Abstract
In addition to genetic abnormalities such as chromosomal translocations and somatic mutations that have been widely acknowledged in the leukemogenesis of acute myeloid leukemia (AML), epigenetic modifications also play a vital role in this process. MicroRNA (miRNA) regulation is emerging as a new layer of epigenetic regulation besides DNA methylation and histone modifications. Amongst the miRNAs first identified to be specifically expressed in hematopoietic cells, the miR-181 family has been implicated in regulating the differentiation of B cells, T cells and natural killer cells during normal hematopoiesis, and has been linked tightly to the pathogenesis and prognosis of AML. Accumulating evidence indicates that miR-181 acts as a tumor suppressor in the pathogenesis of AML and exhibits a significant impact on the survival of patients with AML. Here we review the role of miR-181 as a diagnostic marker and prognostic predictor in AML and discuss the potential use of miR-181 as a therapeutic target for AML.
Keywords: miR-181, acute myeloid leukemia, diagnostic marker, prognostic predictor, tumor suppressor
Introduction
Acute myeloid leukemia (AML) is a hematopoietic stem cell disorder characterized by the rapid growth and abnormal accumulation of granulocyte or monocyte precursors in bone marrow and blood that eventually interferes with the production of normal blood cells [1]. Recurring chromosomal aberrations and gene mutations are frequently found in AML and contribute greatly to the pathogenesis of the disease [2]. Cytogenetic abnormalities, such as t(8;21), t(15;17), t(9;11) chromosomal translocations and inversion of chromosome 16 (inv16), have been found in more than 50% of AML patients and well recognized as key driving forces of leukemogenesis for the corresponding subtypes of AML [1, 3, 4]. In patients with no detectable chromosomal abnormalities, referred to as cytogenetically normal AMLs (CN-AMLs), mutations in specific genes that play crucial roles in leukemogenesis, such as FLT3, NPM1, CEBPA, DNMT3a, TET2 and IDH1/2, have been observed to occur alone or in combination [5-7]. In addition to these genetic changes, epigenetic modifications including DNA methylation and histone modifications have been frequently reported in AML [2]. These aberrant epigenetic changes can be caused by the somatic mutations of epigenetics-modifying genes (e.g., DNMT3a, TET2 and IDH1/2) or by the chimeric fusion proteins generated by chromosomal translocations (e.g., AML1-ETO by t(8;21), PML-RARA by t(15;17), MLL-AF9 by t(9;11)). On the other hand, the effect of the epigenetic machinery may be achieved through these fusion proteins, thereby forming a complicated regulatory network in AML cells.
During the past decade, another layer of complexity has been added to the understanding of cancer including AML, with the accumulating evidence that microRNAs (miRNAs), a class of short (∼22 nucleotides in length) non-coding RNAs play critical roles in the initiation, progression and relapse of various types of cancers [2, 8-11]. Mature miRNAs are generated from primary miRNA transcripts with the sequential processing by two ribonuclease III enzymes, namely Drosha and Dicer [12]. By base-pairing with the 3′ untranslated region (UTR) of target mRNAs, one miRNA can regulate the expression of multiple target genes through mRNA degradation and/or translational inhibition [2]. Deregulation of miRNAs has been widely observed and associated with the development of hematopoietic malignancies by directly or indirectly modulating the expression of genes crucial for hematopoietic development and leukemogenesis [2, 13, 14]. As miRNA regulation is a kind of post-transcriptional regulation that is not involving changes in genomic DNA, regulation mediated by miRNAs can be viewed as a new layer of epigenetic regulation besides DNA methylation and histone modifications.
The miR-181 family, including miR-181a, miR-181b, miR-181c and miR-181d, are among the miRNAs that were first identified to be specifically expressed in hematopoietic cells [13]. Human miR-181a and miR-181b genes are clustered together and located on chromosome 1 (miR-181a1 and miR-181b1) as well as chromosome 9 (miR-181a2 and miR-181b2), whereas miR-181c and miR-181d genes are clustered together on chromosome 19. The members of the miR-181 family are evolutionarily conserved across almost all vertebrates, suggesting that those miRNAs are functionally important. Indeed, accumulating data prove the critical role of miR-181 family members as regulators of normal cell differentiation, especially for cells of hematopoietic origin, including T-cells, B-cells, natural killer cells and megakaryocytes [13, 15, 16]. At the same time, a growing number of evidence has linked miR-181 to leukemia, particularly to AML.
