Abstract
Translation by the ribosome occurs by a complex mechanism involving the coordinated interaction of multiple nucleic acid and protein ligands. Here we have used zero-mode waveguides (ZMWs) and sophisticated detection instrumentation to allow real-time observation of translation at physiologically-relevant (μM) ligand concentrations. Translation at each codon is monitored by stable binding of tRNAs – labeled with distinct fluorophores – to translating ribosomes, allowing direct detection of the identity of tRNA molecules bound to the ribosome, and therefore, the underlying mRNA sequence. We observe the transit of tRNAs on single translating ribosomes and have determined the number of tRNA molecules simultaneously bound to the ribosome, at each codon of an mRNA. Our results show that ribosomes are only briefly occupied by two tRNAs and that release of deacylated tRNA from the E site is uncoupled from binding of A-site tRNA and occurs rapidly after translocation. The methods outlined here have broad application to the study of mRNA sequences, and the mechanism and regulation of translation.
During translation, the ribosome progressively coordinates the dynamic interplay of transfer RNA (tRNA) and protein factors to decipher individual codons of a messenger RNA (mRNA) and synthesize protein. The ribosome contains three tRNA binding sites corresponding to three adjacent codons1. As it elongates, the ribosome repetitively selects aminoacylated tRNA at the A site, orienting them for peptide bond formation with peptidyl tRNA positioned in the P site. Peptidyl transfer is followed by the coordinated movement of the A and P-site tRNAs into the P and E (exit) sites, respectively, thus preparing the deacylated tRNA for dissociation from the ribosome. During this translocation step, which is catalyzed by the GTPase EF-G, the ribosome simultaneously steps along the mRNA, positioning the next codon in the A site and preparing to select another aminoacyl tRNA.
Although dynamic changes in ligand occupancy and positioning in the A, P and E sites are intimately tied to the mechanism of translation2-5, the timing and relation of aminoacyl tRNA arrival at the A site, as a ternary complex (TC) with EF-Tu•GTP, and dissociation of deacylated tRNA from the E site remains unknown. Single-molecule fluorescence methods have recently probed dynamics during translation, such as the selection of tRNA during elongation and ribosomal conformational changes (reviewed in 6). However, traditional single-molecule fluorescence techniques only permit observation of fluorescent ligands in the nanomolar (nM) range, well below the physiological concentration (μM) of most components of the translational apparatus.
Real-time translation in zero-mode waveguides
Zero-mode waveguides (ZMWs, Fig. 1a) are nanophotonic confinement structures consisting of circular holes of 50-200nm diameter in a metal cladding film deposited on a solid, transparent substrate7. In conjunction with laser-excited fluorescence, ZMWs provide observation volumes on the order of zeptoliters (10-21 L), three to four orders of magnitude smaller than far-field excitation volumes. This drastically reduces the background signal from freely-diffusing fluorescent molecules, permitting the observation of fluorescent ligands in the μM range. Advances in fabrication8, surface chemistry9, and detection instrumentation10 have permitted direct monitoring of DNA polymerization in ZMWs11. The binding of labeled ligands to an enzyme immobilized in a ZMW is detected as a pulse of fluorescent light. Here we adapt this instrumentation to the study of translation. Using ZMWs, we observe real-time selection and transit of fluorescently-labeled tRNAs at μM concentration (Fig. 1b) on single ribosomes during multiple rounds of translation elongation. tRNA binding on single ribosomes was tracked using tRNAs that were specifically dye-labeled at their elbow positions without affecting their function12,13. Ribosomes were immobilized in ZMWs as 70S initiation complexes – containing fMet-(Cy3)tRNAfMet – assembled on biotinylated mRNAs, which were tethered to the biotin-PEG-derivatized bottom of ZMWs through neutravidin-biotin linkages; mRNAs contained 5’-UTR and Shine-Dalgarno sequences from T4 gene 32, an initiation codon and coding sequence of 3-12 codons, terminated by a stop (UAA) codon followed by four phenylalanine codons (Fig. 2a). Cy3 fluorescence from an immobilized complex confirmed the presence of initiator tRNA and marked a properly assembled and immobilized ribosome in a ZMW. The number of ribosome complexes immobilized per individual ZMW surfaces increased at higher ribosomal complex concentrations, obeying Poisson statistics, and, as expected, could be blocked by addition of free biotin (Fig. S1). Ellipsometry and ZMW experiments in the absence of ribosomes confirmed minimal nonspecific surface adsorption of translational components (100 μM tRNA, 1μM EF-Tu and EF-G)(Fig. S2).
