains a unique hydrophobic target binding site; 3) profilaggrin participates in protein interactions with stratifin, annexin II and hsp27; and 4) the S100 fused-type CABD and B domain both contribute to target binding selectivity. Furthermore, we propose a model whereby calcium promotes aggregation of profilaggrin into keratohyalin storage granules, and in which the identified binding partners associate with profilaggrin or the cleaved, soluble N-terminal peptide, regulating both its nuclear role in keratinocyte terminal differentiation and incorporation into the functionally important cornified cell envelope (Figure 4c).
Profilaggrin is stored within keratohyalin (KH) granules in a calcium-rich environment; molecular driving forces for KH granule assembly are not fully known. It has been proposed that both extensive phosphorylation of filaggrin units and calcium binding by the N-terminus are required for KH granule formation (Dale et al., 1994). The PF-CABD structure offers several mechanisms of how this domain may contribute to KH granule assembly: 1) homodimerization facilitates aggregation of profilaggrin as there are 50% less free molecules; 2) calcium-bound PF-CABD adopts an open conformation with exposed hydrophobic patches; 3) hydrophobic patches, through self-aggregation or target binding, offer a protein-protein interaction mechanism for assembly. Hsp27, for example, co-localized with profilaggrin to KH granules and loss of hsp27 is associated with hyperkeratinization and misprocessing of profilaggrin (O'Shaughnessy et al., 2007).
Similar to most S100 proteins (Hermann et al., 2012; Isobe et al., 1981), profilaggrin CABD formed homodimers. PF-NT (PF-CABD+B domain), the physiologically relevant proteolytic product, also formed homodimers. In the Y2H system, however, PF-CABD alone did not form homodimers (Figure S5). The likely explanation for this discrepancy is the shorter human PF-CABD protein, but not the longer PF-CABD+B, is prevented from forming homodimers in yeast by the attached yeast GAL4 transcription factor domains. Despite one inconsistent experiment, all other biochemical, structural, and Y2H studies supported dimerization of PF-NT mediated by the S100 domain. Additional studies are needed to determine whether full-length profilaggrin dimerizes; for PF-CABD, dimerization potentially enables binding of two similar or different macromolecules simultaneously. This may impart stability to the stratum corneum; for example, some PF-NT is retained in the stratum corneum and may be crosslinked as part of the cornified envelope (Presland et al., 1997).
Identification of annexin II, stratifin, and hsp27 as target proteins for PF-NT demonstrates human profilaggrin may be an epidermal signaling molecule or involved in protein trafficking, in addition to its role in cytokeratin reorganization and tissue moisturization (Figure 4c). We previously found PF-NT binds loricrin, a component of the cornified cell envelope (Yoneda et al., 2012). Similar to loricrin, stratifin (epithelial specific member of the 14-3-3 protein family also known as 14-3-3sigma) interacted with PF-NT and not with the CABD domain alone. 14-3-3 proteins including stratifin bind phosphorylated proteins, often to R-X-X-pS sequences (Yaffe et al., 1997). Human and mouse PF-NT contain several RXXS sequences (5 in human, 6 in mouse; Table S4) located in the B domain, which may function as stratifin binding sites. Whereas both the S100 and B domains cooperatively bound stratifin in a calcium-independent manner, annexin II bound only to the S100 domain in a calcium-dependent manner. Y2H experiments showed the N-terminal 14 amino acids of annexin II mediated the interaction with PF-CABD. Analysis of the PF-CABD structure bound to the N-terminal 11 residues of annexin II using in silico molecular docking suggests six annexin II (Thr2, Val3, Ile6, Leu7, Lys9, Leu10) and seven profilaggrin (Gln42, Phe40, Met52, Phe56, Met76, Leu80, Tyr84) residues mediate the interaction (Figure S10). There is debate whether Y2H experiments can discriminate between calcium-regulated versus calcium-independent interactions (Deloulme et al., 2003), however, profilaggrin is a fused-type S100 protein distinct from isolated S100 domains, with evidence that downstream protein domains (e.g. B domain) influence target binding affinity, likely through domain cooperativity. To confirm identified protein interactions, multiple techniques were utilized besides Y2H.
In summary, we determined the crystal structure of the N-terminal S100 calcium-binding domain of human profilaggrin, analyzed its suspected target-binding site in molecular detail, and identified protein binding partners for each domain of the cleaved N-terminus. This work enhances our understanding of profilaggrin as a multifunctional epidermal protein; it is a precursor to stratum corneum structure and function, engages in a protein interaction network in human epidermis, and may be active in epidermal protein-protein signaling and protein trafficking.