MiR-181 as a diagnostic marker of AML
With the development of various methods for high-throughput quantification of miRNA expression, miRNA expression profiles have been determined in different sets of leukemia samples. Particularly, with the finding that miRNA expression signature could be used to distinguish AML from other subtypes of leukemia and to discriminate different molecular subtypes of AML [17, 18], it is becoming more clear that miRNAs can be used as diagnostic markers for leukemia, including AML.
In an early study of miRNA expression in 30 AML samples from peripheral blood or bone marrow of patients with normal karyotype, Debernardi et al. used Taqman Realtime qPCR and found that expression of miR-181a correlates with the leukemic morphological subtypes as classified based on the French-American-British (FAB) morphological phenotype [19]. Specifically, an elevated expression of miR-181a was observed in the M1 and M2 subtype samples as compared to those with the M4 and M5 subtypes [19]. This was confirmed later in another study by Isken et al. using 14 normal karyotype AML samples [20]. These studies implied that down-regulation of miR-181a is a hallmark of the M4 and M5 subtypes of AML, although further studies are needed to understand the mechanism by which this deregulation occurs and the targets of miR-181a that might be derepressed and therefore drive the relevant leukemic phenotype.
Beyond morphologically defined subtype distinctions, miR-181 expression has also been examined across cytogenetically normal AML (CN-AML) and cytogenetically abnormal AML (CA-AML) patient samples. In CN-AML, Marcucci et al. reported that all members from the miR-181 miRNA family were up-regulated in patients with CEBPA mutations and therefore characterized this type of AML [21]. Research from the same group also identified the decreased expression of miR-181 in a high-risk subgroup of CN-AML with FLT3-ITD or wild-type NPM1 or both [22]. By use of bead-based miRNA expression profiling assay, Li et al. showed an increased expression of miR-181a, miR-181b, miR-181c, and miR-181d in CA-AML carrying t(8;21), inv(16), and t(15;17) than in patients harboring mixed lineage leukemia (MLL) rearrangements [18]. Therefore, miR-181 expression correlates well with cytogenetic and molecular subtypes of AML and has the potential to be used as a diagnostic marker for AML.
MiR-181 as a prognostic predictor of AML
There is a growing body of evidence to demonstrate that the expression signature of some miRNAs is associated with the clinical outcome of AML patients [23-25], which suggests that miRNAs could serve as prognostic markers and thereby guide the clinical treatment of AML.
Expression of miR-181 family has been consistently reported to positively correlate with favorable prognosis in AML patients (Table 1). Marcucci et al. showed that expression of miR-181a and miR-181b was positively associated with the clinical outcome in molecular high-risk CN-AML (with FLT3-ITD and/or wild-type NPM1) and inversely associated with the risk of an event, such as failure to achieve complete remission (CR), relapse, or death [22]. This correlation remained significant in their multivariable analysis after adjusting for other clinical factors, such as white blood cell count. Based on the inverse correlation between miR-181 expression and their putative target genes such as TLR4, CARD8, CASP1, IL1B, SLC11A1, MSR1, and FGR1A, the authors hypothesized that miR-181 was functionally involved in toll-like receptor (TLR) and interleukin-1β-regulated innate immune response pathways [22]. In addition, they reported that upregulation of members of the miR-181 family in CN-AML with CEBPA mutations was associated with favorable clinical outcomes [21]. They further showed that the truncated C/EBPα-p30 isoform encoded by N-terminal CEBPA mutations bound directly to miR-181a promoter and promoted its expression, suggesting that miR-181a is a direct target of CEBPA mutants [26].
Table 1. Expression patterns and clinical significance of miR-181 family members in AML.