Figure 1. Translation in zero-mode waveguides.
a. Schematic of experimental setup. ZMWs are cylindrical nanostructures with varying diameters (~50-200 nm). The aluminum side wall and quartz bottom surfaces are derivatized to allow specific biotin-streptavidin interactions on the quartz surface and to block non-specific interactions of molecules with ZMWs9,11. Ribosomal complexes are specifically immobilized in the bottom of derivatized ZMWs using biotinylated mRNAs. Ternary complexes Cy5-labeled Phe-tRNAPhe -EF-Tu(GTP) and Cy2-labeled Lys-tRNALys -EF-Tu(GTP), along with EF-G(GTP), are delivered to a ZMW surface-immobilized, initial ribosome complex containing Cy3-labeled fMet-tRNAfMet. Fluorescence is excited by illumination at 488, 532 and 642 nm, and Cy2, Cy3 and Cy5 fluorescence are simultaneously detected using previously described instrumentation10,11 b. Expected signal sequence. Initiation complexes are detected by fluorescence of fMet-(Cy3)tRNAfMet bound at an initiation codon. Fluorescent tRNAs are delivered as TCs. Arrival of Phe-(Cy5)tRNAPhe or Lys-(Cy2)tRNALys at the ribosomal A site is marked by red or blue fluorescent pulse. At low TC concentration, tRNA arrival times are slow (>>1 s), and Cy5- or Cy2-labeled tRNAs can photobleach on the ribosome while waiting for translocation. In the absence of photobleaching, the length of a pulse represents the transit time of that tRNA on the ribosome. At high TC concentrations, tRNA arrival times are fast (<<1 s), and fluorescent pulses are overlapped, which indicates simultaneous occupancy by 2 tRNAs. The tRNA occupancy count is shown below the schematic trace.
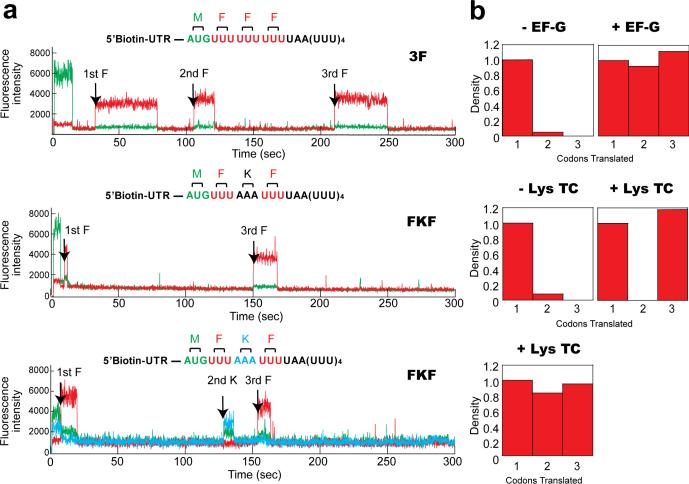
Figure 2. Monitoring translation via fluorescent tRNA binding events.
a. Representative single-ZMW traces of ribosomes translating MFFF mRNA (top) and MFKF mRNA (bottom) in the presence of 30 nM EF-G and 30 nM TC. b. The number of fluorescent pulses observed in ZMWs depends on the presence of EF-G and TC. Event histograms for the three experiments in the absence (n=341) and presence (n=304) of 30 nM EF-G (top), and in the absence (n=278) and presence (n=297) of 30 nM unlabeled Lys-tRNALys TC (middle) and presence (n=355) of 30 nM Lys-(Cy2)-tRNALys TC (bottom). Histograms are normalized by the number of ribosomes showing single events.