Materials & Methods
Profilaggrin, annexin II (p36), and stratifin constructs
Design and production of constructs are detailed in Supplemental Materials and Methods and Table S5.
Antibodies
Primary antibodies were directed against annexin II (Invitrogen, Carlsbad, CA), stratifin (Upstate Biotechnology, Lake Placid, NY), p21 (Cell Signaling Technology, Danvers, MA) and the A (PF-CABD) and B domains of the human profilaggrin N-terminus (Presland et al., 1997).
Protein production and purification
N-terminal calcium binding domain of human profilaggrin (PF-CABD) was produced and purified (detailed in Table S6).
Multi-angle light scattering
Apo-PF-CABD (5.2 mg/ml) in 50mM Tris-HCl buffer (pH 7.8) containing 0.5M NaCl and 1mM EDTA was applied at 0.5 mL/min to Superdex 75 gel filtration column in-line with DAWN HELEOS II light scattering instrument (Wyatt Technology) [laser wavelength 658.0nm]. Data collection/analysis used Astra software (Wyatt Technology) version 5.3.4.20. After preparation by overnight dialysis against 50mM Tris-HCl buffer (pH 7.8) containing 0.5M NaCl and either 5mM or 15mM CaCl2, calcium-bound samples of PF-CABD were analyzed similar to apo state.
Fluorescence spectroscopy
Fluorescence measurements were made using a SLM Aminco spectrofluorometer (SLM Instruments, Inc., Urbana, IL) following established protocol (Bunick et al., 2004).
Crystallization and x-ray data collection
PF-CABD (4.6 mg/ml) in 50mM Tris-HCl buffer (pH 7.8) containing 0.5M NaCl and 1mM EDTA was crystallized by sitting drop vapor diffusion using reservoir solution of 40% PEG 4000, 50mM Tris-HCl buffer (pH 8.5), and 0.1M CaCl2. Drops were prepared by mixing 1μL PF-CABD with 1μL reservoir solution. Crystals grew over 24-48h at 25°C. PF-CABD crystals were soaked 1-3 min in cryoprotectant solution containing 10% PEG 400 in mother liquor prior to flash-cooling of the crystal by direct immersion into a crystal puck storage system maintained under liquid nitrogen. A native data set on a single crystal maintained at ∼100K was collected using the X-29 beamline (λ=1.075Å) at National Synchrotron Light Source in Brookhaven National Laboratory. PF-CABD crystallized in orthorhombic space group P212121 (cell dimensions: a=40.49Å, b=85.28Å, c=96.90Å, α=β=γ=90°) with a unit cell solvent content of 34%. Diffraction data was processed using HKL2000 (HKL Research, Inc., Charlottesville, VA).
Structure determination, refinement, and analysis
PF-CABD structure was determined by molecular replacement with MOLREP (Vagin and Teplyakov, 2010) using calcium-bound S100A12 (PDB 1E8A) as the search model. A simulated annealing composite omit map generated in PHENIX (Adams et al., 2010) was necessary to overcome initial model bias. The PF-CABD structure underwent iterative rounds of model building [Coot (Emsley and Cowtan, 2004)] and refinement (PHENIX). Final model of AU contained four PF-CABD molecules (two homodimers), 8 calcium ions, 120 water molecules, and two PEG 400 molecules. Final Ramachandran statistics: residues in favorable regions, 95.1%; in allowed regions, 4.0%; in outlier regions, 0.9%. Structural analyses were performed with Coot, UCSF Chimera (Resource for Biocomputing, Visualization, and Informatics, University of California, San Francisco), INTERHLX (K. Yap, University of Toronto), and PDBePISA (The European Bioinformatics Institute, European Molecular Biology Laboratory, UK). Electrostatics calculated using PDB2PQR (Dolinsky et al., 2004) and Adaptive Poisson-Boltzmann Software (APBS) (Baker et al., 2001). Figures prepared using UCSF Chimera or PyMOL Molecular Graphics System (Version 1.5.0.4, Schrödinger, LLC).
Yeast two-hybrid assays, expression analysis, and profilaggrin protein interactions
Details for yeast experiments, immunoprecipitation and mass spectrometry studies, and immunofluorescence microscopy are in Supplemental Materials and Methods.
Statement on use of human materials
Normal human neonatal and adult skin, utilized for immunofluorescence microscopy and immunoprecipitation studies, were obtained through the Division of Dermatology with approval from the University of Washington Institutional Review Board.