| AML classification* | miR-181 expression | Clinical significance | References |
|---|---|---|---|
| FAB M1, M2 | miR-181a ↑ | Down-regulation of miR-181a characterizes FAB M4 and M5 subtypes | [19, 20] |
| FAB M4, M5 | miR-181a ↓ | ||
| AML with MLD | miR-181a ↓ | Decreased expression of miR-181a is associated with unfavorable response to therapy | [23] |
| CN-AML with CEBPA mutations | miR-181a ↑ | Increased expression of miR-181a is associated with favorable clinical outcomes | [21] |
| CN-AML with FLT3-ITD and/or wild-type NPM1 | miR-181a/b ↓ | Decreased expression of miR-181a is associated with the risk of failure to achieve CR, relapse, or death | [22, 27] |
| CA-AML with favorable cytogenetic abnormalities such as t(8;21), inv(16), t(15;17) | miR-181a/b ↑ | Increased expression of miR-181a is associated with favorable overall survival | [28] |
| CA-AML with unfavorable cytogenetic abnormalities such as MLL-rearrangements | miR-181a/b ↓ | Decreased expression of miR-181a is associated with unfavorable prognosis | [28] |
AML, acute myeloid leukemia; FAB, French-American-British classification system; MLD, multilineage dysplasia (MLD); CN-AML, cytogenetically normal AML; CA-AML, cytogenetically abnormal AML.
The prognostic significance of miR-181 expression in CN-AML was further confirmed by Schwind et al., who demonstrated that higher expression of a single miRNA, miR-181a, predicted better outcome as shown by a higher CR rate, longer overall survival and longer disease-free survival, especially in CN-AML patients with FLT3-ITD and/or NPM1 wild-tpye [27]. This statistically significant prognostic impact of miR-181a expression was maintained in multivariable analysis considering other molecular prognostic factors [27].
The prognostic potential of miR-181 is not restricted to CN-AML. Li et al. recently demonstrated that increased expression levels of miR-181a and miR-181b are associated with favorable overall survival in CA-AML patients using either univariate or multivariate analysis [28]. The authors also showed that four homeobox superfamily genes (i.e., HOXA7, HOXA9, HOXA11, and PBX3) were targeted by miR-181 family, particularly by miR-181b [28] (also see Figure 1). In fact, miR-181a, miR-181b, miR-181c and miR-181d were expressed at a relatively higher level in patients carrying translocations with favorable prognosis, such as t(8;21), inv(16) and t(15;17), compared to patients with intermediate or poor prognostic translocations, such as those with MLL rearrangements [28]. As a result, expression signatures, not only of miR-181 genes but also of their 4 target genes as a whole, have valuable potential to be employed as independent prognostic factors of clinical outcomes of patients with CA-AML [28].
Figure 1. The miR-181 target gene network in normal and malignant hematopoiesis.
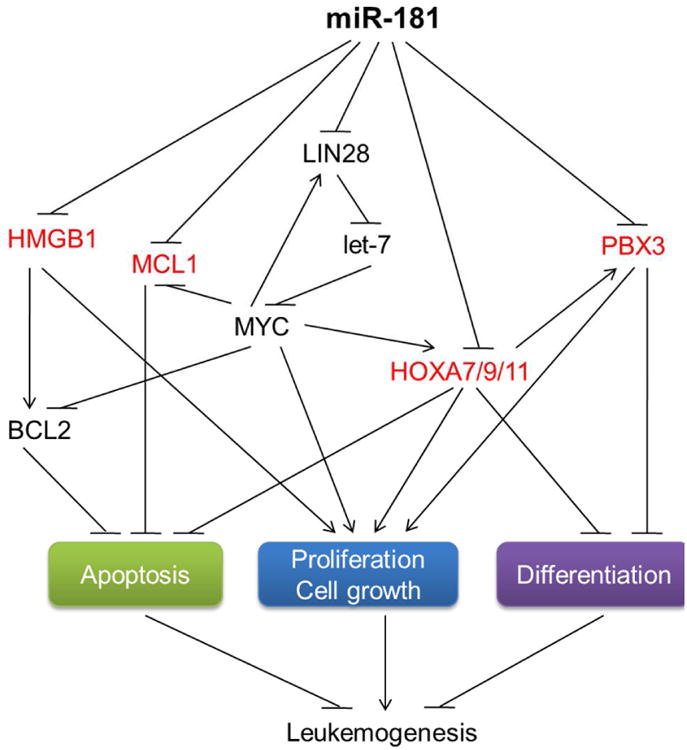
The reported direct targets of miR-181 in AML include PBX3, HOXA7, HOXA9, HOXA11, MCL1, and HMGB1 (in red). Targeting of HMGB1 and MCL1 by miR-181 in AML cells and inhibition of apoptosis by HMGB1 were reported by Lu F et al. [36]. HOXA7/9/11 and PBX3 as direct targets of miR-181 was reported by Li ZJ et al. [28]. Inhibition of apoptosis and promotion of cell growth and transformation of AML cells by HOXA9, as well as action of PBX3 downstream of HOXA9, was reported by Faber J et al. [40]. Inhibition of HOXA9 on neutrophil and macrophage differentiation was reported by Calvo KR et al. [41]. Targeting the LIN28-let-7 regulatory loop by miR-181 was reported by Li X et al. [16]. Regulation of MYC on MCL1, BCL2, and HOXA genes was reported elsewhere [42, 43].