To confirm the utility of ZMWs for investigating translation, we used fluorescence resonance energy transfer (FRET) – a sensitive distance indicator – to observe the path of incoming A-site tRNA accommodation on the ribosome. We have previously used traditional single-molecule total internal reflection fluorescence (TIRF) methods to detect FRET between fMet-(Cy3)tRNAfMet in the P site and Phe-(Cy5)tRNAPhe in the A site12,13. We repeated these experiments using ZMW-immobilized ribosome complexes and excitation at 532 nm. Using FRET values calibrated in ZMWs , we matched the values and timescales observed in prior tRNA-tRNA FRET experiments at tRNA concentrations up to 600nM, more than 20-fold higher than previously measured (Fig. S3, S4). Consistent with previous single-molecule studies, bimolecular arrival rates of TC to surface-bound ribosomes were decreased by an order of magnitude compared to bulk rates, but unimolecular rates were unaffected. Decreased association rates are likely due to steric and surface effects14, but ribosomal function is clearly maintained. These results confirmed the functionality of ZMW-immobilized ribosomes and their ability to detect fluorescent tRNA binding events on the ribosome at >100nM TC concentrations.
Direct detection of tRNA binding to the ribosome during translation
The basic steps of translation were then observed through direct detection of fluorescently-labeled tRNA binding on single ribosomes immobilized in ZMWs. We monitored the binding of ternary complexes Phe-(Cy5)tRNAPhe•EF-Tu•GTP and unlabeled Lys-tRNALys•EF-Tu•GTP or labeled Lys- (Cy2)tRNALys•EF-Tu•GTP to ribosomes programmed by mRNAs encoding 4 amino acids (MFFF or MFKF), (Fig. 2a, b); ZMWs were illuminated simultaneously with 488, 532 and 642nm excitation. Initiated ribosomes were identified by the presence of fMet(Cy3)-tRNAfMet; subsequent real-time arrival and occupation of tRNAs on translating ribosomes were detected as fluorescent pulses of appropriate color10.
Each tRNA pulse marks the arrival and accommodation of that tRNA within the ribosomal A site. The arrival time of the first elongator tRNA encoded by the mRNA marks the transition of ribosomes into elongation. The time between subsequent tRNA pulse arrivals delineates one round of translational elongation and arrival of tRNA at the next codon (Fig. 1b). This time should depend on the concentration of EF-G, which controls the rate of translocation to the next codon. The duration of each tRNA pulse represents the transit time of that tRNA through the A, P, and E sites, followed by dissociation from the ribosome. At the low (<50 nM) factor and tRNA concentrations normally used for single-molecule experiments, photobleaching of ribosome-bound tRNA can also terminate a pulse. Since each pulse describes binding of one tRNA on the ribosome, full translation is detected as a series of pulses corresponding to the number and sequential identity of codons in the mRNA.
To define the order and identity of these pulses, initial experiments were performed at 30 nM EF-G/30 nM TC concentrations, with translation times ~20s per codon, which allows P-site tRNAs to photobleach (lifetime 13.7 s) prior to the arrival of A-site tRNA. In the absence of EF-G, translation of the MFFF message stalls upon arrival of the first tRNAPhe (Fig. 2a). Cy3 fluorescence is followed only by a single red pulse, indicating binding of a single Phe-(Cy5)tRNAPhe and no subsequent translocation (Fig. 2b). In the presence of 30 nM EF-G, three distinct red fluorescent pulses are observed (Fig. 2a, b). The number of fluorescent tRNA pulses is similarly sensitive to the identity of the A-site codon and the presence of correct TC. Translation of the MFKF mRNA in the presence of EF-G but absence of Lys-tRNALys TC results in a single fluorescence pulse following the Cy3 signal, whereas 2 red pulses are observed upon addition unlabeled Lys-tRNALys TC ; finally, two red pulses separated by a blue pulse are observed upon inclusion of fluorescently labeled Lys-(Cy2)tRNALys TC. In all traces, brief single-frame (100 ms) bursts in fluorescence can be observed (Fig. 2b). These events were only observed in the presence of 70S ribosomal complexes, and likely represent non-cognate TC sampling at an A-site codon (see below). These results confirm our ability to track translation through sequential stable tRNA binding events.
Real-time monitoring of translation through sequential fluorescent-tRNA binding events
These tRNA binding signals were then used to observe full translation of distinct heteropolymeric mRNAs encoding 13 amino acids (M(FK)6 and M(FKK)4). The sequence of the mRNA is readily distinguished from the pattern of fluorescent pulses (Fig. 3a). The number of events observed relates the number of codons translated on each mRNA. At 200nM TC and 500nM EF-G, ribosomes translate the entire mRNA (Fig. 3b). The duration of most tRNA pulses is not limited by photobleaching at these high concentrations suggesting that the lifetime of each tRNA signal provides a signal for its transit time on the ribosome (see below). Addition of erythromycin, which binds to the exit tunnel of the ribosome15, blocks translation at 6-8 amino acids16, as expected. These data strongly support the direct link between the pattern of tRNA pulses observed in the ZMW and translation.