Supplementary Material
Figure S1 -- Unbiased Fo-Fc electron density of calcium ions. Profilaggrin crystal asymmetric unit (top; two dimers) in light blue with the eight bound calcium ions as green spheres. Depicted in green wire mesh is the Fo-Fc electron density contoured at 3σ. Pictured at the bottom right within the red zoom box is the canonical EF-hand in the C subunit, showing classic canonical EF-hand calcium coordination (residues labeled) and the Fo-Fc electron density in green (contoured at 3σ).
Figure S2 -- Calcium coordination. Profilaggrin, belonging to the S100 fused-type protein family, contains two types of EF-hands: an N-terminal pseudo/S100 (non-canonical) calcium binding loop (top) and a C-terminal canonical calcium binding loop (bottom). The profilaggrin sequence for each calcium binding loop in the N-terminal calcium binding domain is depicted, along with a position number. The type of calcium coordination for each position within the calcium binding loop is described for both types of calcium binding sites (pseudo/S100/non-canonical vs canonical). Images from the x-ray structure depict the pseudo/S100 (top) and canonical (bottom) calcium coordination for profilaggrin's N-terminus.
Figure S3 -- Inter-molecular interactions between helices IV (top) and helices III (bottom) stabilize the crystallized tetrameric aggregate. (a) Ribbon view of crystal AU, highlighting the helix IV interface between two PF-CABD dimers; helix IV for each subunit (A through D) is colored and labeled (A, red-orange; B, gray; C, blue; D, magenta). (b) Panel ‘a’ has been rotated 90° upward to show a side view of the tetrameric aggregate (helix I from the A and B subunits has been removed for clarity). (c) Focused view of the helix 4 interface, with residues most important for interface stablization shown as stick model. (d) Ribbon view of crystal AU, highlighting the helix III interface between two PF-CABD dimers; the same labeling and color scheme is used from the prior panels, but the colored helix is the 3rd helix, not helix IV as in panels a-c. (e) Focused view (rotated ∼ 180° from panel d) of helix III interactions, with residues most important for interface stablization shown as stick model. The combined buried surface area is ∼ 700 Å2 for the helix IV interface and ∼ 1000 Å2 for the helix III interface. Calcium ions are depicted as green spheres.
Figure S4, related to Figure 2. Stereo image of the electron density for the four aromatic ring cluster present in the N-terminus of profilaggrin. Tyr85 (red) of one monomer (dark blue) stacks against several phenylalanine residues (magenta) derived from the opposing dimer molecule (cyan): Phe14, Phe70, Phe73. The electron density is contoured at 0.8σ.
Figure S5. The human profilaggrin N-terminus forms homodimers in yeast. Human profilaggrin constructs containing either the S100 calcium-binding A domain, B domain or complete N-terminus (A + B domains) prepared in the bait (DNA binding domain) vector (green) or prey (activation domain) (pink) were transformed into yeast, and cells grown on either histidine, leucine and tryptophan (HLT)-deficient media containing 3-amino 1,2,4-triazole (3-AT) (a, c) or leucine and tryptophan(LT)-deficient media as a control (b). Panel (a), bait and prey combinations grown on HLT-deficient containing 5 mM 3-AT. Only the complete terminus (hAB) and the control protein S100A2 formed homodimers in yeast under these selective conditions. Neither human profilaggrin domain A (hA, amino acids 1-91) or B (hB, amino acids 91-293) formed homodimers in yeast. The human AB protein also did not interact with either human S100A2 or yeast MEC3 used as controls. Panel (b), the same bait and prey combinations grown on LT-deficient media, showing robust growth and colony formation. Panel (c), bait and prey human profilaggrin terminus constructs grown on HLT-deficient media with 5 mM 3-AT. Note that the only profilaggrin combinations that interacted in yeast were the AB-293 homodimer containing the complete N-terminus, and the AB-293 in combination with a slightly truncated binding partner, AB-259. The number indicates the number of residues contained in the bait or prey fusion protein. All of the construct combinations grew to confluence on control (LT-deficient) media.
Figure S6. Schematics of constructs used for yeast-two-hybrid analysis of profilaggrin N-terminus homodimerization and N-terminus-annexin II interaction.
Profilaggrin A constructs contain just the S100 domain (residues 1-92), while full-length AB constructs include the complete N-terminus (residues 1-293); additional AB constructs included various truncations made within the B domain, e.g. at residues -98, -120, -140, -160, -218, and -259.
Annexin II (ANXA2) consists of 4 annexin repeats of 70 amino acids (light blue boxes) flanked by N-terminal and C-terminal domains (unshaded boxes). The N-terminal domain of 32 residues includes amino acids 1-14 (shaded black box) that interact with the profilaggrin N-terminus and S100A10.
