Summary
Type I interferons (IFNs) are critical mediators of antiviral defense, but their elicitation by bacterial pathogens can be detrimental to hosts. Many intracellular bacterial pathogens, including Mycobacterium tuberculosis, induce type I IFNs following phagosomal membrane perturbations. Cytosolic M. tuberculosis DNA has been implicated as a trigger for IFN production, but the mechanisms remain obscure. We report that the cytosolic DNA sensor, cyclic GMP-AMP synthase (cGAS), is required for activating IFN production via the STING/TBK1/IRF3 pathway during M. tuberculosis and L. pneumophila infection of macrophages, whereas L. monocytogenes short-circuits this pathway by producing the STING agonist, c-di-AMP. Upon sensing cytosolicDNA, cGAS also activates cell-intrinsic antibacterial defenses, promoting autophagic targeting of M. tuberculosis. Importantly, we show that cGAS binds M. tuberculosis DNA during infection, providing direct evidence that this unique host-pathogen interaction occurs in vivo. These data uncover a mechanism by which IFN is likely elicited during active human infections.
Introduction
Innate immune cells discriminate pathogens from non-pathogens at the earliest stages of infection and tailor their responses to match the level of the threat (Vance et al., 2009). A fundamental way this is achieved is through sensing membrane perturbations mediated by bacterial virulence factors, either directly or via the recognition of specific bacterial molecules in the cytosol (Manzanillo et al., 2012; Thurston et al., 2012; Vance et al., 2009). Cytosolic detection leads to activation of three potent antimicrobial effector pathways – the inflammasome (Lamkanfi and Dixit, 2011), autophagy (Watson et al., 2012), and the cytosolic surveillance pathway (CSP) characterized by elicitation of type I interferons (IFNs) (Monroe et al., 2010; O'Riordan et al., 2002). While type I IFNs (IFN-α, IFN-β) are potent antiviral signaling molecules, they inhibit antibacterial signaling pathways (Mayer-Barber et al., 2011; Teles et al., 2013) and promote infection of many intracellular bacteria including Mycobacterium tuberculosis (Manca et al., 2001; Manzanillo et al., 2012) and Listeria monocytogenes (Auerbuch et al., 2004; Carrero et al., 2004; O'Connell et al., 2004), suggesting that bacterial pathogens have evolved mechanisms to activate this antiviral pathway for their benefit. Likewise, clinical studies have shown that elevated levels of type I IFNs are a biomarker of active TB disease in humans (Berry et al., 2010), indicating that this pathway is engaged during bacterial replication in vivo. Elicitation of IFN-β requires the pore-forming toxin listeriolysin O during L. monocytogenes infection (O'Riordan et al., 2002), and the membrane-disrupting activity of the ESX-1 secretion system during M. tuberculosis infection (Manzanillo et al., 2012). L. monocytogenes actively secretes the bacterial second messenger cyclic diadenylate monophosphate (c-di-AMP) that binds to the host protein STING and activates the STING/TBK1/IRF3 signaling axis to promote a signature transcriptional response that includes IFN-β (Burdette et al., 2011; Sauer et al., 2011). In contrast to L. monocytogenes, M. tuberculosis and another intracellular bacterial pathogen, Legionella pneumophila, appear to activate this same STING-dependent pathway (Lippmann et al., 2008; Manzanillo et al., 2012; Monroe et al., 2010) via recognition of pathogen-derived nucleic acids, although the evidence for this is indirect.
Surprisingly, the STING pathway also potently activates autophagy, a degradative pathway implicated in resistance to intracellular pathogens (Birmingham et al., 2006; Deretic and Levine, 2009; Zhao et al., 2008). Activation of STING and the kinase TBK1 leads to targeting of bacteria and cytosolic DNA to the ubiquitin-mediated selective autophagy pathway in macrophages (Watson et al., 2012), and ATG5, a core autophagy protein, is crucial for limiting M. tuberculosis growth during infection. Despite our growing understanding of the links between the CSP and selective autophagy, the nature of the nucleic acid ligand and the host receptor proteins involved remain major unanswered questions.
Recent studies have identified a DNA sensor, cyclic GMP-AMP synthase (cGAS), as the central cytoplasmic DNA sensor upstream of STING during viral infection (Gao et al., 2013; Schoggins et al., 2014; Sun et al., 2013). Upon binding dsDNA, cGAS synthesizes the secondary messenger cGAMP, which in turn binds to and activates STING, leading to the production of IFN-β through IRF3. Despite some controversy over the involvement of a variety of other DNA receptors (Burdette and Vance, 2013), cGAS is absolutely required for interferon induction upon both DNA transfection and viral infection, and is likely the major cytosolic DNA receptor (Li et al., 2013). cGAS is required for IFN-β induction during Chlamydia trachomatis, but its functional role during bacterial infections is largely unknown (Zhang et al., 2014).
Here, we report two different mechanisms by which bacterial pathogens activate the CSP: M. tuberculosis and L. pneumophila activate cGAS during infection, while L. monocytogenes bypasses cGAS and directly activates STING via c-di-AMP. Furthermore, we show that recognition of M. tuberculosis by cGAS is a critical host-pathogen interaction as it promotes the delivery of bacilli to the ubiquitin-mediated selective autophagy pathway during macrophage infection, and has an unexpected role in cell-autonomous bacterial control. Importantly, we also show that cGAS binds M. tuberculosis genomic DNA during macrophage infection, providing evidence of direct interactions between an M. tuberculosis ligand and a host sensor in vivo.