Figure 3. Real-time translation at near physiological concentrations.
a. Two heteropolymeric mRNAs encoding 13 amino acids were used: M(FK)6 and M(FKK)4. Translation was observed in the presence of 200 nM Phe-(Cy5)tRNAPhe, 200 nM Lys-(Cy2)tRNALys TC and 500 nM EF-G as a series of fluorescent pulses that mirror the mRNA sequence. A long Cy2 pulse is observed upon arrival of the ribosome at the stop codon. Brief sampling pulses (<100 ms) of both Lys-(Cy2)tRNALys and Phe-(Cy5)tRNAPhe TC are observed after arrival at the stop codon. b. Event histograms for translation of M(FK)6 showing translation out to 12 elongation codons (red, n=381). In the presence of 1 μM erythromycin, translation (blue, n=201) is stalled at codon 8 of the mRNA. c. Analysis of translation rates at each codon in M(FK)6. Mean times (avg. ± s. d.) between tRNA arrival events are plotted for translation in the presence of 200nM TC and 30, 100 and 500 nM EF-G. d. Overall tRNA transit times (avg. ± s. d.) for codons 2-12 at (from left) 200 nM TC and 30, 100, or 200 nM EF-G; 500 nM TC and 500 nM EF-G; and 200 nM TC/500 nM EF-G in the presence of 1 μM fusidic acid. e. Cumulative translation times (avg. ± s. d.) for each codon in M(FK)6 at 200 nM TC and 30, 100, or 200 nM EF-G; 500 nM TC and 500 nM EF-G, and 200 nM TC/500 nM EF-G in the presence of 1 μM fusidic acid.
The arrival of tRNAs at single ribosomes tracks the dynamic composition of the translational apparatus in real time. First tRNA arrival events are fast, as they do not depend on translocation. As predicted, the time between subsequent tRNA arrivals decreases with increasing EF-G concentrations between 30 and 500 nM (Fig. 3c). For codons 2-12, the tRNA transit time is also strongly dependent on EF-G, as it represents at least two rounds of peptide bond formation and translocation (Fig. 3d). Inhibition of EF-G by fusidic acid, which stabilizes EF-G•GDP on the ribosome post-translocation17, lengthens the transit time by 3.3-fold (Fig. 3d). Arrival of the ribosome at the UGA stop codon after translation of 12 codons leads to a long pulse from the remaining tRNA in the P-site of the stalled ribosome. The dwell time for this last tRNA is 4.9-fold longer than for preceding pulses, underscoring that photobleaching of the P-site tRNA is not a significant problem using our approach at high factor concentrations: at 500 nM EF-G, the mean lifetime (4.1 s) of tRNAs bound to the ribosome is significantly shorter than the photobleaching lifetime (17.3 s) observed in lower concentration experiments. While paused on the stop codon, tRNA sampling events are observed with short lifetimes (~ 50 ms for Phe-(Cy5)tRNAPhe or Lys-(Cy2)tRNALys,)(Fig. 4a, b), which are clearly distinguishable from real tRNA transit events of > 1 s. These sampling events are consistent with non-cognate TC interaction4 with the A site and their frequency is proportional to TC concentration (Fig. 4c). All trends discussed above were independent of mRNA sequence.
Figure 4. A-site sampling on ribosomes stalled at the stop codon.
a. Fast sampling events at the stop codon position of the M(FK)6 template were observed in the presence of 200 nM Phe-(Cy5)tRNAPhe, 200 nM Lys-(Cy2)tRNALys TC and 500 nM EF-G . b. Dwell time histograms individual sampling pulses of Phe-(Cy5)tRNAPhe (left) and Lys-(Cy2)tRNALys (right). Both histograms are well approximated by a single exponential fit. c. The frequency of fast sampling (avg. ± s. d.) increased linearly with TC concentration (left), while sampling dwell time (avg. ± s. d.) did not depend on TC concentration (right).
