Summary
This article deals with jointly modeling a large number of geographically referenced outcomes observed over a very large number of locations. We seek to capture associations among the variables as well as the strength of spatial association for each variable. In addition, we reckon with the common setting where not all the variables have been observed over all locations, which leads to spatial misalignment. Dimension reduction is needed in two aspects: (i) the length of the vector of outcomes, and (ii) the very large number of spatial locations. Latent variable (factor) models are usually used to address the former, although low-rank spatial processes offer a rich and flexible modeling option for dealing with a large number of locations. We merge these two ideas to propose a class of hierarchical low-rank spatial factor models. Our framework pursues stochastic selection of the latent factors without resorting to complex computational strategies (such as reversible jump algorithms) by utilizing certain identifiability characterizations for the spatial factor model. A Markov chain Monte Carlo algorithm is developed for estimation that also deals with the spatial misalignment problem. We recover the full posterior distribution of the missing values (along with model parameters) in a Bayesian predictive framework. Various additional modeling and implementation issues are discussed as well. We illustrate our methodology with simulation experiments and an environmental data set involving air pollutants in California.
Keywords: Bayesian inference, Factor analysis, Gaussian predictive process, Linear model of coregionalization, Low-rank spatial modeling, Multivariate spatial processes, Spatial misalignment
1. Introduction
With enhanced capabilities in collecting, storing, and accessing geographically referenced data sets, spatial analysts today frequently encounter data comprising a large number of variables across a very large number of locations. In several instances, inference focuses upon three major aspects: (i) estimate associations among the variables, (ii) estimate the strength of spatial association for each variable, and (iii) predict the outcomes at arbitrary locations.
Modeling multiple geographically referenced outcomes proceeds from two different premises. One approach (Royle and Berliner, 1999; Gelfand et al., 2004) considers a conditional regression-like approach where the marginal distribution of the first outcome is specified, followed by the conditional distribution of the second outcome given the first, and so on. This approach is more suitable when there is a natural “ordering” of the outcomes suggesting the sequence for constructing the conditional distributions.
Settings that lack such information prefer joint modeling for the set of outcomes to avoid the explosion in models emerging from alternate ordering schemes. Conditional inference can subsequently proceed from the joint distribution by conditioning on the relevant variables. The challenge in the joint modeling approach is to specify valid multivariate spatial processes using matrix-valued, cross-covariance functions ([e.g., Ch. 7]; Banerjee, Carlin, and Gelfand 2004). Gelfand et al. (2004) offer a detailed comparison of both approaches for multivariate spatial data. Here we focus upon the joint modeling approach.
The linear model of coregionalization (LMC) proposed by Matheron (1982) is among the most general models for multivariate spatial data analysis. Here, the spatial behavior of the outcomes is assumed to arise from a linear combination of the independent latent processes operating at different spatial scales (Chilés and Delfiner, 1999). The idea resembles latent factor analysis (FA) models for multivariate data analysis (e.g., Anderson, 2003) except that in the LMC the number of latent processes is usually taken to be the same as the number of outcomes. Then, an m × m covariance matrix has to be estimated for each spatial scale (see, e.g., Lark and Papritz, 2003; Castrignanó et al., 2005; Zhang, 2007; Finley et al., 2008), where m is the number of outcomes. When m is large (e.g., m ≥ 5 and 300 spatial locations), obtaining such estimates is expensive. Schmidt and Gelfand (2003) and Gelfand et al. (2004) associate only an m × m triangular matrix with the latent processes. However, high dimensional outcomes are still computationally prohibitive for these models.
When the number of independent latent processes in an LMC is taken to be fewer than the number of outcomes (e.g., Zhang, 2007), we obtain a spatial factor model. Wang and Wall (2003) studied multivariate indicators of cancer risk in Minnesota using one spatial factor. Liu, Wall, and Hodges (2005) deployed multiple factors with each underlying factor explaining its own unique set of observed/measured variables. Christensen and Amemiya (2002) developed semiparametric latent variable (shift-factor) models for rectangular grids. Hogan and Tchernis (2004) fitted a one-factor spatial model and compared the results using different forms of spatial dependence through the single factor. Minozzo and Fruttini (2004) applied log-linear spatial FA to geo-referenced frequency counts adopting the classical proportional covariance model to the latent factors.
Our current work is similar to the aforementioned articles in its use of independent latent spatial processes as underlying factors. However, we propose three methodological innovations within this framework. First, we use a multivariate low-rank spatial process to achieve dimension reduction over space. Critically, we need to work with irregularly spaced locations that do not necessarily lie on a grid, nor can they be easily projected onto the same. Although there are several choices here, we deploy the multivariate Gaussian predictive process (Banerjee et al., 2008, 2010). The method is closely related to kernel-convolutions, splines and low-rank kriging, (see, e.g., Wikle and Cressie, 1999; Kamman and Wand, 2003; Ver Hoef et al., 2004; Paciorek, 2007; Cressie and Johannesson, 2008), all of which attempt to facilitate computation through lower dimensional process representations. The predictive process can be applied to any spatial correlation function and maintain the richness of desired hierarchical spatial modeling specifications using a set of locations (or knots).
Second, we do not fix the number of factors but model it stochastically. We do so differently from some existing approaches. For example, Lopes and West (2004) addressed this problem by constructing proposals using the results of a preliminary Markov chain Monte Carlo (MCMC) run under each model. Such an approach has high computational demands, becoming infeasible as the sample size and potential number of factors increase. Dunson (2006) introduced a model averaging method for FA, but the construction of factor selection is inconvenient for spatial models. Chen and Dunson (2003) proposed a Bayesian random effect selection method which is similar to what we propose but, instead of selecting random variables, we choose the underlying latent processes that capture spatial dependence. We avoid complex computational strategies such as reversible jump algorithms and build our adaptive models by utilizing some key identifiability results, hitherto largely unaddressed, to construct hierarchical models.
Third, we reckon with spatial misalignment in the context of spatial FA. Misalignment occurs frequently in spatial data when not all variables have been observed across all locations. Put differently, the sets of observed locations for the different outcomes are not identical (either because they are missing or have been collected by different monitoring sets). Assuming that all covariates are available at a location, we want to estimate the functional relationship between the covariates and the outcomes at that location—even if all the outcomes have not been observed there. We also seek to predict the outcomes at any arbitrary location in the domain, thereby estimating the response surfaces for each outcome.
Our motivating application pertains to ambient air quality assessment in California. The deleterious impact of air pollution upon health and quality of life is widely recognized as a major environmental issue (e.g., Dominici et al., 2006). Spatial interpolation of air pollutants plays a crucial role in assessing and monitoring ambient air quality and modeling multiple pollutants can capture associations within and across different locations, which can enhance predictive performance. Our data set includes five commonly encountered pollutants observed over 300 monitoring stations across California and is spatially misaligned in the aforementined manner. Estimating fully specified joint models for such data will be exorbitant, which is why multivariate spatial modeling has rarely been undertaken in such frameworks.
The remainder of this article is organized as follows. Section 2 describes the features of the LMC and discusses the model construction, identifiability issues, stochastic selection, and prior specification. Section 3 outlines the proposed class of low-rank adaptive spatial factor models, how we handle misaligned data, and carry out inference. Section 4 illustrates the analysis for two simulated data sets and one air quality monitoring data. Finally, Section 5 concludes the paper with a summary and an eye toward future work.