Results
cGAS is required to induce the CSP during intracellular bacterial infection
To test the role of cGAS during M. tuberculosis infection, primary murine bone marrow-derived macrophages (BMDMs) were isolated from cGas-/- and Sting-/- mice. These cells were unresponsive to transfection with interferon-stimulatory DNA (ISD), a 45 bp dsDNA sequence sufficient for activating cGAS (Stetson and Medzhitov, 2006), as measured by quantitative PCR of IRF3 targets (IFN-β and IFIT1 mRNAs, Figure S1A), and by monitoring IRF3 phosphorylation (Figure S1G). Delivery of the STING ligand cGAMP via transfection bypassed the cGAS requirement (Figure S1B), in agreement with previous reports in other cell types establishing that cGAS functions upstream of STING (Sun et al., 2013).
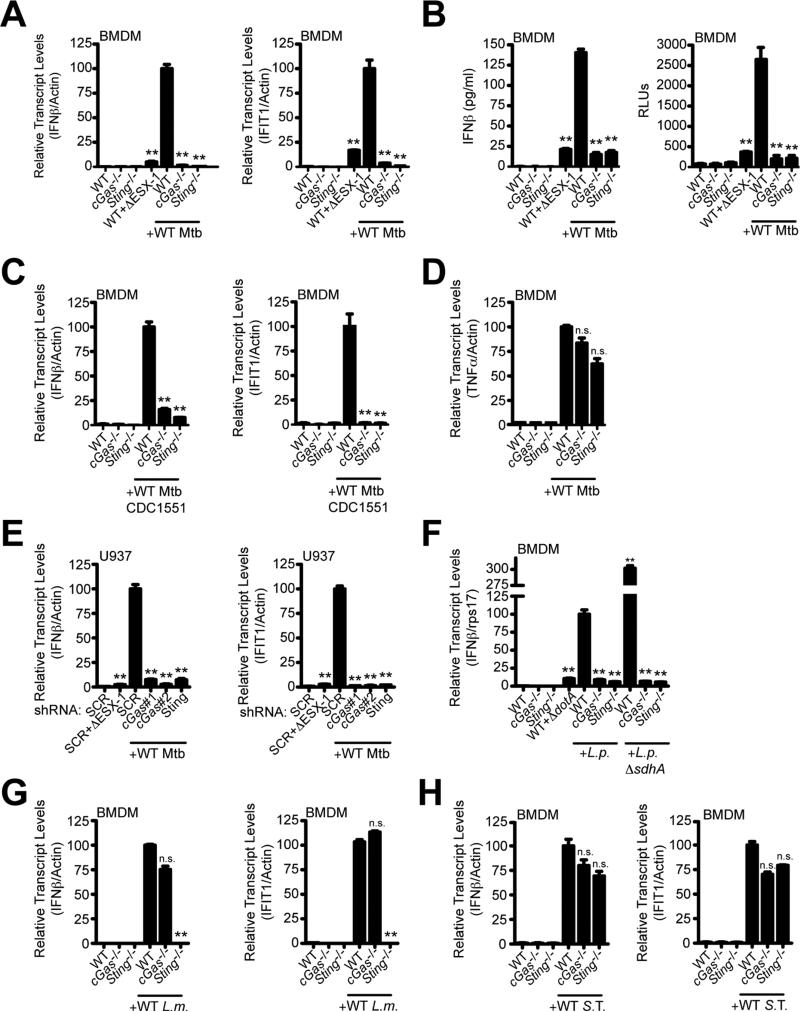
Importantly, infection with the Erdman strain of M. tuberculosis led to robust CSP activation in wild-type BMDMs as measured by both protein and transcript levels. This response was completely blocked in cGas-/- and Sting-/- macrophages (Figures 1A and 1B), similar to infection of wild-type macrophages with an ESX-1 mutant, which fails to induce the CSP (Manzanillo et al., 2012). We observed similar results using a clinical isolate of M. tuberculosis (CDC1551) which, unlike the Erdman strain, has the capacity to produce the STING agonist cdi-GMP (Figure 1C) (Manzanillo et al., 2012). This response is specific to the STING/TBK1/IRF3 pathway as the mRNA level of TNFα, which is not regulated by IRF3, was generally unaffected by these mutations (Figure 1D). Independent shRNA knockdown of cGAS in the RAW 264.7 murine macrophage cell line (Figure S1C), and in the U937 human macrophage cell line (Figure S1D) confirmed the key role of this receptor in responding to cytosolic DNA (Figures S1E and S1F) and M. tuberculosis infection (Figures 1E and S1H). These data demonstrate that cGAS is the major sensor that activates the CSP during M. tuberculosis infection of macrophages. These results also provide strong evidence that CSP activation by wild-type M. tuberculosis is due to exposure of DNA in the cytosol, and support previous findings that endogenous levels of bacterial-produced cyclic dinucleotides are not a major contributor in triggering this response during macrophage infection (Dey et al., 2015; Manzanillo et al., 2012).
Figure 1. cGAS is essential for induction of the cytosolic surveillence response during intracellular bacterial infection.
(A) BMDMs were infected with WT or ΔESX-1 M. tuberculosis (Erdman) for 4 h, and IFN-β and IFIT1 transcripts were measured by RT-qPCR. mRNA levels are expressed as percentages relative to infected WT cells.
(B) Same as (A) but IFN-β protein was measured by ELISA (left) or with ISRE-luciferase cells (right) 24 h post-infection.
(C) Same as (A) but M. tuberculosis strain CDC1551.
(D) Same as (A) but TNFα.
(E) Same as (A) but with U937 knockdown cell lines. mRNA levels are expressed as percentage relative to infected scramble control cells.
(F-H) BMDMs were infected for 4 h and transcript levels were measured during infection with L. pneumophila ΔflaA (L.p.) or ΔflaAΔsdhA (L.p. ΔsdhA) (F), L. monocytogenes (L.m.) (G), or S. Typhimurium (S. T.) (H). n.s., not significant; **p < 0.005 by two-tailed t-test comparing to WT infected with WT bacteria. See also Figure S1.