Figure S7 -- PEG 400 binds to the PF-CABD hydrophobic pocket and alters linker conformation.
(Top left) 24 residues (orange) comprise the hydrophobic pocket of one PF-CABD protein, with bound PEG 400 shown as ball and stick model (carbon, white; oxygen, red).
(Top right) Ile43 and Leu44 (side chains orange) contact PEG 400 (carbon, white; oxygen, red), resulting in a folded conformation of inter-EF-hand linker (L). Wire diagram of PF-CABD monomer (gray) with inter-EF-hand linker (orange).
Bound PEG 400 (by-product of cryocrystallography) alludes to the CABD functioning as a target-binding site. Of 24 residues comprising the hydrophobic pocket, 16 interacted with PEG 400 based on analysis of both the “A” and “C” PF-CABD subunits from the crystal AU (Table S2). The “B” and “D” subunits did not contain PEG. Interestingly, only “A” and “C” subunits have linkers between Helix II and Helix III in a “folded” rather than “extended” conformation. Ile43 and Leu44 are positioned centrally in this linker and in the structure contact PEG 400 (above right), suggesting this flexible linker functions to stabilize target interactions. Linker folding likely explains the slightly more “closed” Helix I/II orientation for the “A” and “C” subunits (Table S1).
Figure S8.
Clustal Omega multiple sequence alignment (top) of the N-termini of 7 epidermal S100 fused-type proteins as well as calbindin D9k, calmodulin, S100B and S100A12. h = human; m = mouse; b = bovine. Completely conserved residues are colored purple and denoted by (*). Highly conserved residues are colored green and denoted (:). Moderately conserved residues are colored orange and denoted by (.). The numbering across the top represents the amino acid sequence numbering for the crystal structure of profilaggrin N-terminus, not positioning for all aligned sequences. The 24 amino acid residues contributing accessible surface area to profilaggrin's N-terminal hydrophobic pocket have a yellow-shaded background; the shading is extended across the multiple sequence alignment to illustrate differences among the protein sequences.
Swiss-Model structural alignment (bottom) between the crystal structure of the N-terminus of human profilaggrin (PDB Code 4PCW) and human S100A12(PDB Code 1E8A), the latter being the model used for molecular replacement. For the profilagrin structure, helices are labeled and colored blue, whereas calcium-binding loops are labeled and colored red.
Figure S9, related to Table 2. Stratifin interacts with profilaggrin N-terminus in yeast. Combinations of bait (green) and prey (pink) fusion proteins were transformed into yeast and grown on HLT-deficient media containing 10 mM 3-AT. Human stratifin (SFN) interacts with full length human profilaggrin N-terminus (hum AB), but not the human A domain (hum A), while SFN interacts with the mouse A and B domains (mou A, B and AB). SFN also gave a similar interaction signal with a mutant human profilaggrin N-terminus protein carrying selected mutations within the calcium-binding domain (hum mut AB). All of the construct combinations grew to confluence on control (LT-deficient) media. Please also see Supplemental Discussion related to this figure.
Figure S10. Structural model of PF-CABD binding to annexin II N-terminus from in silico molecular docking. The N-terminal 11 residues of Gallus gallus annexin II [sequence 1-STVHEILSKLS-11] was molecularly docked onto the PF-CABD dimer structure using the Hex Protein Docking server (http://hexserver.loria.fr/). The structure of the annexin II N-terminus used for docking was dervied from Protein Data Bank Code 1BT6. The sequence differs from human annexin II by a cysteine (human) to serine (chicken) substitution at position 8. Pictured at the bottom is the PF-CABD dimer (light and dark blue) with the annexin II N-terminal helix colored red (the six residues highlighted in orange are the annexin II residues that interact with the PF-CABD binding pocket. In the zoomed in image (top), the seven PF-CABD residues within the hydrophobic pocket that interact with annexin II are highlighted yellow and labeled.
Table S1. Characterization of the interhelical angles and interhelical dimerization contacts of human profilaggrin N-terminal calcium binding domain (PF-CABD).
Table S2, related to Figure 2. Amino Acid Residues Surrounding the Target Binding Pocket and associated Accessible Surface Area. The asterisks (*) denote residues contacting the bound PEG 400 molecule in either the “A” or “C” subunit from the crystal asymmetric unit. The five residues conserved across the S100 fused-type protein family are denoted (italics, #). A, B, C, D = subunits from the crystal asymmetric unit.
Table S3, related to Table 2. Identification of three human profilaggrin N-terminus–associated proteins by immunoprecipitation and mass spectrometrya
Table S4. Potential stratifin binding sites at R-X-X-S motifs within the N-terminus of human and mouse profilaggrin. The amino acid sequence is numbered excluding the N-terminal methionine (leading serine = 1).