The total translation time for different mRNAs is characterized by the arrival times of tRNAs at different codons. As expected, translation rates depend on the concentrations of TC and EF-G. For the M(FK)6 mRNA (Fig. 3e), translation rates increased from 0.08 to 0.4 s-1 for EF-G concentrations ranging from 30 and 500 nM (200 nM TC). Likewise, increasing the concentration of TC also increases the overall translation rate. At the highest concentrations of TC and EF-G, the translation rate was nearly 1 codon · s-1, approaching that obtained using cell extracts in vitro18. When 30S pre-initiation complexes are immobilized in ZMWs and translation is initiated by addition of 50S subunits, TC, and EF-G – requiring initiation prior to protein synthesis (Fig. S5) – the overall translation rate is unaffected, except for a delay (12 s) in arrival of the first tRNA. This delay is consistent with the timescale of 50S subunit joining during initiation19, prior to progression to elongation.
Our approach allows analysis of translational rates at each codon of an mRNA. Arrival of the first tRNA is independent of EF-G concentration, as expected. However, the first EF-G catalyzed translocation of the ribosome may become the slowest step in elongation, as revealed by the time between tRNA arrivals (Fig. 3e). This first EF-G-catalyzed translocation step is about 2-fold slower than subsequent translocation events, at all EF-G concentrations and for all mRNAs tested (Fig. S5). A similar trend in elongation rate was previously observed by following the global conformation of single translating ribosomes (Aitken & Puglisi, submitted). Codons after position 3 are all translated with similar rates in M(FK)6 until the long stall at the final stop codon. Slight differences in overall translation rates are observed for distinct mRNA sequences, with MF12 translated most slowly and M(FKK)4 most rapidly (Fig. S5). The hydrophobic character of the poly(phe) peptide may inhibit translation of the MF12 mRNA.
To define the mechanism linking tRNA arrival at the A site and release from the E site, we used these signals to measure the real-time tRNA occupancy of the ribosome during translation. While at least two tRNAs must occupy the ribosome during peptide bond formation2-6, the ribosome contains three tRNA-binding sites (A,P,E) and stable tRNA occupancy in the E site after translocation would cause accumulation of 3 tRNAs on the ribosome. The arrival of tRNA in the A site may signal tRNA departure from the E site, or dissociation from the E site may occur spontaneously upon translocation. In our single-molecule traces, overlapping fluorescence pulses report on the number of tRNAs simultaneously bound to the ribosome, while appearance and departure of fluorescence indicates tRNA arrival and dissociation (Fig. 1b). During translation of the M(FK)6, at 200 nM Phe-(Cy5)tRNAPhe and Lys-(Cy2)tRNALys TC and 500 nM EF-G, most (82.2%) consecutive Cy5 and Cy2 pulses are overlapping, indicating that two tRNAs occupy the ribosome simultaneously during translation.
Dynamic tRNA occupancy on the ribosome during translation
To determine the real-time occupancy of the ribosome at each codon, we post-synchronized 381 traces according to the arrival of aminoacylated tRNA at each codon (Fig. 5a). In this formulation, two-dimensional color plots reveal the time-dependent tRNA occupancy of hundreds of single ribosomes during each elongation cycle along the mRNA.
Figure 5. Monitoring the dynamic tRNA occupancy of translating ribosomes.
a. Post-synchronization plots for time-resolved tRNA occupancy at codons 2-6 during translation of M(FK)6. Two-dimensional histograms are post-synchronized in time with respect to each tRNA transit event (1st F~5th F) at 500, 100, and 30 nM EF-G, and at 500 nM EF-G in the presence of fusidic acid. b. tRNA occupancy time (avg. ± s. d.) at 500, 100, and 30 nM EF-G, and at 500 nM EF-G in the presence of fusidic acid.