We next tested the role of cGAS during infection with another intracellular bacterial pathogen, L. pneumophila, which elicits type I IFNs by accessing the cytosol through its type IV secretion system (T4SS) (Stetson and Medzhitov, 2006). L. pneumophila infection failed to induce the CSP in cGas-/- BMDMs (Figure 1F). In addition, sdhA mutant L. pneumophila cells, which enter into the cytoplasm more readily than wild-type bacteria (Creasey and Isberg, 2012), induced significantly higher levels of IFN-β mRNA (Monroe et al., 2009; Vance et al., 2009), and this too was dependent on cGAS (Figure 1F). The absolute requirement for cGAS is consistent with the notion that DNA is exposed to the cytosol during L. pneumophila infection via vacuolar perforations from its T4SS (Monroe et al., 2010; Stetson and Medzhitov, 2006).
In contrast with M. tuberculosis, CSP activation by L. monocytogenes infection was mostly independent of cGAS (Figure 1G). In cGas-/- BMDMs there was only a slight reduction in IFN-β and no decrease in IFIT1 mRNA levels compared to infected wild-type BMDMs, whereas the response was completely blocked in Sting-/- BMDMs (Sauer et al., 2011). The absolute requirement of STING but not cGAS indicates that L. monocytogenes successfully short-circuits the CSP pathway by providing its own second messenger, whereas M. tuberculosis and L. pneumophila generate the signal via DNA binding to cGAS.
Finally, infection with Salmonella enterica serovar Typhimurium led to normal levels of IFN-β and IFIT1 mRNAs in both cGas-/- and Sting-/- BMDMs, indicating that this pathogen predominantly activates type I IFNs via a CSP-independent mechanism, likely via the TLR4/TRIF pathway (Figure 1H) (Kawai and Akira, 2010; Zughaier et al., 2005). Taken together, these data suggest that intracellular bacterial pathogens have evolved the ability to activate type I IFNs via different mechanisms.
cGAS targets cytosolic DNA and M. tuberculosis to the selective autophagy pathway
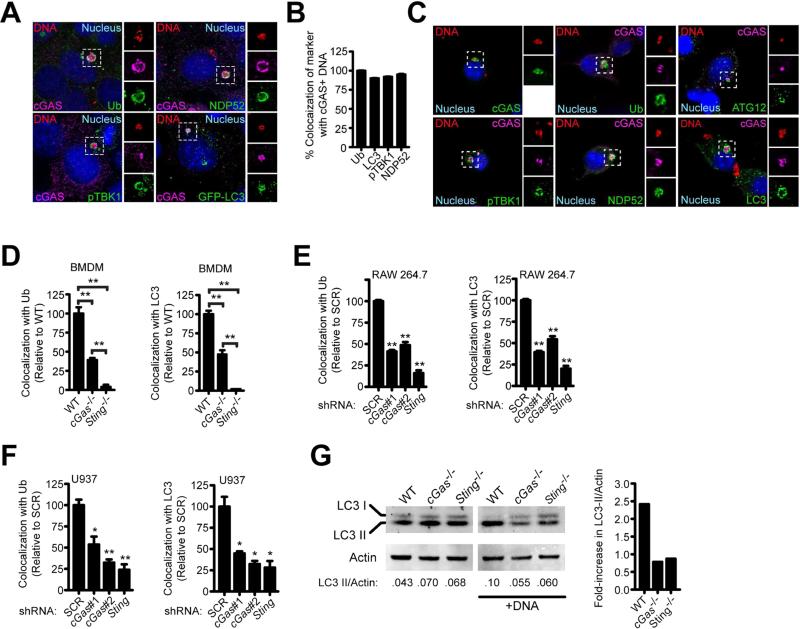
Newly-recognized connections between DNA sensing and autophagy suggests that cGAS activation may also promote bacterial clearance (Liang et al., 2014; Manca et al., 2001; Manzanillo et al., 2012; Watson et al., 2012). To test the role of cGAS in autophagic targeting, we transfected murine embryonic fibroblasts (MEFs) with Cy3-labeled dsDNA and observed that a similar percentage of DNA puncta colocalized with cGAS as with the selective autophagy markers LC3, ubiquitin, NDP52, and activated phospho-TBK1 (pTBK1) (Figures S2A and S2B). Multicolor fluorescence microscopy revealed that the large majority of cGAS+ dsDNA structures contained all of these selective autophagy markers in both transfected MEFs and RAW 264.7 cells (Figures 2A-2C). Importantly, in cGas-/- macrophages, colocalization of the autophagy targeting components, ubiquitin and LC3, with transfected DNA was reduced by approximately 50% (Figure 2D). Curiously, we consistently observed a stronger reduction in selective autophagy marker colocalization with DNA in Sting-/- macrophages compared to cGas-/-cells, suggesting that other factors, perhaps additional DNA sensors including IFI204 (Manzanillo et al., 2012), work upstream of STING to target cytosolic DNA to autophagy. The requirement for STING in selective autophagic targeting is consistent with our previous finding that TBK1 is also required for this effect, whereas IRF3 is only required for the transcriptional output of this pathway (Watson et al., 2012). ShRNA-mediated knockdown of cGAS or STING in human or murine macrophage cell lines also led to decreased recruitment of selective autophagy markers to dsDNA 4 h post-transfection (Figures 2E and 2F), though we did not observe the cGAS-independent contribution to this effect. As an additional method of testing the role of cGAS in targeting dsDNA to the selective autophagy pathway, we measured the levels of LC3-II, the lipidated form of LC3 generated during activation of autophagy, in wild-type, cGas-/-, and Sting-/- BMDMs after transfection with dsDNA. Western blot analysis revealed that while wild-type BMDMs had increased LC3-II levels after dsDNA transfection, cGas-/- and Sting-/- BMDMs failed to induce LC3-II conversion (Figure 2G). Importantly, the effect of cGAS on autophagy is not due to general defects in bulk autophagy as starvation of macrophages led to LC3-II conversion and degradation regardless of genotype (Figure S2C).