2. Model Construction
2.1 LMC Model Structure and Specification
LMC consists of decomposing the set of original second-order random stationary outcomes into a set of reciprocally orthogonal regionalized factors. Suppose, for a study region D ⊆ ℝd, an m × 1 process Z(s) = (Z1(s), …, Zm (s))′ is a second-order stationary process. Then, for all s, h ∈ ℝd and i, j = 1, …, m, we have E[Zi (s)] = μi and cov{Zi (s), Zj (s + h)} = Cij (h). The matrix-valued function C(h) = {Cij (h)} is called the multivariate cross-covariance. Generally for i ≠ j, a change in the order of the variables or a change in the sign of the separation vector h changes the values of the cross-covariances. If both the sequence and the sign are changed, we would have the same value Cij (h) = Cj i (−h), which implies C(h) = C(−h)′. C(h) must also be a positive definite function. That is, for any finite set of spatial locations s1, …, sn ∈ D and any vectors ai ∈ ℝm, i = 1, …, n, . Specifying a valid cross-covariance function is less straightforward because of this constraint, but several spectral and constructive approaches (Ver Hoef and Barry, 1998; Chilés and Delfiner, 1999) have been proposed and used in multivariate spatial analysis.
A straightforward cross-covariance is the so-called intrinsic specification, C(h) = ρ(h)T, where T is an m × m positive definite matrix and ρ(h) is a univariate correlation function. The limitation here is that each Zi (s) has the same spatial parameters, so each outcome would have the same strength in spatial association over the domain. An extension (Wackernagel, 2003) specifies , where for each k, Tk is a rank-one positive semidefinite matrix and ρk (h; ϕk) is a correlation function that depends upon additional parameters ϕk. Here, r is the total number of different spatial correlation functions in the multivariate cross-covariance.
The spectral decomposition yields , where uk is the normalized (i.e., ‖uk‖ = 1) eigenvector of Tk corresponding to the only positive eigenvalue ξk of Tk. Because Tk is symmetric with rank one, all its other eigenvalues are zero. This implies that there is an m × 1 vector, , which is the square root of Tk such that . Although ‖uk‖ = 1, it still has two possible directions. To build a one-to-one transformation between Tk and λk, we need to impose some further constraints on λk by, for instance, restricting the first element of λk to be positive.
Let wk (s), k = 1, …, r, be independently distributed univariate Gaussian processes, each with unit variance and a parametric correlation function. We write wk (s) ~ GP(0, ρk (·; ϕk)) with var{wk (s)} = 1, cov{wk (s), wk (t)} = ρk (s, t; ϕk), and cov{wk (s), wl (t)} = 0 whenever k ≠ l for all s and t (even when s = t). Here ρk (·; ϕk) is a correlation function associated with wk (s), and ϕk includes the spatial decay and smoothness parameters. We can easily see that the multivariate cross-covariance function for the process wk (s)λk is ρk (h; ϕk)Tk and hence, for , the function is .
We often assume that the mean of the outcomes arises linearly in the predictors so that the mean of the jth outcome Yj (s) is modeled as xj (s)′ βj, where xij (s) is a pj × 1 vector of predictors, assumed to be known at location s, and βj is the corresponding pj × 1 vector of slopes. Let Y(s) be the m × 1 vector of outcomes with jth element Yj (s) and let X(s)′ be an m × p block diagonal matrix, where and the jth diagonal block is given by xj(s)′ and is a p × 1 vector of slopes. Then, the spatial factor model is
| (1) |
where Λ = (λ1, …, λr) is a m × r matrix with kth column λk, w(s) = (w1(s), …, wr (s))′, and ε(s) is an m × 1 vector of measurement errors distributed as N (0, Ψ). The measurement error variance Ψ can be any m × m positive definite matrix but is usually assumed diagonal with elements for j = 1, …, m along the diagonal.
2.2 Identifiability in Spatial Factor Model
Model (1) is similar to FA models with the factor-loading matrix Λ and latent factor w(s), except that w(s) is now a multivariate stochastic process positing spatial dependence. As is well known (see, e.g., Anderson, 2003), orthogonal FA models must be further constrained to ensure identifiability. A widely used approach is to fix certain elements of Λ to constant values, usually to zeroes, such as restricting Λ to be an upper or lower triangular matrix with strictly positive diagonal elements (Lopes and West, 1999).
In LMCs, (see, e.g., Schmidt and Gelfand, 2003; Gelfand et al., 2004; Finley et al., 2008), a lower triangular Λ with positive diagonal elements identifies the covariances among the outcomes within a location because C(0) = ΛΛ′. But what we really seek to model is C(h). Finding proper identifiability constraints on Λ is equivalent to finding transformations that retain the statistical properties of the model.
Let P be an r × r orthogonal matrix, so that P′P = PP′ = Ir. The random effect term in (1) can be written as Λw(s) = ΛPP′w(s) = Λ̄ w̄(s), where Λ̄ = ΛP and w̄(s) = P′w(s). For nonspatial or traditional FA, the elements of w(s) are uncorrelated and the model is invariant to any orthogonal transformation because w(s) and w̄(s) have identical cross-covariances and ΛΛ′ = Λ̄Λ̄′. One can, therefore, obtain an infinite number of equivalent matrices of factor loadings by simply applying orthogonal transformations.
Matters are subtly different when w(s) is a spatial process. The cross-covariance matrix cov{w(s), w(t)} = Γ(s, t; ϕ), is now diagonal (and not an identity matrix as in traditional FA) with the kth diagonal element ρk (s, t; ϕk). Orthogonal transformations can, therefore, alter the distribution of w̄(s). To be precise, now cov{w̄(s), w̄(t)} = P′Γ(s, t; ϕ)P, which is neither necessarily diagonal nor equal to Γ(s, t; ϕ). Therefore, spatial factor models are not necessarily invariant to any orthogonal transformation.
Our primary motivation for using a Λ without any pre-specified 0 element is stochastic modeling for the number of factors (see Section 2.3). Such a model would automatically identify the latent spatial processes and, hence, the corresponding columns of Λ that are retained in (1). Specifying Λ to be lower triangular is now problematic because it is unlikely that such a structure will be retained throughout the stochastic selection process. On the other hand, using a Λ that is identifiable but that does not restrict certain elements to be zero will avoid such awkwardness.
In fact, we argue (see Web Appendix A for details) that only two groups of orthogonal transformations lead to nonidentifiability in spatial factor models. The first transformation P satisfies P′Γ(s, t; ϕ)P = Γ(s, t; ϕ). Such a P must be diagonal with 1’s and −1’s only. It is a special reflector (i.e., P2 = I) and is obtained as a product of elementary reflectors of the form , where ei is the ith Euclidean standard coordinate vector. The orthogonal matrix P alters the sign of the columns in Λ. To identify Λ (or have a one-to-one relationship between Λ and ΛΛ′), we need to specify one element in each column of Λ as positive or negative. Without losing generality, we could set the first row of Λ to be positive.
Permutation matrices constitute the second group of orthogonal transformations that lead to nonidentifiability in spatial factor models. A permutation matrix switches the elements of w(s) and the corresponding columns of Λ simultaneously, so the distributions of Λ̄w̄(s) and Λw(s) are the same. To address such identifiability issues, we impose some constraints on ρk (h; ϕk), more specifically on ϕk (Zhang, 2007). For simplicity, we consider the exponential correlation function ρk (h; ϕk) = exp(−ϕk ‖h‖), which has a spatial decay parameter ϕk as the only unknown parameter. We require the range parameter ϕk, k = 1, …, r, to be ordered as ϕ1 < ϕ2 ⋯ < ϕr or ϕ1 > ϕ2 ⋯ > ϕr. Simulation studies (see Sections 4.1 and 4.2) reveal that without this constraint, parameter estimation becomes problematic.
2.3 Adaptive Bayesian Factor Model
Determining the number of spatial factors (r) is challenging because of the lack of rigorous theoretical results that hint at the data’s ability to inform about r. Generally, previous work Webster, Atteias, and Duboiss (1994) employing the LMC assumes r < m is fixed. The choice of the number of spatial processes and their respective scales is a critical point in geostatistical models. In the applications of LMC to multivariate spatial analysis (see, e.g., Webster et al., 1994; Castrignanó et al., 2005; Buttafuoco et al., 2010), the spatial correlation functions ρk (h; ϕk), the parameters ϕk, and r are obtained from empirical estimates of the auto- and cross-variograms before any modeling. This approach ignores the uncertainty in the estimates of the spatial parameters and may yield dubious inference.