This analysis shows that EF-G driven translocation controls the number of tRNAs on the ribosome. At 30 nM EF-G, the two-tRNA state lasts ~6.3 s at each codon, consistent with the estimated time for translocation. Increasing concentrations of EF-G to 500 nM shorten the lifetime of the 2-tRNA bound state of the ribosome from 6.3 s to 1.5 s (Fig. 5b). These trends are observed at different codons and for distinct mRNA sequences (Fig. S6), confirming the generality of the conclusions. The 2-tRNA state is not followed by a 3-tRNA state; even at high concentrations of both EF-G and TC, ribosomes occupied by three tRNA are almost never observed (1.7%), which would be consistent with decreased E-site affinity upon A site tRNA arrival. However departure of E-site tRNA, is not linked to arrival of the next tRNA at the A site in our experiments, as correlation analysis shows no connection between E-site tRNA departure events and A-site arrival events (r = 0.04). Instead, these results suggest that EF-G binding and subsequent GTP hydrolysis drives the tRNA from the A/P P/E hybrid states to the P and E sites, at which point the E-site tRNA rapidly dissociates. Consistent with this model, fusidic acid-stalled EF-G (GDP) in the A site inhibits arrival of the next tRNA, and inhibits each round of elongation, but does not affect the rate of tRNA dissociation from the E site (Fig 5b).
The results presented here demonstrate that translation can be observed in real time using single ribosomes immobilized in ZMWs. The application of ZMWs to the observation of translating ribosomes permits the sensitivity and precision of single-molecule measurements at near physiological concentrations (μM) of both tRNA and protein factors (Fig. S7). By using specifically dye-labeled tRNAs, long-lived binding events to mRNA-programmed ribosomes are readily observed and distinguished from transient sampling. The sequence of tRNA binding events reveals the encoding mRNA sequence. Full translation requires the presence of both EF-G and the appropriate TCs. Ribosome-directed antibiotics interfere with translation as predicted by their mechanism of action: fusidic acid blocks release of EF-G from the ribosome, slowing elongation, whereas erythromycin blocks elongation beyond 7 amino acids. The dynamics of tRNA binding events at each codon revealed slow initiation and long pauses upon encountering the stop codon; sampling of TC at the stop codon of stalled ribosomes is observed. Translation at μM concentrations of factors and ligands is efficient and rapid, avoiding limitations of dye photobleaching, and allows correlation of bimolecular binding events on single ribosomes.
The mechanism by which tRNAs transit through the ribosome during decoding, peptide bond formation and translocation was explored using our approach. Various models for interplay of the A and E site have been proposed. Recent dynamic and structural studies suggest that EF-G interaction within the A-site may control the conformation of the E site20. The ability to probe tRNA dynamics on the ribosome at high TC and factors concentrations in ZMWs allowed us to determine the time-dependent composition of the ribosome at each codon during translation. These results show unambiguously that tRNA release from the E site is rapid once translocation has occurred and is uncorrelated to arrival of the next tRNA. This is consistent with a model of transient E site occupancy after translocation21,22.
Three tRNAs are rarely observed on translating ribosomes, and only at high concentrations of TC (μM) where arrival of the third tRNA in the A site is rapid. These data agree with dynamic investigations of the E site that show coupling of E site opening, and in particular the L1 stalk23-25, to translocation and E-site occupancy, and are consistent with the rapid rates required for efficient elongation. Three tRNA occupancy occurs when a slow E-site dissociation event is coincident with rapid tRNA arrival in the A site (Fig S7). Slow E-site dissociation delays subsequent rounds of elongation, and may be important in rare translational events, such as frameshifting26.
Future perspectives
The real-time system outlined here has broad application to the study of translation. The dynamic events underlying translational fidelity and ribosomal movement are probed directly at each codon during translation, allowing rare translational events to be uncovered. Time-resolved analysis of compositional changes in the ribosome can be extended to initiation, elongation and release factor binding and can be merged with FRET signals to correlate ligand binding and ribosomal conformational changes. Eukaryotic translational systems can be readily substituted to probe the dynamics of translational control and regulation. This approach allows the direct detection of mRNA coding sequence, and may permit the observation of translational events involved in the regulation of protein synthesis, such as frameshifting.
Full Methods
Instrumentation
Instrumentation10 and chips containing 3,000 individual ZMWs 8 were used and prepared as described previously11. Specific immobilization of ribosome complexes in ZMWs was achieved by surface passivation using polyphosphonate with Biotin-PEG–silane9. ZMW diameters were in the range of 120-135 nm. Cy2, Cy3 and Cy5 fluorescence was detected upon simultaneous excitation at 488, 532 and 642 nm. Each laser power for three excitations was 0.5 μW/μm2 for all three color experiments, but 2.5μW/μm2 for two color experiments (Fig. 2a). Each dye lifetime were 17.3 s for Phe-(Cy5)tRNAfMet in P site, 22.5 s for fMet-(Cy3)tRNAfMet in P site and for 16.5 s fMet-(Cy2)tRNAfMet in P site.