Figure 2. cGAS is required to target cytosolic DNA to the ubiquitin-mediated selective autophagy pathway.
(A) MEFs expressing 3×FLAG-tagged cGAS transfected with Cy3-labeled plasmid DNA for 4 h and immunostained for 3×FLAG or indicated markers.
(B) Quantification of cGAS+ Cy3-DNA co-stained with indicated marker from (A). Differences are not statistically significant.
(C) RAW 264.7 cells stably expressing 3×FLAG-cGAS transfected with Cy3-DNA for 4 h and immunostained for 3×FLAG and indicated markers.
(D) Quantification of ubiquitin and LC3 colocalization with Cy3-DNA 4 h post-transfection in BMDMs.
(E) Same as (D) but RAW 264.7 knockdown cell lines.
(F) Same as (D) but U937 knockdown cell lines.
(G) BMDMs were transfected with interferon-stimulatory DNA (ISD) for 2 h, and LC3-II conversion was analyzed by quantitative Western blot (left) and expressed as a fold-increase in the ratio of LC3-II/Actin. Results are representative of at least three independent experiments. *p < 0.05, **p < 0.005 by two-tailed t-test comparing to SCR unless otherwise indicated. See also Figure S2.
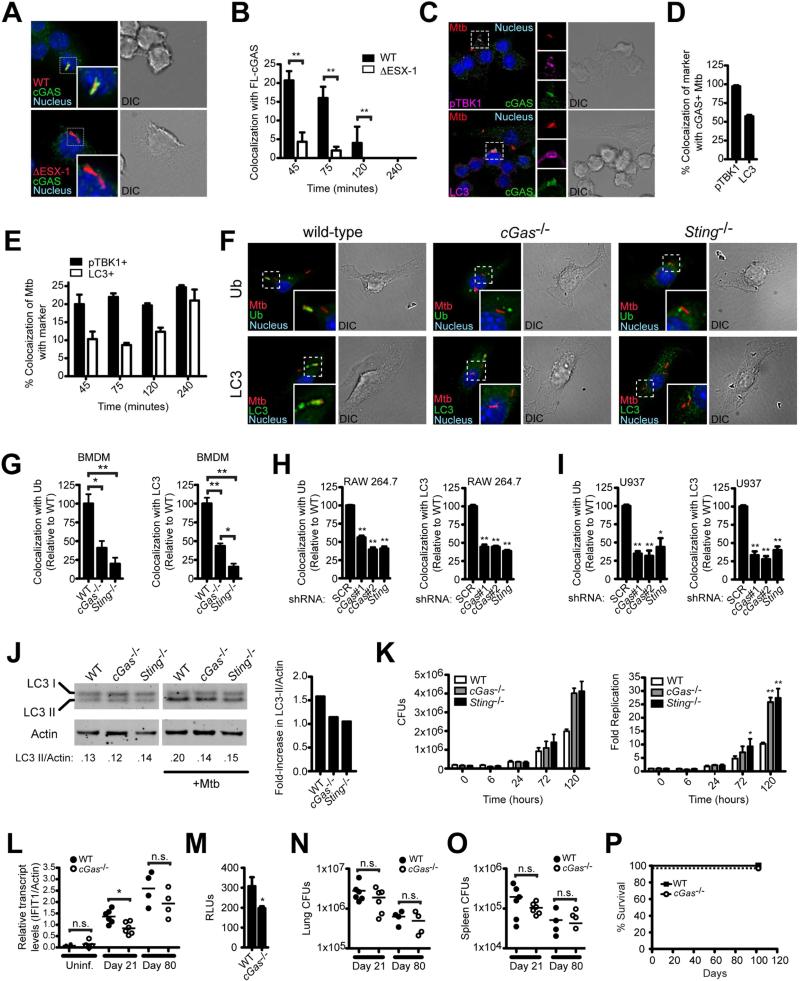
To begin to test the role of cGAS in autophagic targeting of M. tuberculosis, we infected RAW 264.7 cells expressing epitope-tagged cGAS with mCherry-expressing M. tuberculosis and monitored cGAS localization. Early in infection, cGAS colocalized with approximately 20% of wild-type M. tuberculosis, and this colocalization decreased at later time points (Figures 3A and 3B). Importantly, we observed significantly decreased colocalization of cGAS with ESX-1 mutant bacteria at all time points, confirming that recognition requires ESX-1-mediated cytosolic access. As observed with transfected DNA, the majority of cGAS+ bacilli also colocalized with pTBK1 and LC3, suggesting that cGAS is important for recruiting these selective autophagy markers to intracellular M. tuberculosis (Figures 3C and 3D). Time course studies showed that the appearance of cGAS on bacilli coincided with pTBK1 recruitment but preceded the recruitment of the terminal autophagy marker LC3 (Figure 3E), which is consistent with the notion that cGAS is a proximal sensor of bacteria that leads to subsequent targeting.
Figure 3. cGAS is required to target Mycobacterium tuberculosis to the ubiquitin-mediated selective autophagy pathway.
(A) RAW 264.7 cells stably expressing 3×FLAG-cGAS infected with mCherry M. tuberculosis for 45 min and immunostained for 3×FLAG.
(B) Quantification of (A) during the indicated time course.
(C) RAW 264.7 cells stably expressing 3×FLAG-cGAS infected with mCherry M. tuberculosis for 45 min and immunostained for 3×FLAG and indicated markers.
(D) Quantification of (C).
(E) Quantification of (C) during the indicated time course.
(F) BMDMs infected with mCherry M. tuberculosis and immunostained for ubiquitin and LC3 4 h post-infection.