In addition, in AML with multilineage dysplasia (MLD) that is often associated with unfavorable cytogenetic profile and response to therapy, miR-181a is the only miRNA that has been shown to be downregulated [23], further supporting its role as a prognostic predictor in AML.
MiR-181 in therapeutic development for AML
The aim of research in cancer is to translate basic scientific findings into better clinical strategies that would benefit patients. MiRNAs serve as such a type of ideal targets from this point of view as they can act as either oncogenes or tumor suppressors or both. The potential of using miRNA inhibitors or miRNA mimics has been studied to evaluate the feasibility of interfering with the function of oncogenic miRNAs or restoring the expression/function of tumor-suppressor miRNAs in the treatment of cancers [2, 10, 29, 30]. Moreover, combination of miRNA inhibitors or mimics with chemotherapeutic drugs has also proven to be more efficacious in cancer treatment and thus represents a very attractive therapeutic strategy [31, 32].
Although an oncogenic role of miR-181 has been reported in other types of cancer [33, 34], the role of miR-181 as a tumor suppressor in AML has been consistently suggested based on the correlation between miR-181 expression and the overall survival of AML patients [21, 22, 27, 28]. Specifically, Li et al. studied in detail the tumor suppressive role of miR-181 in AML and showed that ectopic expression of miR-181b significantly inhibited cell viability and promoted apoptosis of MLL-rearranged AML cells, and inhibited MLL-fusion-mediated cell transformation and leukemogenesis [28]. This study sheds new light on the application of miR-181b for clinical treatment of MLL-rearranged AML.
Hickey et al. have shown that lenalidomide, a drug approved by U.S. Food and Drug Administration (FDA) for the treatment of myelodysplastic syndromes and multiple myeloma, can induce endogenous expression of miR-181a through enhancing translation of the C/EBPα-p30 isoform [26]. In xenograft mouse models, either forced expression of miR-181a or lenalidomide treatment could significantly inhibit tumor growth of AML [26]. Thus, inducing endogenous expression of tumor-suppressor miRNAs by therapeutic agents represents a new therapeutic strategy.
Recently, studies on combining miR-181 with chemotherapeutic drugs have been conducted, with the hope of providing better strategy for AML therapy. MiR-181a expression was shown to be reduced in the HL-60/Ara-C subline that was derived from HL60 with Ara-C resistance, and restoration of this miRNA sensitized the cells to Ara-C treatment [35]. Similarly, lenalidomide can sensitize AML cells to conventional Ara-C therapy through increasing endogenous expression of miR-181a [26]. In another study, Lu et al. showed that miR-181b was down-regulated in human multidrug-resistant leukemia cells and relapsed/refractory AML patient samples; when overexpressed, miR-181b sensitized these leukemia cells to cytotoxic chemotherapeutic drugs by targeting HMGB1 [36]. These studies provided preliminary evidence for the combined therapy of AML using miR-181 and chemotherapeutic drugs.
Discussion
With increasing studies of miRNA expression profiling and functions in AML, our understanding of the functions and clinical implications of miRNAs in the pathogenesis and prognosis of AML is expanding. In addition to their prognostic value, miRNA expression signatures also have tremendous potential to translate into clinical therapeutics. For example, in unfavorable subtypes of cytogenetically normal and abnormal AML with low expression of miR-181a/b, replacement therapies through nanoparticle delivery of miR-181a/b oligos and/or drug (e.g., lenalidomide) treatment can restore the normal expression/function of miR-181a/b and thereby inhibit leukemia progression. The optimal therapeutic effect on treating poor-prognosis AML could be achieved by the combination of miR-181 replacement therapy with other therapies such as standard chemotherapy.