Table S5. Primers used to generate constructs used in yeast two-hybrid studies.
Table S6. Profilaggrin Protein Production and Purification Protocol.
Acknowledgments
We thank Joe Watson and Bill Eliason for assistance with purification and light scattering experiments, respectively; Dr. Ivan Lomakin for ANS fluorescence assistance; Drs. Daniel Eiler and Jimin Wang for crystallographic discussions; Rebecca Hjorten for performing immunohistochemistry experiments; Jessica Huard and Bradley Sainsbury for Y2H assistance. We thank Walter J. Chazin for critical manuscript review. Work was supported by the Dermatology Foundation through a Dermatologist Investigator Research Fellowship and a Career Development Award (to C.G.B.), the National Institutes of Health/NIAMS Dermatology Training Grant to Yale (PI: Richard Edelson) T32 AR007016 (to C.G.B.), and by grant R01 AR49183 from the NIH (to R.B.P). D.J.P was partially supported by grant P01 AM21557 from the NIH.
Abbreviations used
- ANS
8-anilinonaphthalene-1-sulfonic acid
- ASA
accessible surface area
- AU
asymmetric unit of crystal
- CABD
calcium-binding domain
- EDTA
ethylenediaminetetraacetic acid
- Da
dalton
- KH
keratohyalin
- MW
molecular weight
- PEG
polyethylene glycol
- PF-CABD
N-terminal S100 fused-type calcium-binding domain of human profilaggrin
- PF-NT
human profilaggrin N-terminus containing CABD plus B domain
- RMSD
root mean square deviation
- Y2H
yeast-two-hybrid
Footnotes
Accession Number. Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 4PCW.
Conflict of Interest: The authors state no conflict of interest.
Author Contributions: C.G.B. designed and conducted the crystallography study, including purification and crystallization of the protein; collection, processing, and refinement of the crystallographic data; structural analysis, gel filtration and light scattering studies; wrote the manuscript. Yeast two-hybrid, immunoprecipitation, mass spectrometry, and cell biology studies were performed by O.T.L., D.J.P., and R.B.P; L.M.M. and T.A.S. aided in experimental design, analyzed the data, and provided guidance. All authors contributed to the writing of the manuscript, and approved the final version.
References
- Adams PD, Afonine PV, Bunkóczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aho S, Harding CR, Lee JM, et al. Regulatory role for the profilaggrin N-terminal domain in epidermal homeostasis. J Invest Dermatol. 2012;132:2376–85. doi: 10.1038/jid.2012.174. [DOI] [PubMed] [Google Scholar]
- Akiyama M. FLG mutations in ichthyosis vulgaris and atopic eczema: spectrum of mutations and population genetics. Br J Dermatol. 2010;162:472–7. doi: 10.1111/j.1365-2133.2009.09582.x. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, et al. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj A, Bydoun M, Holloway R, et al. Annexin A2 heterotetramer: structure and function. Int J Mol Sci. 2013;14:6259–305. doi: 10.3390/ijms14036259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Bunick CG, Chazin WJ. Target selectivity in EF-hand calcium binding proteins. Biochim Biophys Acta. 2004;1742:69–79. doi: 10.1016/j.bbamcr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Brown SJ, Kroboth K, Sandilands A, et al. Intragenic copy number variation within filaggrin contributes to the risk of atopic dermatitis with a dose-dependent effect. J Invest Dermatol. 2012;132:98–104. doi: 10.1038/jid.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunick CG, Nelson MR, Mangahas S, et al. Designing sequence to control protein function in an EF-hand protein. J Am Chem Soc. 2004;126:5990–8. doi: 10.1021/ja0397456. [DOI] [PubMed] [Google Scholar]
- Dale B, Resing K, Presland R. Keratohyalin granules. In: Leigh I, Lane E, Watt F, editors. The Keratinocyte Handbook. Cambridge University Press; 1994. pp. 323–50. [Google Scholar]
- Dale BA, Lonsdale-Eccles JD, Holbrook KA. Stratum corneum basic protein: an interfilamentous matrix protein of epidermal keratin. Curr Probl Dermatol. 1980;10:311–25. doi: 10.1159/000396298. [DOI] [PubMed] [Google Scholar]