(G) Quantification of (F).
(H) Quantification of ubiquitin- and LC3-positive M. tuberculosis in RAW 264.7 knockdown cell lines.
(I) Same as (H) but U937 knockdown cell lines.
(J) BMDMs were infected with M. tuberculosis for 2 h, and LC3-II conversion was analyzed by quantitative Western blot (left) and expressed as a fold-change in the ratio of LC3-II/Actin. Results are representative of at least three independent experiments.
(K) CFUs from BMDMs infected with M. tuberculosis at 6, 24, 72, and 120 h (left) and normalized to CFUs at 0 h (right).
(L-P) WT and cGas-/- mice were infected with ~100 aerosolized M. tuberculosis CFUs (n = 4 or 6 per group).
(L) IFIT1 transcripts in lungs of uninfected and infected mice.
(M) Serum IFN-β levels 21 days post-infection as measured by ISRE-luciferase reporter cells.
(N-O) CFUs in lungs (N) and spleens (O) 21 and 80 days post-infection.
(P) Survival of mice monitored for 102 days (n = 6 WT and 3 cGas-/-). *p < 0.05, **p < 0.005, n.s. not significant by two-tailed t-test comparing to SCR or WT unless otherwise indicated. See also Figure S3.
We next determined the requirement of cGAS for targeting M. tuberculosis to the selective autophagy pathway. Consistent with our previous observations, Sting-/- BMDMs were defective in recruiting ubiquitin, LC3, NDP52, pTBK1, and Atg12, giving rise to an approximately 75-80% decrease in colocalization of any of these markers with bacteria compared with wild-type (Figures 3F, 3G, and S3A-C) (Leber et al., 2008; Watson et al., 2012; Woodward et al., 2010). Importantly, cGas-/- BMDMs were also defective for autophagic targeting of M. tuberculosis, with a reduction in colocalization of ~50%, which mirrors our observations with transfected cytosolic dsDNA. Stable shRNA knockdowns of cGAS or STING in human and mouse macrophage cell lines led to similar results (Figures 3H and 3I). Consistent with the recruitment of selective autophagy markers to M. tuberculosis, wild-type BMDMs had increased levels of LC3-II conversion, while cGas-/- and Sting-/- BMDMs were partially defective for LC3-II conversion after infection (Figure 3J). Together, these data indicate that DNA sensing by cGAS is required for full targeting of M. tuberculosis to the ubiquitin-mediated selective autophagy pathway, and indicate that a cGAS-independent pathway is also at play.
Previous work demonstrated that ATG5 is important for controlling intracellular M. tuberculosis replication in vivo (Gutierrez et al., 2004; Watson et al., 2012). In contrast, IRF3-driven type I IFNs appear to work in a cell extrinsic fashion promoting infection in vivo (Manca et al., 2001; Manzanillo et al., 2012). Given the potential positive and negative roles of these two outputs of STING/TBK1 activation during infection, we sought to determine the overall role of cGAS in M. tuberculosis pathogenesis. Infection of cGas-/- mice led to partially decreased type I IFN levels when compared with wild-type mice, indicating that while this pathway is activated by M. tuberculosis during infection, other pathways can stimulate IFN in vivo (Figures 3L and 3M). Moreover, these animals had no overt defect in overall resistance to M. tuberculosis as we observed similar bacterial burdens in the tissues of these mice at both early and late time points after infection (Figures 3N and 3O), and none of the cGas-/- succumbed to infection during the 100-day experiment (Figure 3P). Consistent with these observations, a parallel study by Collins et al. using the same M. tuberculosis strain but different cGas-/- mice (Li et al., 2013), also found that M. tuberculosis-infected cGas-/- mice have similar bacterial loads and cytokine levels compared to wild-type mice through an extended time-course of infection. In these studies, Collins et al. report a small increase in susceptibility of cGas-/- mice to M. tuberculosis manifest after the 100-day time point, a phenotype that was not observed in Sting-/- mice. Although we do not understand the basis for these small discrepancies, both studies support the overall conclusion that removing cGAS from the context of an intact immune system is insufficient to dramatically alter host resistance to M. tuberculosis. This is in stark contrast to both IRF3-/- and myeloid-specific ATG5-/- mice (Manzanillo et al., 2012; Watson et al., 2012). Since we observed a significant reduction in autophagic targeting in cGas-/- macrophages, we sought to determine the cell-intrinsic contribution of cGAS in controlling M. tuberculosis replication in macrophages. Consistent with cGAS targeting M. tuberculosis to the selective autophagy pathway for destruction by lysosomes, cGas-/- and Sting-/- BMDMs were permissive for M. tuberculosis growth, resulting in three-fold higher bacterial numbers compared to wild-type macrophages five days post-infection (Figure 3K). Thus, while cGAS appears to be dispensable for overt bacterial resistance in a mouse model, this receptor is required for cell-intrinsic bacterial killing in macrophages. While we do not understand why the susceptibility of cGas-/- macrophages does not translate to a similar phenotype during infection of the entire mouse, we suspect that compensatory factors manifest only in vivo, such as the involvement of additional host DNA sensors or functionally redundant autophagic targeting pathways, may be at play. Alternatively, the decrease of pro-bacterial type I IFNs in cGas-/- mice during M. tuberculosis infection may be balanced by the reduced antibacterial capacity of selective autophagy in the macrophages of cGas-/- mice. Finally, while cGAS activation leads to both interferon and autophagic targeting in macrophages, specific functions of IRF3 and ATG5 (Choi et al., 2014; Zhao et al., 2008) independent of their roles in interferon production and autophagic targeting, respectively, may still be active in cGas-/- mice.