We adapt earlier work by Kuo and Mallick (1998) and Chen and Dunson (2003) to propose an approach for selecting spatial processes corresponding to different spatial scales using a Bayesian hierarchical model. Indicator variables δ = {δ1, …, δr}, where each δk is supported at two points 1 and 0, are introduced in model (1) to yield:
| (2) |
When δk = 1, we include the kth spatial random process wk (s) and the corresponding λk in the model. Otherwise, we omit the kth spatial scale. This yields 2r submodels. We call (2) the adaptive Bayesian factor model, in which the unknown parameters are estimated together with which factors will be retained.
2.4 Prior Specification
We place a multivariate normal prior on the slope parameter β with mean μβ and variance Σβ. Often, a flat prior is used. Each diagonal element of Ψ is assigned an Inverse Gamma (IG) distribution. With , the prior for Ψ becomes with hyperparameters a and b. A customary choice is to use a = 2, which suggests a distribution with infinite variance and a mean of b (often gleaned from a semivariogram).
For simplicity, we assume independent priors for δ and Λ (Kuo and Mallick, 1998). The indicator variables δk, k = 1, …, r, are taken a priori to be independent, with p(δk = 1 | ω) = ω. We regard ω as unknown and assume that it has a uniform prior on (0, 1). Imposing constraints on Λ, only requires a minor modification in the derivation of the full conditional distribution. Here, we take independent priors such as . The first row of Λ is restricted to be positive, that is, for k = 1, …, r, where I(·) is the indicator function. Berger and Pericchi (2001) suggest caution in the use of such priors in generic hierarchical models because the outcome of the model selection process can be quite sensitive to their vagueness. However, this seems to be less of an issue in dynamic factor models once the loading matrix is made identifiable (Lopes and West, 2004), where-upon inference and model selection were robust to the prior specifications. Similar results are obtained in random effect selection models (Chen and Dunson, 2003; Cai and Dunson, 2006). In our current context, restricting the first row of Λ to be positive, hence assigning a truncated normal to each element of the first row of Λ, ensures robust model selection and related inference to .
One also needs to assign priors on . The prior for ϕ depends upon the choice of correlation functions. Quite remarkably, the spatial process parameters are not consistently estimable and the effect of the prior does not disappear with increasing amounts of data (Zhang, 2004). Hence, prior information becomes an even more delicate issue. Typically, we set prior distributions for the range parameters relative to the size of their domains. In this article, an exponential correlation function is used and the prior for the range parameter is specified on a support with upper and lower limits denoted as ϕu = −log(0.01)/dmin and ϕl = −log(0.05)/dmax, where dmin and dmax are the minimum and maximum distances across all the locations (Wang and Wall, 2003). Because of identifiability issues discussed in Section 2.2, we construct a joint distribution for the ϕk’s to ensure ordering. In particular, we set
where π(ϕ1) is a uniform density with support (ϕl, ϕu) and subsequently, for k = 2, 3, …, r,
| (3) |
Here the hyperparameters ck > 0 controls the shape of the distribution and the separation of the ϕk’s. In all our subsequent analyses, we fix ck = 2k as a reasonable choice that delivers robust inference. For any finite domain this prior is proper (i.e., integrable). To offer some additional insight, let ϕk − ϕk−1 ∈ (0, hck) and h = 4. Numerical integration yields p(ϕk − ϕk−1 < ck /2 | ϕk−1) < 0.01. This implies that ϕk is unlikely to appear in (ϕk−1, ϕk−1 + ck /2) unless there is a strong mode from the likelihood. So (3) could efficiently separate the ϕk’s. On the other hand, , which indicates that π(ϕk | ϕk−1) becomes informative when ϕk−1 and ϕk are away from each other.
An alternative specification is π(ϕ) ∝ I (ϕl < ϕ1) I (ϕ1 < ϕ2) ⋯ I (ϕr < ϕu). This is somewhat simpler than (3), but encounters problems in practical implementation. Here, the posteriors for ϕk and ϕk +1, although they theoretically obey ϕk < ϕk +1, can become very close to each other. In that case, one of the two latent processes becomes redundant, yet stochastic factor selection keeps both processes. The specification in (3) avoids this situation.
3. Predictive Process Factor Models
Although FA is a powerful tool for summarizing multivariate outcomes and conducting dimension reduction on the number of outcomes, the spatial FA is prohibitive with a large number of locations. A popular model-based approach for dimension reduction over space uses low- or fixed-rank representations for w(s). Their likelihood resembles linear-mixed models and can be estimated using standard algorithms with some minor adaptations.
Here we consider one such representation that projects the spatially associated latent factors w(s) onto a lower-dimensional subspace determined by a partial realization of the process w(s) over a manageable set of locations called “knots.” Unlike several other low-rank methods, the predictive process does not introduce additional parameters or kernels. Also, some approaches require empirical estimates of the data’s covariance structure. This may be challenging here as the variability in the “data” is assumed to be a sum of unobserved factors. Therefore, we cannot use variograms on the data to isolate empirical estimates of its spatial covariance structure from that of the individual factors.
3.1 Model Construction
A Gaussian predictive process (see, e.g., Banerjee et al., 2008; Finley et al., 2009; Banerjee et al., 2010) uses a set of fixed “knots” , n* ≤ n which are usually fixed and may, but need not, form a subset of the observed locations 𝒮 = {s1, …, sn}. The Gaussian process defined in Section 2.1 implies that follows , where is the n* × n* covariance matrix whose (i, j)th element is . The spatial interpolation or “kriging” function at a site s0 is given by , where dk (s0; ϕk) is an n* × 1 vector whose ith element is . This defines the predictive process w̃k (s) ~ GP(0, ρ̃k (·; ϕk)) derived from the parent process wk (s), where .
The predictive process underestimates the variance of the parent process wk (s0) at any location s0 because var{wk (s0)} − var{w̃k (s0)} = var{wk (s0) − w̃k (s0)} ≥ 0. The veracity of this is an immediate consequence of the definition of w̃k (s) as a conditional expectation. This means that the estimated Ψ from the predictive process model roughly captures the same amount of variability as the estimated Ψ + Λ(Ir − E[Γ̃(s0)])Λ′ from (1), where Γ̃(s0) is an r × r diagonal matrix with kth diagonal element ρ̃k (s0, s0; ϕk).
Defining , where , we easily obtain var{fk (s)} = var{wk (s)} = 1, as desired. Replace w(s) with f(s) = (f1(s), …, fr (s))′ in (2) and define fk = (fk (s1), …, fk (sn))′, F = (f1, …, fr) and . This yields the posterior distribution p(F, w*, β, Ψ, ϕ, Λ, δ, ω |Y) proportional to
| (4) |
where Dk (ϕk) = (dk (s1), …, dk (sn))′ and Σfk is an n × n diagonal matrix with ith diagonal element . π(ω) is uniformly distributed on (0, 1) and π(ϕ) is defined in (3). Estimation of (4) proceeds using MCMC sampling (see Web Appendix B for details).
We offer some remarks on knot selection. With evenly distributed locations, knots on a uniform grid (see, e.g., Diggle and Lophaven, 2006) may suffice. With irregular locations, space-covering designs (e.g., Royle and Nychka, 1998) yield a more representative set. In general, different knots selection methods still produce robust results in predictive process models (see, e.g., Finley et al., 2009; Banerjee et al., 2010). And the modified predictive process model fk (s) is even less sensitive than w̃k (s) to such choices. Here, we use the K-means clustering algorithm (Hartigan and Wong, 1979) to arrive at a set of knots.