Data collection & analysis
Data were collected on a highly parallel confocal fluorescence detection instrument, using prism-based dispersion optics and an EMCCD camera. Fluorescence traces were recorded at a rate of 30 frames per second for 3 min, with the exception experiments using MFF, MFKF mRNAs and those at 30 nM EF-G for M(FK)6 mRNA, which were recorded at a rate of 100 frames per second for 5 min. Using custom software written in Matlab (MathWorks), fluorescence traces that displayed Cy3 fluorescence corresponding to an fMet-(Cy3)tRNAfMet molecule and colocalized with the arrival of labeled ternary complex upon delivery were analyzed to extract individual tRNA transit times, time between tRNA arrivals and tRNA occupancy within single ribosomes. Data from individual molecules (n > 300 molecules for all experiments) were accumulated into statistical distributions to extract mean estimates for the above-described values.
Supplementary Material
Acknowledgement
Supported by NIH GM51266 (JDP). We thank T. Funatsu at University of Tokyo, and A. Tsai & A. Petrov (Stanford) for encouragement and stimulating discussions. We thank J. Gray for performing ellipsometry experiments.
Footnotes
Supplementary Materials are available in the online version of the paper at www.nature.com/nature.
References
- 1.Green R, Noller HF. Ribosomes and translation. Annu. Rev. Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 2.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 3.Hausner TP, Geigenmüller U, Nierhaus KH. The allosteric three –site model for the ribosomal elongation cycle. New insights into the inhibition mechanism of aminoglycosides, thiostrepton, and viomycin. J. Biol. Chem. 1988;263:13103–13111. [PubMed] [Google Scholar]
- 4.Rodnina MW, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome : kinetic and structural mechanisms. Annu. Rev. Biochem. 2001;70:415–435. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 5.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous racheting of the ribosome. Mol. Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annu. Rev. Biochem. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- 7.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 8.Foquet M, et al. Improved fabrication of zero-mode waveguides for single-molecule detection. J. Appl. Phys. 2008;103:034301. [Google Scholar]
- 9.Korlach J, et al. Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures. Proc. Natl Acad. Sci. USA. 2008;105:1176–1181. doi: 10.1073/pnas.0710982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lundquist PM, et al. Parallel confocal detection of single molecules in real time. Opt. Lett. 2008;33:1026–1028. doi: 10.1364/ol.33.001026. [DOI] [PubMed] [Google Scholar]
- 11.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 12.Blanchard SC, Gonzalez RL, Jr., Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 13.Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc. Natl Acad. Sci. USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan V, Graves DJ, McKenzie SE. The biophysics of DNA hybridization with immobilized oligonucleotide probes. Biophys. J. 1995;69:2243–2255. doi: 10.1016/S0006-3495(95)80095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlunzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 16.Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 2003;330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 17.Bodley JW, Godtfredsen WO. Studies on translocation. XI. Structure-function relationships of the fusidane-type antibiotics. Biochem. Biophys. Res. Commun. 1972;46:871–877. doi: 10.1016/s0006-291x(72)80222-5. [DOI] [PubMed] [Google Scholar]
- 18.Underwood KA, Swartz JR, Puglisi JD. Quantitative polysome analysis identifies limitations in bacterial cell-free protein synthesis. Biotechnol. Bioeng. 2005;91:425–435. doi: 10.1002/bit.20529. [DOI] [PubMed] [Google Scholar]
- 19.Marshall RA, Aitken CE, Puglisi JD. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol. Cell. 2009;35:37–47. doi: 10.1016/j.molcel.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lill R, Wintermeyer W. Destabilization of codon-anticodon interaction in the ribosomal exit site. J. Mol. Biol. 1987;196:137–148. doi: 10.1016/0022-2836(87)90516-x. [DOI] [PubMed] [Google Scholar]
- 22.Semenkov YP, Rodnina MV, Wintermeyer W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc. Natl Acad. Sci. USA. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornish PV, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl Acad. Sci. USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc. Natl Acad. Sci. USA. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders CL, Curran JF. Genetic analysis of the E site during RF2 programmed frameshifting. RNA. 2007;13:1483–1491. doi: 10.1261/rna.638707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.