- Deloulme JC, Gentil BJ, Baudier J. Monitoring of S100 homodimerization and heterodimeric interactions by the yeast two-hybrid system. Microsc Res Tech. 2003;60:560–8. doi: 10.1002/jemt.10298. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, et al. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–7. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Hermann A, Donato R, Weiger TM, et al. S100 calcium binding proteins and ion channels. Front Pharmacol. 2012;3:67. doi: 10.3389/fphar.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida-Yamamoto A, Takahashi H, Presland RB, et al. Translocation of profilaggrin N-terminal domain into keratinocyte nuclei with fragmented DNA in normal human skin and loricrin keratoderma. Lab Invest. 1998;78:1245–53. [PubMed] [Google Scholar]
- Isobe T, Ishioka N, Okuyama T. Structural relation of two S-100 proteins in bovine brain; subunit composition of S-100a protein. Eur J Biochem. 1981;115:469–74. doi: 10.1111/j.1432-1033.1981.tb06225.x. [DOI] [PubMed] [Google Scholar]
- Kam E, Resing KA, Lim SK, et al. Identification of rat epidermal profilaggrin phosphatase as a member of the protein phosphatase 2A family. J Cell Sci. 1993;106(Pt 1):219–26. doi: 10.1242/jcs.106.1.219. [DOI] [PubMed] [Google Scholar]
- Kizawa K, Takahara H, Unno M, et al. S100 and S100 fused-type protein families in epidermal maturation with special focus on S100A3 in mammalian hair cuticles. Biochimie. 2011;93:2038–47. doi: 10.1016/j.biochi.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Lonsdale-Eccles JD, Haugen JA, Dale BA. A phosphorylated keratohyalin-derived precursor of epidermal stratum corneum basic protein. J Biol Chem. 1980;255:2235–8. [PubMed] [Google Scholar]
- O'Shaughnessy RF, Welti JC, Cooke JC, et al. AKT-dependent HspB1 (Hsp27) activity in epidermal differentiation. J Biol Chem. 2007;282:17297–305. doi: 10.1074/jbc.M610386200. [DOI] [PubMed] [Google Scholar]
- Palmer CN, Irvine AD, Terron-Kwiatkowski A, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- Pearton DJ, Dale BA, Presland RB. Functional analysis of the profilaggrin N-terminal peptide: identification of domains that regulate nuclear and cytoplasmic distribution. J Invest Dermatol. 2002;119:661–9. doi: 10.1046/j.1523-1747.2002.01831.x. [DOI] [PubMed] [Google Scholar]
- Presland RB, Haydock PV, Fleckman P, et al. Characterization of the human epidermal profilaggrin gene. Genomic organization and identification of an S-100-like calcium binding domain at the amino terminus. J Biol Chem. 1992;267:23772–81. [PubMed] [Google Scholar]
- Presland RB, Kimball JR, Kautsky MB, et al. Evidence for specific proteolytic cleavage of the N-terminal domain of human profilaggrin during epidermal differentiation. J Invest Dermatol. 1997;108:170–8. doi: 10.1111/1523-1747.ep12333356. [DOI] [PubMed] [Google Scholar]
- Presland RB, Rothnagel JA, Lawrence OT. Profilaggrin and the Fused S100 Family of Calcium-Binding Proteins. In: Elias PM, Feingold KR, editors. Skin Barrier. New York: Taylor & Francis Group; 2006. pp. 111–40. [Google Scholar]
- Resing KA, Thulin C, Whiting K, et al. Characterization of profilaggrin endoproteinase 1. A regulated cytoplasmic endoproteinase of epidermis. J Biol Chem. 1995;270:28193–8. doi: 10.1074/jbc.270.47.28193. [DOI] [PubMed] [Google Scholar]
- Sandilands A, O'Regan GM, Liao H, et al. Prevalent and rare mutations in the gene encoding filaggrin cause ichthyosis vulgaris and predispose individuals to atopic dermatitis. J Invest Dermatol. 2006;126:1770–5. doi: 10.1038/sj.jid.5700459. [DOI] [PubMed] [Google Scholar]
- Smith FJ, Irvine AD, Terron-Kwiatkowski A, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–42. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–5. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–71. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Yoneda K, Nakagawa T, Lawrence OT, et al. Interaction of the profilaggrin N-terminal domain with loricrin in human cultured keratinocytes and epidermis. J Invest Dermatol. 2012;132:1206–14. doi: 10.1038/jid.2011.460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 -- Unbiased Fo-Fc electron density of calcium ions. Profilaggrin crystal asymmetric unit (top; two dimers) in light blue with the eight bound calcium ions as green spheres. Depicted in green wire mesh is the Fo-Fc electron density contoured at 3σ. Pictured at the bottom right within the red zoom box is the canonical EF-hand in the C subunit, showing classic canonical EF-hand calcium coordination (residues labeled) and the Fo-Fc electron density in green (contoured at 3σ).