3.2 Handling Missing Observations
Missing outcomes arise frequently in many environmental applications. The outcomes may be missing for a variety of unintended reasons: nonresponse, equipment failure, lack of collection, and so on. Bayesian analysis often proceeds from data augmentation (Tanner, 1993) or multiple imputation (Rubin, 1976) to handle the incomplete data problem. Both methods impute the missing values within a Gibbs step and then use the complete data set. Instead, here we condition on the latent factors, which simplifies the MCMC algorithm and reduces the computational burden. Conditional on the latent factor f(s), the outcomes are independent of each other, which yields a likelihood depending only upon observed data. The missing values can, then, be recovered from the posterior predictive distribution.
Let Y(s) be the m × 1 vector of measured and unmeasured outcomes at site s. Suppose the measured and unmeasured elements of Y(s) have indices i1, …, ids and ids+1, …, im, respectively, where ds is the number of observed outcomes at s. Let vj, j = 1, …, m, be an m × 1 vector whose jth element is 1 and the rest are all 0. We can then construct matrices R1(s)ds×m and R2(s)(m−ds)×m, which can extract the observed and unobserved elements when multiplied by Y(s). More precisely, R1(s) = (vi1, …, vids)′ and R2(s) = (vids+1, …, vim)′.
Observe that Yo (s) = R1(s)Y(s) and Yu (s) = R2(s)Y(s) consist of the observed and unobserved elements, respectively. Multiplying both sides of model (2) by R1(s) reveals that the likelihood p(Yo (s) | f(s), γ, β, Λ, Ψ) follows a multivariate normal distribution with variance R1(s)ΨR1(s)′ and mean . Note that R1(s)ΨR1(s)′ is a ds × ds diagonal matrix with lth diagonal element for l = 1, …, dS. This ensures that R1(s)ΨR1(s)′ is a valid covariance matrix, and so is R2(s)ΨR2(s)′ for similar reasons. See Web Appendix B for implementation details.
3.3 Prediction and Predictive Model Comparison
Spatial analysis for data sets with missing values customarily entail three types of inference: (i) recover missing values, (ii) predict outcome Y(t) = (Y1(t), …, Ym (t))′ at any arbitrary location t, and (iii) generate replicates for the observed data for use in model assessment and selection (Gelfand and Ghosh, 1998).
To recover the missing values, we use composition sampling to draw Yu from p(Yu|Yo), where Yo = (Yo (s1)′, …, Yo (sn)′)′ and Yu = (Yu (s1)′, …, Yu (sn)′)′. Note that
where Λ̃ = (δ1λ1, …, δrλr). Using the samples {F(l), θ(l)} from p(F, θ |Yo), the missing value at s is recovered by drawing for each, l = 1, …, L, from an (m − ds) × 1 multivariate normal distribution with mean R2(s)[X(s)′ β(l) + Λ̃(l) f(l) (s)] and variance R2(s)Ψ(l)R2(s)′.
Prediction of the outcomes at unsampled or ungauged sites is often a major study objective. Using notations defined earlier, we have
where and . Here, Gk (ϕk) is the variance of fk, which equals . For prediction, we would first sample fk (t)(l) from using posterior samples and . Then, Y(t)(l) is drawn from an m ×1 multivariate normal distribution with mean X(t)′β(l) + Λ̃(l) f(t)(l) and variance Ψ(l).
Finally we turn to generating replicated observations for Yo. This is easily achieved by first sampling from p(w*, θ |Yo). The replicate is generated from a ds × 1 multivariate normal distribution with mean R1(s)[X(s)′β(l) + Λ̃(l) w̃(l)(s)] and variance R1(s)[Ψ(l) + Λ̃(l) Σf(s) (ϕ(l))Λ̃(l)′]R1(s)′, where Σf(s) is a diagonal matrix with kth diagonal element . Then and can be calculated respectively, where i indexes the observed data. Gelfand and Ghosh (1998) present a posterior predictive criterion that balances goodness-of-fit and predictive variance under a squared error loss function. This assigns a score to each model that is the sum of two terms, P and G, where is an error sum of squares and represents goodness-of-fit, whereas represents predictive variance and acts as a penalty term. Therefore, complexity is penalized and a parsimonious choice with lower D = G + P indicates preferred models.
4. Illustrations
We executed our adaptive spatial factor models using the R programming language. The most demanding model (12 × 1 vector of outcomes across 1, 000 locations) took approximately 48 hours to deliver its entire inferential output from 20, 000 MCMC iterations, including 4, 000 samples for burn-in, on a 3.10-GHz Intel i5–2400 processor with 4.0 Gbytes of RAM. The statistics of Gelman and Rubin (1992) was used to assess chain convergence alongside visual inspection of the trace plots and empirical autocorrelation functions.
4.1 Simulation Study One
The objective of this simulation study is to explore different specifications for Λ and demonstrate model identifiability. Adaptive factor modeling and predictive processes are not employed here. The data generating model uses a 2 × 2 factor loading matrix Λ and a spatial range parameter ϕ whose first element is smaller than the second. A synthetic data set comprising 250 locations within a [0, 50] × [0, 50] square was generated from (1) with r = m = 2. An isotropic exponential correlation function, ρk (s − t; ϕk) = exp(−ϕk ‖s − t‖) for k = 1, 2, was used to produce a spatially dependent bivariate random field. The bivariate outcomes, Y(s), were simulated with the following parameters:
where β is the mean vector for Y(s) and . Note the strong negative cross-correlation between the two spatial processes. Also, the correlation decays six times faster in the second process than in the first, yielding spatial ranges −log(0.05)/ϕ1 ≈ 30 and −log(0.05)/ϕ2 ≈ 5 units for the first and second process, respectively.
Three models are estimated and compared. The first restricts Λ to be lower-triangular, as in the LMC setting, but does not order the ϕi’s a priori. The other two models use factor loadings with positive elements in the first row of Λ and use the prior in (3) for ϕ. One of these two models assumes ϕ1 < ϕ2, reflecting the true ordering, although the other reverses the inequality and results in a misspecified model a priori. However, because the factor loadings are now identifiable, the estimated Λ contains permuted columns according to the order of the ϕi’s. This is attractive in practice—one need not ascertain the correct ordering of the range parameters a priori. We arrange them in the same order as the true values although presenting estimates in Table 1.
Table 1.