cGAS binds M. tuberculosis genomic DNA in vivo in an ESX-1 dependent manner
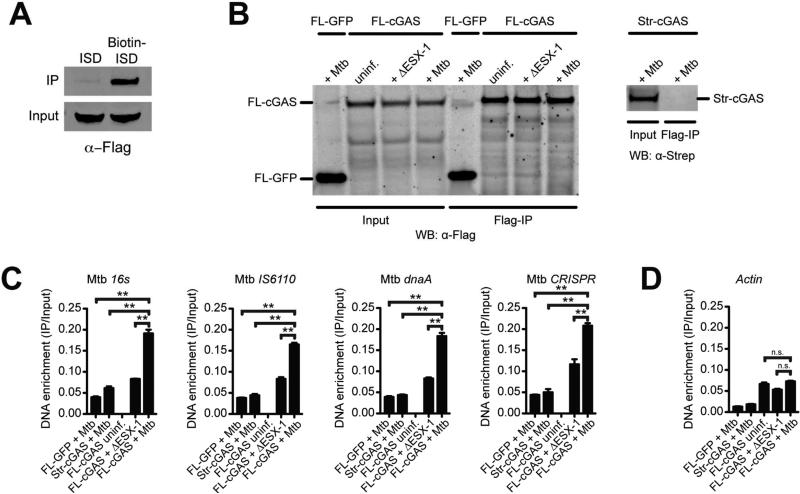
Our finding that cGAS is activated by M. tuberculosis supports an interaction between a bacterial-derived molecule (DNA) and a host cytosolic sensor in vivo. However, surprisingly few bona fide physical interactions have been identified between M. tuberculosis and macrophages in the context of infection. This prompted us to explore the molecular mechanism by which M. tuberculosis activates cGAS by detecting a physical association of cGAS and DNA within macrophages. Consistent with previous findings, we were able to efficiently co-precipitate cGAS following transfection with biotin-labeled ISD in macrophages (Figure 4A) (Liang et al., 2014; Sun et al., 2013). To perform the reciprocal experiment using unlabeled DNA, we adapted a chromatin-IP protocol to capture DNA bound by cGAS in live cells. We first transfected macrophages with ISD, and after formaldehyde crosslinking and immunoprecipitating with α-FLAG beads (Figure S4A), we measured the abundance of ISD by qPCR. Importantly, we identified an enrichment of ISD only in immunoprecipitates from FLAG-cGAS-expressing cells transfected with ISD, but not in untransfected cells or cells expressing cGAS with a different epitope tag (Strep-cGAS, Figure S4B).
Figure 4. cGAS binds to M. tuberculosis-derived DNA during infection.
(A) Western blot of 3×FLAG-cGAS after streptavidin pulldown of cell lysates from 3×FLAG-cGAS RAW 264.7 cells transfected with interferon-stimulatory DNA (ISD) or biotinylated-ISD.
(B) Western blot of inputs and FLAG IPs from WT or ΔESX-1 M. tuberculosis infected RAW 264.7 cells stably expressing 3×FLAG-GFP or 3×FLAG-cGAS (left panel), or Strep-cGAS as a negative control (right panel). Whole cell lysates (Input) or IPs (Flag-IP) were visualized by Western blot using anti-FLAG (left) or anti-Strep (right) antibodies.
(C) qPCR of M. tuberculosis-derived sequences from DNA isolated from IPs in (B). Quantities were normalized to inputs.
(D) Same as (C) but mouse Actin. n.s., not significant, **p < 0.005 by two-tailed t-test. See also Figure S4.
We next utilized this ChIP-like methodology to determine if cGAS binds directly to M. tuberculosis genomic DNA during infection. We infected cGAS-expressing macrophages with M. tuberculosis, crosslinked and immunoprecipitated cGAS (Figure 4B), and measured the abundance of several M. tuberculosis DNA sequences, including the IS6110 transposon and the CRISPR repeat, two repetitive elements in the M. tuberculosis genome (Figure 4C). These M. tuberculosis-derived sequences were significantly enriched in FLAG-cGAS immunoprecipitates as compared to controls (FLAG-GFP and Strep-cGAS). Importantly, we observed an enrichment of mycobacterial DNA bound by cGAS in cells infected with wild-type M. tuberculosis compared to cells infected with ESX-1 mutant bacteria, which cannot actively access the cytosol (Figure 4C). The M. tuberculosis DNA sequences that were detected in the ESX-1 mutant-infected cells likely arise from nonspecific phagosomal rupture that can occur when macrophages undergo necrotic cell death during high MOI infections (Lee et al., 2006). Furthermore, we did not observe any change in the abundance of host-derived DNA sequences in the immunoprecipitates after M. tuberculosis infection (Figure 4D), although the low levels of host sequences that were detected may be due to low levels of nonspecific binding of cGAS to nuclear DNA during normal cell processes (Schoggins et al., 2014). Together, these data show that bacterial-derived DNA exposed to the cytosol is bound by cGAS during infection.
Discussion
Our data, as well as the work presented in the accompanying papers (Collins et al., 2015; Wassermann et al., 2015), support a model in which M. tuberculosis triggers the STING/TBK1 pathway using the ESX-1 secretion system to disrupt phagosomal membranes, allowing bacterial DNA access to cGAS in the cytosol . Subsequent activation of TBK1 likely couples a number of cellular events, including the IRF3-dependent IFN response and, as described here, antibacterial autophagic targeting. Although indirect evidence had implicated the role of cytosolic bacterial DNA in triggering these pathways (Manzanillo et al., 2012; Watson et al., 2012), here we provide direct evidence that bacterial DNA is a bona fide “pathogen-associated molecular pattern” sensed by M. tuberculosis-infected cells. The mechanism by which DNA is liberated from M. tuberculosis and how ESX-1 permeabilizes phagosomal membranes to allow cytosolic access are currently unknown. However, we suspect that the nucleic acid may be sensed by cGAS at the bacterial cell surface, providing important spatial cues for the selective recruitment of autophagic vesicles.