However, special care must be taken when employing these data for guidance of clinical application. Although almost all the clinical data available support miR-181 as a tumor suppressive miRNA family in AML, there was still some controversial evidence on the function of miR-181 in leukemia. For example, miR-181b was shown to be down-regulated in acute promyelocytic leukemia (APL) patients and cell lines treated with all-trans-retinoic acid (ATRA) [37]; in addition, overexpression of miR-181a was shown to prevent 1,25-dihydroxyvitamin D(3)-induced monocytic differentiation [38], ATRA-induced granulocytic differentiation, or phorbol myristate acetate (PMA)-induced macrophage-like differentiation of human AML cells [39]. It is better to rule out the genetic and epigenetic causes of these discrepancies before considering any potential clinical applications. Additionally, it is essential for us to further clarify the molecular mechanisms underlying the regulation of miR-181 expression in different subtypes of AML and the impact of miR-181 expression signatures on the prognosis of patients with AML. Importantly, more bona fide targets of miR-181 need to be identified in AML, although several target genes have been reported in normal and malignant hematopoiesis (Figure 1). It is only after we have a more comprehensive and in-depth understanding of the regulatory mechanisms and pathological functions of miR-181 in AML that we can develop the optimal miR-181-based therapies and apply them to treat currently therapy-resistant AML.
Acknowledgments
This work was supported in part by the National Institutes of Health (NIH) R01CA182528 and R01CA178454 (to JC), American Cancer Society (ACS) Research Scholar grant (to JC), Leukemia & Lymphoma Society (LLS) Translational Research grant (to JC), Innovation Award of Alex's Lemonade Stand Foundation for Childhood Cancer (to JC), and the Spastic Paralysis Foundation of the Illinois, Eastern Iowa Branch of Kiwanis International (to JC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heerema-McKenney A, Arber DA. Acute myeloid leukemia. Hematol Oncol Clin North Am. 2009;23:633–54. doi: 10.1016/j.hoc.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Fatica A. Noncoding RNAs in Acute Myeloid Leukemia: From Key Regulators to Clinical Players. Scientifica (Cairo) 2012;2012:925758. doi: 10.6064/2012/925758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 6.Mrozek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–48. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer SC, Levine RL. Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol. 2014;15:e382–e94. doi: 10.1016/S1470-2045(14)70008-7. [DOI] [PubMed] [Google Scholar]
- 8.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120:953–60. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classifyhuman cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 13.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 14.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–8. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 15.Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187:6171–5. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Zhang J, Gao L, McClellan S, Finan MA, Butler TW, Owen LB, Piazza GA, Xi Y. MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ. 2012;19:378–86. doi: 10.1038/cdd.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander SK, Le Beau MM, Larson RA, Golub TR, Rowley JD, Chen J. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:19971–6. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD, Chen J. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105:15535–40. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–6. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 20.Isken F, Steffen B, Merk S, Dugas M, Markus B, Tidow N, Zuhlsdorf M, Illmer T, Thiede C, Berdel WE, Serve H, Muller-Tidow C. Identification of acute myeloid leukaemia associated microRNA expression patterns. Br J Haematol. 2008;140:153–61. doi: 10.1111/j.1365-2141.2007.06915.x. [DOI] [PubMed] [Google Scholar]
- 21.Marcucci G, Maharry K, Radmacher MD, Mrozek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus CD, Liu CG, Ruppert AS, Powell BL, Carroll AJ, Caligiuri MA, Kolitz JE, Larson RA, Bloomfield CD. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J Clin Oncol. 2008;26:5078–87. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, Garzon R, Croce CM, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–28. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 23.Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, Flomenberg N, Marcucci G, Calin GA, Kornblau SM, Kantarjian H, Bloomfield CD, Andreeff M, Croce CM. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–9. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Y, Kasar S, Underbayev C, Prakash S, Raveche E. MicroRNAs in Acute Myeloid Leukemia and Other Blood Disorders. Leuk Res Treatment. 2012;2012:603830. doi: 10.1155/2012/603830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Li Z, He C, Wang D, Yuan X, Chen J, Jin J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44:191–7. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickey CJ, Schwind S, Radomska HS, Dorrance AM, Santhanam R, Mishra A, Wu YZ, Alachkar H, Maharry K, Nicolet D, Mrozek K, Walker A, Eiring AM, Whitman SP, Becker H, Perrotti D, Wu LC, Zhao X, Fehniger TA, Vij R, Byrd JC, Blum W, Lee LJ, Caligiuri MA, Bloomfield CD, Garzon R, Marcucci G. Lenalidomide-mediated enhanced translation of C/EBPalpha-p30 protein up-regulates expression of the antileukemic microRNA-181a in acute myeloid leukemia. Blood. 2013;121:159–69. doi: 10.1182/blood-2012-05-428573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwind S, Maharry K, Radmacher MD, Mrozek K, Holland KB, Margeson D, Whitman SP, Hickey C, Becker H, Metzeler KH, Paschka P, Baldus CD, Liu S, Garzon R, Powell BL, Kolitz JE, Carroll AJ, Caligiuri MA, Larson RA, Marcucci G, Bloomfield CD. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:5257–64. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, Radmacher MD, Maharry K, Elkahloun A, Yang X, He C, He M, Zhang Z, Dohner K, Neilly MB, Price C, Lussier YA, Zhang Y, Larson RA, Le Beau MM, Caligiuri MA, Bullinger L, Valk PJ, Delwel R, Lowenberg B, Liu PP, Marcucci G, Bloomfield CD, Rowley JD, Chen J. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–24. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol. 2011;29:475–86. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 31.Sugimura K, Miyata H, Tanaka K, Hamano R, Takahashi T, Kurokawa Y, Yamasaki M, Nakajima K, Takiguchi S, Mori M, Doki Y. Let-7 expression is a significant determinant of response to chemotherapy through the regulation of IL-6/STAT3 pathway in esophageal squamous cell carcinoma. Clin Cancer Res. 2012;18:5144–53. doi: 10.1158/1078-0432.CCR-12-0701. [DOI] [PubMed] [Google Scholar]
- 32.Weng H, Huang H, Dong B, Zhao P, Zhou H, Qu L. Inhibition of miR-17 and miR-20a by Oridonin Triggers Apoptosis and Reverses Chemoresistance by Derepressing BIM-S. Cancer Res. 2014;74:4409–19. doi: 10.1158/0008-5472.CAN-13-1748. [DOI] [PubMed] [Google Scholar]
- 33.Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123:150–63. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji D, Chen Z, Li M, Zhan T, Yao Y, Zhang Z, Xi J, Yan L, Gu J. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer. 2014;13:86. doi: 10.1186/1476-4598-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai H, Cao Z, Deng C, Zhou L, Wang C. miR-181a sensitizes resistant leukaemia HL-60/Ara-C cells to Ara-C by inducing apoptosis. J Cancer Res Clin Oncol. 2012;138:595–602. doi: 10.1007/s00432-011-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu F, Zhang J, Ji M, Li P, Du Y, Wang H, Zang S, Ma D, Sun X, Ji C. miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int J Oncol. 2014;45:383–92. doi: 10.3892/ijo.2014.2390. [DOI] [PubMed] [Google Scholar]
- 37.Garzon R, Pichiorri F, Palumbo T, Visentini M, Aqeilan R, Cimmino A, Wang H, Sun H, Volinia S, Alder H, Calin GA, Liu CG, Andreeff M, Croce CM. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–57. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Gocek E, Liu CG, Studzinski GP. MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle. 2009;8:736–41. doi: 10.4161/cc.8.5.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su R, Lin HS, Zhang XH, Yin XL, Ning HM, Liu B, Zhai PF, Gong JN, Shen C, Song L, Chen J, Wang F, Zhao HL, Ma YN, Yu J, Zhang JW. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene. 2014 doi: 10.1038/onc.2014.274. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 40.Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, Zwaan CM, Kung AL, Armstrong SA. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–85. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calvo KR, Knoepfler PS, Sykes DB, Pasillas MP, Kamps MP. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc Natl Acad Sci U S A. 2001;98:13120–5. doi: 10.1073/pnas.231115398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beverly LJ, Varmus HE. MYC-induced myeloid leukemogenesis is accelerated by all six members of the antiapoptotic BCL family. Oncogene. 2009;28:1274–9. doi: 10.1038/onc.2008.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, Chen P, He C, You D, Zhang S, Wang J, Arnovitz S, Elkahloun A, Price C, Hong GM, Ren H, Kunjamma RB, Neilly MB, Matthews JM, Xu M, Larson RA, Le Beau MM, Slany RK, Liu PP, Lu J, Zhang J, Chen J. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22:524–35. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]