Posterior percentiles (50%, 2.5%, and 97.5%) estimated for the parameters in different model specifications
| True parameter |
Lower triangular setting |
General factor loadings | |
|---|---|---|---|
| Increasing ϕk | Decreasing ϕk | ||
| β1 = 5 | 4.77 (1.22, 7.49) | 5.00 (3.29, 6.98) | 5.58 (3.77, 7.85) |
| β2 = 10 | 10.38 (8.65, 12.73) | 10.15 (8.38, 11.60) | 9.68 (7.73, 11.16) |
| C(0)1,1 = 10 | 8.59, (4.76, 15.08) | 10.82 (6.53, 18.54) | 10.65 (6.23, 17.66) |
| C(0)2,1 = −4 | −5.64 (−9.99, −2.93) | −5.34 (−10.84, −1.82) | −5.11 (−9.88, −1.67) |
| C(0)2,2 = 8 | 12.17 (6.76, 17.54) | 12.99 (7.15, 19.43) | 12.83 (7.15, 18.42) |
| Ψ1 = 2 | 3.65 (2.61, 4.88) | 2.30 (1.19, 3.66) | 2.35 (1.15, 3.55) |
| Ψ2 = 5 | 3.27 (0.97, 8.38) | 3.33 (1.06, 7.99) | 3.21 (0.99, 7.99) |
| ϕ1 = 0.1 | 0.06 (0.05, 0.12) | 0.07 (0.05, 0.13) | 0.07 (0.05, 0.14) |
| ϕ2 = 0.6 | 0.95 (0.25, 2.15) | 1.17 (0.52, 2.58) | 1.15 (0.49, 2.44) |
| Λ1,1 = 3 | 2.93 (2.18, 3.88) | 3.11 (2.32, 4.15) | 3.07 (2.31, 4.06) |
| Λ1,2 = −2 | −1.93, (−2.80, −1.17) | −2.53 (−3.42, −1.79) | −2.48 (−3.33, −1.68) |
| Λ2,1 = 1 | Fixed at 0 | 1.02 (0.56, 1.68) | 1.01 (0.55, 1.78) |
| Λ2,2 = 2 | 2.89, (1.84, 3.47) | 2.57 (1.33, 3.22) | 2.60 (1.34, 3.23) |
In the first model Λ21 = 0, ϕi’s are assigned uniform priors over (ϕl, ϕu) without any further restrictions. Priors for the two other models are assigned as discussed in Section 2.4 with hyperparameters μβ = 0, , ϕl = 0.047, ϕu = 25, a = 2, and b = 5. Posterior inferences for the latter two models are consistent. This indicates the importance of ordering constraints; without them, the posterior distributions for ϕ and Λ can produce multiple modes as the MCMC chain could jump between the possible specifications (Lopes and West, 2004). The parameter Λ21 (boldface in Table 1), which was set as 0 in the lower triangular setting, is significantly different from 0 in both the models using general factor loadings. Thus, we can capture the underlying cross-covariance structure without restricting the loading matrix, but by imposing an ordering on the spatial decay parameters. In addition, our proposed models have a lower posterior predictive model comparison score (D = 14, 202) than the lower triangular setting (D = 14, 941). This, again, demonstrates a preference for using general factor loadings.
4.2 Simulation Study Two
Now we demonstrate the adaptive spatial factor model using a simulated data set with missing values. We generated 1, 000 locations in a [0, 30] × [0, 30] square. At each location, we simulated a 12 × 1 vector of outcomes, Y(s), from (1) with three factors and outcome-specific intercepts as the only regressors. Each spatial process, wk (s), was generated from an isotropic exponential correlation function. The parameters are defined as:
After the data was generated, we allowed a quarter of the locations to retain all the outcomes, although randomly omitting 2/3 of the outcomes from the remaining locations. This produced a data set with approximately 50% missingness.
We applied the Gibbs sampling algorithm described in Section 3 for estimation. For the predictive process, we selected 200 knots using a K-means clustering algorithm (Hartigan and Wong, 1979). The maximum number of latent factors, r, was taken to be 5. For comparison, we also fit spatial factor models using a fixed number of latent factors for the same data set. The prior distributions resemble those in Section 4.1, except that now ϕl = 0.07 and ϕu = 310 to better reflect the domain. Also, for the adaptive model we used independent Bernoulli priors for δ with p(δk = 1| ω) = ω, k = 1, …, r and ω ~ U(0, 1).
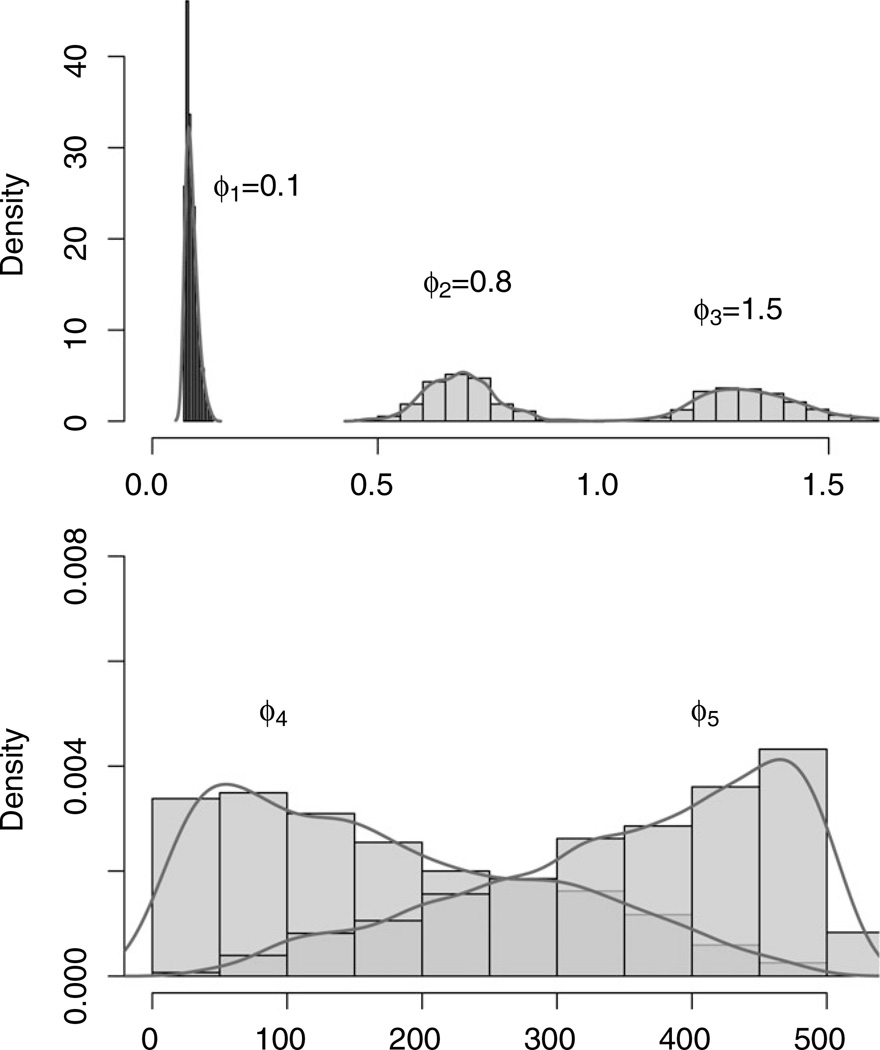
Stochastic selection produced the final model with three latent factors based upon the highest posterior probabilities. Figure 1 illustrates the posterior distributions for ϕ. The spatial parameters in the active processes are ϕ1, ϕ2, ϕ3 (corresponding to δk = 1) and their posteriors are well identified. The true values are all included in the central 95% credible intervals. The posteriors for ϕ4 and ϕ5 only reflect prior information. The wide range of their posteriors suggest that there is very little posterior learning and the MCMC sampler is sampling from the prior. The marginal posterior density for ω reveals substantial posterior learning. The credible intervals of β and Ψ contain the true values, as does most of the elements of Λ, suggesting sound estimative performance.
Figure 1.
Histograms of the posterior distributions for ϕ.
Spatial factor models with fixed number of latent factors equaling 1, 2, 4, and 5 are also fitted. The posterior predictive criterion discussed in Section 3.3 is presented for model assessment and the scores are compared in Table 2. The adaptive spatial factor model with D = 99740 (boldface in Table 2) seems to be the winner here. There is overwhelming evidence that the regression model with random processes f1(s), f2(s), and f3(s) is optimal. This, pleasantly, agrees with the true specification.
Table 2.
Simulation study two: Model comparison criteria
| Number of factors | |||||
|---|---|---|---|---|---|
| Criterion | r = 1 | r = 2 | Adaptive factor | r = 4 | r = 5 |
| G | 69567 | 53893 | 45982 | 46011 | 46036 |
| P | 71698 | 60173 | 53758 | 53880 | 53939 |
| D | 141265 | 114066 | 99740 | 99891 | 99975 |
4.3 Air Monitor Value Data
Our current application involves data compiled from the Environmental Protection Agency across 316 air monitors in California involving five pollutants: carbon monoxide (CO), nitrogen dioxide (NO2), ozone (O3), particulate matter with diameter < 2.5 micrometers (PM2.5), and particulate matter with diameter < 10 micrometers (PM10). Concentrations (ppm) measured by monitoring equipments are used for CO, NO2, and O3 and (µg/m3) for PM2.5 and PM10. Here we use the annual average of the monitor values recorded in 2008. All the variables were standardized to mean 0 and variance 1.