It is tempting to speculate that M. tuberculosis has evolved to use its own chromosomal DNA to elicit type I IFNs, as these cytokines are produced throughout in vivo mouse infections (Manzanillo et al., 2012) as well as in active human disease (Berry et al., 2010). While the modest decreases in type I IFNs and similar CFU counts we observed during cGas-/- mouse infection implicate a role for other host sensor molecules, this work provides a detailed view of one of the molecular interactions that occurs during M. tuberculosis infection of macrophages. Indeed, the situation in vivo is certainly much more complicated, and we cannot exclude the possible role of additional DNA sensors which may work in concert with cGAS to coordinate an innate immune response, or the potentially crucial role of signaling in other cell types at different stages of infection. Although these other factors remain uncharacterized, we suspect that cGAS contributes to the robust type I IFN signature associated with active disease in humans (Berry et al., 2010; Novikov et al., 2011).
The cGAS/STING/TBK1 pathway is a major pathway by which innate immune cells recognize both viral and bacterial pathogens that access the cytosol, but different bacteria have evolved diverse mechanisms to initiate this response. We suspect that this primarily antiviral pathway has been co-opted by bacterial pathogens, perhaps primarily to elicit type I IFNs. Indeed, this pathway may be an easy target for bacterial pathogens as the potential for evolution of bacteria to elicit this response would, in theory, be relatively straightforward as it simply requires exposure of existing metabolites such as nucleic acids or cyclic dinucleotides to the inside of infected cells. However, activation also comes at a cost to bacterial replication as cGAS/STING/TBK1 activation simultaneously induces autophagic targeting. The ability of some pathogens, like L. pneumophila (Choy et al., 2012) and L. monocytogenes (Gao et al., 2013; Sun et al., 2013; Tattoli et al., 2013), to inhibit autophagy may be a common mechanism by which pathogens shift the balance of cytosolic detection to promoting infection. Understanding how to shift this balance instead toward autophagic targeting of bacterial pathogens represents an attractive host-directed therapeutic strategy to combat infection in humans.
Experimental Procedures
Cell lines and cell culture
Bone marrow derived macrophages (BMDMs) were generated from wild-type C57BL/6 (The Jackson Laboratory), cGas-/- (Schoggins et al., 2014) (or (Li et al., 2013) for experiments with L. pneumophila), and Stinggt/gt (referred to as Sting-/-) (Sauer et al., 2011) mice that were 8-12 weeks old. All mice were housed and treated using procedures described in animal care protocols approved by the Institutional Animal Care and Use Committee of UC San Francisco and Washington University St. Louis.
Bacterial strains
The wild-type M. tuberculosis strain used in these studies was the Erdman strain except when CDC1551 was used as mentioned. The 10403S strain of L. monocytogenes, the SL1344 strain S. enterica serovar Typhimurium, and the LPO2 ΔflaA strain of L. pneumonphila were used as wild type strains.
Bacterial infections
For M. tuberculosis infections, cells were infected at a multiplicity of infection (MOI) of 10 for RNA and cytokine analysis and 1 for CFUs and microscopy studies. For L. monocytogenes, S. Typhimurium, and L. pneumophila infections, BMDMs were infected at an MOI of 10, 5, and 1, respectively. Cells were harvested 4 or 6 h post-infection.
RNA isolation and qPCR analysis
For RNA analysis, cells were harvested in Trizol 4 h post-treatment, and RNA was isolated using PureLink RNA Mini Kits (Invitrogen). cDNA was synthesized with iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed using Taq DNA Polymerase (NEB) and SYBR Green I (Sigma). Both the averages and the standard deviations of the raw values were normalized to the average of the treated wild-type sample, which was set at 100%.
Colocalization of markers with M. tuberculosis and cytosolic DNA
Cytosolic DNA puncta or bacterium-containing phagosomes were visualized directly by fluorescence microscopy. A series of images were captured and analyzed by counting the number of cell-associated DNA puncta or bacteria that colocalized with the corresponding marker. At least one hundred events were analyzed per cover slip, and each condition was performed with triplicate cover slips. For experiments in knockout or knockdown cells, genotypes were blinded throughout.
Mouse infection
All procedures involving animals were conducted following NIH guidelines in accordance with an approved IACUC protocol at Washington University in St. Louis School of Medicine. cGas+/+ and cGas-/- mice (Schoggins et al., 2014) (C57BL/6 background, male and female littermates, approximately 8-10 weeks old; see Supplemental Experimental Procedures) were infected with ~100 M. tuberculosis (Erdman strain) using an inhalation exposure system (Glas-Col). Samples were collected for CFUs, RNA, or serum analysis at the indicated time points.
cGAS pulldown and DNA analysis
RAW 267.4 cells were infected at an MOI of 20 or transfected with 25 g ISD and fixed in 4% PFA after 2 h. Tagged constructs were immunoprecipated using M2 FLAG Magnetic Beads (Sigma) eluted with FLAG peptide (synthesized by Bioneer, Inc., Alameda, CA). IP efficiency was confirmed by Western blot analysis. Samples were analyzed by qPCR to measure the abundance of fragments from the M. tuberculosis or mouse genome.
Statistics
Statistical analysis of data was performed using GraphPad Prism software (Graphpad, San Diego, CA). Two-tailed unpaired Student's t-tests were used for statistical analyses, and unless otherwise noted, all results are representative of at least two independent biological experiments and are reported as the mean ± SD (n = 3 per group).
Supplementary Material
Highlights.