Figure S2 -- Calcium coordination. Profilaggrin, belonging to the S100 fused-type protein family, contains two types of EF-hands: an N-terminal pseudo/S100 (non-canonical) calcium binding loop (top) and a C-terminal canonical calcium binding loop (bottom). The profilaggrin sequence for each calcium binding loop in the N-terminal calcium binding domain is depicted, along with a position number. The type of calcium coordination for each position within the calcium binding loop is described for both types of calcium binding sites (pseudo/S100/non-canonical vs canonical). Images from the x-ray structure depict the pseudo/S100 (top) and canonical (bottom) calcium coordination for profilaggrin's N-terminus.
Figure S3 -- Inter-molecular interactions between helices IV (top) and helices III (bottom) stabilize the crystallized tetrameric aggregate. (a) Ribbon view of crystal AU, highlighting the helix IV interface between two PF-CABD dimers; helix IV for each subunit (A through D) is colored and labeled (A, red-orange; B, gray; C, blue; D, magenta). (b) Panel ‘a’ has been rotated 90° upward to show a side view of the tetrameric aggregate (helix I from the A and B subunits has been removed for clarity). (c) Focused view of the helix 4 interface, with residues most important for interface stablization shown as stick model. (d) Ribbon view of crystal AU, highlighting the helix III interface between two PF-CABD dimers; the same labeling and color scheme is used from the prior panels, but the colored helix is the 3rd helix, not helix IV as in panels a-c. (e) Focused view (rotated ∼ 180° from panel d) of helix III interactions, with residues most important for interface stablization shown as stick model. The combined buried surface area is ∼ 700 Å2 for the helix IV interface and ∼ 1000 Å2 for the helix III interface. Calcium ions are depicted as green spheres.
Figure S4, related to Figure 2. Stereo image of the electron density for the four aromatic ring cluster present in the N-terminus of profilaggrin. Tyr85 (red) of one monomer (dark blue) stacks against several phenylalanine residues (magenta) derived from the opposing dimer molecule (cyan): Phe14, Phe70, Phe73. The electron density is contoured at 0.8σ.
Figure S5. The human profilaggrin N-terminus forms homodimers in yeast. Human profilaggrin constructs containing either the S100 calcium-binding A domain, B domain or complete N-terminus (A + B domains) prepared in the bait (DNA binding domain) vector (green) or prey (activation domain) (pink) were transformed into yeast, and cells grown on either histidine, leucine and tryptophan (HLT)-deficient media containing 3-amino 1,2,4-triazole (3-AT) (a, c) or leucine and tryptophan(LT)-deficient media as a control (b). Panel (a), bait and prey combinations grown on HLT-deficient containing 5 mM 3-AT. Only the complete terminus (hAB) and the control protein S100A2 formed homodimers in yeast under these selective conditions. Neither human profilaggrin domain A (hA, amino acids 1-91) or B (hB, amino acids 91-293) formed homodimers in yeast. The human AB protein also did not interact with either human S100A2 or yeast MEC3 used as controls. Panel (b), the same bait and prey combinations grown on LT-deficient media, showing robust growth and colony formation. Panel (c), bait and prey human profilaggrin terminus constructs grown on HLT-deficient media with 5 mM 3-AT. Note that the only profilaggrin combinations that interacted in yeast were the AB-293 homodimer containing the complete N-terminus, and the AB-293 in combination with a slightly truncated binding partner, AB-259. The number indicates the number of residues contained in the bait or prey fusion protein. All of the construct combinations grew to confluence on control (LT-deficient) media.
Figure S6. Schematics of constructs used for yeast-two-hybrid analysis of profilaggrin N-terminus homodimerization and N-terminus-annexin II interaction.
Profilaggrin A constructs contain just the S100 domain (residues 1-92), while full-length AB constructs include the complete N-terminus (residues 1-293); additional AB constructs included various truncations made within the B domain, e.g. at residues -98, -120, -140, -160, -218, and -259.
Annexin II (ANXA2) consists of 4 annexin repeats of 70 amino acids (light blue boxes) flanked by N-terminal and C-terminal domains (unshaded boxes). The N-terminal domain of 32 residues includes amino acids 1-14 (shaded black box) that interact with the profilaggrin N-terminus and S100A10.
Figure S7 -- PEG 400 binds to the PF-CABD hydrophobic pocket and alters linker conformation.
(Top left) 24 residues (orange) comprise the hydrophobic pocket of one PF-CABD protein, with bound PEG 400 shown as ball and stick model (carbon, white; oxygen, red).