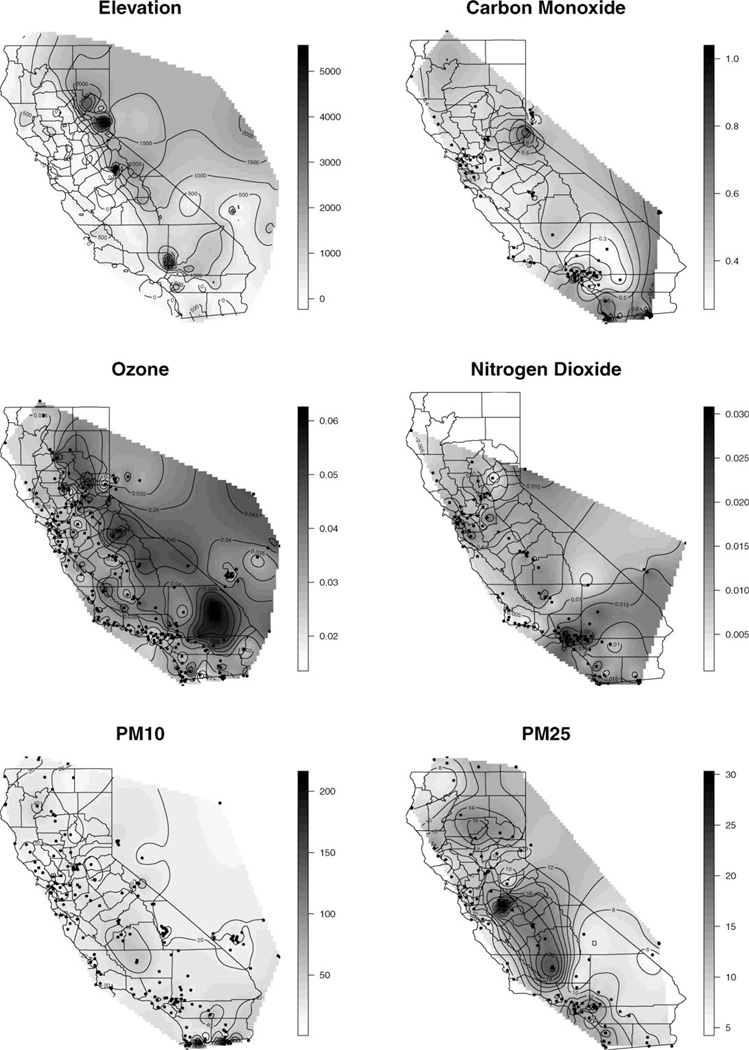
For the modeling, each pollutant is assigned its own intercept term. Elevation (in kilometers) is the only predictor and is depicted as a contoured image in the top-left panel of Figure 2. The instruments only monitor some of the five pollutants at most of the sites, so the data set has about 53% of observations missing. The longitude and latitude of the monitors are transformed to Easting and Northing in kilometer units. For the predictive process, 50 knots were selected using a K-means clustering algorithm. Interpolation for the air pollutants and the locations of monitor sites (·) appear in Figure 2.
Figure 2.
Interpolation of air pollutants measured on monitor sites across California.
Visually, the air pollutants exhibit spatially varying concentrations as well as the associations among them at a given location. We seek to model both kinds of dependence. It is not clear whether it is appropriate to assume a single factor underlying the different air pollutants or if additional factors need to be introduced. To address this question, we repeated the approach described in Section 3 using the exact model (4). The priors are the same as in Section 4.2 except for ϕ, which has distribution (3) with ϕl = 0.002 and ϕu = 136.
The model is implemented in the same manner as in Section 4.2. The maximum possible number of factors r is taken to be 3. We decide to use 4,000 iterations for a burn-in period, and then a further 16,000 iterations for posterior inference.
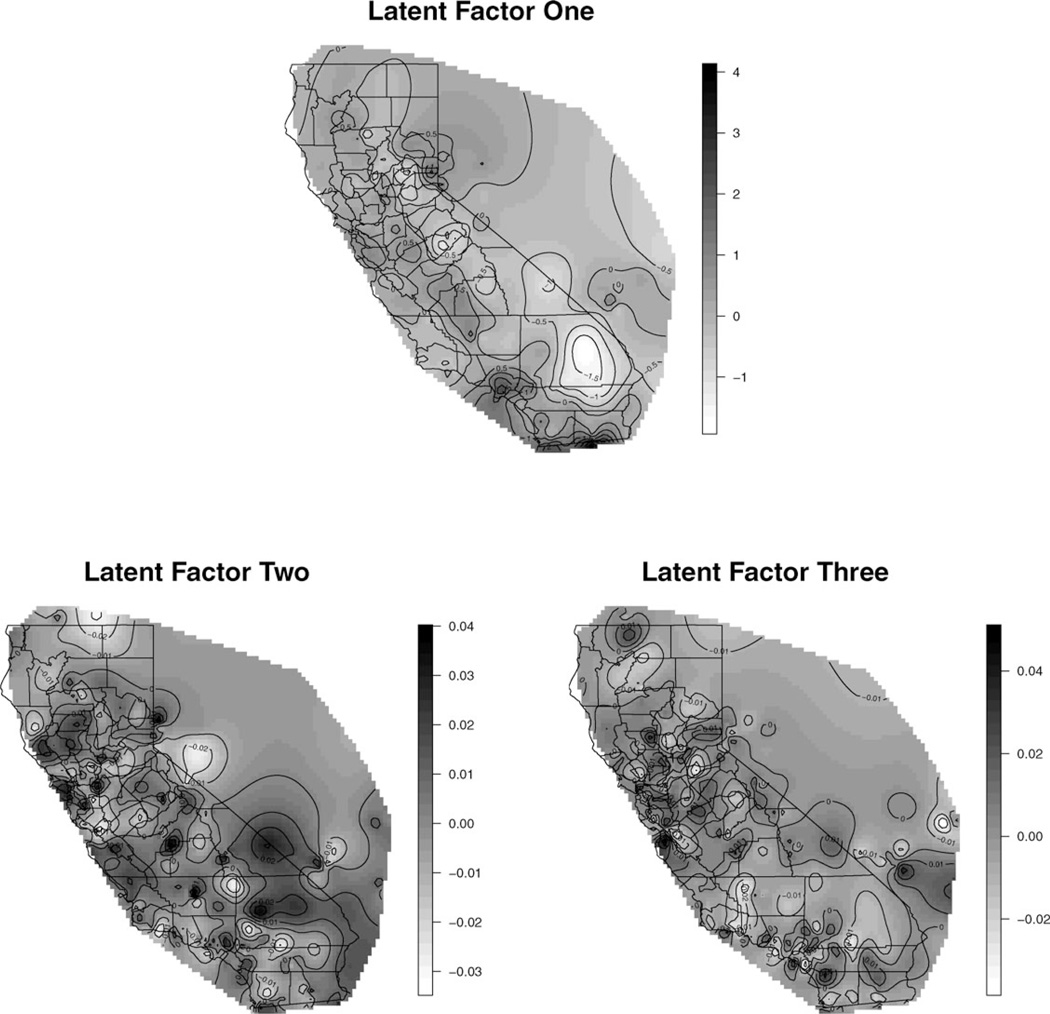
The probability assigned to the model with one latent factor is 1, suggesting that one factor is sufficient. The interpolation of the latent spatial processes are displayed in Figure 3. The panel at the top depicts the only active process (δ1 = 1), which presents some characteristics for the spatial concentrations of the air pollutants. The panels at the bottom do not show significant association with the data and only demonstrate the prior information. For each air pollutant Yj (s), the percentage of the variance explained by the spatial factor fk (s), is simply , j = 1, …, 5. Overall, about 50% of the variation is explained by the first latent factor—NO2 and CO are more closely related to this latent factor, although PM2.5 is weakly explained.