■ cGAS is essential for type I IFN production during M. tuberculosis infection
■ cGAS activation targets M. tuberculosis to the selective autophagy pathway
■ cGAS deficiency leads to increased intracellular M. tuberculosis growth
■ cGAS directly binds to M. tuberculosis genomic DNA during infection
Acknowledgments
The authors thank Z. Chen for providing femurs from cGas-/- mice, T. Parry and B. Penn for sharing reagents, and J. Woodward, K. Patrick, C. Homer, and M. Bell for technical assistance. We are grateful to S. Johnson, K. Thorn, and the Nikon Imaging Center staff at UCSF for training and use of their microscopes. This work was supported by NIH grant PO1 AI063302, NIH grant U19 AI109725, Crohn's and Colitis Foundation of America grant 274415, the Arnold and Mabel Beckman Foundation, HHMI, The Damon Runyon Cancer Research Foundation, NSF Graduate Research Fellowships, and NIGMS Cell and Molecular Biology Training Grant GM007067.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
R.O.W. and S.L.B. contributed equally to this work. Experiments were designed, performed and analyzed primarily by R.O.W., S.L.B., and J.S.C. with contributions from D.A.M., E.J.D., and J.O. Mouse infection was performed by J.M.K. The manuscript was written by R.O.W., S.L.B., and J.S.C., and figures were prepared by R.O.W. and S.L.B. All authors read and commented on the manuscript during preparation.
References
- Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. The Journal of Experimental Medicine. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MPR, Graham CM, Mcnab FW, Xu Z, Bloch SAA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. The Journal of Experimental Medicine. 2004;200:535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, Fujita N, Saitoh T, Akira S, Yoshimori T, et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity. 2014;40:924–935. doi: 10.1016/j.immuni.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey EA, Isberg RR. The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proceedings of the National Academy of Sciences. 2012;109:3481–3486. doi: 10.1073/pnas.1121286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host & Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B, Dey RJ, Cheung LS, Pokkali S, Guo H, Lee J-H, Bishai WR. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat Med. 2015;21:401–406. doi: 10.1038/nm.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP Synthase Is an Innate Immune Sensor of HIV and Other Retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Modulation of inflammasome pathways by bacterial and viral pathogens. J Immunol. 2011;187:597–602. doi: 10.4049/jimmunol.1100229. [DOI] [PubMed] [Google Scholar]
- Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, Portnoy DA. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Remold HG, Ieong MH, Kornfeld H. Macrophage apoptosis in response to high intracellular burden of Mycobacterium tuberculosis is mediated by a novel caspase-independent pathway. J Immunol. 2006;176:4267–4274. doi: 10.4049/jimmunol.176.7.4267. [DOI] [PubMed] [Google Scholar]
- Li X-D, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Seo GJ, Choi YJ, Kwak M-J, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner K-P, Ha T, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host & Microbe. 2014;15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann J, Rothenburg S, Deigendesch N, Eitel J, Meixenberger K, van Laak V, Slevogt H, N'guessan PD, Hippenstiel S, Chakraborty T, et al. IFNbeta responses induced by intracellular bacteria or cytosolic DNA in different human cells do not require ZBP1 (DLM-1/DAI). Cell Microbiol. 2008;10:2579–2588. doi: 10.1111/j.1462-5822.2008.01232.x. [DOI] [PubMed] [Google Scholar]
- Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE3, Freedman VH, Kaplan G. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc Natl Acad Sci U S A. 2001;98:5752–5757. doi: 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host & Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber KD, Andrade BB, Barber DL, Hieny S, Feng CG, Caspar P, Oland S, Gordon S, Sher A. Innate and adaptive interferons suppress IL-1α and IL-1β production by distinct pulmonary myeloid subsets during Mycobacterium tuberculosis infection. Immunity. 2011;35:1023–1034. doi: 10.1016/j.immuni.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe KM, McWhirter SM, Vance RE. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog. 2009;5:e1000665. doi: 10.1371/journal.ppat.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe KM, McWhirter SM, Vance RE. Induction of type I interferons by bacteria. Cell Microbiol. 2010;12:881–890. doi: 10.1111/j.1462-5822.2010.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikov A, Cardone M, Thompson R, Shenderov K, Kirschman KD, Mayer-Barber KD, Myers TG, Rabin RL, Trinchieri G, Sher A, et al. Mycobacterium tuberculosis triggers host type I IFN signaling to regulate IL-1β production in human macrophages. J Immunol. 2011;187:2540–2547. doi: 10.4049/jimmunol.1100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. The Journal of Experimental Medicine. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan M, Yi CH, Gonzales R, Lee K-D, Portnoy DA. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc Natl Acad Sci USA. 2002;99:13861–13866. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer J-D, Sotelo-Troha K, Moltke, von J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D, Medzhitov R. Recognition of Cytosolic DNA Activates an IRF3-Dependent Innate Immune Response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I, Sorbara MT, Yang C, Tooze SA, Philpott DJ, Girardin SE. Listeria phospholipases subvert host autophagic defenses by stalling pre-autophagosomal structures. Embo J. 2013;32:3066–3078. doi: 10.1038/emboj.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles RMB, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, Komisopoulou E, Kelly-Scumpia K, Chun R, Iyer SS, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science. 2013;339:1448–1453. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TLM, Wandel MP, Muhlinen, von N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host & Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yeruva L, Marinov A, Prantner D, Wyrick PB, Lupashin V, Nagarajan UM. The DNA Sensor, Cyclic GMP-AMP Synthase, Is Essential for Induction of IFN-β during Chlamydia trachomatis Infection. J Immunol. 2014;193:2394–2404. doi: 10.4049/jimmunol.1302718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host & Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zughaier SM, Zimmer SM, Datta A, Carlson RW, Stephens DS. Differential induction of the toll-like receptor 4-MyD88-dependent and -independent signaling pathways by endotoxins. Infect Immun. 2005;73:2940–2950. doi: 10.1128/IAI.73.5.2940-2950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.