(Top right) Ile43 and Leu44 (side chains orange) contact PEG 400 (carbon, white; oxygen, red), resulting in a folded conformation of inter-EF-hand linker (L). Wire diagram of PF-CABD monomer (gray) with inter-EF-hand linker (orange).
Bound PEG 400 (by-product of cryocrystallography) alludes to the CABD functioning as a target-binding site. Of 24 residues comprising the hydrophobic pocket, 16 interacted with PEG 400 based on analysis of both the “A” and “C” PF-CABD subunits from the crystal AU (Table S2). The “B” and “D” subunits did not contain PEG. Interestingly, only “A” and “C” subunits have linkers between Helix II and Helix III in a “folded” rather than “extended” conformation. Ile43 and Leu44 are positioned centrally in this linker and in the structure contact PEG 400 (above right), suggesting this flexible linker functions to stabilize target interactions. Linker folding likely explains the slightly more “closed” Helix I/II orientation for the “A” and “C” subunits (Table S1).
Figure S8.
Clustal Omega multiple sequence alignment (top) of the N-termini of 7 epidermal S100 fused-type proteins as well as calbindin D9k, calmodulin, S100B and S100A12. h = human; m = mouse; b = bovine. Completely conserved residues are colored purple and denoted by (*). Highly conserved residues are colored green and denoted (:). Moderately conserved residues are colored orange and denoted by (.). The numbering across the top represents the amino acid sequence numbering for the crystal structure of profilaggrin N-terminus, not positioning for all aligned sequences. The 24 amino acid residues contributing accessible surface area to profilaggrin's N-terminal hydrophobic pocket have a yellow-shaded background; the shading is extended across the multiple sequence alignment to illustrate differences among the protein sequences.
Swiss-Model structural alignment (bottom) between the crystal structure of the N-terminus of human profilaggrin (PDB Code 4PCW) and human S100A12(PDB Code 1E8A), the latter being the model used for molecular replacement. For the profilagrin structure, helices are labeled and colored blue, whereas calcium-binding loops are labeled and colored red.
Figure S9, related to Table 2. Stratifin interacts with profilaggrin N-terminus in yeast. Combinations of bait (green) and prey (pink) fusion proteins were transformed into yeast and grown on HLT-deficient media containing 10 mM 3-AT. Human stratifin (SFN) interacts with full length human profilaggrin N-terminus (hum AB), but not the human A domain (hum A), while SFN interacts with the mouse A and B domains (mou A, B and AB). SFN also gave a similar interaction signal with a mutant human profilaggrin N-terminus protein carrying selected mutations within the calcium-binding domain (hum mut AB). All of the construct combinations grew to confluence on control (LT-deficient) media. Please also see Supplemental Discussion related to this figure.
Figure S10. Structural model of PF-CABD binding to annexin II N-terminus from in silico molecular docking. The N-terminal 11 residues of Gallus gallus annexin II [sequence 1-STVHEILSKLS-11] was molecularly docked onto the PF-CABD dimer structure using the Hex Protein Docking server (http://hexserver.loria.fr/). The structure of the annexin II N-terminus used for docking was dervied from Protein Data Bank Code 1BT6. The sequence differs from human annexin II by a cysteine (human) to serine (chicken) substitution at position 8. Pictured at the bottom is the PF-CABD dimer (light and dark blue) with the annexin II N-terminal helix colored red (the six residues highlighted in orange are the annexin II residues that interact with the PF-CABD binding pocket. In the zoomed in image (top), the seven PF-CABD residues within the hydrophobic pocket that interact with annexin II are highlighted yellow and labeled.
Table S1. Characterization of the interhelical angles and interhelical dimerization contacts of human profilaggrin N-terminal calcium binding domain (PF-CABD).
Table S2, related to Figure 2. Amino Acid Residues Surrounding the Target Binding Pocket and associated Accessible Surface Area. The asterisks (*) denote residues contacting the bound PEG 400 molecule in either the “A” or “C” subunit from the crystal asymmetric unit. The five residues conserved across the S100 fused-type protein family are denoted (italics, #). A, B, C, D = subunits from the crystal asymmetric unit.
Table S3, related to Table 2. Identification of three human profilaggrin N-terminus–associated proteins by immunoprecipitation and mass spectrometrya
Table S4. Potential stratifin binding sites at R-X-X-S motifs within the N-terminus of human and mouse profilaggrin. The amino acid sequence is numbered excluding the N-terminal methionine (leading serine = 1).
Table S5. Primers used to generate constructs used in yeast two-hybrid studies.
Table S6. Profilaggrin Protein Production and Purification Protocol.