Figure 3.
Interpolation of latent spatial factors across California.
Table 3 presents the posterior inferences of β, Ψ, ϕ, Λ, and the correlation ρ = {ρi,j}, i, j = 1, …, 5, j > i, among air pollutants, which is estimated from the variance estimator ΛΛ′ + Ψ. Only CO and NO2 reveal significant (negative) intercept coefficients. The slope parameters seem to suggest that elevation has a significant impact only upon Ozone among the five pollutants. The posterior inference for ϕ1 has median 0.018, which corresponds to the effective range (i.e., the distance at which the correlation drops to 0.05) roughly 160 kilometers. The posterior inferences for ϕ2 and ϕ3 indicate weak spatial dependence. So we only present the first column of Λ and omit the rest in Table 3. The Λj1’s reflect the influence of the common factor on the corresponding outcome variables. Because the residual variances, , differ across j, the Λj1’s measure covariances on different scales and cannot be directly compared to assess strength of association between outcomes. The parameters can be interpreted as independent measurement error variances. The covariances of the outcomes collected within one set are , where λ1 = (Λ11, …, Λ51)′. The corresponding correlations among the outcomes are also presented in Table 3.
Table 3.
The posterior credible intervals estimated for the parameters in air pollutants data set. The subscripts 1–5 in β, Ψ, ρ, and the row index of λ1 refer to CO, NO2, O3, PM10, and PM25, respectively. Subscripts on ϕ refer to the three spatial range parameters
| Parameter | 50% (2.5%, 97.5%) | Parameter | 50% (2.5%, 97.5%) | Parameter | 50% (2.5%, 97.5%) | |
|---|---|---|---|---|---|---|
| β1,0 | −0.76 (− 1.37, −0.29) | Λ11 | 0.85, (0.58, 1.39) | ρ1,2 | 0.60 (0.44, 0.80) | |
| β2,0 | −0.61 (− 1.23, −0.14) | Λ21 | 0.88 (0.64, 1.47) | ρ1,3 | −0.45 (− 0.70, −0.27) | |
| β3,0 | 0.04 (− 0.24, 0.41) | Λ31 | −0.57 (− 0.98, −0.36) | ρ1,4 | 0.50 (0.33, 0.68) | |
| β4,0 | −0.30 (− 0.69, 0.05) | Λ41 | 0.68, (0.49, 1.02) | ρ1,5 | 0.39 (0.19, 0.64) | |
| β5,0 | −0.27 (− 0.72, 0.07) | Λ51 | 0.55 (0.27, 1.00) | ρ2,3 | −0.48 (− 0.73, −0.28) | |
| β1,E lev | −0.02 (− 0.50, 0.45) | 0.52 (0.35, 0.77) | ρ2,4 | 0.52, (0.37, 0.70) | ||
| β2,E lev | 0.33 (− 0.24, 0.91) | 0.44 (0.30, 0.63) | ρ2,5 | 0.41 (0.20, 0.66) | ||
| β3,E lev | 0.40 (0.21, 0.58) | 0.59 (0.44, 0.79) | ρ3,4 | −0.39 (− 0.60, −0.23) | ||
| β4,E lev | 0.02 (− 0.28, 0.33) | 0.63 (0.45, 0.85) | ρ3,5 | −0.30 (− 0.55, −0.14) | ||
| β5,E lev | −0.12 (− 0.43, 0.16) | 0.86 (0.62, 1.19) | ρ4,5 | 0.33 (0.15, 0.55) | ||
| ϕ1 | 0.018 (0.004, 0.039) | |||||
| ϕ2 | 43(5.1, 110) | |||||
| ϕ3 | 101(33, 136) |
5. Summary
We have addressed the problem of modeling large multivariate spatial data, where dimension reduction is sought both in the number of outcomes and in the number of spatial locations. The former is achieved with fewer number of factors, although the latter is achieved using a knot-based predictive process. Model-based strategies (Guhaniyogi et al., 2011) exist for choosing knots but recent findings, including our own explorations here, suggests that simple space-covering or clustering algorithms usually deliver robust inference. Our adaptive model lets the number of factors be stochastic and allows the data to drive the inference.
One can use either the predictive process w̃k (s) or its “modification” fk (s) in the adaptive spatial factor models but fk (s) adapts better to the data and, hence, is less sensitive to the knots. Although the substantive inference is quite robust to both these processes, some subtle differences are seen in the estimation algorithm with regard to transition among models. This is related to the strength of the spatial random field and is explained in Web Appendix C.
Given the widespread use of R as a statistical language, we presented computational benchmarks on R running on fairly standard architectures. In multivariate spatial analysis, “large” refers to the size of m × n, where m is the number of outcomes and n is the number of locations. With R running on standard architectures, mn > 1, 000 is usually deemed exorbitant without dimension reduction. Substantial computational gains accrue from using lower-level languages (C/C++) on shared memory systems.
Alternative approaches for modeling large spatial data sets include an SPDE/GMRF approach proposed by Lindgren, Rue, and Lindström (2011) that uses explicit Markov representations of the Matérn covariance family using a class of stochastic partial differential equations. Rather than MCMC, they use a faster Integrated Nested Laplace Approximation algorithm for Bayesian inference. Another approach, termed covariance tapering (Furrer, Genton, and Nychka 2006), relies upon compactly supported correlation functions to produce sparse covariance matrices containing only a moderate number of nonzero elements. How effective these alternative approaches will be with dynamic spatial factor models is yet to be ascertained.
Space-time or dynamic factor modeling using low-rank processes can also be envisioned. Misalignment can now occur both over space and over time to yield data matrices that are highly irregular. Our formulation can, nevertheless, be easily adapted to such settings. Also, although we attended only to point-referenced data here, adaptive space-time factor models could be used for multivariate regionally aggregated data (Jin, Banerjee, and Carlin 2007) as well. Here, usually the number of regions is not onerous, so dimension reduction is relevant over the number of outcomes (FA) and time (a temporal predictive process).
Supplementary Material
Acknowledgements
The authors thank the referees, associate editor, and editor for valuable comments resulting in substantial improvements to the manuscript. This work was supported by NIH grant 1-RC1-GM092400-01 and NSF grant DMS-1106609.
Footnotes
The Web Appendices referenced in Sections 2.2, 3.1, 3.2, and 5, the ambient air quality data analyzed in Section 4.3, and the R code implementing our models are available with this paper at the Biometrics website on Wiley Online Library.
References
- Anderson TW. An Introduction to Multivariate Statistical Analysis. 3rd edition. New York, NY, USA: Wiley; 2003. [Google Scholar]
- Banerjee S, Carlin BP, Gelfand AE. Hierarchical Modeling and Analysis for Spatial Data. Boca Raton, Florida: CHAPMAN & HALL/CRC; 2004. [Google Scholar]
- Banerjee S, Finley AO, Waldmann P, Ericsson T. Hierarchical spatial process models for multiple traits in large genetic trials. Journal of the American Statistical Association. 2010;105:506–521. doi: 10.1198/jasa.2009.ap09068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Gelfand AE, Finley AO, Sang H. Gaussian predictive process models for large spatial data sets. Journal of the Royal Statistical Society Series B Statistical Methodology. 2008;70:825–848. doi: 10.1111/j.1467-9868.2008.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JO, Pericchi LR. Objective Bayesian methods for model selection: Introduction and comparison. Lecture Notes- Monograph Series. 2001;38:135–207. [Google Scholar]
- Buttafuoco G, Castrignanò A, Colecchia AS, Ricca N. Delineation of management zones using soil properties and a multivariate geostatistical approach. The Italian Journal of Agronomy. 2010;4:323–332. [Google Scholar]
- Cai B, Dunson DB. Bayesian covariance selection in generalized linear mixed models. Biometrics. 2006;62:446–457. doi: 10.1111/j.1541-0420.2005.00499.x. [DOI] [PubMed] [Google Scholar]
- Castrignanó A, Cherubini C, Giasi CI, Castore M, Mucci GD, Molinari M. ISTRO Conference Brno. Brno, Czech Republic: ISTRO; 2005. Using multivariate geostatistics for describing spatial relationships among some soil properties. In; pp. 383–390. [Google Scholar]
- Chen Z, Dunson DB. Random effects selection in linear mixed models. Biometrics. 2003;59:762–769. doi: 10.1111/j.0006-341x.2003.00089.x. [DOI] [PubMed] [Google Scholar]
- Chilés J, Delfiner P. Geostatistics: Modeling Spatial Uncertainty. New York, USA: John Wiley; 1999. [Google Scholar]
- Christensen WF, Amemiya Y. Latent variable analysis of multivariate spatial data. Journal of the American Statistical Association. 2002;97:302–317. [Google Scholar]
- Cressie NAC, Johannesson G. Fixed rank kriging for very large spatial data sets. Mathematical Geology. 2008;70:209–226. [Google Scholar]
- Diggle P, Lophaven S. Bayesian geostatistical design. Scandinavian Journal of Statistics. 2006;33:53–64. [Google Scholar]
- Dominici F, Peng R, Bell M, Pham L, McDermott A, Zeger S, Samet J. Fine particulate air pollution and hospital admission for cardiovascular and respiratory diseases. Journal of the American Medical Association. 2006;295:1127–1134. doi: 10.1001/jama.295.10.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunson DB. Isds discussion paper. Duke University; 2006. Efficient Bayesian model averaging in factor analysis. [Google Scholar]
- Finley AO, Banerjee S, Ek AR, McRoberts RE. Bayesian multivariate process modeling for prediction of forest attributes. Journal of Agricultural, Biological, and Environmental Statistics. 2008;1:60–83. [Google Scholar]
- Finley AO, Sang H, Banerjee S, Gelfand AE. Improving the performance of predictive process modeling for large datasets. Computational Statistics & Data Analysis. 2009;53:2873–2884. doi: 10.1016/j.csda.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrer R, Genton MG, Nychka D. Covariance tapering for interpolation of large spatial datasets. Journal Of Computational And Graphical Statistics. 2006;15:502–523. [Google Scholar]
- Gelfand AE, Ghosh SK. Model choice: A minimum posterior predictive loss approach. Biometrika. 1998;85:1–11. [Google Scholar]
- Gelfand AE, Schmidt AM, Banerjee S, Sirmans CF. Nonstationary multivariate process modeling through spatially varying coregionalization. TEST. 2004;13:263–312. [Google Scholar]
- Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Statistical Science. 1992;7:457–472. [Google Scholar]
- Guhaniyogi R, Finley AO, Banerjee S, Gelfand AE. Adaptive gaussian predictive process models for large spatial datasets. Environmetrics. 2011;22:997–1007. doi: 10.1002/env.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan JA, Wong MA. A k-means clustering algorithm. Applied Statistics. 1979;28:100–108. [Google Scholar]
- Hogan JW, Tchernis R. Bayesian factor analysis for spatially correlated data, with application to summarizing area-level material deprivation from census data. Journal of the American Statistical Association. 2004;99:314–324. [Google Scholar]
- Jin X, Banerjee S, Carlin BP. Order free co-regionalized areal data models with application to multiple-disease mapping. Journal Of The Royal Statistical Society Series B. 2007;69:817–838. doi: 10.1111/j.1467-9868.2007.00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamman E, Wand M. Geoadditive models. Applied Statistics. 2003;52:1–18. [Google Scholar]
- Kuo L, Mallick B. Variable selection for regression models. Sankhya, Series B, The Indian Journal of Statistics. 1998;60:65–81. [Google Scholar]
- Lark R, Papritz A. Fitting a linear model of coregionalization for soil properties using simulated annealing. Geoderma. 2003;115:245–260. [Google Scholar]
- Lindgren F, Rue H, Lindström J. An explicit link between Gaussian fields and Gaussian Markov random fields: The stochastic partial differential equation approach. Journal of the Royal Statistical Society - Series B: Statistical Methodology. 2011;73:423–498. [Google Scholar]
- Liu X, Wall MM, Hodges JS. Generalized spatial structural equation models. Biostatistics. 2005;6:539–557. doi: 10.1093/biostatistics/kxi026. [DOI] [PubMed] [Google Scholar]
- Lopes HF, West M. Model uncertainty in factor analysis. Technical report, Institute of Statistics and Decision Sciences, Duke University; 1999. [Google Scholar]
- Lopes HF, West M. Bayesian model assessment in factor analysis. Statistica Sinica. 2004;14:41–67. [Google Scholar]
- Matheron G. Centre de Géostatistique. Fontainebleau, France: E.N.S.M.P; 1982. Pour une analyse krigeante des données régionalisées. Report N-732. [Google Scholar]
- Minozzo M, Fruttini D. Loglinear spatial factor analysis: An application to diabetes mellitus complications. Environmetrics. 2004;15:423–434. [Google Scholar]
- Paciorek CJ. Computational techniques for spatial logistic regression with large datasets. Computational Statistics and Data Analysis. 2007;51:3631–3653. doi: 10.1016/j.csda.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle JA, Berliner LM. A hierarchical approach to multivariate spatial modeling and prediction. Journal of Agricultural Biological and Environmental Statistics. 1999;4:29–56. [Google Scholar]
- Royle JA, Nychka D. An algorithm for the construction of spatial coverage designs with implementation in splus. Computers & Geosciences. 1998;24:479–488. [Google Scholar]
- Rubin DB. Inference and missing data. Biometrika. 1976;63:581–592. [Google Scholar]
- Schmidt AM, Gelfand AE. A Bayesian coregionalization approach for multivariate pollutant data. Journal of Geophysical Research. 2003;108:8783–8788. [Google Scholar]
- Tanner MA. Tools for Statistical Inference. Methods for the Exploration of Posterior Distributions and Likelihood Functions. New York, USA: Springer-Verlag; 1993. [Google Scholar]
- Ver Hoef JM, Barry RP. Consructing and fitting models for cokriging and multivariable spatial prediction. Journal of Statistical Planning and Inference. 1998;69:275–294. [Google Scholar]
- Ver Hoef JM, Cressie N, Barry RP. Flexible spatial models based on the fast fourier transform (fft) for cokriging. Journal of Computational and Graphical Statistics. 2004;13:265–282. [Google Scholar]
- Wackernagel H. Multivariate Geostatistics: An Introduction With Applications. Berlin: Springer-Verlag Telos; 2003. [Google Scholar]
- Wang F, Wall MM. Generalized common spatial factor model. Biostatistics. 2003;4:569–582. doi: 10.1093/biostatistics/4.4.569. [DOI] [PubMed] [Google Scholar]
- Webster R, Atteias O, Duboiss JP. Coregionalization of trace metals in the soil in the swiss jura. European Journal of Soil Science. 1994;45:205–218. [Google Scholar]
- Wikle C, Cressie N. A dimension-reduced approach to space-time Kalman filtering. Biometrika. 1999;86:815–829. [Google Scholar]
- Zhang H. Inconsistent estimation and asymptotically equal interpolations in model-based geostatistics. Journal of the American Statistical Association. 2004;99:250–261. [Google Scholar]
- Zhang H. Maximum-likelihood estimation for multivariate spatial linear coregionalization models. Environmetrics. 2007;18:125–139. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.